Abstract
HSD17B1 is an important candidate gene in breast cancer via its role in converting estrone to estradiol. A non-synonymous G-to-A transition (rs605059) and an intronic C-to-A (rs676387) SNP, which captured most common variation in HSD17B1, were evaluated in several breast cancer studies with inconclusive results. We followed-up these findings in the Polish Breast Cancer Study (1,995 cases, 2,296 controls) and the British SEARCH study (4,470 cases, 4,560 controls). Meta-analyses of published data and our own were also conducted among Caucasian women. Consistent with previous reports, we found little to no association with overall risk for heterozygotes and minor allele homozygotes, compared to major allele homozygotes, for rs605059 (summary ORs (95% CIs): 0.93 (0.87-0.99) for GA and 0.96 (0.85-1.08), based on 11,762 cases and 14,329 controls from 10 studies) and for rs676387 (summary ORs (95% CIs): 1.04 (0.97-1.12) and 1.12 (0.99-1.27), based on analyses of 11,074 cases and 13,605 controls from 8 studies). Data from the Polish (n=586 ER-negative cases) and British (n=407) studies did not support the previous findings that ER-negative tumors were inversely associated with rs676387 AA genotype and positively associated with rs605059 GG genotype, based on sub-analyses in 5 prospective cohorts with 354 ER-negative cases. In conclusion, it is unlikely that common genetic variation in HSD17B1 is associated with a moderate modulation in breast cancer risk overall; however, we cannot exclude the possibility of a very weak effect. Associations between HSD17B1 genotypes and risk of ER-negative breast cancer were inconsistent across studies and should be studied further.
Introduction
Genetic variation in the estrogen synthesis pathway may affect breast cancer risk by increasing lifetime exposure to the cell proliferation- and genotoxic-effects of estrogen (1). 17β-hydroxysteroid dehydrogenase 1 (HSD17B1) has been suggested to be an important candidate gene in breast carcinogenesis because its coding enzyme, 17HSD1, catalyzes the conversion of estrone to estradiol, the final step of estrogen synthesis (2, 3). In conjunction with 17HSD1, 17HSD2, which catalyzes the reverse estradiol to estrone, predominates the control of available active estrogen (4). However, 17HSD1 is over-expressed in malignant breast tissue of pre- and post-menopausal women (5) and in breast tumors with lower estrogen receptor (ER) expression (6).
A non-synonymous G-to-A (rs605059) single nucleotide polymorphism (SNP) in HSD17B1 has been examined in relationship to breast cancer risk in eight studies with inconclusive results (7-11). In a recent report of five U.S. and European cohorts (5,370 cases, 7,480 controls) (12), additional genetic variation in the gene and its flanking regions, as designated by 4 haplotype-tagging SNPs (htSNPs; rs676387, rs605059, rs598126, and rs2010750), was not found to be associated with breast cancer risk. However, among a subset of 2,091 Caucasian cases with estrogen receptor (ER) data and 2,982 controls, risk of ER-negative tumors (n=354) was lower among women with at least one copy of the variant A allele of rs676387, and higher among those with minor alleles of 3 strongly-correlated htSNPs – rs605059, rs598126, and rs2010750. We followed-up these findings in two studies, in which we genotyped the same 4 htSNPs in the Polish Breast Cancer Study (PBCS; 1,995 cases, 2,296 controls), and rs676387 and rs605059 in the Studies of Epidemiology and Risk Factors in Cancer Heredity (SEARCH; 4,470 cases, 4,560 controls).
Methods
The present analysis is based on data from two case-control studies conducted in Poland (13) and England (14) that have been previously described in detail. Both studies received approval from their respective institutional review committees and all study respondents provided informed consent.
Study Populations
The PBCS study was conducted between 2000 and 2003 among women residing in Warsaw and Lodz, Poland (13). Eligible cases were women aged 20 to 74 years who were newly diagnosed with either histologically or cytologically confirmed in situ or invasive breast cancer. Study personnel identified cases through a rapid identification system and cancer registries to ensure complete case ascertainment. Controls with no history of breast cancer were randomly selected through a database of all Polish residents. Controls were frequency-matched to cases by city and age in 5-year categories. A total of 1,995 cases (65% of eligible cases identified) and 2,296 controls (63% of eligible controls identified) provided a personal interview on known and suspected risk factors and donated a venous blood sample. Medical records of cases were obtained and extracted for tumor characteristics and initial breast cancer treatment regimens. Hormone receptor status was available for 76% of cases, and determined by immunohistochemical assays (91% of cases) and biochemical methods (9%).
In SEARCH, cases were drawn from the East Anglian Cancer Registry. All patients diagnosed with invasive breast cancer before age 55 years since 1991 and still alive in 1996 (prevalent cases, median age 48 years), together with all those diagnosed <70 years between 1996 and the present (incident cases, median age 54 years), were eligible to take part. Sixty-seven percent of eligible breast cancer patients returned a questionnaire and 64% provided a blood sample for DNA analysis. Female controls were randomly selected from the Norfolk component of the European Prospective Investigation of Cancer (EPIC) in approximate order of recruitment through general practice age-sex registers. EPIC is a prospective study of diet and cancer being carried out in nine European countries. The EPIC-Norfolk cohort comprises 25,000 residents of Norfolk, East Anglia – the same region from which the cases were recruited. Controls were not matched to cases, but were broadly similar with respect to age (range= 42-81 years). This analysis is based on a subset of 4,470 cases and 4,560 controls all of whom completed an epidemiological questionnaire and provided a blood sample for DNA analysis. Tumor characteristics, including ER status, were extracted from tumor registry data. ER data were available for 52% of cases in SEARCH.
Genotyping
htSNPs were selected based on the efforts of the Breast and Prostate Cohort Consortium (BPC3) (15), as previously described (12, 16). Given the close proximity of the adjacent genes, including N-acetylglucosaminidase-α (NAGLU) gene and a pseudogene for HSD17B1 (HSD17BP1) in the 5′ direction and CoA synthase (COASY) and transcription factor-like 4 (TCFL4) in the 3′ direction, the location of htSNPs for HSD17B1 extended into these neighboring genes. Four htSNPs (rs676387, rs605059, rs598126, and rs2010750) were selected, that capture a majority of common haplotype diversity among European-Americans (RH2 = 0.82).
Methods for genotype assays for rs676387, rs605059, rs598126, and rs2010750 performed on PBCS samples are described on-line: http://snp500cancer.nci.nih.gov (17). A total of 100 duplicate DNA pairs were interspersed throughout the PBCS DNA samples. All SNPs were >97% concordant for duplicate pairs and >99% complete for all samples. Genotype frequencies were in Hardy-Weinberg equilibrium (HWE) among controls (p-value >0.44).
In the SEARCH DNA samples, only rs605059 and rs676387 were genotyped, because there was a strong correlation between rs605059, rs598126, and rs2010750 in the PBCS and in previous studies (12). Genotyping was performed using a fluorescent 5′ exonuclease assay (Taqman) and the ABI PRISM 7900 Sequence Detection Sequence (PE Biosystems). Cases and controls were arrayed together in twelve 384-well plates and a 13th plate contained 8 duplicate samples from each of the 12 plates. All SNPs were 100% concordant for duplicate pairs and >99% complete for all samples. Genotype frequencies were in HWE among controls (p-value >0.33).
Statistical Analyses
Unconditional logistic regression models were used to estimate odds ratios (ORs) and 95% confidence intervals (CIs), adjusted for study site (Warsaw, Lodz, or Norfolk), for the association between individual SNPs and breast cancer, using STATA (version 10.0). Genotypes were evaluated using indicator variables. We assumed an additive mode of inheritance to calculate the p for trend. We evaluated effect modification by age at baseline (defined as age at diagnosis for cases and age at interview for controls; age categories: <45, 45-55, >55) and menopausal status (premenopausal and postmenopausal for PBCS only) comparing the −2 log likelihood of models with and without interaction terms. Breast cancer cases were further defined by ER (ER+, ER−) and PR (PR+, PR−) status of the breast tumor, excluding cases with unknown hormone receptor status. Polytomous logistic regression models were used to estimate ORs and 95% CIs for two different case groups and controls. To test for differences in genotype associations between case groups, we evaluated case-only logistic regression models, adjusted for age and study, with ER status (positive/ negative) or PR status (positive/ negative) as the outcome variable and genotype as the explanatory variable.
Meta-analysis
Study-specific ORs (95% CI) were extracted for the risk of invasive breast cancer overall and by ER status among the Caucasian participants of the five cohorts previously involved in a pooling project (i.e., CPS-II=American Cancer Society Cancer Prevention Study II (9), EPIC=European Prospective Investigation into Cancer and Nutrition cohort, NHS=Nurses’ Health Study (8), MEC=Hawaii-Los Angeles Multiethnic Cohort, and WHS=Women’s Health Study) (study-specific ORs and 95% CIs provided in Supplemental Table 2 in reference (12)), as well as those for PBCS and SEARCH. Additional published studies were found in PubMed searches conducted using the key words “HSD17B1” or “17β-hydroxysteroid dehydrogenase 1” and “breast cancer risk”. Four studies (7, 10, 11, 18) were identified that published data on rs605059 and breast cancer risk prior to June 2008; however, we excluded two studies conducted among Asian women (7, 18). Studies, conducted in Germany (referred to as the GENICA=Gene Environment Interaction and Breast Cancer in Germany (19)) and Finland (10), were included in our meta-analysis. ORs (95% CIs) of breast cancer risk overall (risk estimates by ER status were not available) were extracted from these publications. Summary ORs (95% CIs) were calculated using fixed-effect and random-effect models with each study result weighted by the within-study and between-study variance (20). A two-sided p-value <0.05 was deemed statistically significant. The presence of between-study heterogeneity was assessed by the Q test (20). Given the small number of studies included in our meta-analysis, as well as the low power of the aforementioned test (21-23), a two-sided p-value of <0.10 was considered statistically significant and that of <0.20, borderline significant. Statistical analyses were performed in STATA10 (StataCorp, College Station, TX).
Results
Description of Study Populations
The average age of the PBCS controls under study was 55.7 (± 10.0) years and all were Caucasian. The SEARCH controls were slightly older (mean age=57.8 ± 10.7) and >98% were self-reported Caucasian. The SEARCH cases tended to be diagnosed with ER+ tumors (79.1%), while slightly less PBCS cases were diagnosed with ER+ tumors (65.1%). The Polish cases were on average older (55.8 ± 10.0 years) than the SEARCH cases (51.7 ± 8.7 years).
Although rs598126 and rs2010750 were also genotyped in PBCS, only results for rs605059 are presented as results were similar for these three htSNPs given the high correlation between them in Polish controls [rs605059-rs598126 (r2 =0.98), rs605059-rs2010750 (r2 =0.80), and rs2010750-rs598126 (r2 =0.82)]. The minor allele frequencies in controls were 22% in PBCS and 27% in SEARCH for rs676387 and 48% and 44%, respectively, for rs605059.
Main effect associations between HSD17B1 htSNPs and breast cancer risk
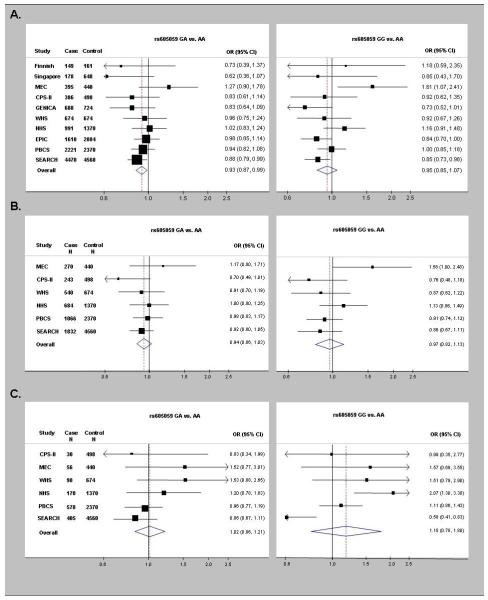
Compared with the AA genotype, the GA and GG genotypes of rs605059 were associated with weak decreases in breast cancer risk in SEARCH (OR=0.88, 95% CI 0.79-0.99 and OR=0.85, 95% CI 0.73-0.98, respectively). However, neither genotype was associated with risk in PBCS (OR=0.94, 95% CI 0.82-1.08 and OR=1.00, 95% CI 0.85-1.18, respectively) (Table 1). The summary ORs (95% CIs), based on 11,762 cases and 14,329 controls from 10 studies, were 0.93 (0.87-0.99) for the GA genotype and 0.96 (0.85-1.08) for the GG genotype (Figure 1 and Supplemental Table 1). Significant between-study heterogeneity was detected for the summary ORs for the GG genotype (p-value for between-study heterogeneity=<0.0001), but not for the AG genotype (p-value for between-study heterogeneity=0.02). In models restricted to the 6 studies (i.e., MEC, CPS-II, WHS, NHS, PBCS, and SEARCH) included in the ER-specific breast cancer risk analyses below (i.e., 5,982 invasive Caucasian cases and 9,912 Caucasian controls), the summary OR (95% CI) for rs605059 GG genotype was 0.98 (0.86-1.11) with evidence of significant between-study heterogeneity (p-value=0.04), similar to the findings based on all studies (Figure 1 and Supplemental Table 1).
Table 1. Age- and study-adjusted odds ratios (OR) and 95% confidence intervals (CI) for the associations between HSD17B1 htSNPs (rs605059 and rs676387) and breast cancer, overall and stratified by estrogen receptor (ER) expression, from 2 independent studies.
| SNP | Cases | Controls | Cases vs. Controls | ER+ Cases | ER+ vs. Controls | ER− Cases | ER− vs. Controls | p- value2 |
|---|---|---|---|---|---|---|---|---|
| N1 | N1 | OR (95% CI) | N1 | OR (95% CI) | N1 | OR (95% CI) | ||
| rs6050593 (Ex6+220 G>A) | ||||||||
| Polish Breast Cancer Study (PBCS) 4 | ||||||||
| AA | 622 | 641 | 1.00 | 302 | 1.00 | 160 | 1.00 | |
| AG | 1,046 | 1,155 | 0.94 (0.82, 1.08) | 537 | 0.98 (0.83, 1.17) | 274 | 0.96 (0.77, 1.19) | 0.75 |
| GG | 510 | 527 | 1.00 (0.85, 1.18) | 227 | 0.91 (0.74, 1.12) | 144 | 1.11 (0.86, 1.43) | 0.18 |
| SEARCH Study 5 | ||||||||
| AA | 1,444 | 1,417 | 1.00 | 596 | 1.00 | 145 | 1.00 | |
| AG | 2,074 | 2,168 | 0.88 (0.79, 0.99) | 902 | 0.92 (0.80, 1.05) | 207 | 0.86 (0.67, 1.11) | 0.57 |
| GG | 800 | 880 | 0.85 (0.73, 0.98) | 334 | 0.85 (0.71, 1.01) | 53 | 0.58 (0.41, 0.83) | 0.01 |
| rs676387 (IVS4-150 C>A) | ||||||||
| Polish Breast Cancer Study (PBCS) 4 | ||||||||
| CC | 1,305 | 1,442 | 1.00 | 613 | 1.00 | 363 | 1.00 | |
| CA | 780 | 805 | 1.07 (0.95, 1.21) | 413 | 1.21 (1.04, 1.41) | 181 | 0.89 (0.73, 1.09) | 0.01 |
| AA | 136 | 123 | 1.22 (0.94, 1.57) | 62 | 1.19 (0.86, 1.63) | 42 | 1.34 (0.92, 1.93) | 0.56 |
| SEARCH Study 5 | ||||||||
| CC | 2,225 | 2,407 | 1.00 | 944 | 1.00 | 187 | 1.00 | |
| CA | 1,765 | 1,787 | 1.11 (1.00, 1.24) | 763 | 1.13 (1.00, 1.29) | 176 | 1.34 (1.06, 1.70) | 0.58 |
| AA | 372 | 352 | 1.16 (0.96, 1.40) | 142 | 1.04 (0.82, 1.31) | 44 | 1.53 (1.03, 2.27) | 0.16 |
Ns may not add up to total because of missing genotype data
p-value for case heterogeneity by ER status
In PBCS (1,995 cases, 2,296 controls), results for rs598126 and rs2010750 were similar to those for rs605059 because the SNPs were highly correlated (r2>0.8)
The PBCS includes 1,995 case and 2,296 controls. Of 1,677 cases with ER data, 1,089 had ER-positive tumors and 588 had ER-negative tumors
The SEARCH study includes 4,470 cases and 4,560 controls. Of 2,256 cases with ER data, 1.849 had ER-positive tumors and 407 had ER-negative tumors.
Figure 1.
Forest plots of study-specific odds ratios (OR) and 95% confidence intervals (CI) for the risk of breast cancer overall (A) and for risk of estrogen receptor (ER)-positive breast tumors (B) and risk of ER-negative breast tumors (C) associated with genotypes of rs605059. Studies are weighted and ranked according to the inverse of the variation of the log OR. The size of the boxes indicates the variance of the log OR. The solid line represents an OR of one and the dotted line represents the summary OR of all studies. The acronyms stand for the study names: CPS-II=American Cancer Society Cancer Prevention Study II, EPIC=European Prospective Investigation into Cancer and Nutrition cohort, GENICA=Gene Environment Interaction and Breast Cancer in Germany, NHS=Nurses’ Health Study, MEC=Hawaii-Los Angeles Multiethnic Cohort, Poland=Polish Breast Cancer Study, SEARCH=Studies of Epidemiology and Risk Factors in Cancer Heredity, and WHS=Women’s Health Study.
Overall, breast cancer risk for rs676387 was slightly elevated for AA genotype, compared to the CC genotype, in both PBCS (OR=1.22, 95% CI 0.94-1.57) and SEARCH (OR=1.16, 95% CI 0.96-1.40), although neither OR was statistically significant. The summary OR (95% CI), based on 11,074 cases and 13,605 controls in 7 studies, for the AA genotypes, compared to the CC genotypes, was 1.12 (95% CI 0.99-1.27; p-value for between-study heterogeneity=0.30).
Associations between HSD17B1 htSNPs and ER-specific breast cancer risk
While there was no significant difference in OR estimates for rs605059 in the PBCS between ER-positive and ER-negative breast tumors (p-values for case heterogeneity=0.75 for AG vs. AA, and 0.18 for homozygote GG vs. AA; Table 1), SEARCH results suggested a stronger decreased risk of the GG genotype for ER-negative (OR=0.58, 95% CI 0.41-0.83) than ER-positive (OR=0.85, 95% CI 0.71-1.01; p-value for case heterogeneity=0.01; Table 1) tumors. For our meta-analysis, ER data were available for cases from 6 studies (i.e., MEC, CPS-II, WHS, NHS, PBCS, and SEARCH), including 4,635 ER-positive cases and 1,347 ER-negative cases. No significant associations were found between rs605059 and ER-positive breast cancers (Figure 1 and Supplemental Table 1): summary ORs (95% CIs) were 0.94 (0.86-1.03) for the GA genotype and 0.97 (0.84-1.11) for the AA genotype with no evidence of between-study heterogeneity (p-values=0.48 and 0.16, respectively). There was also no significant association between the GG genotype of rs605059 and risk of ER-negative tumors (OR=1.18, 95% CI 0.78-1.80), but these ORs were not consistent across studies (p-value for between-study heterogeneity=0.001). To determine whether an individual study was the source of between-study heterogeneity, we systematically dropped each study and recalculated the summary OR for the GG genotype of rs605059. Exclusion of each study with the exception of SEARCH did not affect the test of between-study heterogeneity nor the random effect estimates. When we excluded SEARCH, the summary OR (95% CI) for the GG genotype of rs605059 was 1.39 (1.04-1.87) and p-value for the test of between-study heterogeneity was 0.20.
In PBCS and SEARCH, no significant difference between risk of ER-positive breast cancer and risk of ER-negative breast cancer was found for the AA genotype of rs676387 (p-values=0.56 and 0.16, respectively). However, risk of ER-negative tumors were elevated for rs676387 AA genotype in PBCS (OR=1.34, 95% CI 0.92-1.93) and SEARCH (OR=1.53, 95% CI 1.03-2.27). These findings were not consistent with summary OR (95% CI), based on 1,347 ER-negative cases and 9,912 controls that suggested no association (OR=1.02, 95% CI 0.70-1.50) between the AA genotype of rs676387 and risk of ER-negative tumors, although there was weak evidence of between-study heterogeneity in this estimate (p-value for between-study heterogeneity=0.11). None of the individual studies found an association between rs676387 AA genotype and ER-positive breast cancer risk (p-value for between-study heterogeneity=0.95); the summary OR (95% CI) was 1.02 (0.70-1.50).
Interactions by menopausal status and age
Menopausal status did not modify any of the genotype associations in the PBCS (p for interactions > 0.42; no additional data shown). Data on menopausal status was not available for SEARCH; therefore, we used age at baseline as a proxy of menopausal status. We also did not observe modification by age of the association between breast cancer risk and rs676387 or rs605059 (Supplemental Table 3).
Discussion
Data from two independent studies in Poland and the UK totaling 6,465 cases and 6,856 controls showed weak to null associations with overall breast cancer risk for two SNPs (rs605059 or rs676387) that captured most of the known common variation in HSB17B1. Meta-analysis results including data from these two studies and previously published data (up to 11,762 cases and 14,329 controls in total) were consistent with these findings.
Previous analyses of 2,091 cases and 2,982 controls in 5 prospective cohort studies found ER-negative tumors (n=354) were negatively associated with the rs676387 AA genotype and positively associated with the rs605059 GG genotype (12). However, data from the Polish (n=586 ER-negative cases) and British (n=407 ER-negative cases) studies did not support these findings, and meta-analyses including previously published data demonstrated significant evidence for heterogeneous relative risk estimates for ER-negative disease across studies. Factors that could explain the observed heterogeneity include unidentified biases, a true effect modified by a factor with varying prevalences across study populations, or chance. All of the study participants were Caucasian and had similar minor allele frequencies, therefore ethnic differences are unlikely to explain the observed heterogeneity. Furthermore, the differences in main effect estimates across studies were minimal, and confounding by population stratification is unlikely to more strongly affect subgroup analyses, such as those for ER-negative tumors.
Studies differed in the distribution of age and menopausal status: study subjects in the previous report (12) were all postmenopausal women, while PBCS and SEARCH included both premenopausal and postmenopausal women. Differences in study population characteristics may explain between-study heterogeneity. However, we did not observe evidence of modification of genotype associations by menopausal status in PBCS or by age at baseline in SEARCH or PBCS. Thus, differences in age distributions between studies are unlikely to appreciably contribute to disparate results across studies.
Although it is unlikely that genotype is related to selection or participation in case-control or cohort studies, other study-specific biases may influence observed relative risks. Bias may have been introduced by the restriction of cases to those with information on hormone receptor status. In PBCS and SEARCH, genotype frequencies were similar for cases with and without ER data. In the previous report (12), one of the largest studies (1,610 cases, 2,884 controls) included in analyses of the association between genotypes and overall breast cancer risk was not included in the analyses by hormone receptor data. Interestingly, estimates from this study showed an overall 33% risk increase for rs676387 (12), which is inconsistent with the lower risk of ER-negative tumors and lack of association with ER-positive tumors observed in the other cohorts (12). If individual studies had specific biases, it is unlikely that they would be in the same direction for all studies; this may explain some of the heterogeneity across studies.
In conclusion, it does not appear that common genetic variation in HSD17B1 is associated with a moderate modulation in breast cancer risk overall; however, we cannot exclude the possibility of a very weak effect. Additional data for HSD17B1 polymorphisms and breast cancer risk from studies with well characterized tumors is needed to clarify the findings for ER-negative tumors.
Supplementary Material
Acknowledgements
The authors are grateful to Drs. Witold Zatonski, and Neonila Szeszenia-Dabrowska for their contributions to the Polish Breast Cancer study, and to Shahana Ahmedand and the entire SEARCH breast study team. The Polish study is funded by Intramural Research Program of the National Cancer Institute, NIH, Department of Health and Human Services, USA. SEARCH is funded by a grant from Cancer Research UK.
References
- 1.Thompson PA, Ambrosone C. Molecular epidemiology of genetic polymorphisms in estrogen metabolizing enzymes in human breast cancer. J Natl Cancer Inst Monogr. 2000;(27):125–34. doi: 10.1093/oxfordjournals.jncimonographs.a024235. [DOI] [PubMed] [Google Scholar]
- 2.Vihko P, Harkonen P, Soronen P, et al. 17 beta-hydroxysteroid dehydrogenases--their role in pathophysiology. Mol Cell Endocrinol. 2004;215(1-2):83–8. doi: 10.1016/j.mce.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 3.Miettinen M, Mustonen M, Poutanen M, et al. 17Beta-hydroxysteroid dehydrogenases in normal human mammary epithelial cells and breast tissue. Breast Cancer Res Treat. 1999;57(2):175–82. doi: 10.1023/a:1006217400137. [DOI] [PubMed] [Google Scholar]
- 4.Jansson A, Gunnarsson C, Stal O. Proliferative responses to altered 17{beta}-hydroxysteroid dehydrogenase (17HSD) type 2 expression in human breast cancer cells are dependent on endogenous expression of 17HSD type 1 and the oestradiol receptors. Endocr Relat Cancer. 2006;13(3):875–84. doi: 10.1677/erc.1.01181. [DOI] [PubMed] [Google Scholar]
- 5.Gunnarsson C, Ahnstrom M, Kirschner K, et al. Amplification of HSD17B1 and ERBB2 in primary breast cancer. Oncogene. 2003;22(1):34–40. doi: 10.1038/sj.onc.1206078. [DOI] [PubMed] [Google Scholar]
- 6.Oduwole OO, Li Y, Isomaa VV, et al. 17beta-hydroxysteroid dehydrogenase type 1 is an independent prognostic marker in breast cancer. Cancer Res. 2004;64(20):7604–9. doi: 10.1158/0008-5472.CAN-04-0446. [DOI] [PubMed] [Google Scholar]
- 7.Wu AH, Seow A, Arakawa K, Van Den Berg D, Lee HP, Yu MC. HSD17B1 and CYP17 polymorphisms and breast cancer risk among Chinese women in Singapore. Int J Cancer. 2003;104(4):450–7. doi: 10.1002/ijc.10957. [DOI] [PubMed] [Google Scholar]
- 8.Setiawan VW, Hankinson SE, Colditz GA, Hunter DJ, De Vivo I. HSD17B1 gene polymorphisms and risk of endometrial and breast cancer. Cancer Epidemiol Biomarkers Prev. 2004;13(2):213–9. doi: 10.1158/1055-9965.epi-03-0241. [DOI] [PubMed] [Google Scholar]
- 9.Feigelson HS, McKean-Cowdin R, Coetzee GA, Stram DO, Kolonel LN, Henderson BE. Building a multigenic model of breast cancer susceptibility: CYP17 and HSD17B1 are two important candidates. Cancer Res. 2001;61(2):785–9. [PubMed] [Google Scholar]
- 10.Mannermaa A, Peltoketo H, Winqvist R, et al. Human familial and sporadic breast cancer: analysis of the coding regions of the 17 beta-hydroxysteroid dehydrogenase 2 gene (EDH17B2) using a single-strand conformation polymorphism assay. Hum Genet. 1994;93(3):319–24. doi: 10.1007/BF00212030. [DOI] [PubMed] [Google Scholar]
- 11.Justenhoven C, Hamann U, Schubert F, et al. Breast cancer: a candidate gene approach across the estrogen metabolic pathway. Breast Cancer Res Treat. 2008;108(1):137–49. doi: 10.1007/s10549-007-9586-8. [DOI] [PubMed] [Google Scholar]
- 12.Feigelson HS, Cox DG, Cann HM, et al. Haplotype analysis of the HSD17B1 gene and risk of breast cancer: a comprehensive approach to multicenter analyses of prospective cohort studies. Cancer Res. 2006;66(4):2468–75. doi: 10.1158/0008-5472.CAN-05-3574. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Closas M, Brinton LA, Lissowska J, et al. Established breast cancer risk factors by clinically important tumour characteristics. Br J Cancer. 2006;95(1):123–9. doi: 10.1038/sj.bjc.6603207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cebrian A, Pharoah PD, Ahmed S, et al. Tagging single-nucleotide polymorphisms in antioxidant defense enzymes and susceptibility to breast cancer. Cancer Res. 2006;66(2):1225–33. doi: 10.1158/0008-5472.CAN-05-1857. [DOI] [PubMed] [Google Scholar]
- 15.The National Cancer Institute Breast and Prostate Cancer Cohort Consortium A candidate gene approach to searching for low-penetrance breast and prostate cancer genes. Nat Rev Cancer. 2005;5(12):977–85. doi: 10.1038/nrc1754. [DOI] [PubMed] [Google Scholar]
- 16.Kraft P, Pharoah P, Chanock SJ, et al. Genetic variation in the HSD17B1 gene and risk of prostate cancer. PLoS Genet. 2005;1(5):e68. doi: 10.1371/journal.pgen.0010068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Packer BR, Yeager M, Burdett L, et al. SNP500Cancer: a public resource for sequence validation, assay development, and frequency analysis for genetic variation in candidate genes. Nucleic Acids Res. 2006;34:D617–21. doi: 10.1093/nar/gkj151. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakoda LC, Blackston C, Doherty JA, et al. Polymorphisms in steroid hormone biosynthesis genes and risk of breast cancer and fibrocystic breast conditions in chinese women. Cancer Epidemiol Biomarkers Prev. 2008;17(5):1066–73. doi: 10.1158/1055-9965.EPI-07-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Justenhoven C, Hamann U, Schubert F, et al. Breast cancer: a candidate gene approach across the estrogen metabolic pathway. Breast Cancer Res Treat. 2007 doi: 10.1007/s10549-007-9586-8. [DOI] [PubMed] [Google Scholar]
- 20.Laird NM, Mosteller F. Some statistical methods for combining experimental results. Int J Technol Assess Health Care. 1990;6(1):5–30. doi: 10.1017/s0266462300008916. [DOI] [PubMed] [Google Scholar]
- 21.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–101. [PubMed] [Google Scholar]
- 22.Poole C, Greenland S. Random-effects meta-analyses are not always conservative. Am J Epidemiol. 1999;150(5):469–75. doi: 10.1093/oxfordjournals.aje.a010035. [DOI] [PubMed] [Google Scholar]
- 23.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Bmj. 1997;315(7109):629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.