Abstract
Parasitic helminth neuromuscular function is a proven target for chemotherapeutic control. Although neuropeptide signaling plays a key role in helminth motor function, it has not yet provided targets for known anthelmintics. The majority of biologically active neuropeptides display a C-terminal amide (NH2) motif, generated exclusively by the sequential action of two enzymes, peptidylglycine α-hydroxylating monooxygenase (PHM) and peptidylglycine α-amidating lyase (PAL). Further to our previous description of a monofunctional PHM enzyme (SmPHM) from the human blood fluke Schistosoma mansoni, here we describe a cDNA encoding S. mansoni PAL (SmPAL). SmPAL is a monofunctional enzyme which, following heterologous expression, we find to have functionally similar catalytic activity and optimal pH values, but key catalytic core amino acid substitutions, when compared to other known PALs including those found in humans. We have used in situ hybridization to demonstrate that in adult schistosomes, SmPAL mRNA (Sm-pal-1) is expressed in neuronal cell bodies of the central nervous system, consistent with a role for amidated neuropeptides in S. mansoni neuromuscular function. In order to validate SmPAL as a putative drug target we applied published RNA interference (RNAi) methods in efforts to trigger knockdown of Sm-pal-1 transcript in larval schistosomula. Although transcript knock-down was recorded on several occasions, silencing was variable and inconsistent and did not associate with any observable aberrant phenotype. The inconsistent outcomes of RNAi suggest that there may be tissue-specific differences in the applicability of RNAi methods for S. mansoni, with neuronal targets proving more difficult or refractory to knockdown. The key role played by schistosome amidating enzymes in neuropeptide maturation make them appealing as drug targets; their validation as such will depend on the development of more robust reverse genetic tools to facilitate efficient neuronal gene function studies.
Keywords: Peptidylglycine α- amidating lyase, Neuropeptide amidation, Schistosoma mansoni, In-situ hybridisation, RNA interference
1. Introduction
Diseases caused by helminth parasites remain one of the most significant public health problems facing society. The most prevalent and chronic flatworm infection of humans, resulting in a major, poverty-related problem in the developing world is schistosomiasis. Schistosoma mansoni, designated a Neglected Tropical Disease, is a major cause of human schistosomiasis, having a global prevalence of over 200 million in an estimated 74 developing countries [1]. The difficult situation facing health organisations involved in the reduction of morbidity due to schistosomiasis is aggravated by over-reliance on a single drug, praziquantel (PZQ). Although resistance does not appear to be widespread, it has been generated in the laboratory [2], and has occurred transiently in field situations [3, 4]. These observations, coupled with increasing distribution and usage of PZQ, provide circumstances known to foster the development of drug resistance. As a result of this dependence and the consequent risk of inadvertently selecting for drug-resistant parasites, there is a pressing need to exploit and develop new drug targets and to develop novel chemotherapeutic agents for the treatment and control of schistosomiasis [5].
The neuropeptidergic system is central to flatworm motor control - neuropeptide signalling molecules appear to be involved in the modulation of a range of biological processes throughout the phylum Platyhelminthes [6–11]. So far, we have identified 18 distinct amidated peptides in S. mansoni [12, 13], several of which have been localised to the CNS and PNS of larval and adult schistosomes [13–19]. Some of these peptides have also been shown to exert potent myoexcitatory effects on isolated muscle fibres from adult schistosomes, and as such are thought to be directly involved in muscle function [20]. Consequently, the neuropeptidergic component of the S. mansoni nervous system represents an attractive repository of novel drug targets.
The majority of neuropeptides require a carboxy-terminal amide motif to confer biological activity, as demonstrated by the reduced activity of most non-amidated glycine-extended precursors (<10 %) in comparison to their α-amidated counterparts [21–24]. Two key enzymes act sequentially in the only known mechanism for secretory peptide amidation: peptidylglycine α-hydroxylating monooxygenase (PHM), a copper, oxygen and ascorbate dependant enzyme which catalyses hydroxylation of the C-terminal glycine present on a peptide precursor; and peptidylglycine α-amidating lyase (PAL), a zinc dependant enzyme which catalyses dealkylation of the hydroxylated intermediate to form the α-amidated neuropeptide and glyoxylate [21, 23]. This process occurs throughout the Metazoa, the importance of which is demonstrated in larval Drosophila melanogaster mutants, where the deletion of the PHM gene results in embryonic death [25]. In higher organisms PHM and PAL are commonly encoded by the same transcript, generating a bifunctional protein designated peptidylglycine α-amidating monooxygenase (PAM) [24]. In stark contrast, the genomes of a number of invertebrates including the flatworm Dugesia japonica, encode single copies of monofunctional PHM and PAL [26], while others possess multiple monofunctional enzymes which can be membrane associated, soluble or even inactive [24, 27]. Crucially, adult S. mansoni express monofunctional PHM (SmPHM), which is functionally divergent from the mammalian homologue [28]. These organisational and functional differences between the schistosome and mammalian amidating enzymes encourage the characterisation of schistosome amidating enzymes and their exploration as potential drug targets.
Even though neuropeptide signalling is a core component of schistosome neural function and neuromuscular modulation, it has not been a major focus for novel drug target discovery and validation in schistosomes, largely hindered by the absence of known neuropeptides and their cognate receptors. Recent proliferations in EST datasets and publication of the S. mansoni genome sequence [29] have fuelled the recent discovery of SmPHM and eighteen schistosome neuropeptides, making neuropeptide signalling ripe for more detailed interrogation and validation as a source of suitable targets for schistosome control [13, 28, 29]. Further, genome level bioinformatics has uncovered a set of at least 24 putative schistosome neuropeptide receptors [29], which are appealing because of the proven druggability of many known G-protein coupled receptors (GPCRs) and their suitability for high throughput screens. We hypothesise that neuropeptide activation/processing provides an alternative and potentially powerful set of drug targets. Since the combined activities of PHM and PAL are the only known conduit to neuropeptide amidation/activation and we already know schistosomes possess multiple amidated peptide messengers, these enzymes have much appeal as drug target candidates. Previously we demonstrated that one half of the neuropeptide amidating pathway in S. mansoni is functionally distinct from that seen in vertebrates and, indeed, other invertebrates, such that we now switch focus to the second half of the amidation pathway, PAL. Here we report the identification, localization, cloning and functional characterisation of a S. mansoni cDNA encoding a novel, monofunctional PAL enzyme. Further, we attempt to validate its candidature as a drug target through the application of RNA interference (RNAi).
2. Materials and methods
2.1. Schistosoma mansoni culture
Schistosome infected mice supplied by Dr Fred Lewis, Biomedical Research Institute, Rockville MD, USA, were maintained and processed at Iowa State University (ISU). Adult schistosomes recovered from infected mice were either processed for whole-mount in situ hybridisation (WISH) experiments as previously described [30], or stored in RNAlater to allow subsequent RNA extraction, before being shipped to Queen’s University Belfast (QUB) on dry ice. Schistosomes used in the RNA interference (RNAi) experiments were maintained at QUB in Biomphalaria glabrata snails; cercariae were shed from infected snails by photostimulus, and mechanically transformed to schistosomula by vortexing, schistosome bodies were isolated from tails by centrifugation over a solution of 30% Percoll in water (500 g for 15 min at 4°C), protocol adapted from [53]. Schistosomula were maintained in vitro in complete RPMI media [31] containing 20% serum, at 37°C in a 5% CO2 atmosphere.
2.2. Bioinformatics
BLAST methodology [32] was employed to uncover novel putative PAL encoding transcripts from S. mansoni EST and genomic datasets. BLASTn and tBLASTn tools were used at the National Centre for Biotechnology Information (NCBI) BLAST server (http://blast.ncbi.nlm.nih.gov/Blast) as well as the Schistosoma mansoni genome database, SmGeneDB (http://www.genedb.org/genedb/smansoni). Searches were performed on the ‘est_others’ database, had an expect value of >1000, and were limited by the query ‘Schistosoma mansoni’. Known PAL genes [25] were used as query sequences. Returns were translated in all six reading frames (http://www.expasy.org/tools/dna.html), and examined for both the presence of PAL specific motifs and similarity to other PAL genes/proteins (http://www.ebi.ac.uk/Tools/InterProScan) [33]. Sequences were also analysed for the presence of an N-terminal secretory signal peptide, and putative N-glycosylation sites, using the online SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) [34], and NetNGlyc servers (http://www.cbs.dtu.dk/services/NetNGlyc/) respectively.
2.3. RNA extraction and PCR analyses
Messenger RNA was extracted from between 10–50 mg of S. mansoni tissue using Dynabeads mRNA Direct™ kit (Invitrogen) according to the manufacturer’s instructions. mRNA eluted in Tris-HCl was quantified using a NanoDrop™ 1000 spectrophotometer. Separate populations of 5′ and 3′ RACE-ready cDNAs were generated using the SMART™ RACE cDNA Amplification kit (Clontech/Takara), using ≥1 μg mRNA per synthesis according to manufacturer’s instructions. RACE cDNA was stored at −20°C until use.
Gene specific primers (GSP) designed against the putative S. mansoni PAL (SmPAL) ESTs (GenBank accession numbers AM047261 and AM043268) were used in 50 μl PCR reactions (94°C for 2 min; 40 cycles of : 94°C 1 min, 55°C 1 min, and 72°C 1 min; 72°C for 7 min) to confirm the presence of SmPAL in the following reaction mixture: 5 μl 10x PCR buffer (Invitrogen), 3 μl MgCl2 (50 mM, Invitrogen), 1 μl dNTP mix (10 mM, Promega), 1 μl of primer SmPAL-F1 and SmPAL-R1 (20 μM; Table 1), 1 μl cDNA template, 0.3 μl Platinum® Taq DNA Polymerase (5 U/μl, Invitrogen), and ddH2O to 50 μl. Reaction products were TOPO-TA cloned (Invitrogen) and at least 3 plasmids were sequence verified.
Table 1.
Primer sequences
| Primer Name | Primer sequence 5′-3′ | |
|---|---|---|
| SmPAL-F1 | CGGCACGAGGGAACCAT CATACC | |
| SmPAL-R1 | ACTACT ACGTCCCGTTGGT TTAC | |
| SmPAL-Xba1-F | TCTAGAAT GTACTTTACATTAATAATC | |
| SmPAL-Bgl2-R | AGATCTATAATCGTATTCAA AGTTAAAACG | |
| SmPAL-Nhe1-F | GCTAGCATCGTAT ATGATTTCC | |
| SmPAL-Hpa1-R | GAATTCGTTAACTCACG CAGG | |
| SmPAL-ISH-T7-F1 | TAATACGACTCACTATAGGGCATGTATCGTATATGATTTCC | |
| SmPAL-ISH-R1 | GAGATTTAAATCATATGGTCC | |
| SmPAL-ISH-F2 | CATGTATCGTATATGATTTCC | |
| SmPAL-ISH-T7-R2 | TAATACGACTCACTAT AGGGGAGATTTAAATCATATGGTCC | |
| SmPAL-RNAi-T7-F1 | TAATAC GACTCACTATAGGGCATGTATCGTATATGATTTCC | |
| SmPAL-RNAi-R1 | GTATTT TCATCCCATACTCG | |
| SmPAL-RNAi-F2 | CATGTATCGTATA TGATTTC | |
| SmPAL-RNAi-T7-R2 | TAATACGACTCACTATAGGGGTATTTTCATCCC ATACTCG | |
| SmPHM-RNAi-T7-F1 | TAATACGACTCACTATAGGGCGTATTCCAGGAGTGACAAC | |
| SmPHM-RNAi-R1 | GACTCAGATTCACGACATATAG | |
| SmPHM-RNAi-F2 | CGTATTCCAGGAGTGACAAC | |
| SmPHM-RNAi-T7-R2 | TAATACGACTCACTATAGGGGACTCAGATTCACGACATATAG | |
| SmPAL-RT S1 | GCTTTAAACCTGGTTCAAGC | 1322 bp RT-PCR product |
| SmPAL-RT AS1 | ATAATCGTATTCAAAGTTAAAACG | |
| SmAP-RT S1 | GCCATCCGACAAGGAATATAAGTGT | 98 bp RT-PCR product |
| SmAP-RT AS1 | GGTCCATTGAAAAAGGAGGATATGAGA | |
T7 RNA polymerase promoter sequences are underlined
2.4. Functional SmPAL expression
The SmPAL open reading frame (1236 bp) was PCR-amplified using sense primer SmPAL-Xba1-F and antisense primer SmPAL-Bgl2-R (see Table 1) incorporating 5′ Xba1 and Bgl2 restriction sites respectively. Products were TOPO-TA cloned (Invitrogen) and sequence verified. Using Xba1, Bgl2, and pBS.rhodopsin, the rhodopsin tag (11 amino acids) was appended to the C-terminus of SmPAL. pCIS.SmPAL-rhod was then created using Xba1 and Hpa1. pBS.SmPAL-rhod was used as a template to PCR-amplify SmPAL-rhod without the native S. mansoni signal peptide (aa 1–18) using primers SmPAL-Nhe1-F and SmPAL-Hpa1-R (see Table 1). The SmPAL-rhod reaction product was placed after the rat PAM signal peptide and a poly histidine tag in pCIS.sig-His6 yielding pCIS.sig-His6-SmPAL-rhod. pCIS.SmPAL-rhod was used in functional expression assays.
2.5. Cell culture, transfection, and PAL enzyme assay
Functional expression and characterisation of SmPAL enzymatic activity were carried out as described [35]. SmPAL, a control vector encoding cytosolic Enhanced Green Fluorescent Protein (EGFP; pEGFP-N2, Clontech, Palo Alto, CA, USA), and Drosophila PAL (dPAL2) [35] were expressed in pEAK RAPID cells (a derivative of hEK-293 cells, Edge Biosystems, Gaithersburg, MD, USA). Cells were transiently transfected using Lipofectamine 2000 (2 μg/30 mm well). 24 h after transfection, medium was replaced with serum-free medium, and the cells were incubated for a further 24 h before cells and spent media were harvested for analysis.
Cell extracts and spent medium were assayed for catalytic activity and Western blots were carried out to evaluate secretion efficiency; protease inhibitor cocktail and phenylmethanesulfonylfluoride (PMSF) were added to cell extracts and to spent medium prior to analysis. For comparison, cells and spent medium containing expressed dPAL2 [34] were analysed under the same experimental conditions as SmPAL. pEAK RAPID cells expressing SmPAL were extracted in a low ionic strength buffer with detergent and protease inhibitors. Spent medium from pEAK RAPID cells was centrifuged to remove debris. For Western blot analysis, media were denatured by boiling in Laemmli sample buffer. For enzyme assays, media were diluted into assay diluents (20 mM Na TES (pH 7.4), 10 mM mannitol, 1.0 mg/mL bovine serum albumin, and1% Triton X-100) [39].
Aliquots of cell extract and spent medium were fractionated on 4–15% polyacrylamide gels (Invitrogen), transferred to PVDF membranes and subjected to Western blot analysis using a monoclonal rhodopsin antibody. Secreted, rhodopsin tagged SmPAL was assessed for catalytic activity, using 0.5 μM αN-acetyl-Tyr-Val-α-hydroxyglycine (mixed with a trace amount of [125I]-labelled peptide), prepared using recombinant PHM. SmPAL was diluted in 20 mM Na TES (pH 7.4, 100mM Tris, 100mM EDTA and 2% SES), 10 mM mannitol, 1.0 mg/ml bovine serum albumin, and 1% Triton X-100 (Pierce). The reaction volume (40 μl) typically contained 0.4 μl of spent medium (diluted to 5 μl), 100–150 mM Na MES (pH 4.5), 1 mM CdCl2 and 0.05% Thesit (Boehringer Mannheim). The reaction mixture was incubated for 30 min to 1 h at 37 °C, following which the substrate and product were separated using ethyl acetate phase separation [39]. SmPAL dependence upon pH was analysed using the catalytic activity assay conditions described above and altering pH using 150 mM Na MES buffers that ranged in pH from 3.0 to 7.5. In all activity assays and Western blots, pEGFP-transfected cells were used as a blank.
2.6. Sm-pal-1 probe generation and WISH
Sm-pal-1 WISH probe templates were generated by PCR (94 °C 2 min; 40 cycles of: 94 °C 1 min, 55 °C 1 min, 72 °C 1 min; 72 °C for 7 min) using GSPs flanked by T7 RNA polymerase promoter sequences (sense labelled T7 template: sense primer SmPAL-ISH-T7-F1 and antisense primer SmPAL-ISH-R1; antisense T7 labelled template: sense primer SmPAL-ISH-F2 and antisense primer SmPAL-ISH-T7-R2; see Table 1) in a 50 μl reaction mixture as described above. Probe templates were cloned into pCR2.1-TOPO and at least 3 plasmids were sequence verified. Sm-pal-1 digoxigenin (DIG)-labelled single stranded RNA (ssRNA) probes with sense and antisense polarity were generated from their cDNA templates, and WISH was carried out according to the methods described [30, 36]. Hybridised probes were detected with substrate (5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium tablet; BCIP/NBT, Sigma-Aldrich) [30]. Specimens were photographed using a Leica DFC300FX camera and Leica FW4000 V 1.2 software with a Leica DMR light microscope.
2.7. dsRNA generation
A 226 bp fragment of Sm-pal-1 was generated by PCR (94 °C 2 min; 40 cycles of: 94 °C 1 min, 55 °C 1 min, 72 °C 1 min; 72 °C for 7 min) using GSP flanked by T7 RNA polymerase promoter sequences (sense T7 labelled template: sense primer SmPAL-RNAi-T7-F1 and antisense primer SmPAL-RNAi-R1; antisense T7 labelled template: sense primer SmPAL-RNAi-F2 and antisense primer SmPAL-RNAi-T7-R2; see Table 1), in a 50 μl reaction mixture as described above. In addition a 224 bp fragment of the SmPHM transcript [28] was amplified by PCR (94 °C 2 min; 40 cycles of: 94 °C 1 min, 55 °C 1 min, 72 °C 1 min; 72 °C for 7 min) using GSP flanked by T7 RNA polymerase promoter sequences (sense T7 labelled template: sense primer SmPHM-RNAi-T7-F1 and antisense primer SmPHM-RNAi-R1; antisense T7 labelled template: sense primer SmPHM-RNAi-F2 and antisense primer SmPHM-RNAi-T7-R2; see Table 1), in a 50 μl reaction mixture. Sm-phm-1 dsRNA was used in Sm-pal-1 RNAi experiments as a non-target control dsRNA. Products were TOPO-TA cloned and at least 3 plasmids were sequence verified. The Sm-pal-1 construct displayed no similarity to any other S. mansoni sequence as shown by BLASTp and tBLASTn searches of schistosome sequences on NCBI BLAST server.
Separate populations of ssRNA were generated from T7 labelled templates with sense or antisense polarity, with the MEGAshortscript™ High Yield Transcription kit (Ambion) at 37 °C for 4 h according to the manufacturer’s instructions. 1 μl of DNase I (New England Biolabs) was incubated with ssRNA populations at 37 °C for 15 min, equimolar amounts of sense and antisense ssRNA were mixed and annealed at 75 °C for 5 min and cooled at room temperature for 30 min to generate dsRNA. 4 μl of RNase If (New England Biolabs) was added to dsRNA and incubated at 37 °C for 20 min to remove any ssRNA. dsRNA was recovered by phenol/chloroform extraction and ethanol precipitation, re-suspended in 100 μl H2O, quantified using the NanoDrop 1000 spectrophotometer, and stored at −80 °C until use.
2.8. dsRNA delivery
Sm-pal-1 and Sm-phm-1 dsRNA delivery by electroporation was carried out as described [37]. 200, 10 day old schistosomula were electroporated in 50 μl of serum-free RPMI culture medium supplemented with 1 mg/ml Sm-pal-1 dsRNA. During each experiment 200 schistosomula were electroporated in serum-free RPMI without the addition of dsRNA (−dsRNA), as a negative control. Schistosomula were observed immediately following electroporation to assess damage, in all cases worms appeared normal following exposure. Schistosomula were cultured overnight in 100 μl complete RPMI culture medium, which was replaced with fresh complete RPMI (100 μl) each day for two days after which gene expression was measured by reverse-transcriptase (RT)-PCR. Schistosomula were observed once a day, throughout the post-dsRNA delivery culture period, for the appearance of any aberrant phenotypes associated with altered movement. dsRNA delivery by soaking was also attempted in this study. Following the 10 day transformation period, approximately 200 schistosomula were transferred into 100 μl of serum-free RPMI supplemented with 0.1 mg/ml Sm-pal-1 and Sm-phm-1 dsRNA. Each day 50 μl of RPMI was replaced with fresh RPMI supplemented with 0.1 mg/ml dsRNA. One batch of schistosomula was cultured for the duration of each soaking experiment in serum-free RPMI in the absence of dsRNA (−dsRNA), as a negative control. Soaking was originally carried out for 7 days; however, a high death rate after 5–6 days forced a shorter soaking time such that experiments were reduced to 5 days. Each day, dsRNA treated and untreated schistosomula were observed visually for the presence of any aberrant phenotypes, including differences in general appearance, morphology and movement (number and speed of contractions). After 5 days gene expression was measured by RT-PCR.
2.9. Reverse-transcriptase PCR (RT-PCR)
mRNA was extracted from schistosomula using Dynabeads mRNA Direct™ kit (Invitrogen) according to the manufacturer’s instructions. Any remaining DNA was removed by DNase digestion using DNase I (New England Biolabs), following which the mRNA was purified using RNeasy Mini Elute Cleanup kit (Qiagen), and quantified using the NanoDrop 1000 spectrophotometer. Specific gene products were amplified from 12 ng of mRNA per RT-PCR reaction using GSP (Table 1) at a final concentration of 0.6 μM, and the QIAGEN OneStep RT-PCR kit. In addition to GSPs designed against the experimental target gene Sm-pal-1, GSPs designed against a 98 bp fragment of the S. mansoni alkaline phosphatase gene [31], were employed in RT-PCR reactions to enable the comparison of the relative expression levels of Sm-pal-1 and Sm-ap. Prior to use in RT-PCR reactions, all primer sets were used in RT-PCR optimisation experiments to ensure that they amplified specific gene products consistently. Each primer set (SmPAL and SmAP) was used in RT-PCR reactions on mRNA extracted from schistosomula treated with Sm-pal-1 dsRNA, Sm-phm-1 dsRNA (non-target control dsRNA) or buffer without dsRNA (negative control). Cycling conditions were as follows: 50 °C for 30 min, 95 °C for 15 min, followed by 40 cycles of 94 °C 30 sec, 50 °C 45 sec, and 72 °C for 2 min, and a final extension step of 72 °C for 10 min. RT-PCR products were visualised by agarose gel electrophoresis (as previously described), images were taken, and transcript abundance quantified relative to standardised DNA size markers using Quantity One 1-D Analysis software (BioRad).
3. Results
3.1. Identification and characterisation of schistosome PAL (SmPAL)
Interrogation of the schistosome EST database with the Drosophila PAL1 sequence (GenBank accession number ACJ13179) identified two ESTs encoding a putative SmPAL (GenBank accession numbers AM047261 and AM043286). Subsequent computational analysis showed that these ESTs could represent a single SmPAL enzyme with significant similarity to the PAL domain of PAM proteins and to monofunctional PAL enzymes. Highest homology (identified via tBLASTn searches) was to the partial ORF of a putative Macrostomum lignano PAL (GenBank accession number EG951302, 34.0 % identity). Primers designed for PCR confirmation of the corresponding full length SmPAL open reading frame generated a 1236 nucleotide cDNA sequence (GenBank accession number FJ668386), encoding a 412 amino acid protein. SmPAL has a predicted molecular mass of 48 kDa, and includes an 18 amino acid (aa) signal peptide [38]. The coding sequence of SmPAL is flanked by a 333 bp 5′ untranslated region (UTR) and a 107 bp 3′ UTR with a polyadenylation signal (AATATA) 25 bp upstream of the poly(A)+ tail. The sequence of SmPAL suggests that adult schistosomes express a gene encoding an active monofunctional PAL enzyme.
A number of amino acids common to all eukaryotic PAL proteins and essential for catalysis are conserved [39–41] (Fig. 1). The six-bladed β-propeller structure of the rat PAL catalytic core (rat PALcc) includes a catalytic Zn2+ and a structural Ca2+. The three His residues that bind the catalytic Zn2+ (His585, His690, His786 in rat PALcc) are conserved, suggesting that SmPAL and rat PAL may bind Zn2+ in a similar manner. In rat PALcc, the Ca2+ is bound by two main chain carbonyls (Val520, Leu587) and a carboxylate (Asp787). While Leu587 is conserved, Val520 is replaced by Ala and Asp787 is replaced by Ser in SmPAL; although the interactions of Ca2+ with the main chain carbonyls would be conserved in SmPAL, its interaction with Ca2+ may be altered. The four cysteine residues that form disulfide bonds positioning key active site residues in rat PALcc are conserved in SmPAL, which has four additional cysteine residues not common to rat PALcc; three of these additional cysteine residues are located in what is predicted to form the sixth β-propeller of SmPAL. All of the key active site residues in rat PALcc (Arg533, Tyr654 and Arg706) are completely conserved [41].
Fig. 1.
ClustalW alignment of schistosome PAL (SmPAL) with the catalytic core of other eukaryotic PAL domains/proteins. Text boxed in yellow denotes perfectly conserved residues, text boxed in blue denotes partially conserved residues. Four conserved cysteine residues are marked by black circles, while four additional SmPAL specific cysteine residues are highlighted in red. Three conserved His residues involved in binding Zn2+ are marked by black triangles. Residues in rat PALcc that bind Ca2+ (Val520, Leu587, Asp787) are not perfectly conserved and are marked by black boxes. Conserved key active site residues in rat PALcc (Arg533, Tyr654, Arg706) are marked by asterisks. Rat PALcc Met784, which interacts with the peptide substrate, and is replaced by Asp in SmPAL is marked by a black rectangle. Four possible SmPAL N-glycosylation sites are highlighted in magenta. GenBank accession numbers: Rat PAL (RatPALcc, P14925), Human PAL (HsPALcc, AAA36414), Drosophila melanogaster PAL 1 (dPAL1cc, ACJ13179), Drosophila melanogaster PAL 2 (dPAL2cc, AAF47043), Lymnaea stagnalis PAL (LsPALcc, AAD42259), and Caenorhabditis elegans (CePALcc, 172108).
3.2. Functional analysis of SmPAL
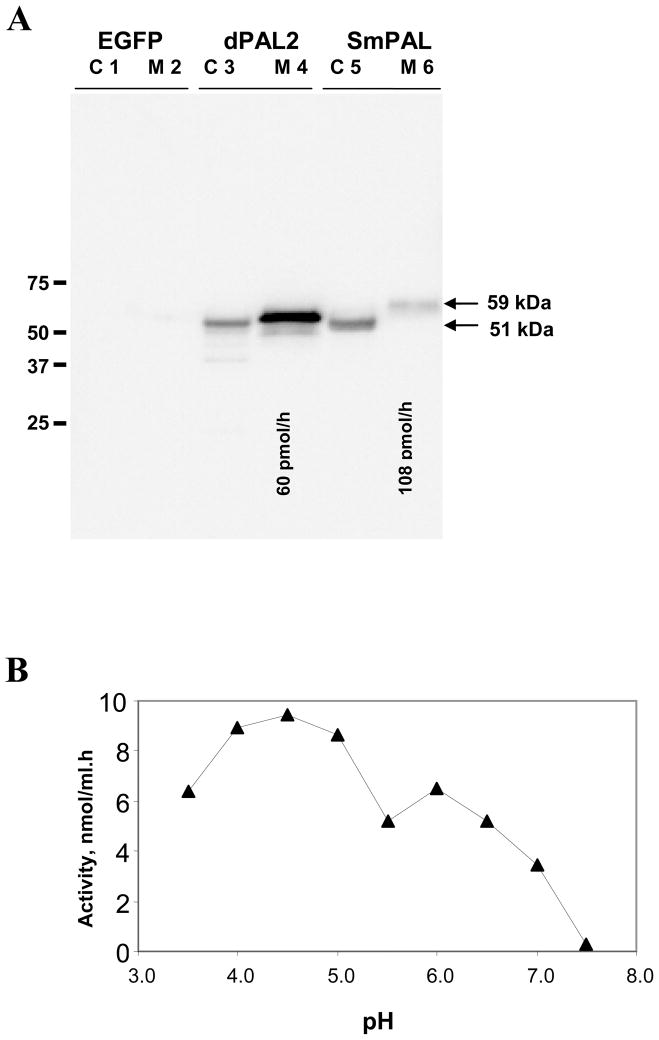
In order to assess the catalytic activity of SmPAL, it was expressed transiently in mammalian cells. To ensure efficient secretion and detection, the signal sequence of SmPAL was replaced by the signal sequence of rat PAM; further, an epitope tag (rhodopsin) was appended to the C-terminus of SmPAL. Drosophila PAL (dPAL2) was previously analyzed in a similar manner [35] and was expressed at the same time for comparison. Spent medium and cell extracts were analysed by Western blot, revealing the presence of rhodopsin-tagged SmPAL in cell extracts and in spent medium (Fig. 2A); transient expression of EGFP served as a control. While the SmPAL and dPAL2 present in cell extracts were similar in mass (51 kDa), the SmPAL recovered from spent medium was 59 kDa larger than secreted dPAL2. The predicted mass of His6-SmPAL.rho is 48 kDa, the SmPAL sequence includes four potential N-linked glycosylation sites (N-X-S/T) (Fig. 1). The potential N-glycosylation sites closest to the N- and C-termini of SmPAL fall outside of the β-propeller structure observed for rat PALcc and, therefore, should be accessible to oligosaccharide transferase [41]; the remaining sites may also be accessible since they would be expected to occur in the loops connecting the β1/β2 strands of propeller 1 and the β3/β4 strands of propeller 2. The mass of the rhodopsin-tagged SmPAL protein in cell extracts (51 kDa) makes it unlikely that more than one or two N-linked oligosaccharide chains are present. The size increase observed upon secretion of SmPAL suggests the presence of additional modifications such as the addition of sialic acid residues to N-linked oligosaccharides or O-linked glycosylation.
Fig. 2.
In vitro expression of SmPAL. Western blot analysis of the steady-state distribution of transfected EGFP (control), dPAL2 and SmPAL. Cell extracts (C; 4% of total amount cell extract) and spent medium (M; 0.8% of total amount spent medium) are shown; monoclonal antibody to rhodopsin was used to visualize the transiently expressed proteins. PAL activity was quantified in aliquots of spent medium; the total amount of activity loaded into the lanes containing medium is indicated below lanes M4 and M6. The apparent molecular masses of SmPAL and dPAL2 were compared; Western blot analysis yielded an apparent molecular mass of 51 kDa for cellular SmPAL (C5) and 59 kDa for secreted SmPAL (M6), dPAL2 is shown for comparison (C3 and M4). (B) Graph showing analysis of the pH dependence of SmPAL activity, indicating optimal activity at pH 4.5, with an additional peak of activity at pH 6.
Rhodopsin-tagged SmPAL and dPAL2 were both secreted; the amount of medium analyzed represents the amount secreted in approximately 5 h. Equivalent amounts of cell extract and spent medium were analyzed for dPAL2 and SmPAL. Since misfolded proteins are not efficiently secreted, this result indicates that SmPAL, like dPAL2, folds properly in mammalian cells. Although SmPAL and dPAL2 were expressed at similar levels (Fig. 2A), SmPAL was secreted less efficiently. In order to avoid any contribution from newly synthesized, immature enzyme, activity assays were carried out on spent medium. Cells transiently expressing EGFP were analyzed as a control and the low level of activity detected in their medium was subtracted as background. SmPAL enzyme activity was readily detected in the spent medium (Fig. 2A). The enzymatic activity of recombinant rhodopsin-tagged dPAL2 was previously characterised [35], and activity was readily detected in the spent medium. Since both proteins were detected using antibody to rhodopsin, their specific activities can be compared. Under the assay conditions used, the specific activity of SmPAL is substantially higher than that of dPAL2.
In mammalian cells, PAL must function in the acidic environment of the secretory granule. To assess the effect of assay pH on SmPAL activity, aliquots of spent medium from cells expressing SmPAL were diluted into buffers ranging in pH from 3.5 to 7.5. Optimal activity was detected at pH 4.5, with a second clear peak of SmPAL activity apparent at pH 6 (Figure 2B). No activity was detectable above pH 7.0. Based on the crystal structure of rat PALcc, a single residue contacts the amino acid that is to be amidated; the Met784 that serves this function in rat PALcc is replaced by Asp in SmPAL (Fig. 1). If titration of the side chain carboxylate affects the binding of substrate to SmPAL, this may explain the presence of two distinct pH optima.
3.3. Sm-pal-1 transcript expression
Sm-pal-1 in situ hybridisation experiments were carried out on whole mount preparations of a mixed sex population of adult S. mansoni. Antisense probes were used as experimental probes to identify the distribution of Sm-pal-1 transcript, visualised by a purple/brown staining pattern, while sense probes were used in time- and condition-matched experiments as a negative control. Colour deposition could be visualised within 2 h of incubation with the BCIP/NBT substrate and continued to develop overnight.
Sm-pal-1 gene expression was localised to neuronal cell bodies in the CNS of adult schistosomes. Female schistosomes display a marked deposition of Sm-pal-1 staining in the cerebral ganglia and longitudinal nerve cords of the CNS in the anterior fore-body (Fig. 3A and B). Transcript localisation is notable in the longitudinal nerve cords surrounding the oral sucker, and running towards the posterior, parallel to the oesophagus and the bifurcated gut (Fig. 3A and B). The oesophagus and gut intestinal caecae appear dark brown in colour due to non-specific interaction of BCIP/NBT with endogenous phosphatses (also evident in sense-strand treated control worms, where no CNS staining was observed, see Fig. 3). The longitudinal nerve cords in the mid body of the female worm also demonstrate Sm-pal-1 expression, where they can be seen in close proximity to the uterus (Fig. 3C). Interestingly, the longitudinal nerve cords display Sm-pal-1 gene expression in a distinct, but increasingly more diffuse pattern than noted in the bilobed cerebral ganglia. This irregular staining pattern was also observed in the tail of male worms, where Sm-pal-1 gene expression is evident in several discrete accumulations of staining (Fig. 3D). There was no evidence of Sm-pal-1 gene expression in the PNS – likely due to the limited access of antibodies and staining reagents in the absence of flat-fixing.
Fig. 3.
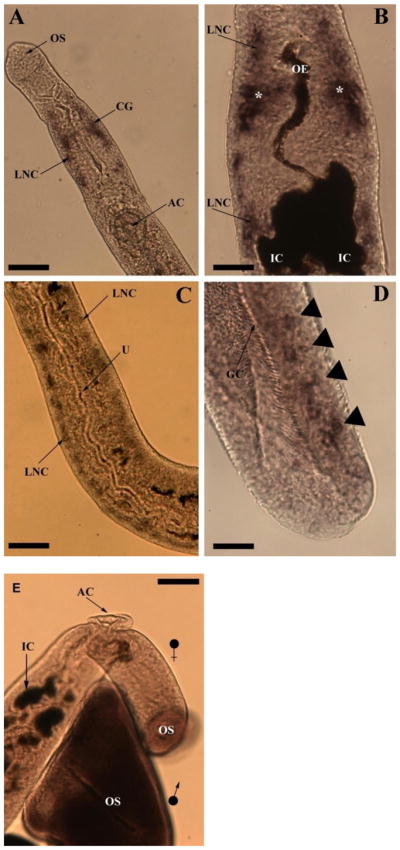
Light microscopy images of whole mount in situ hybridisation in adult Schistosoma mansoni showing Sm-pal-1 gene expression. (A) A female S. mansoni showing Sm-pal-1 localisation in the cerebral ganglia (CG) and longitudinal nerve cords (LNC). The muscular oral sucker (OS) and acetabulum (AC) are also labelled. (B) A female worm with Sm-pal-1 gene expression in the cerebral ganglia (asterisk, *) and LNC running in an anterior and posterior direction. The oesophagus (OE) and intestinal caecae (IC) are also shown. (C) A female specimen showing Sm-pal-1 gene expression in the LNCs running parallel to the uterus (U). (D) The tail of a male S. mansoni showing Sm-pal-1 transcript distribution (arrow heads) close to the posterior end of the gynaecophoric canal (GC). (E) A S. mansoni negative control, treated with Sm-pal-1 sense probes. An adult male and female pair can be seen to lack Sm-pal-1 staining. The oral sucker (OS), acetabulum (AC) and intestinal caecae (IC) of the female worm are shown, with the OS of the male positioned below the female anterior fore-body. Scale bars: A=60 μm; B=20 μm; C=40 μm; D=27 μm; E= 50 μm.
Sense probes were employed in negative control experiments to identify any non-specific staining. Specimens showed no observable staining in control experiments. Note that dark brown staining of the oesophagus and intestinal caecae was observed in female negative control specimens as in experimental worms, demonstrating the non-specific staining of these elements by the BCIP/NBT substrate; no CNS staining was evident (Fig. 3E).
3.4. Sm-pal-1 RNAi
Following SmPAL dsRNA delivery by electroporation, and 2 day post-dsRNA delivery schistosomula culture, RT-PCRs revealed no measurable changes in Sm-pal-1 transcript levels (data not shown). dsRNA delivery by electroporation was attempted twice with two separate batches of dsRNA, and on both occasions was unsuccessful, thus longer-term soaking as a means of dsRNA delivery was explored.
In total, six soaking experiments were carried out, of which only two experiments revealed diminished transcript abundance of Sm-pal-1. In experiments where transcript knock-down was detected, dsRNA delivery by soaking induced a marked and comparable reduction in Sm-pal-1 transcript levels as measured by endpoint detection (71–90 %; Fig. 4). Levels of Sm-pal-1 were unaffected in Sm-phm-1 dsRNA treated worms (Fig. 4). The alkaline phosphatase control gene Sm-ap showed no measurable difference in expression levels in dsRNA treated worms (Fig. 4). These observations suggest a specific reduction in the quantity of Sm-pal-1 transcript. All RT-PCR no template controls were negative.
Figure 4.
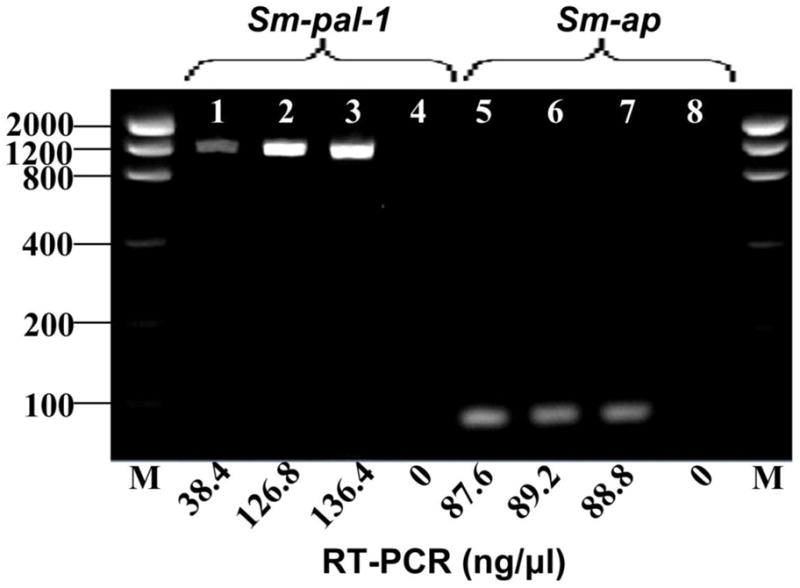
The effects of RNA interference on the expression of Schistosoma mansoni PAL (Sm-pal-1) and S. mansoni alkaline phosphatase (Sm-ap) in larval schistosomula. Representative RT-PCR results showing marked reduction in Sm-pal-1 transcript levels (~71 %, lane 1) following treatment with double stranded (ds) RNA for Sm-pal-1. Transcript levels of Sm-ap are unaffected (lanes 5, 6 and 7). (Lanes: 1, SmPAL-dsRNA treated; 2, SmPHM-dsRNA treated; 3, untreated (− dsRNA) control; 4, no template control; 5, SmPAL-dsRNA treated; 6, SmPHM-dsRNA treated; 7, - dsRNA control; 8, no template control).
On each of four further attempts to diminish Sm-pal-1 transcript levels in schistosomula, RT-PCR did not show any measurable difference in transcript abundance. In all experiments, whether or not Sm-pal-1 transcript levels were detectably reduced, schistosomula appeared normal and did not display any observable aberrant phenotypes from untreated control experiments.
4. Discussion
Carboxy-terminal α-amidation is an essential post-translational modification in the generation of biologically active neuropeptides. Various studies, involving the amidating enzymes of eukaryotes, report emerging disparities in amidating enzyme gene organisation [see 24 for review]. For the most part, PHM and PAL are expressed as separate domains of a bifunctional PAM protein in higher organisms such as Rattus norvegicus [42], and Xenopus laevis [43]. In contrast, an increasingly complicated picture of PHM and PAL expression is emerging from invertebrates, as a number of species including Lymnaea stagnalis [44], Aplysia californica [45], Calliactis parasitica [46], Drosophila [25, 35], Caenorhabditis elegans [47], and the planarian Dugesia japonica [26] encode single or multiple copies of one or both enzymes, expressed as bifunctional or monofunctional proteins.
PHM has previously been characterised from S. mansoni and identified as a monofunctional enzyme [28]. In the same study the use of degenerate primers and BLAST analysis of the GenBank EST dataset, failed to identify an S. mansoni PAL cDNA [28]. Capitalizing on more recent S. mansoni EST datasets, we identified a cDNA encoding a monofunctional schistosome PAL. Interestingly, extended interrogation of schistosome EST and genome datasets confirmed the absence of an EST representing a bifunctional PAM enzyme in S. mansoni.
Monofunctional SmPAL is a soluble, secreted enzyme, similar in sequence to other PALs such as the secreted Drosophila enzyme, dPAL2 [35], retaining four conserved cysteines for the maintenance of secondary structure, plus four cysteine residues unique to SmPAL, and a tyrosine residue known to be essential for functionality [39, 40]. Yet there are key amino acid substitutions in SmPAL, in the Ca2+ binding site, and where the amino acid preceding the α-hydroxyglycine and the PAL active site interact, which set it apart from the host enzyme. The latter of the two appears particularly significant as it is the Met784 in rat PALcc, substituted by Asp in SmPAL (Fig. 1), which recognizes the carbonyl group of the peptide bond linking the amino acid to be amidated to α-hydroxyglycine [41].
Rat PAL is completely N-glycosylated at a single consensus site located in the loop connecting β3/β4 in propeller five [48]. Although it is unclear what dictates the efficiency with which N-glycosylation sites are used [49], it is probable that only two of the four candidate N-linked glycosylation sites within SmPAL are glycosylated. The most N- and most C-terminal sites are not included in the β-propeller structure; while both sites may be accessible, the termination of translation may limit N-glycosylation of the most C-terminal site. The other two possible sites are in loops and could also be used (Figure 1). The size increase observed upon secretion of SmPAL may be a result of further post-translational modifications such as the addition of sialic acid residues or O-linked glycosylation, which occur at a later stage in protein processing [49].
The conversion of α-N-acetyl-Tyr-Val-α-hydroxyglycine, to α-N-acetyl-Tyr-Val-α-NH2, a reaction mechanism specific to PAL enzymes, revealed SmPAL catalytic properties within the range of other eukaryote enzymes. The pH optimum of SmPAL coincides with the reported characteristics of other PALs [35, 44, 50, 51], and as expected, is less acidic than the pH optimum of SmPHM [28]. The low pH optimum and high Km (44 μM) of SmPHM, which deviate from the catalytic properties of the human host enzyme, make SmPHM a potential novel drug target [28]. Although the functional characteristics of SmPAL revealed in this study do not display the same degree of divergence from the schistosome host enzyme as SmPHM [51], it is the degree of structural divergence evident from the crystallography of rat PALcc [41] which may provide opportunity for chemotherapeutic intervention. Regardless, the expression of a catalytically active monofunctional SmPAL, unequivocally confirms the existence of monofunctional amidating enzymes in schistosomes which act independently of each other, in contrast to the mammalian host enzyme.
Although SmPAL transcript localisation is consistent with the immunolocalisation patterns of the amidating co-enzyme SmPHM in the schistosome CNS, SmPHM is also localised extensively in the PNS of adult schistosomes [28]. Although currently there is no molecular or bioinformatic evidence supporting the possibility of a second SmPAL enzyme, the enzyme complement of other eukaryotes [24], and the restricted Sm-pal-1 expression pattern presented here, does not totally diminish the possibility of another active schistosome amidating lyase. One possible reason for the absence of SmPAL staining in the PNS is that transcript abundance was below our detection limit. Another reason could be that most of the PNS cell bodies reside close to the CNS such that transcript does not physically associate with much of the PNS and as such is identified as CNS staining by in situ hybridisation. Clearly, more reliable comparisons could be made with immunocytochemical approaches using a SmPAL antiserum.
The expression of Sm-pal-1 in the CNS of adult schistosomes, coupled with the extensive immunolocalisation of SmPHM throughout the nervous system of adult schistosomes [28] is consistent with the hypothesis that SmPAL and SmPHM are neuronally expressed enzymes, involved in neuropeptide biosynthesis. The widespread localisation of neuropeptide amidating enzymes in the schistosome nervous system is supported by a recent study which exploited available transcriptomic and genomic datasets for flatworms, including S. mansoni, and reported seventeen novel amidated neuropeptides from S. mansoni [13]. A transcript encoding schistosome NPFF (PQRFamide motif), a peptide similar to vertebrate neuropeptide FF and not previously reported in invertebrates, was identified, along with several RFamides, an additional NPF, and novel L/Iamides and PWamides [13]. Thus our recent bioinformatic data go some way to support and explain the localisation pattern of sm-pal-1 and the extensive staining patterns reported for SmPHM.
RNAi has quite recently become popular for the functional analysis of schistosome genes and a growing number of publications report successful gene silencing, of non-neuronal transcripts, in larval parasites following dsRNA/siRNA delivery by electroporation and soaking [31, 37, 52–64]. Our attempts to knockdown schistosomula Sm-pal-1 gene transcripts were somewhat less successful than previous publications of schistosomula gene knockdown. Importantly however, none of the genes previously targeted in schistosomes have been exclusively involved in neuronal processes, and are most commonly genes such as the cathepsins B and D, and alkaline phosphatase, which are highly expressed in tissues relatively more accessible to external dsRNA than neuronal cells [31, 37, 53, 57, 60].
In the studies mentioned above, electroporation has been reported to induce >85 % knockdown of some non-neuronal transcripts in schistosomula [31]. However, in this study electroporation did not induce measurable gene knockdown of our PAL transcript on either of two attempts. Some reports detail concentration dependant responses to dsRNA [55, 57] such that it is likely, in the case of Sm-pal-1, that future electroporation experiments may benefit from the optimisation of dsRNA concentration to produce measurable and consistent gene silencing.
DsRNA delivery by electroporation is reported to produce much more dramatic gene suppression (>10 fold) than dsRNA delivery by soaking [31, 57]. In this study however, soaking did successfully and specifically reduce Sm-pal-1 transcript abundance by up to 90 % on two occasions out of six. While these results do indicate that some, if not all, schistosome neuronal genes are susceptible to RNAi, the question remains as to why neuronal gene silencing in this case does not appear to be consistently reproducible. Tissue types are known to differ in their degree of penetrability by dsRNA constructs, resulting in varying degrees of gene suppression between different tissues [65]. This may present one possible reason for Sm-pal-1 knockdown unreliability, as dsRNA constructs may have limited access to deeper tissues such as neuronal cell bodies, where target transcripts are expressed.
Further, dsRNA delivery by soaking may have proven more successful if larval parasites were exposed to dsRNA during cercarial transformation. It is thought that schistosome intra-molluscan life stages are more susceptible to RNAi during transformation when tegumental membranes are undergoing reorganisation [52]; some reports suggest that this may also be the case during cercarial transformation [53]. In contrast, other studies involving dsRNA delivery during cercarial transformation disagree, suggesting that dsRNA uptake is no more efficient when external membranes are in flux, than when schistosomula are fully formed [57]. The exploration of alternative dsRNA delivery methods, or the application of smaller siRNAs may prove beneficial to the induction of neuronal RNAi in schistosomes.
The recent discovery of a rich flatworm neuropeptide complement provides strong support for the importance of neuropeptide signaling in flatworm neurobiology. One particularly appealing feature of these discoveries is the fact that the majority of putative neuropeptides discovered in the flatworm datasets are novel peptides with no obvious homologues in other animal phyla. Clearly, such differences are a preferred feature of parasite drug targets and underscore the possibility of exploiting flatworm neuropeptide signaling processes for parasite control. The mechanisms and processes associated with neuropeptide signaling such as peptide amidation represent attractive targets for chemotherapeutic intervention. The presence of a widely expressed PHM enzyme in the nervous system of adult schistosomes [28], and the identification by this study of a functionally active, monofunctional schistosome PAL enzyme provides target enzymes for consideration as drug target candidates. Unfortunately, RNAi has yet to be optimised for neuronally-expressed transcripts in schistosomes such that validating neuronal gene products as drug targets remains a challenge. Although we only observed Sm-pal-1 RNAi knockdown intermittently, the fact that knockdown occurred on a couple of occasions gives hope that further optimization of RNAi methods could facilitate robust neuronal RNAi.
Acknowledgments
This work was supported by a Department of Employment and Learning studentship grant (LA), NIH Grant AI49162 (T.A.D, A.G.M), and DK-32949 (B.A.E).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hotez PJ, Brindley PJ, Bethony JM, et al. Helminth infections: the great neglected tropical diseases. J Clin Invest. 2008;118:1311–21. doi: 10.1172/JCI34261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fallon PG, Doenhoff MJ. Drug-resistant schistosomiasis: resistance to praziquantel and oxamniquine induced in Schistosoma mansoni in mice is drug specific. Am J Trop Med Hyg. 1994;51:83–8. doi: 10.4269/ajtmh.1994.51.83. [DOI] [PubMed] [Google Scholar]
- 3.Geerts S, Gryseels B. Anthelmintic resistance in human helminths: a review. Trop Med Int Health. 2001;6:915–21. doi: 10.1046/j.1365-3156.2001.00774.x. [DOI] [PubMed] [Google Scholar]
- 4.Botros S, Sayed H, Amer N, et al. Current status of sensitivity to praziquantel in a focus of potential drug resistance in Egypt. Int J Parasitol. 2005;35:787–91. doi: 10.1016/j.ijpara.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Caffrey CR, Rohwer A, Oellien F, et al. A comparative chemogenomics strategy to predict potential drug targets in the metazoan pathogen, Schistosoma mansoni. PLoS One. 2009;4:e4413. doi: 10.1371/journal.pone.0004413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halton DW, Gustafsson MK. Functional morphology of the platyhelminth nervous system. Parasitology. 1996;113:S47. [Google Scholar]
- 7.Gustafsson MK, Halton DW, Kreshchenko ND, et al. Neuropeptides in flatworms. Peptides. 2002;23:2053–61. doi: 10.1016/s0196-9781(02)00193-6. [DOI] [PubMed] [Google Scholar]
- 8.Maule AG, Mousley A, Marks NJ, et al. Neuropeptide signaling systems - potential drug targets for parasite and pest control. Curr Top Med Chem. 2002;2:733–58. doi: 10.2174/1568026023393697. [DOI] [PubMed] [Google Scholar]
- 9.Mousley A, Marks NJ, Halton DW, et al. Arthropod FMRFamide-related peptides modulate muscle activity in helminths. Int J Parasitol. 2004;34:755–68. doi: 10.1016/j.ijpara.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Mousley A, Marks NJ, Maule AG. Neuropeptide signalling: a repository of targets for novel endectocides? Trends Parasitol. 2004;20:482–7. doi: 10.1016/j.pt.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 11.Kreshchenko ND. Functions of flatworm neuropeptides NPF, GYIRF and FMRF in course of pharyngeal regeneration of anterior body fragments of planarian, Girardia tigrina. Acta Biol Hung. 2008;59 (Suppl):199–207. doi: 10.1556/ABiol.59.2008.Suppl.29. [DOI] [PubMed] [Google Scholar]
- 12.Humphries JE, Kimber MJ, Barton YW, et al. Structure and bioactivity of neuropeptide F from the human parasites Schistosoma mansoni and Schistosoma japonicum. J Biol Chem. 2004;279:39880–5. doi: 10.1074/jbc.M405624200. [DOI] [PubMed] [Google Scholar]
- 13.McVeigh P, Mair GR, Atkinson L, et al. Discovery of multiple neuropeptide families in the phylum Platyhelminthes. Int J Parasitol. 2009;39:1243–52. doi: 10.1016/j.ijpara.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skuce PJ, Johnston CF, Fairweather I, et al. A confocal scanning laser microscope study of the peptidergic and serotoninergic components of the nervous system in larval Schistosoma mansoni. Parasitology. 1990;101 (Pt 2):227–34. doi: 10.1017/s0031182000063277. [DOI] [PubMed] [Google Scholar]
- 15.Skuce PJ, Johnston CF, Fairweather I, et al. Immunoreactivity to the pancreatic polypeptide family in the nervous system of the adult human blood fluke, Schistosoma mansoni. Cell Tissue Res. 1990;261:573–81. doi: 10.1007/BF00313537. [DOI] [PubMed] [Google Scholar]
- 16.Solis-Soto JM, De Jong Brink M. Immunocytochemical study on biologically active neurosubstances in daughter sporocysts and cercariae of Trichobilharzia ocellata and Schistosoma mansoni. Parasitology. 1994;108 ( Pt 3):301–11. doi: 10.1017/s0031182000076149. [DOI] [PubMed] [Google Scholar]
- 17.Fairweather I, Skuce PO, Brownlee DJ, et al. Light and electron microscopic immunocytochemistry of FMRFamide and neuropeptide F immunoreactivities in the human blood fluke, Schistosoma mansoni. Acta Biol Hung. 1995;46:211–20. [PubMed] [Google Scholar]
- 18.Marks NJ, Halton DW, Maule AG, et al. Comparative analyses of the neuropeptide F (NPF)- and FMRFamide-related peptide (FaRP)-immunoreactivities in Fasciola hepatica and Schistosoma spp. Parasitology. 1995;110 ( Pt 4):371–81. doi: 10.1017/s0031182000064714. [DOI] [PubMed] [Google Scholar]
- 19.Mair GR, Maule AG, Shaw C, et al. Muscling in on parasitic flatworms. Parasitol Today. 1998;14:73–6. doi: 10.1016/s0169-4758(97)01182-4. [DOI] [PubMed] [Google Scholar]
- 20.Day TA, Maule AG, Shaw C, et al. Platyhelminth FMRFamide-related peptides (FaRPs) contract Schistosoma mansoni (Trematoda: Digenea) muscle fibres in vitro. Parasitology. 1994;109 ( Pt 4):455–9. doi: 10.1017/s0031182000080707. [DOI] [PubMed] [Google Scholar]
- 21.Eipper BA, Stoffers DA, Mains RE. The biosynthesis of neuropeptides: peptide alpha-amidation. Annu Rev Neurosci. 1992;15:57–85. doi: 10.1146/annurev.ne.15.030192.000421. [DOI] [PubMed] [Google Scholar]
- 22.Bolkenius FN, Ganzhorn AJ. Peptidylglycine alpha-amidating mono-oxygenase: neuropeptide amidation as a target for drug design. Gen Pharmacol. 1998;31:655–9. doi: 10.1016/s0306-3623(98)00192-x. [DOI] [PubMed] [Google Scholar]
- 23.Kulathila R, Merkler KA, Merkler DJ. Enzymatic formation of C-terminal amides. Nat Prod Rep. 1999;16:145–54. doi: 10.1039/a801346b. [DOI] [PubMed] [Google Scholar]
- 24.Prigge ST, Mains RE, Eipper BA, et al. New insights into copper monooxygenases and peptide amidation: structure, mechanism and function. Cell Mol Life Sci. 2000;57:1236–59. doi: 10.1007/PL00000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolhekar AS, Roberts MS, Jiang N, et al. Neuropeptide amidation in Drosophila: separate genes encode the two enzymes catalyzing amidation. J Neurosci. 1997;17:1363–76. doi: 10.1523/JNEUROSCI.17-04-01363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asada A, Orii H, Watanabe K, et al. Planarian peptidylglycine-hydroxylating monooxygenase, a neuropeptide processing enzyme, colocalizes with cytochrome b561 along the central nervous system. FEBS J. 2005;272:942–55. doi: 10.1111/j.1742-4658.2004.04528.x. [DOI] [PubMed] [Google Scholar]
- 27.Eipper BA, Milgram SL, Husten EJ, et al. Peptidylglycine alpha-amidating monooxygenase: a multifunctional protein with catalytic, processing, and routing domains. Protein Sci. 1993;2:489–97. doi: 10.1002/pro.5560020401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mair GR, Niciu MJ, Stewart MT, et al. A functionally atypical amidating enzyme from the human parasite Schistosoma mansoni. FASEB J. 2004;18:114–21. doi: 10.1096/fj.03-0429com. [DOI] [PubMed] [Google Scholar]
- 29.Berriman M, Haas BJ, LoVerde PT, et al. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460:352–8. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dillon GP, Illes JC, Isaacs HV, et al. Patterns of gene expression in schistosomes: localization by whole mount in situ hybridization. Parasitology. 2007;134:1589–97. doi: 10.1017/S0031182007002995. [DOI] [PubMed] [Google Scholar]
- 31.Ndegwa D, Krautz-Peterson G, Skelly PJ. Protocols for gene silencing in schistosomes. Exp Parasitol. 2007;117:284–91. doi: 10.1016/j.exppara.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 33.Zdobnov EM, Apweiler R. InterProScan--an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–8. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 34.Emanuelsson O, Brunak S, von Heijne G, et al. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–71. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 35.Han M, Park D, Vanderzalm PJ, et al. Drosophila uses two distinct neuropeptide amidating enzymes, dPAL1 and dPAL2. J Neurochem. 2004;90:129–41. doi: 10.1111/j.1471-4159.2004.02464.x. [DOI] [PubMed] [Google Scholar]
- 36.Pownall ME, Tucker AS, Slack JM, et al. eFGF, Xcad3 and Hox genes form a molecular pathway that establishes the anteroposterior axis in Xenopus. Development. 1996;122:3881–92. doi: 10.1242/dev.122.12.3881. [DOI] [PubMed] [Google Scholar]
- 37.Correnti JM, Brindley PJ, Pearce EJ. Long-term suppression of cathepsin B levels by RNA interference retards schistosome growth. Mol Biochem Parasitol. 2005;143:209–15. doi: 10.1016/j.molbiopara.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 38.Nielsen H, Engelbrecht J, Brunak S, et al. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 39.Kolhekar AS, Bell J, Shiozaki EN, et al. Essential features of the catalytic core of peptidyl-alpha-hydroxyglycine alpha-amidating lyase. Biochemistry. 2002;41:12384–94. doi: 10.1021/bi0260280. [DOI] [PubMed] [Google Scholar]
- 40.De M, Bell J, Blackburn NJ, et al. Role for an essential tyrosine in peptide amidation. J Biol Chem. 2006;281:20873–82. doi: 10.1074/jbc.M513886200. [DOI] [PubMed] [Google Scholar]
- 41.Chufan EE, De M, Eipper BA, et al. Amidation of bioactive peptides: the structure of the lyase domain of the amidating enzyme. Structure. 2009;17:965–73. doi: 10.1016/j.str.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ouafik LH, Stoffers DA, Campbell TA, et al. The multifunctional peptidylglycine alpha-amidating monooxygenase gene: exon/intron organization of catalytic, processing, and routing domains. Mol Endocrinol. 1992;6:1571–84. doi: 10.1210/mend.6.10.1448112. [DOI] [PubMed] [Google Scholar]
- 43.Iwasaki Y, Shimoi H, Saiki H, et al. Tissue-specific molecular diversity of amidating enzymes (peptidylglycine alpha-hydroxylating monooxygenase and peptidylhydroxyglycine N-C lyase) in Xenopus laevis. Eur J Biochem. 1993;214:811–8. doi: 10.1111/j.1432-1033.1993.tb17984.x. [DOI] [PubMed] [Google Scholar]
- 44.Spijker S, Smit AB, Eipper BA, et al. A molluscan peptide alpha-amidating enzyme precursor that generates five distinct enzymes. FASEB J. 1999;13:735–48. doi: 10.1096/fasebj.13.6.735. [DOI] [PubMed] [Google Scholar]
- 45.Fan X, Spijker S, Akalal DB, et al. Neuropeptide amidation: cloning of a bifunctional alpha-amidating enzyme from Aplysia. Brain Res Mol Brain Res. 2000;82:25–34. doi: 10.1016/s0169-328x(00)00173-x. [DOI] [PubMed] [Google Scholar]
- 46.Williamson M, Hauser F, Grimmelikhuijzen CJ. Genomic organization and splicing variants of a peptidylglycine alpha-hydroxylating monooxygenase from sea anemones. Biochem Biophys Res Commun. 2000;277:7–12. doi: 10.1006/bbrc.2000.3629. [DOI] [PubMed] [Google Scholar]
- 47.C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–8. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 48.Yun HY, Johnson RC, Mains RE, et al. Topological switching of the COOH-terminal domain of peptidylglycine alpha-amidating monooxygenase by alternative RNA splicing. Arch Biochem Biophys. 1993;301:77–84. doi: 10.1006/abbi.1993.1117. [DOI] [PubMed] [Google Scholar]
- 49.Kolhekar AS, Quon AS, Berard CA, et al. Post-translational N-glycosylation of a truncated form of a peptide processing enzyme. J Biol Chem. 1998;273:23012–8. doi: 10.1074/jbc.273.36.23012. [DOI] [PubMed] [Google Scholar]
- 50.Eipper BA, Perkins SN, Husten EJ, et al. Peptidyl-alpha-hydroxyglycine alpha-amidating lyase. Purification, characterization, and expression. J Biol Chem. 1991;266:7827–33. [PubMed] [Google Scholar]
- 51.Husten EJ, Eipper BA. Purification and characterization of PAM-1, an integral membrane protein involved in peptide processing. Arch Biochem Biophys. 1994;312:487–92. doi: 10.1006/abbi.1994.1336. [DOI] [PubMed] [Google Scholar]
- 52.Boyle JP, Wu XJ, Shoemaker CB, et al. Using RNA interference to manipulate endogenous gene expression in Schistosoma mansoni sporocysts. Mol Biochem Parasitol. 2003;128:205–15. doi: 10.1016/s0166-6851(03)00078-1. [DOI] [PubMed] [Google Scholar]
- 53.Skelly PJ, Da’dara A, Harn DA. Suppression of cathepsin B expression in Schistosoma mansoni by RNA interference. Int J Parasitol. 2003;33:363–9. doi: 10.1016/s0020-7519(03)00030-4. [DOI] [PubMed] [Google Scholar]
- 54.Correnti JM, Pearce EJ. Transgene expression in Schistosoma mansoni: introduction of RNA into schistosomula by electroporation. Mol Biochem Parasitol. 2004;137:75–9. doi: 10.1016/j.molbiopara.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 55.Cheng GF, Lin JJ, Shi Y, et al. Dose-dependent inhibition of gynecophoral canal protein gene expression in vitro in the schistosome (Schistosoma japonicum) by RNA interference. Acta Biochim Biophys Sin (Shanghai) 2005;37:386–90. doi: 10.1111/j.1745-7270.2005.00058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dinguirard N, Yoshino TP. Potential role of a CD36-like class B scavenger receptor in the binding of modified low-density lipoprotein (acLDL) to the tegumental surface of Schistosoma mansoni sporocysts. Mol Biochem Parasitol. 2006;146:219–30. doi: 10.1016/j.molbiopara.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 57.Krautz-Peterson G, Radwanska M, Ndegwa D, et al. Optimizing gene suppression in schistosomes using RNA interference. Mol Biochem Parasitol. 2007;153:194–202. doi: 10.1016/j.molbiopara.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 58.Nabhan JF, El-Shehabi F, Patocka N, et al. The 26S proteasome in Schistosoma mansoni: bioinformatics analysis, developmental expression, and RNA interference (RNAi) studies. Exp Parasitol. 2007;117:337–47. doi: 10.1016/j.exppara.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 59.Krautz-Peterson G, Skelly PJ. Schistosome asparaginyl endopeptidase (legumain) is not essential for cathepsin B1 activation in vivo. Mol Biochem Parasitol. 2008;159:54–8. doi: 10.1016/j.molbiopara.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morales ME, Rinaldi G, Gobert GN, et al. RNA interference of Schistosoma mansoni cathepsin D, the apical enzyme of the hemoglobin proteolysis cascade. Mol Biochem Parasitol. 2008;157:160–8. doi: 10.1016/j.molbiopara.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao ZR, Lei L, Liu M, et al. Schistosoma japonicum: inhibition of Mago nashi gene expression by shRNA-mediated RNA interference. Exp Parasitol. 2008;119:379–84. doi: 10.1016/j.exppara.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 62.de Moraes Mourao M, Dinguirard N, Franco GR, et al. Phenotypic Screen of Early-Developing Larvae of the Blood Fluke, Schistosoma mansoni, using RNA Interference. PLoS Negl Trop Dis. 2009;3:e502. doi: 10.1371/journal.pntd.0000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rinaldi G, Morales ME, Alrefaei YN, et al. RNA interference targeting leucine aminopeptidase blocks hatching of Schistosoma mansoni eggs. Mol Biochem Parasitol. 2009;167:118–26. doi: 10.1016/j.molbiopara.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beckmann S, Buro C, Dissous C, et al. The Syk kinase SmTK4 of Schistosoma mansoni is involved in the regulation of spermatogenesis and oogenesis. PLoS Pathog. 2010:6. doi: 10.1371/journal.ppat.1000769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–12. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]