Abstract
Normal development of the mammary gland proceeds via interactions between the epithelium and the mesenchyme that start during embryogenesis and continue during pubertal outgrowth and differentiation. The function of specific peptide growth factors that bind members of the receptor tyrosine kinase family and the cytokine receptor family are required at each stage. In many cases the peptides are produced in one compartment and act on receptors in the other compartment. One of the striking differences between normal development and cancer is the loss of this cross-talk. Mammary tumor cells often produce a peptide and express the receptor on the same cell leading to autocrine activation of signaling pathways, a mechanism that is characteristic for cancer cells. We will discuss different peptides in the context of normal development and cancer in this review.
Growth factors signal between the epithelium and mesenchyme during mammary gland development. This crosstalk can be lost in cancer, in which the same cell often expresses both a growth factor and its receptor.
Development of the mouse mammary gland begins at embryonic day 10.5 with formation of the milk line, a thickening of the ectoderm that extends from the anterior to the posterior limb buds. By E11.5 five placode pairs invaginate into the surrounding stroma and undergo limited branching morphogenesis. At birth a rudimentary ductal tree is present in a stromal fat pad. At the onset of puberty ovarian hormones induce ductal elongation until the fat pad is filled with a system of ducts and side branches. During pregnancy the glands fill with secretory alveolar units that on parturition produce copius amounts of milk for the offspring. As the pups are weaned, these alveoli are removed by programmed cell death coupled with extensive tissue remodeling, an event termed involution. All these processes are under the control of a wide variety of hormonal and peptide factors. In this review, which is concentrated on peptide growth factors, we will discuss those factors that have important roles in normal development and have been implicated in breast cancer.
RECEPTOR TYROSINE KINASES AND PEPTIDE LIGANDS IN NORMAL DEVELOPMENT AND CANCER
The superfamily of receptor tyrosine kinases (RTK) includes approximately 60 members divided into 19 subfamilies. Each receptor has an extracellular ligand binding region, a single membrane-spanning region, and a cytoplasmic tyrosine kinase-containing domain. In this review we will discuss the epidermal growth factor receptor (EGFR) or ErbB family and the EGF-related ligands, the fibroblast growth factor receptor (FGFR), and FGFs as well as insulin-like growth factors (IGFs) and their receptors, mentioning their involvement in normal development of the mammary gland and in breast cancer.
EGF FAMILY OF GROWTH FACTORS AND THEIR RECEPTORS
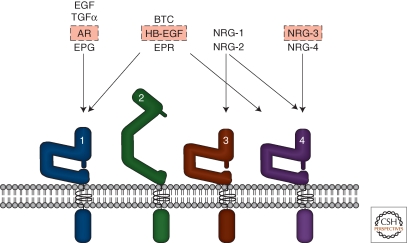
There are four members of the ErbB RTK family: EGFR, ErbB2, ErbB3, and ErbB4. Peptides in the EGF family bind ErbB receptors and based on their receptor specificity are divided into three groups: The first includes EGF, transforming growth factor (TGF)-α, amphiregulin (AR), and epigen (EPG), which bind EGFR; the second includes betacellulin (BTC), heparin-binding EGF (HB-EGF), and epiregulin (EPR), which bind both EGFR and ErbB4; the third group, the neuregulins (NRGs), forms two subgroups based on their capacity to bind ErbB3 and ErbB4 (NRG-1 and NRG-2) or ErbB4 (NRG-3 and NRG-4) (Hynes and Lane 2005) (Fig. 1).
Figure 1.
EGF-family peptides and ErbB Receptors. The four ErbB receptors are shown: (1) EGFR, (2) ErbB2, (3) ErbB3, and (4) ErbB4. The EGF-family peptides bind specific ErbB receptors: EGF, TGF-α,AR, and EPG bind EGFR; BTC, HB-EGF, EPR have dual specificity, binding both EGFR and ErbB4; NRG-1 and NRG-2 bind ErbB3 and ErbB4; NRG-3 and NRG-4 bind ErbB4 (Hynes and Lane 2005). Various roles for specific ligands (boxed in red) and all the ErbB receptors have been described in normal mammary gland development. In breast cancer, overexpressed ErbB2 together with ErbB3 function as a unit to drive proliferation of specific subsets of tumors (Holbro et al. 2003). EGFR is overexpressed in 30%–60% of basal-like breast cancers. See text for more information.
Ligand binding initiates signaling by causing specific homo- or heterodimeric receptor formation and activation of the cytoplasmic kinase domain that phosphorylates tyrosines in the tail region of each receptor. Phosphorylation triggers the association of specific signaling molecules whose binding initiates downstream signaling events. Of note, ErbB3 has impaired kinase activity and only acquires signaling potential, i.e., tyrosine phosphorylation, when dimerized with another receptor. Furthermore, ErbB2 does bind any of the EGF-family ligands, however, it is activated via heterdimerization with the other ligand-bound receptors (Hynes and Lane 2005) and is their preferred heterodimerization partner (Graus-Porta et al. 1997).
FGF FAMILY OF PEPTIDE GROWTH FACTORS AND RECEPTORS
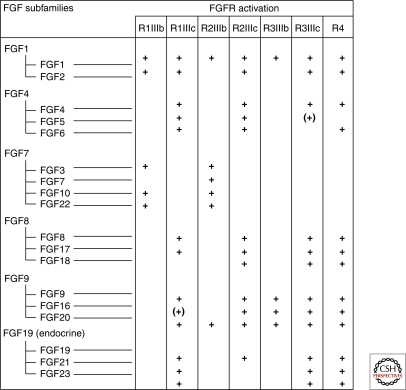
Members of the FGF ligand family and their receptors form a complex network with four FGFR genes that undergo alternative splicing to yield minimally seven FGFRs. Alternative splicing of FGFR 1–3 in the D3 region (III) of the ectodomain yields, e.g., FGFR1-IIIb and FGFR1-IIIc, that bind different FGFs and are expressed in epithelial or mesenchmal compartments, respectively. The 18 FGFs are divided into six subfamilies; the FGF 11-14 subfamily has been renamed fibroblast homologous factors because their core structure is similar to other FGFs, however, several key residues for receptor binding are missing and they do not bind FGFRs (reviewed in Itoh and Ornitz 2004; Beenken and Mohammadi 2009).
Each FGF ligand binds a specific FGFR (Fig. 2), which enforces receptor dimerization, kinase activation and autophosphorylation of specific tyrosine residues in the cytoplasmic domain (Mohammadi et al. 1996; Lew et al. 2009). In addition to the receptor, FRS2, a major adaptor linking FGFRs to the ERK and PI3K pathways becomes phosphorylated on specific tyrosine residues (Kouhara et al. 1997; Ong et al. 2001).
Figure 2.
Members of the FGF family are divided into six subgroups of closely related peptides. Each ligand binds to a specific subset of the FGFRs. () refers to the fact that there is still uncertainty regarding the binding. The table summarizes results from many laboratories and some relevant publications are as follows: (Ornitz et al. 1996; Zhang et al. 2006; Kurosu et al. 2006, 2007; Suzuki et al. 2008; Schwertfeger 2009).
INSULIN, IGFs, AND THEIR RECEPTORS
The peptides insulin and IGF1 bind to various members of the insulin receptor (IR)-IGF1 receptor (IGF1R) family. The receptors are tetrameric structures composed of half receptors each of which has an extracellular ligand-binding α chain and a transmembrane spanning β chain with cytoplasmic tyrosine kinase activity. IGF1 binds IGF1R as well as a hybrid composed of an IR and an IGF1R. Moreover, the bioavailability of IGF1 is controlled by IGF binding proteins (IGFBP) (reviewed in De Meyts 2004). As for other RTKs, ligand binding stimulates multiple intracellular pathways. One specific characteristic of the IR-IGF1R family is their dependence on insulin responsive substrate (IRS) family members, in particular IRS-1/2, for intracellular signaling.
ROLES FOR EGF-FAMILY PEPTIDES, FGFs, AND IGF1 AND THEIR RECEPTORS IN MAMMARY GLAND DEVELOPMENT
Embryonic Development of the Mammary Gland
Development of the mouse mammary gland begins in at embryonic day E10.5 and by E11.5, five placode pairs invaginate into the surrounding stroma and undergo branching morphogenesis. FGF10 and its receptor FGFR2-IIIb have important roles during embryonic mammary gland development. Starting at E10.5, somatic-expressed FGF10 acts on FGFR2-IIIb in the ectoderm to initiate induction and positioning of the mammary line and placode formation. The ligand and the receptor are both essential for induction of four of the five placode pairs (Mailleux et al. 2002; Veltmaat et al. 2006). Considering upstream regulators of the pathway, it has been shown that FGF10 levels are reduced in somites lacking the transcription factor GLI3, and the mammary line and placode three fail to form in its absence (Veltmaat et al. 2006).
NRG3 was discovered as a hypomorphic allele causing hypoplastic mammary buds in ska mutant mice (Howard et al. 2005). NRG3 is expressed in the mesenchyme adjacent to sites where placodes form and acts to promote mammary morphogenesis. ErbB4, the NRG3 receptor, as well as the other ErbB receptors, are expressed at sites of epithelial–mesenchymal interactions during morphogenesis of the embryonic mammary gland (Wansbury et al. 2008). Intriguingly, embryonic mammary placode formation appears to be normal in ErbB4 null mice (Tidcombe et al. 2003). Indeed, embryos deleted for each of the four ErbB receptors appear to form normal mammary placodes (Wansbury et al. 2008). This suggests that while NRG3 is limiting in the process of placode formation, the ability of an individual ErbB receptor to form a heterodimer with the other family members provides sufficient signaling to allow embryonic placode formation.
The IGF/IGF1R axis has an essential role in embryonic bud development. Intriguingly, it was observed that embryos deficient in P190-B, a member of the RhoGTPase activating (RhoGAP) family that interacts with integrins, had smaller mammary buds. The defect was in both compartments, because the epithelial bud proliferation was lower and the underlying mesenchyme was aberrant. A similar phenotype was observed in embryonic mammary buds lacking IRS-1/2 (Heckman et al. 2007). Taken together, the results suggest that IGF1/IGFR1 signaling through a RhoGAP and the IRS effector proteins has an essential role in the epithelial mesenchymal interactions needed for proper mammary bud morphogenesis.
Ductal Outgrowth and Differentiation
The rudimentary epithelial tree in the mammary fat pad remains quiescent until puberty onset when ovarian steroid hormone production commences and induces ductal outgrowth. Bulbous structures on the tips of the ducts, the terminal end buds (TEBs) are highly proliferative and penetrate into the fat pad as the ducts elongate. TEBs bifurcate and secondary branches form during this process, until the entire fat pad is filled with a network of branched ducts. During repeated estrous cycles the ductal network increases in complexity and side-branches grow under progesterone control. Under control of prolactin, alveolar structures bud off the ductal system during pregnancy, and these differentiate into milk-producing structures. Steroid hormones, growth hormone (GH), and prolactin are the master regulators of mammary growth and pregnancy-induced differentiation, whereas peptide factors of the EGF and FGF family, and IGF-1 have specific roles during these processes.
During ductal outgrowth multiple FGFs: FGF1, FGF2, FGF4, FGF7, and FGF10, as well as FGFR1 and FGFR2 are expressed (reviewed in Schwertfeger 2009). TEBs have high levels of FGFR2, which is essential at this stage because FGFR2 null glands penetrate the fat pad more slowly and show fewer branch points compared with WT controls (Lu et al. 2008). Indeed, it has been shown that the epithelial specific FGFR2-IIIb isoform is responsible for ductal outgrowth (Parsa et al. 2008). During pregnancy and lactation some FGFs are expressed (Coleman-Krnacik and Rosen 1994), however, to our knowledge a specific role for the FGF/FGFR signaling network at this developmental stage has not been described.
The pituitary GH is essential for pubertal mammary development (reviewed in Hovey et al. 2002); IGF1, which is produced in the mammary gland, acts as the local effector of GH (reviewed in Wood et al. 2000; Kleinberg and Ruan 2008; Kleinberg et al. 2009). During postnatal development of the gland, IGF1 is produced in both stromal and epithelial cells (Loladze et al. 2006) and, in its absence, TEBs fail to develop correctly and the glands show limited outgrowth potential (Ruan and Kleinberg 1999). These results, together with data showing that embryonic IGF1R null mammary buds transplanted into WT hosts also possess limited outgrowth potential and show defects in TEB proliferation (Bonnette and Hadsell 2001), illustrate the importance of the IGF1 axis for mammary development. The RhoGAP, p190-B mentioned previously, also modulates the IGF1/IGF1R axis during pubertal outgrowth. In p190-B heterozygous females the ductal outgrowth rate is slower and decreased levels of IRS-1/2 were observed in TEBs (Chakravarty et al. 2003). Interestingly, pregnancy hormones partially overcome the proliferation defect of the IGF1R null epithelium, potentially due to increased insulin sensitivity (discussed in Bonnette and Hadsell 2001). IGF1 is also required for lobuloalveolar development of the mammary gland (Loladze et al. 2006).
Multiple EGF-family ligands and all of the ErbB receptors are expressed in the mammary gland during ductal outgrowth, pregnancy-induced alveolar differentiation, and in lactation (Schroeder and Lee 1998). EGF-family ligands are produced as transmembrane precursors that on cleavage bind and activate ErbB receptors. Members of the ADAM (a disintegrin and metalloproteinase) family of proteases, as well as matrix metalloproteinases (MMP) cleave and release EGF-related ligands (Sanderson et al. 2006; Bergers and Coussens 2000). During pubertal outgrowth EGFR is strongly expressed in TEBs and adjacent stroma (Coleman et al. 1988) and waved-2 mutants with a kinase-impaired EGFR show less outgrowth (Sebastian et al. 1998). Reciprocal transplant experiments between WT epithelium and EGFR mutant stroma and the reverse, revealed that stromal EGFR is required during this phase (Wiesen et al. 1999).
The evidence pointing to epithelial-produced AR as the EGFR activating ligand at this developmental stage is very strong. Expression of AR, but none of the other EGFR ligands, is very high during puberty (Sternlicht et al. 2005). Moreover, AR appears to be a direct target of estradiol since upon injection of the steroid into 3-wk-old ovariectomized mice, AR expression is strongly up-regulated (Ciarloni et al. 2007). Finally, AR null epithelial grafts do not grow in WT mammary fat pads (Ciarloni et al. 2007). Interestingly, ADAM17 null mice have a phenotype very similar to that of AR null mice and it was shown that in the absence of this protease there is impaired cleavage and release of AR (Sternlicht et al. 2005). Thus, AR is required specifically downstream from estradiol in the epithelial compartment, where it is released by ADAM17 and acts on stromal EGFR to promote ductal elongation during puberty. This suggests a model whereby stromal cells respond to AR and produce soluble factors that act back on the epithelium. Some interesting candidates are expressed at this developmental stage including FGFs, IGF1, and HGF. Because absence of the epithelial FGFR2-IIIb also impairs pubertal outgrowth (Parsa et al. 2008), it is tempting to speculate that stromal-produced FGFs, e.g., FGF7 or FGF10, are the ligands acting on the epithelium. Indeed, it has been shown that FGF2 acts downstream from EGFR and supports growth of EGFR null mammary organoids in culture (Sternlicht et al. 2005). Despite the fact that stromal ErbB2 is also active at puberty (Sebastian et al. 1998), analyses carried out on ErbB2 null mammary epithelium showed that the receptor has a limiting role in the epithelium, not in the stroma. In the absence of epithelial ErbB2, impaired ductal elongation and branching (Jackson-Fisher et al. 2004; Andrechek et al. 2005), as well as alterations in TEB structures were observed (Jackson-Fisher et al. 2004) during pubertal outgrowth. ErbB2 requires a heterodimerization partner for ligand-induced activation and likely candidates for this role are NRG1 and ErbB3. On the one hand, implantation of NRG1 slow-release pellets promoted ductal outgrowth (Jones et al. 1996). On the other hand, transplantation of mammary buds from ErbB3 null embryos into recipient WT fat pads showed that in the absence of ErbB3 pubertal outgrowth was severely impaired and TEBs showed increased apoptosis (Jackson-Fisher et al. 2008).
EGFR, ErbB2, and ErbB3 each have proliferative and survival roles during mammary outgrowth. In contrast, ErbB4 has an essential role late in pregnancy and in lactation (reviewed in Stern 2008). Initial studies showed that during lactation ErbB4 was strongly phosphorylated on tyrosine (Schroeder and Lee 1998), indicative of a role at this developmental stage. A direct examination of ErbB4 null mammary epithelium showed that pubertal outgrowth was normal, however, alveolar differentiation and lactation were impaired (Tidcombe et al. 2003). Moreover, the defect was shown to be due to impaired phosphorylation of the transcription factor Stat5, which has an essential role in mammary gland differentiation and is activated during lactation by ErbB4 (Jones et al. 1999). ErbB4 is the only member of the family that undergoes γ-secretase cleavage, which releases the intracellular cytoplasmic domain of the receptor, allowing its nuclear entry (Ni et al. 2001; Sardi et al. 2006). This is interesting in light of the finding that in lactating mammary glands this domain of ErbB4 is present in the nucleus (Long et al. 2003). The other three ErbB family members have also been detected in the nucleus of cells in various normal tissues, although this has not been described in the mammary gland (reviewed in Wang and Hung 2009).
There are two candidates for the ligand activating ErbB4 during late stages of pregnancy and in lactation. One is NRGα1, which when deleted has a transient effect on alveolar development late in pregnancy and early in lactation (Li et al. 2002). In addition, there is evidence suggesting that HB-EGF might mediate ErbB4 activation during lactation. The heparin-sulfate proteoglycan CD44 has been shown to bind MMP-7 and pro-HB-EGF on the surface of mammary epithelial cells and this binding is required for HB-EGF processing. A lactation defect has been observed in CD44 null females. Moreover, neither HB-EGF processing, nor ErbB4 activation were observed in mammary glands lacking the proteoglycan (Yu et al. 2002).
RTKs AND MAMMARY CANCER
A prevailing theme during the proliferative phase of mammary gland development is paracrine receptor activation by ligands produced in other cells. One of the most striking differences between normal development and cancer is the ability of the estrogen receptor (ER) positive cancer cells to proliferate; in the normal gland the ER positive cells do not divide but respond by producing paracrine factors that act on other cells, AR being a good example. This characteristic appears to be lost in cancer, something that distinguishes the normal mammary gland from breast cancer (Anderson and Clarke 2004). In this section we will briefly review cancer roles for the peptides discussed in normal development.
FGFs AND FGFRs IN CANCER
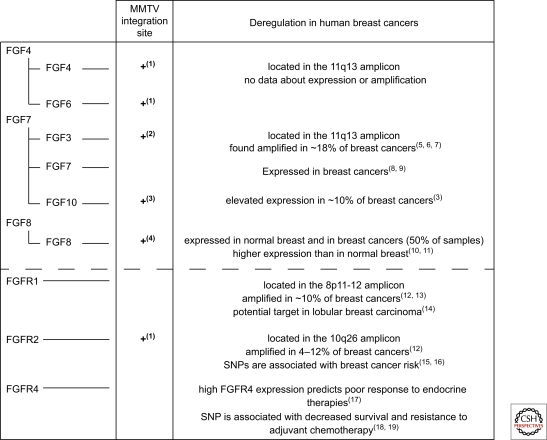
The association between FGF and mammary cancer was first established in MMTV-induced tumors, where proviral insertional mutagenesis led to FGF3 transcriptional activation (Peters et al. 1989). A recent large-scale screen revealed that the FGF pathway was activated in >65% of MMTV-induced tumors. Integration sites were adjacent to FGFR1 and FGFR2 in addition to FGF3, 4, 6, 8, and 10 (Theodorou et al. 2007). Interestingly, FGF8 and FGF10, ligands for FGFR2 (Fig. 2), are also overexpressed in human breast cancer (Marsh et al. 1999; Theodorou et al. 2004).
FGFR1, FGFR2, and FGFR4 have each been implicated in breast cancer development. The 8p11-12 amplicon harboring FGFR1 has been detected in 8%–10% of breast cancers (Adnane et al. 1991; Ray et al. 2004; Reis-Filho et al. 2006) and correlates with poor outcome in patients with ER positive tumors (Elsheikh et al. 2007), suggestive of a role in endocrine resistance. FGFR1 is overexpressed in some, but not all tumors with the amplicon, likely reflecting the complexity of this amplicon. Irrespective of this, elevated levels of the receptor have been detected in lobular breast carcinomas (Reis-Filho et al. 2006; Xian et al. 2009). Genome-wide screens aimed at uncovering breast cancer associated genes identified single-nucleotide polymorphisms (SNP) in introns of FGFR2 (Easton et al. 2007; Hunter et al. 2007). One of the SNPs alters the binding of Oct-1/Runx2 to the DNA and, using an in vitro transcriptional assay it was shown that this SNP increased FGFR2 transcription (Meyer et al. 2008). Primary tumors with this SNP show elevated FGFR2 levels (Nordgard et al. 2007). FGFR2 amplification (10q26) has also been detected in a subgroup of triple negative (negative for ER, PR, and high ErbB2) breast cancers (Turner et al. 2010). High levels of the FGFR4Arg388 allele have been correlated with poor response to chemotherapy (Thussbas et al. 2006) and endocrine therapy (Meijer et al. 2008). Introduction of this allele into a transgenic mammary tumor model led to more rapid tumor development and tumor progression (Seitzer et al. 2010). Other clinical observations on the ligands and receptors are presented in Figure 3.
Figure 3.
Deregulation of FGF ligands and receptors in mammary cancer. MMTV proviral insertional mutagenesis led to the identification of five FGF ligands and the FGFR2 whose expression promoted mammary cancer. Some of these ligands have also been implicated in human breast cancer. FGFR1 and FGFR2 have also been shown to contribute to breast cancer development, whereas FGFR4 has been implicated in therapy response. The references used for this figure are: (1) (Theodorou et al. 2007), (2) (Peters et al. 1989), (3) (Theodorou et al. 2004), (4) (MacArthur et al. 1995), (5) (Naidu et al. 2001), (6) (Fioravanti et al. 1997), (7) (Berns et al. 1995), (8) (Jacquemier et al. 1998), (9) (Bansal et al. 1997), (10) (Zammit et al. 2002), (11) (Marsh et al. 1999), (12) (Adnane et al. 1991), (13) (Ray et al. 2004), (14) (Reis-Filho et al. 2006), (15) (Hunter et al. 2007), (16) (Easton et al. 2007), (17) (Meijer et al. 2008), (18) (Bange et al. 2002).
EGF-FAMILY PEPTIDES AND ErbB RECEPTORS IN CANCER
In contrast to the FGFs that are often activated by MMTV insertional mutagenesis (Theodorou et al. 2007), we are not aware of any reports showing that EGF-family peptides or their receptors are activated by the provirus. However, mammary transgenic models, most driven by the MMTV promoter, have been made with multiple peptides and with the EGFR and ErbB2. MMTV-NRG transgenics show persistent TEBs and mammary cancers arise with a long latency (Krane and Leder 1996). MMTV-EGFR transgenics develop hyperplasias and cancer (Brandt et al. 2000). Mammary tumors arising from transgenic expression of TGFα and ErbB2/Neu have been extensively analyzed (reviewed in Cardiff and Wellings 1999; Mikaelian et al. 2004). Moreover, MMTV-ErbB2/Neu mice are often used to test the importance of particular proteins in tumor outgrowth and metastasis formation (White et al. 2004; Lahlou et al. 2007).
A vast body of evidence suggests that the EGF-family peptides and their receptors have an important role in human breast cancer. ERBB2 gene amplification was the first consistent genetic alteration found in approximately 25% of primary tumors (Slamon et al. 1987; van de Vijver et al. 1987; Berger et al. 1988). Because this was discovered more than 20 yr ago, numerous studies on the role of this receptor in breast cancer and on its potential as a therapeutic target have been published (reviewed in Hynes and Lane 2005). The basal-like subtype of breast cancer, also referred to as triple negative due to lack of ER, PR (progesterone receptor), and ErbB2, has a very aggressive clinical behavior (Nielsen et al. 2004). EGFR is expressed at moderate to high levels in 30%–60% of basal-like breast cancer (Reis-Filho and Tutt 2008; Thike et al. 2009). Clinical trials testing the potential of EGFR tyrosine kinase inhibitors are ongoing in breast cancer (see ClinicalTrials.gov).
IGF-1 AND IGF1R IN CANCER
There are numerous studies suggesting that IGF-1 and its receptor have important roles in breast cancer (Sachdev and Yee 2001; Pollak et al. 2004). The IGF/IGF1R axis impacts on many aspects of the tumor via its ability to act on the Ras/MAPK/ERK proliferative pathway and the PI3K survival pathway. Indeed, even the ratio of IGF-1 to its binding protein IGFBP-3 has been shown to modify breast cancer risk in BRCA mutation carriers (Mitchell et al. 2006). It is also interesting to note that down-regulation of IGFBP-3 leading to up-regulation of IGF1-R signaling and PI3K pathway activation was described as a mechanism underlying resistance to an ErbB inhibitor (Guix et al. 2008). This shows not only the importance of IGF-1/IGF1R signaling in activating the PI3K pathway, but also suggests that in the clinic up-regulation of this pathway should be monitored when ErbB blocking inhibitors are used. Recently, a mouse model of basal-like breast cancer that is dependent on IGF1R has been described and in the future should be useful for testing targeted therapies (Klinakis et al. 2009).
CYTOKINES AND THEIR RECEPTORS IN NORMAL DEVELOPMENT AND CANCER
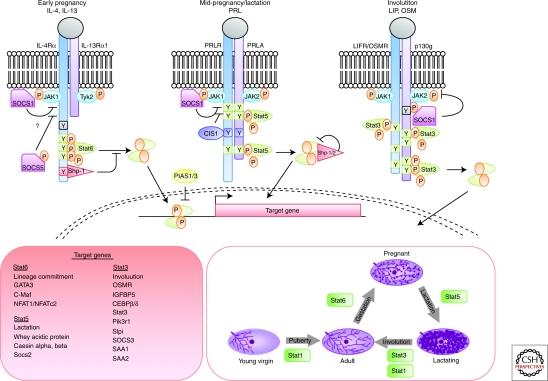
Cytokines comprise a large family of soluble proteins that mediate cell communication and induce potent biological responses often by inducing the expression of receptors, co-receptors, or adaptors. Their receptors can be grouped into families that share similar structures and signal through the Jak/Stat pathway. Notably, unlike the EGF, FGF, and IGF families of receptors, cytokine receptors do not have intrinsic kinase activity. They rely instead on receptor-associated Jak kinases (Haan et al. 2006). In mammals there are four Jaks that are ubiquitously expressed with the exception of Jak3. On ligand binding, the Jaks transphosphorylate each other and the receptor chains to create docking sites for Stats that are then also tyrosine phosphorylated by the Jaks to create active Stat dimers. These subsequently translocate to the nucleus, bind to palindromic motifs (TTCCNGGAA), and activate transcription of sets of target genes that are specific for each Stat (Fig. 4).
Figure 4.
Cytokine signaling through the Jak/Stat pathway in mammary gland development. Sequential activation of Stat6 and Stat5 control alveologenesis during pregnancy. These Stats are activated in response to specific ligands, IL-4 and IL-13 (Stat6) and Prl (Stat5) that bind to receptors of the cytokine family. During involution, cell death is mediated by Stat3, which is activated by LIF and then by OSM, which share the common gp130 receptor. Stat signaling is negatively regulated by the family of SOCS proteins that are direct transcriptional targets of each Stat factor. Once activated, Stats bind to similar consensus sequences but activate transcription of distinct sets of target genes to control mammary gland development.
CYTOKINE RECEPTOR FAMILIES
There are five main receptor families that have different receptor chains (usually two or three) and are activated by different ligands: the interferon (IFN), gp130, γC, IL-3, and single chain families (Schindler and Plumlee 2008). The corresponding ligands are diverse and include the interleukins (IL), leptin, leukemia inhibitory factor (LIF), and prolactin (Prl). Different combinations of JAKs associate with each receptor, for example, Jak1 and Jak2 bind to the IFNγ receptor. The exception is the single chain family where the receptor is a homodimer and Jak2 is invariably the associated kinase. The most relevant member of this family is the Prl receptor (PRLR).
THE STAT FAMILY OF TRANSCRIPTION FACTORS
There are seven mammalian Stats (Stats1–6, 5a, and 5b) that are approximately 91 kDa in size and are distributed in pairs on three different chromosomes. They appear to have arisen from a single primordial gene with the Stat3 and Stat5a/b pair being most closely related to homologs found in Dictyostelium, Caenorhabditis elegans, and Drosophila (Kisseleva et al. 2002).
A variety of biochemical and genetic studies have revealed which Stats are downstream effectors of particular cytokines. Although Stat2 is specific for IFN-I, Stat4 for IL-12, IL-23, and IL-27, and Stat6 for IL-4 and IL-13, a much wider range of cytokines signal through Stat1, Stat3, and Stats5a/b. The IL-6 family (IL-6, IL-11, IL-31, LIF, CNTF, CLC/CLF, NP, CT1, and OSM), the IL-10 family (IL-10, IL-19, IL-20, IL-22, IL-24, and IL-26), and G-CSF, Leptin, and IL-21 all activate Stat3 and the IL-3 family (IL-3, IL-5, and GM-CSF), the single chain family (GH, Prl, Tpo, and Epo), and the γC family (IL-2, IL-7, IL-9, IL-15, and IL-21) activate Stat5a and Stat5b that are 96% identical at the amino acid level. Apart from the response to Prl (Stat5a) and GH (Stat5b) these Stats are not entirely functionally redundant and, interestingly, appear to bind DNA differently as Stat5a preferentially forms teteramers whereas Stat5b favors dimers (Verdier et al. 1998). Stat1 is the only Stat downstream from IFNγ but is also activated by some cytokines of the gp130 family and EGF.
THE ROLES OF CYTOKINES AND STATS IN MAMMARY GLAND DEVELOPMENT
Development of the embryonic mammary gland proceeds unperturbed in mice that are deficient for Stats 1, 2, 4, 5, and 6. Thus, these Stats are unlikely to be important in mammary stem cells or early tissue development. Because deletion of Stat3 results in early embryonic lethality, it has not been possible to determine whether Stat3 is important at this stage of development. However, given that Stat3 is implicated in self-renewal of embryonic stem cells in culture, it would be interesting to determine if Stat3 has a role in establishing or maintaining embryonic mammary stem cells.
The adult mammary gland is quite different and, individual members of the Stat family have important roles. Stats 1 and 3 are constitutively expressed but Stat1 is tyrosine phosphorylated only in virgin gland and in late involution, following remodeling of the gland, while Stat3 activity is highest on the day of birth and during the first 6 days of involution (Watson 2006). Stat6 is expressed also throughout adult mammary gland development, and is activated during pregnancy before Stat5. Thus, there are discrete windows of Stat activity in the adult mammary gland. The expression of cytokines is less well known although a role for LIF in ductal elongation during puberty has been described (Kritikou et al. 2003).
Genetic studies in mice have revealed roles for Stats 3, 5a and b, and 6 in the adult gland during a pregnancy cycle whereas ablation of Stat1 has not been reported to have a major effect. Stat6 is activated by the type-2 cytokines, IL-4 and IL-13, in T cells and is associated with changes in chromatin structure and transcriptional up-regulation of GATA-3, c-Maf, and NFAT1/NFATc2. Surprisingly, in mammary gland, Stat6 is also activated by IL-4 and IL-13 and this correlates with the expression of IL4Rα and GATA-3 in the epithelium (Khaled et al. 2007). Importantly, in the absence of either Stat6 or IL-4 and IL-13, lobuloalveolar development is substantially delayed with numbers of alveoli reduced by about 70% although this defect is resolved by late pregnancy when it is likely that levels of GATA-3 are sufficient to specify and maintain the lineage in the absence of Stat6. GATA-3 is critically important as demonstrated by two similar studies in which Gata3 was conditionally deleted at different stages of mammary development (Kouros-Mehr et al. 2006; Asselin-Labat et al. 2007). Deletion of Gata3 in alveolar cells during pregnancy resulted in a block in alveolar differentiation and failed lactogenesis (Asselin-Labat et al. 2007). Doxycycline-inducible Cre-mediated deletion of Gata3 in late pregnancy resulted also in the disruption of ductal architecture (Kouros-Mehr et al. 2006). Using FACS to isolate subpopulations of epithelial cells revealed that the size of the luminal progenitor pool increases significantly in Gata3-null mice. Because overexpression of GATA3 in null cells induced the expression of the milk proteins β-casein and WAP, in the absence of lactogenic hormone stimulation, it seems reasonable to conclude that GATA-3 promotes the differentiation of lineage-restricted progenitor cells (Asselin-Labat et al. 2007).
Levels of GATA-3 are high also in the mammary epithelium of virgin mice and it is one of the earliest transcription factors to be expressed in mammary gland, being detectable in the embryonic placode. The importance of GATA-3 is illustrated further by the failure to form TEBs and a severe reduction in ductal outgrowth in the absence of GATA-3. A transcriptional link between GATA-3 and FOXA1 was uncovered using microarray and promoter analysis coupled with chromatin immunoprecipitation to identify a functional GATA-3 binding site in the FOXA1 promoter. Interestingly, FOXA1, is required for ERα transcriptional activity. Thus, GATA-3 is required at various stages of mammary gland development, placing this transcription factor earlier in the luminal lineage than Stat5 and Stat6.
It is interesting that phosphorylated Stat5 levels are reduced in Stat6-deficient mammary glands. Whether this is a consequence of reduced levels of IL-5, which is known to activate Stat5, rather than Prl is not known. A hierarchy of signaling from IL-4/IL-13 through Stat6 and GATA3 is thus an important constituent of commitment to the alveolar luminal lineage. Notably, GATA3 is highly expressed in breast cancers of the luminal A subtype, which also express ERα (Sorlie et al. 2003).
Stats 5a and 5b have long been known to be essential mediators of lobuloalveolar development (Miyoshi et al. 2001; Cui et al. 2004). These two Stat5 gene have different roles reflecting their upstream activators Prl (Stat5a) and GH (Stat5b) although they can compensate for each other, as shown by the rescue of impaired alveologenesis and failed lactation in the Stat5a knockout mouse by Stat5b (Liu et al. 1998). It can be concluded that Stats5a and b are essential for normal differentiation and survival of alveolar mammary epithelial cells but that because ductal morphogenesis is unaffected in their absence, this requirement is specific to the alveolar lineage. This distinguishes Stat5 from GATA-3, which is required in both ductal and alveolar lineages. Recently, transplantation studies of FACS-sorted cells suggested that alveolar progenitor cells could not be generated from a presumptive earlier progenitor in the absence of Stats5a and b. Notably, whereas GATA-3 expression is unperturbed in Stat5a/b null cells, the expression of Elf5 is severely diminished (Yamaji et al. 2009).
Once again, there are parallels with the hematopoietic system because Stat5a/b have been shown to control stem and progenitor cell fate and loss of a single allele of Stat5a/b resulted in increased HSC cycling and depletion of the long-term HSC pool (Wang et al. 2009). Furthermore, results obtained with mice completely null for Stat5a/b reveal that these factors are required for the normal development of all lymphoid cells (Yao et al. 2006).
Taken together, the data suggest that there are at least two alveolar lineages: a Prl-Stat5 controlled lineage where Elf5 is an essential regulator and a Stat6-GATA-3 lineage that may be ERα positive. It is interesting that these lineages are not independent of each other because ablation of either Stat5 or GATA-3 results in a loss of differentiated epithelium perhaps indicating essential interactions between cells of these lineages. Because GATA-3 is expressed in virgin gland whereas Stat6 is activated in only a few ductal cells in the virgin, it may be that GATA-3 is switched off in a subset of alveolar cells during pregnancy to be replaced by Elf5.
Stat3 is essential for the initiation of apoptosis and remodeling following forced weaning (Chapman et al. 1999). In the absence of Stat3, involution is dramatically delayed and the reversible phase can be extended for up to 6 d (Humphreys et al. 2002). Stat3 is activated by a number of cytokines. In involuting mammary gland, the upstream ligand is LIF, shown using LIF deficient mice (Kritikou et al. 2003) or by implanting LIF pellets into lactating mammary glands (Schere-Levy et al. 2003). Another upstream regulator of Stat3 activity in involution is TGFβ3 (Nguyen and Pollard 2000). Interestingly, IL-6 does not regulate Stat3 activity in involution (Zhao et al. 2002) although it does in breast cancer cells, suggesting perhaps that different signals downstream from their cognate receptors specifies either a proliferative or death response to specific Stat3-activating cytokines. Recently, it has been shown that the receptor for oncostatin M (OSMR) is required for the activation of Stat3 during the second phase of involution (Tiffen et al. 2008) where it may substitute for LIF which peaks at 12 hours involution. Because OSMR is transcriptionally regulated by Stat3, this provides positive feedback on the pathway enhancing the Stat3 signal in the second phase (Tiffen et al. 2008).
Thus, cytokines and their Stat targets have important roles in lineage commitment, survival, and death of mammary gland epithelium.
CYTOKINE SIGNALING AND BREAST CANCER
As mentioned earlier, GATA-3 expression is strongly correlated with the Luminal A subtype of breast tumors, which have the best prognostic outcome (Sorlie et al. 2003). Low GATA-3 expression is an indicator of poor clinical outcome. In mouse models of breast cancer, GATA-3 suppresses tumor dissemination and maintains differentiation.
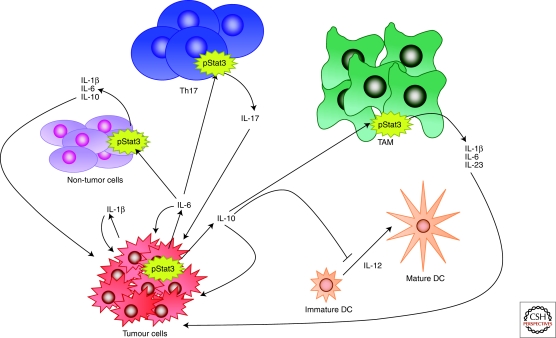
Several Stats, and in particular Stat3, have been shown to be constitutively activated in a wide range of blood and solid tumors and are frequently activated in breast carcinoma (Watson and Miller 1995) (Fig. 5). The apparent paradox between the death-inducing function of Stat3 in involution and its oncogenic and survival function in breast tumors, has prompted extensive investigation. Apart from the different cell types involved, namely fully differentiated alveolar cells in involution compared with ductal cells in most breast carcinomas, Stat3 activation results in the secretion of cytokines. Thus, chronic Stat3 activity in tumor cells results in paracrine signaling to cells in the tumor microenvironment including infiltrating immune cells. It is well established that inflammation has tumor-promoting potential and active Stat3 is often found at the invasive edge of tumors, adjacent to inflammatory cells (Bromberg and Wang 2009). Stat3 is also a key mediator of anti-inflammatory responses downstream from the IL-10 receptor and has recently been shown to enable tumors to evade the immune system by inhibiting the maturation of dendritic cells and macrophages (Wang et al. 2004). Accordingly, inhibition of Stat3 in macrophages induced an antitumor immune response in a rat model of breast cancer (Sun et al. 2006). Recently, using an activated ErbB2 mouse model of breast cancer, it was shown that although Stat3 does not affect the initiation of tumors, it dramatically increases lung metastasis (Ranger et al. 2009). Ablation of Stat3 in this model was associated with a reduction in inflammatory and angiogenic responses. Thus, breast cancers with active Stat3 condition other cells in the tumor microenvironment by secreting cytokines that promote both metastasis and immune evasion.
Figure 5.
Constitutive Stat3 activity in tumors affects the surrounding microenvironment. Chronic Stat3 activation in tumor cells results in secretion of IL-1β, IL-6, and IL-10; these in turn act on nontumor cells, T helper 17 cells (Th17) and tumor associated macrophages (TAM) resulting in further secretion of cytokines, which act in a feedback loop to promote growth and differentiation of the tumor cells. Stat3-directed secretion of IL-10 by the tumor cells also results in inhibition of antitumor immunity, for example by inhibiting the maturation of dendritic cells (DCs). Thus, Stat3 and the cytokines that it induces, promote tumor invasion and metastasis. Diagram adapted from Pensa et al. (2009).
Recently, a role for IL-6 in self-renewal of breast cancer stem cells was proposed (Sansone et al. 2007). Using the so-called mammosphere assay, where stem cells and their derivatives are cultured in floating spheroids, it was shown that breast tumor cells express higher levels of IL-6 mRNA than nontumor cells from the same patient although secretion of IL-6 by these cells was not measured. Furthermore, IL-6 treatment promoted spheroid growth in a Notch-3- dependent manner. IL-6 has been shown also to be a potent growth factor in ERα positive breast tumors (Sasser et al. 2007). Overexpression of IL-6 in MCF-7 breast cancer cells repressed E-cadherin expression and induced an EMT phenotype, and xenografts of these cells showed increased proliferation and advanced tumor grade (Sullivan et al. 2009). Interestingly, it has been shown recently that EMT induces stem cell-like properties (Mani et al. 2008). Cooperation between Stat3 and EGFR has been shown to be involved in EMT by mediating the expression of the TWIST gene (Lo et al. 2007). So, Stat3 could influence tumor spread by directly inducing EMT via up-regulation of TWIST expression and down-regulation of E-cadherin levels and enhancing self-renewal properties.
Stat3 and Stat5 have quite different roles in normal mammary gland development and have been suggested to play opposing roles in cell fate with Stat3 being pro-apoptotic and Stat5 prosurvival (Clarkson et al. 2006). A subset of breast tumors have both Stat3 and Stat5 constitutive activity and a recent study showed that these tumors are more differentiated than tumors with only Stat3 activity and furthermore, have more favorable prognostic characteristics (Walker et al. 2009). These data in breast tumors contrast with those in transgenic mice that develop tumors as a consequence of overexpression of either normal or activated Stat5 although these also have a differentiated phenotype (Iavnilovitch et al. 2004). Recent work has revealed that expression of constitutively active Stat5a suppresses the motility and invasiveness of MCF7 cells while in contrast, constitutively active Stat5b has no effect suggesting that expression of Stat5a, but not Stat5b, could be an indicator of better prognosis in breast cancer (Tang et al. 2010).
Although there is little evidence that Stat6 is oncogenic, Stat6 deficient mice are significantly more resistant to tumors arising from 4T1 mammary adenocarcinoma cells compared with wild-type BALB/c mice (Jensen et al. 2003). Furthermore, ablation of Stat6 results in enhanced survival of mice in which 4T1 primary tumors have been surgically excised (Ostrand-Rosenberg et al. 2002). This has been proposed to result from the polarization of the immune response toward a type 1 cytokine profile because Stat6 KO mice are unable to generate a type 2 response.
Thus, the Stats that are important for normal alveologenesis in mammary gland are also important in tumorigenesis either by promoting tumor growth (Stat5), generating an inflammatory tumor microenvironment (Stat3), or by facilitating immune evasion (Stat6 and Stat3).
ACKNOWLEDGMENTS
We would like to thank Drs. G. MacDonald and J. Dey (Basel) and Dr. J. Wickenden (Cambridge) for useful discussions and help with the figures. Work in the Hynes laboratory is supported by the Novartis Research Foundation. Work in the Watson laboratory is funded by the BBSRC and the Breast Cancer Campaign.
Footnotes
Editors: Mina J. Bissell, Kornelia Polyak, and Jeffrey Rosen
Additional Perspectives on Mammary Gland Biology available at www.cshperspectives.org
REFERENCES
- Adnane J, Gaudray P, Dionne CA, Crumley G, Jaye M, Schlessinger J, Jeanteur P, Birnbaum D, Theillet C 1991. BEK and FLG, two receptors to members of the FGF family, are amplified in subsets of human breast cancers. Oncogene 6: 659–663 [PubMed] [Google Scholar]
- Anderson E, Clarke RB 2004. Steroid receptors and cell cycle in normal mammary epithelium. J Mammary Gland Biol Neoplasia 9: 3–13 [DOI] [PubMed] [Google Scholar]
- Andrechek ER, White D, Muller WJ 2005. Targeted disruption of ErbB2/Neu in the mammary epithelium results in impaired ductal outgrowth. Oncogene 24: 932–937 [DOI] [PubMed] [Google Scholar]
- Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, et al. 2007. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol 9: 201–209 [DOI] [PubMed] [Google Scholar]
- Bange J, Prechtl D, Cheburkin Y, Specht K, Harbeck N, Schmitt M, Knyazeva T, Muller S, Gartner S, Sures I, et al. 2002. Cancer progression and tumor cell motility are associated with the FGFR4 Arg(388) allele. Cancer Res 62: 840–847 [PubMed] [Google Scholar]
- Bansal GS, Cox HC, Marsh S, Gomm JJ, Yiangou C, Luqmani Y, Coombes RC, Johnston CL 1997. Expression of keratinocyte growth factor and its receptor in human breast cancer. Br J Cancer 75: 1567–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken A, Mohammadi M 2009. The FGF family: Biology, pathophysiology and therapy. Nat Rev Drug Discov 8: 235–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MS, Locher GW, Saurer S, Gullick WJ, Waterfield MD, Groner B, Hynes NE 1988. Correlation of c-erbB-2 gene amplification and protein expression in human breast carcinoma with nodal status and nuclear grading. Cancer Res 48: 1238–1243 [PubMed] [Google Scholar]
- Bergers G, Coussens LM 2000. Extrinsic regulators of epithelial tumor progression: Metalloproteinases. Curr Opin Genet Dev 10: 120–127 [DOI] [PubMed] [Google Scholar]
- Berns EM, Foekens JA, van Staveren IL, van Putten WL, de Koning HY, Portengen H, Klijn JG 1995. Oncogene amplification and prognosis in breast cancer: Relationship with systemic treatment. Gene 159: 11–18 [DOI] [PubMed] [Google Scholar]
- Bonnette SG, Hadsell DL 2001. Targeted disruption of the IGF-I receptor gene decreases cellular proliferation in mammary terminal end buds. Endocrinology 142: 4937–4945 [DOI] [PubMed] [Google Scholar]
- Brandt R, Eisenbrandt R, Leenders F, Zschiesche W, Binas B, Juergensen C, Theuring F 2000. Mammary gland specific hEGF receptor transgene expression induces neoplasia and inhibits differentiation. Oncogene 19: 2129–2137 [DOI] [PubMed] [Google Scholar]
- Bromberg J, Wang TC 2009. Inflammation and cancer: IL-6 and STAT3 complete the link. Cancer Cell 15: 79–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardiff RD, Wellings SR 1999. The comparative pathology of human and mouse mammary glands. J Mammary Gland Biol Neoplasia 4: 105–122 [DOI] [PubMed] [Google Scholar]
- Chakravarty G, Hadsell D, Buitrago W, Settleman J, Rosen JM 2003. p190-B RhoGAP regulates mammary ductal morphogenesis. Mol Endo 17: 1054–1065 [DOI] [PubMed] [Google Scholar]
- Chapman RS, Lourenco PC, Tonner E, Flint DJ, Selbert S, Takeda K, Akira S, Clarke AR, Watson CJ 1999. Suppression of epithelial apoptosis and delayed mammary gland involution in mice with a conditional knockout of Stat3. Genes Dev 13: 2604–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarloni L, Mallepell S, Brisken C 2007. Amphiregulin is an essential mediator of estrogen receptor alpha function in mammary gland development. Proc Natl Acad Sci 104: 5455–5460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson RW, Boland MP, Kritikou EA, Lee JM, Freeman TC, Tiffen PG, Watson CJ 2006. The genes induced by signal transducer and activators of transcription (STAT)3 and STAT5 in mammary epithelial cells define the roles of these STATs in mammary development. Mol Endocrinol 20: 675–685 [DOI] [PubMed] [Google Scholar]
- Coleman-Krnacik S, Rosen JM 1994. Differential temporal and spatial gene expression of fibroblast growth factor family members during mouse mammary gland development. Mol Endocrinol 8: 218–229 [DOI] [PubMed] [Google Scholar]
- Coleman S, Silberstein GB, Daniel CW 1988. Ductal morphogenesis in the mouse mammary gland: Evidence supporting a role for epidermal growth factor. Dev Biol 127: 304–315 [DOI] [PubMed] [Google Scholar]
- Cui Y, Riedlinger G, Miyoshi K, Tang W, Li C, Deng CX, Robinson GW, Hennighausen L 2004. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol 24: 8037–8047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meyts P 2004. Insulin and its receptor: Structure, function and evolution. Bioessays 26: 1351–1362 [DOI] [PubMed] [Google Scholar]
- Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, et al. 2007. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 447: 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsheikh SE, Green AR, Lambros MBK, Turner NC, Grainge MJ, Powe D, Ellis IO, Reis-Filho JS 2007. FGFR1 amplification in breast carcinomas: A chromogenic in situ hybridisation analysis. Breast Cancer Res 9: R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fioravanti L, Cappelletti V, Coradini D, Miodini P, Borsani G, Daidone MG, Di Fronzo G 1997. int-2 Oncogene amplification and prognosis in node-negative breast carcinoma. Int J Cancer 74: 620–624 [DOI] [PubMed] [Google Scholar]
- Graus-Porta D, Beerli RR, Daly JM, Hynes NE 1997. ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J 16: 1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guix M, Faber AC, Wang SE, Olivares MG, Song Y, Qu S, Rinehart C, Seidel B, Yee D, Arteaga CL, et al. 2008. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest 118: 2609–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haan C, Kreis S, Margue C, Behrmann I 2006. Jaks and cytokine receptors–an intimate relationship. Biochem Pharmacol 72: 1538–1546 [DOI] [PubMed] [Google Scholar]
- Heckman BM, Chakravarty G, Vargo-Gogola T, Gonzales-Rimbau M, Hadsell DL, Lee AV, Settleman J, Rosen JM 2007. Crosstalk between the p190-B RhoGAP and IGF signaling pathways is required for embryonic mammary bud development. Dev Biol 309: 137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbro T, Beerli RR, Maurer F, Koziczak M, Barbas CF 3rd, Hynes NE 2003. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc Natl Acad Sci 100: 8933–8938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovey RC, Trott JF, Vonderhaar BK 2002. Establishing a framework for the functional mammary gland: From endocrinology to morphology. J Mammary Gland Biol Neoplasia 7: 17–38 [DOI] [PubMed] [Google Scholar]
- Howard B, Panchal H, McCarthy A, Ashworth A 2005. Identification of the scaramanga gene implicates Neuregulin3 in mammary gland specification. Genes Dev 19: 2078–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys RC, Bierie B, Zhao L, Raz R, Levy D, Hennighausen L 2002. Deletion of Stat3 blocks mammary gland involution and extends functional competence of the secretory epithelium in the absence of lactogenic stimuli. Endocrinology 143: 3641–3650 [DOI] [PubMed] [Google Scholar]
- Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A, et al. 2007. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet 39: 870–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes NE, Lane HA 2005. ERBB receptors and cancer: The complexity of targeted inhibitors. Nat Rev Cancer 5: 341–354 [DOI] [PubMed] [Google Scholar]
- Iavnilovitch E, Cardiff RD, Groner B, Barash I 2004. Deregulation of Stat5 expression and activation causes mammary tumors in transgenic mice. Int J Cancer 112: 607–619 [DOI] [PubMed] [Google Scholar]
- Itoh N, Ornitz DM 2004. Evolution of the Fgf and Fgfr gene families. Trends Genet 20: 563–569 [DOI] [PubMed] [Google Scholar]
- Jackson-Fisher AJ, Bellinger G, Breindel JL, Tavassoli FA, Booth CJ, Duong JK, Stern DF 2008. ErbB3 is required for ductal morphogenesis in the mouse mammary gland. Breast Cancer Res 10: R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Fisher AJ, Bellinger G, Ramabhadran R, Morris JK, Lee KF, Stern DF 2004. ErbB2 is required for ductal morphogenesis of the mammary gland. Proc Natl Acad Sci 101: 17138–17143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemier J, Sun ZZ, Penault-Llorca F, Geneix J, Devilard E, Adelaide J, Birnbaum D 1998. FGF7 protein expression in human breast carcinomas. J Pathol 186: 269–274 [DOI] [PubMed] [Google Scholar]
- Jensen SM, Meijer SL, Kurt RA, Urba WJ, Hu HM, Fox BA 2003. Regression of a mammary adenocarcinoma in STAT6-/- mice is dependent on the presence of STAT6-reactive T cells. J Immunol 170: 2014–2021 [DOI] [PubMed] [Google Scholar]
- Jones FE, Jerry DJ, Guarino BC, Andrews GC, Stern DF 1996. Heregulin induces in vivo proliferation and differentiation of mammary epithelium into secretory lobuloalveoli. Cell Growth Differ 7: 1031–1038 [PubMed] [Google Scholar]
- Jones FE, Welte T, Fu XY, Stern DF 1999. ErbB4 signaling in the mammary gland is required for lobuloalveolar development and Stat5 activation during lactation. J Cell Biol 147: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaled WT, Read EK, Nicholson SE, Baxter FO, Brennan AJ, Came PJ, Sprigg N, McKenzie AN, Watson CJ 2007. The IL-4/IL-13/Stat6 signalling pathway promotes luminal mammary epithelial cell development. Development 134: 2739–2750 [DOI] [PubMed] [Google Scholar]
- Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW 2002. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene 285: 1–24 [DOI] [PubMed] [Google Scholar]
- Kleinberg DL, Ruan W 2008. IGF-I, GH, and sex steroid effects in normal mammary gland development. J Mammary Gland Biol Neoplasia 13: 353–360 [DOI] [PubMed] [Google Scholar]
- Kleinberg DL, Wood TL, Furth PA, Lee AV 2009. Growth hormone and insulin-like growth factor-I in the transition from normal mammary development to preneoplastic mammary lesions. Endocr Rev 30: 51–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinakis A, Szabolcs M, Chen G, Xuan S, Hibshoosh H, Efstratiadis A 2009. Igfr1 as a therapeutic target in a mouse model of basal-like breast cancer. Proc Natl Acad Sci 106: 2359–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouhara H, Hadari YR, Spivak-Kroizman T, Schilling J, Bar-Sagi D, Lax I, Schlessinger J 1997. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell 89: 693–702 [DOI] [PubMed] [Google Scholar]
- Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z 2006. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell 127: 1041–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krane IM, Leder P 1996. NDF/heregulin induces persistence of terminal end buds and adenocarcinomas in the mammary glands of transgenic mice. Oncogene 12: 1781–1788 [PubMed] [Google Scholar]
- Kritikou EA, Sharkey A, Abell K, Came PJ, Anderson E, Clarkson RW, Watson CJ 2003. A dual, non-redundant, role for LIF as a regulator of development and STAT3-mediated cell death in mammary gland. Development 130: 3459–3468 [DOI] [PubMed] [Google Scholar]
- Kurosu H, Choi M, Ogawa Y, Dickson AS, Goetz R, Eliseenkova AV, Mohammadi M, Rosenblatt KP, Kliewer SA, Kuro-o M 2007. Tissue-specific expression of βKlotho and fibroblast growth factor (FGF) receptor isoforms determines metabolic activity of FGF19 and FGF21. J Biol Chem 282: 26687–26695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosu H, Ogawa Y, Miyoshi M, Yamamoto M, Nandi A, Rosenblatt KP, Baum MG, Schiavi S, Hu MC, Moe OW, et al. 2006. Regulation of fibroblast growth factor-23 signaling by klotho. J Biol Chem 281: 6120–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahlou H, Sanguin-Gendreau V, Zuo D, Cardiff RD, McLean GW, Frame MC, Muller WJ 2007. Mammary epithelial-specific disruption of the focal adhesion kinase blocks mammary tumor progression. Proc Natl Acad Sci 104: 20302–20307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew ED, Furdui CM, Anderson KS, Schlessinger J 2009. The precise sequence of FGF receptor autophosphorylation is kinetically driven and is disrupted by oncogenic mutations. Sci Signal 2: ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Cleary S, Mandarano MA, Long W, Birchmeier C, Jones FE 2002. The breast proto-oncogene, HRGα regulates epithelial proliferation and lobuloalveolar development in the mouse mammary gland. Oncogene 21: 4900–4907 [DOI] [PubMed] [Google Scholar]
- Liu X, Gallego MI, Smith GH, Robinson GW, Hennighausen L 1998. Functional rescue of Stat5a-null mammary tissue through the activation of compensating signals including Stat5b. Cell Growth Differ 9: 795–803 [PubMed] [Google Scholar]
- Lo HW, Hsu SC, Xia W, Cao X, Shih JY, Wei Y, Abbruzzese JL, Hortobagyi GN, Hung MC 2007. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res 67: 9066–9076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loladze AV, Stull MA, Rowzee AM, Demarco J, Lantry JH 3rd, Rosen CJ, Leroith D, Wagner KU, Hennighausen L, Wood TL 2006. Epithelial-specific and stage-specific functions of insulin-like growth factor-I during postnatal mammary development. Endocrinology 147: 5412–5423 [DOI] [PubMed] [Google Scholar]
- Long W, Wagner KU, Lloyd KC, Binart N, Shillingford JM, Hennighausen L, Jones FE 2003. Impaired differentiation and lactational failure of Erbb4-deficient mammary glands identify ERBB4 as an obligate mediator of STAT5. Development 130: 5257–5268 [DOI] [PubMed] [Google Scholar]
- Lu P, Ewald AJ, Martin GR, Werb Z 2008. Genetic mosaic analysis reveals FGF receptor 2 function in terminal end buds during mammary gland branching morphogenesis. Dev Biol 321: 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur CA, Shankar DB, Shackleford GM 1995. Fgf-8, activated by proviral insertion, cooperates with the Wnt-1 transgene in murine mammary tumorigenesis. J Virol 69: 2501–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailleux AA, Spencer-Dene B, Dillon C, Ndiaye D, Savona-Baron C, Itoh N, Kato S, Dickson C, Thiery JP, Bellusci S 2002. Role of FGF10/FGFR2b signaling during mammary gland development in the mouse embryo. Development 129: 53–60 [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. 2008. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133: 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh SK, Bansal GS, Zammit C, Barnard R, Coope R, Roberts-Clarke D, Gomm JJ, Coombes RC, Johnston CL 1999. Increased expression of fibroblast growth factor 8 in human breast cancer. Oncogene 18: 1053–1060 [DOI] [PubMed] [Google Scholar]
- Meijer D, Sieuwerts AM, Look MP, van Agthoven T, Foekens JA, Dorssers LC 2008. Fibroblast growth factor receptor 4 predicts failure on tamoxifen therapy in patients with recurrent breast cancer. Endocr Relat Cancer 15: 101–111 [DOI] [PubMed] [Google Scholar]
- Meyer KB, Maia AT, O'Reilly M, Teschendorff AE, Chin SF, Caldas C, Ponder BA 2008. Allele-specific up-regulation of FGFR2 increases susceptibility to breast cancer. PLoS Biol 6: e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikaelian I, Blades N, Churchill GA, Fancher K, Knowles BB, Eppig JT, Sundberg JP 2004. Proteotypic classification of spontaneous and transgenic mammary neoplasms. Breast Cancer Res 6: R668–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell G, Antoniou AC, Warren R, Peock S, Brown J, Davies R, Mattison J, Cook M, Warsi I, Evans DG, et al. 2006. Mammographic density and breast cancer risk in BRCA1 and BRCA2 mutation carriers. Cancer Res 66: 1866–1872 [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Shillingford JM, Smith GH, Grimm SL, Wagner KU, Oka T, Rosen JM, Robinson GW, Hennighausen L 2001. Signal transducer and activator of transcription (Stat) 5 controls the proliferation and differentiation of mammary alveolar epithelium. J Cell Biol 155: 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi M, Dikic I, Sorokin A, Burgess WH, Jaye M, Schlessinger J 1996. Identification of six novel autophosphorylation sites on fibroblast growth factor receptor 1 and elucidation of their importance in receptor activation and signal transduction. Mol Cell Biol 16: 977–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidu R, Wahab NA, Yadav M, Kutty MK, Nair S 2001. Detection of amplified int-2/FGF-3 gene in primary breast carcinomas using differential polymerase chain reaction. Int J Mol Med 8: 193–198 [DOI] [PubMed] [Google Scholar]
- Nguyen AV, Pollard JW 2000. Transforming growth factor beta3 induces cell death during the first stage of mammary gland involution. Development 127: 3107–3118 [DOI] [PubMed] [Google Scholar]
- Ni CY, Murphy MP, Golde TE, Carpenter G 2001. γ-Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science 294: 2179–2181 [DOI] [PubMed] [Google Scholar]
- Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, et al. 2004. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res 10: 5367–5374 [DOI] [PubMed] [Google Scholar]
- Nordgard SH, Johansen FE, Alnaes GI, Naume B, Borresen-Dale AL, Kristensen VN 2007. Genes harbouring susceptibility SNPs are differentially expressed in the breast cancer subtypes. Breast Cancer Res 9: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SH, Hadari YR, Gotoh N, Guy GR, Schlessinger J, Lax I 2001. Stimulation of phosphatidylinositol 3-kinase by fibroblast growth factor receptors is mediated by coordinated recruitment of multiple docking proteins. Proc Natl Acad Sci 98: 6074–6079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M 1996. Receptor specificity of the fibroblast growth factor family. J Biol Chem 271: 15292–15297 [DOI] [PubMed] [Google Scholar]
- Ostrand-Rosenberg S, Clements VK, Terabe M, Park JM, Berzofsky JA, Dissanayake SK 2002. Resistance to metastatic disease in STAT6-deficient mice requires hemopoietic and nonhemopoietic cells and is IFN-γ dependent. J Immunol 169: 5796–5804 [DOI] [PubMed] [Google Scholar]
- Parsa S, Ramasamy SK, De Langhe S, Gupte VV, Haigh JJ, Medina D, Bellusci S 2008. Terminal end bud maintenance in mammary gland is dependent upon FGFR2b signaling. Dev Biol 317: 121–131 [DOI] [PubMed] [Google Scholar]
- Pensa S, Watson CJ, Poli V 2009. Stat3 and the inflammation/acute phase response in involution and breast cancer. J Mammary Gland Biol Neoplasia 14: 121–129 [DOI] [PubMed] [Google Scholar]
- Peters G, Brookes S, Smith R, Placzek M, Dickson C 1989. The mouse homolog of the hst/k-FGF gene is adjacent to int-2 and is activated by proviral insertion in some virally induced mammary tumors. Proc Natl Acad Sci 86: 5678–5682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak MN, Schernhammer ES, Hankinson SE 2004. Insulin-like growth factors and neoplasia. Nat Rev Cancer 4: 505–518 [DOI] [PubMed] [Google Scholar]
- Ranger JJ, Levy DE, Shahalizadeh S, Hallett M, Muller WJ 2009. Identification of a Stat3-dependent transcription regulatory network involved in metastatic progression. Cancer Res 69: 6823–6830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray ME, Yang ZQ, Albertson D, Kleer CG, Washburn JG, Macoska JA, Ethier SP 2004. Genomic and expression analysis of the 8p11-12 amplicon in human breast cancer cell lines. Cancer Res 64: 40–47 [DOI] [PubMed] [Google Scholar]
- Reis-Filho JS, Simpson PT, Turner NC, Lambros MB, Jones C, Mackay A, Grigoriadis A, Sarrio D, Savage K, Dexter T, et al. 2006. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin Cancer Res 12: 6652–6662 [DOI] [PubMed] [Google Scholar]
- Reis-Filho JS, Tutt AN 2008. Triple negative tumours: A critical review. Histopathology 52: 108–118 [DOI] [PubMed] [Google Scholar]
- Ruan W, Kleinberg DL 1999. Insulin-like growth factor I is essential for terminal end bud formation and ductal morphogenesis during mammary development. Endocrinology 140: 5075–5081 [DOI] [PubMed] [Google Scholar]
- Sachdev D, Yee D 2001. The IGF system and breast cancer. Endocr Relat Cancer 8: 197–209 [DOI] [PubMed] [Google Scholar]
- Sanderson MP, Dempsey PJ, Dunbar AJ 2006. Control of ErbB signaling through metalloprotease mediated ectodomain shedding of EGF-like factors. Growth Factors 24: 121–136 [DOI] [PubMed] [Google Scholar]
- Sansone P, Storci G, Tavolari S, Guarnieri T, Giovannini C, Taffurelli M, Ceccarelli C, Santini D, Paterini P, Marcu KB, et al. 2007. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J Clin Invest 117: 3988–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardi SP, Murtie J, Koirala S, Patten BA, Corfas G 2006. Presenilin-dependent ErbB4 nuclear signaling regulates the timing of astrogenesis in the developing brain. Cell 127: 185–197 [DOI] [PubMed] [Google Scholar]
- Sasser AK, Sullivan NJ, Studebaker AW, Hendey LF, Axel AE, Hall BM 2007. Interleukin-6 is a potent growth factor for ER-α-positive human breast cancer. FASEB J 21: 3763–3770 [DOI] [PubMed] [Google Scholar]
- Schere-Levy C, Buggiano V, Quaglino A, Gattelli A, Cirio MC, Piazzon I, Vanzulli S, Kordon EC 2003. Leukemia inhibitory factor induces apoptosis of the mammary epithelial cells and participates in mouse mammary gland involution. Exp Cell Res 282: 35–47 [DOI] [PubMed] [Google Scholar]
- Schindler C, Plumlee C 2008. Inteferons pen the JAK-STAT pathway. Semin Cell Dev Biol 19: 311–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C, Levy DE, Decker T 2007. JAK-STAT signaling: From interferons to cytokines. J Biol Chem 282: 20059–20063 [DOI] [PubMed] [Google Scholar]
- Schroeder JA, Lee DC 1998. Dynamic expression and activation of ERBB receptors in the developing mouse mammary gland. Cell Growth Differ 9: 451–464 [PubMed] [Google Scholar]
- Schwertfeger KL 2009. Fibroblast growth factors in development and cancer: Insights from the mammary and prostate glands. Curr Drug Targets 10: 632–644 [DOI] [PubMed] [Google Scholar]
- Sebastian J, Richards RG, Walker MP, Wiesen JF, Werb Z, Derynck R, Hom YK, Cunha GR, DiAugustine RP 1998. Activation and function of the epidermal growth factor receptor and erbB-2 during mammary gland morphogenesis. Cell Growth Differ 9: 777–785 [PubMed] [Google Scholar]
- Seitzer N, Mayr T, Streit S, Ullrich A 2010. A single nucleotide change in the mouse genome accelerates breast cancer progression. Cancer Res 70: 802–812 [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL 1987. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235: 177–182 [DOI] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. 2003. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci 100: 8418–8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DF 2008. ERBB3/HER3 and ERBB2/HER2 duet in mammary development and breast cancer. J Mammary Gland Biol Neoplasia 13: 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternlicht MD, Sunnarborg SW, Kouros-Mehr H, Yu Y, Lee DC, Werb Z 2005. Mammary ductal morphogenesis requires paracrine activation of stromal EGFR via ADAM17-dependent shedding of epithelial amphiregulin. Development 132: 3923–3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan NJ, Sasser AK, Axel AE, Vesuna F, Raman V, Ramirez N, Oberyszyn TM, Hall BM 2009. Interleukin-6 induces an epithelial-mesenchymal transition phenotype in human breast cancer cells. Oncogene 28: 2940–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Yao Z, Liu S, Tang H, Yan X 2006. An oligonucleotide decoy for Stat3 activates the immune response of macrophages to breast cancer. Immunobiology 211: 199–209 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Uehara Y, Motomura-Matsuzaka K, Oki J, Koyama Y, Kimura M, Asada M, Komi-Kuramochi A, Oka S, Imamura T 2008. βKlotho is required for fibroblast growth factor (FGF) 21 signaling through FGF receptor (FGFR) 1c and FGFR3c. Mol Endocrinol 22: 1006–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JZ, Zuo ZH, Kong XJ, Steiner M, Yin Z, Perry JK, Zhu T, Liu DX, Lobie PE 2010. Signal transducer and activator of transcription (STAT)-5A and STAT5B differentially regulate human mammary carcinoma cell behavior. Endocrinology 151: 43–55 [DOI] [PubMed] [Google Scholar]
- Theodorou V, Boer M, Weigelt B, Jonkers J, van der Valk M, Hilkens J 2004. Fgf10 is an oncogene activated by MMTV insertional mutagenesis in mouse mammary tumors and overexpressed in a subset of human breast carcinomas. Oncogene 23: 6047–6055 [DOI] [PubMed] [Google Scholar]
- Theodorou V, Kimm MA, Boer M, Wessels L, Theelen W, Jonkers J, Hilkens J 2007. MMTV insertional mutagenesis identifies genes, gene families and pathways involved in mammary cancer. Nat Genet 39: 759–769 [DOI] [PubMed] [Google Scholar]
- Thike AA, Cheok PY, Jara-Lazaro AR, Tan B, Tan P, Tan PH 2009. Triple-negative breast cancer: Clinicopathological characteristics and relationship with basal-like breast cancer. Mod Pathol [DOI] [PubMed] [Google Scholar]
- Thussbas C, Nahrig J, Streit S, Bange J, Kriner M, Kates R, Ulm K, Kiechle M, Hoefler H, Ullrich A, et al. 2006. FGFR4 Arg388 allele is associated with resistance to adjuvant therapy in primary breast cancer. J Clin Oncol 24: 3747–3755 [DOI] [PubMed] [Google Scholar]
- Tidcombe H, Jackson-Fisher A, Mathers K, Stern DF, Gassmann M, Golding JP 2003. Neural and mammary gland defects in ErbB4 knockout mice genetically rescued from embryonic lethality. Proc Natl Acad Sci 100: 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffen PG, Omidvar N, Marquez-Almuina N, Croston D, Watson CJ, Clarkson RW 2008. A dual role for oncostatin M signaling in the differentiation and death of mammary epithelial cells in vivo. Mol Endocrinol 22: 2677–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N, Lambros MB, Horlings HM, Pearson A, sharpe R, Natrajan R, Geyer FC, van Kouwenhove M, Kreike B, Mackay A, et al. 2010. Integrative molecular profiling of triple negatvie breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene doi: 10.1038/onc.2009.489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Vijver M, van de Bersselaar R, Devilee P, Cornelisse C, Peterse J, Nusse R 1987. Amplification of the neu (c-erbB-2) oncogene in human mammmary tumors is relatively frequent and is often accompanied by amplification of the linked c-erbA oncogene. Mol Cell Biol 7: 2019–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltmaat JM, Relaix F, Le LT, Kratochwil K, Sala FG, van Veelen W, Rice R, Spencer-Dene B, Mailleux AA, Rice DP, et al. 2006. Gli3-mediated somitic Fgf10 expression gradients are required for the induction and patterning of mammary epithelium along the embryonic axes. Development 133: 2325–2335 [DOI] [PubMed] [Google Scholar]
- Verdier F, Rabionet R, Gouilleux F, Beisenherz-Huss C, Varlet P, Muller O, Mayeux P, Lacombe C, Gisselbrecht S, Chretien S 1998. A sequence of the CIS gene promoter interacts preferentially with two associated STAT5A dimers: A distinct biochemical difference between STAT5A and STAT5B. Mol Cell Biol 20: 389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SR, Nelson EA, Zou L, Chaudhury M, Signoretti S, Richardson A, Frank DA 2009. Reciprocal effects of STAT5 and STAT3 in breast cancer. Mol Cancer Res 7: 966–976 [DOI] [PubMed] [Google Scholar]
- Wang S-C, Hung M-C 2009. Nuclear translocation of the epidermal growth factor receptor family membrane tyrosine kinase receptors. Clin Cancer Res 15: 6484–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li G, Tse W, Bunting KD 2009. Conditional deletion of STAT5 in adult mouse hematopoietic stem cells causes loss of quiescence and permits efficient nonablative stem cell replacement. Blood 113: 4856–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Niu G, Kortylewski M, Burdelya L, Shain K, Zhang S, Bhattacharya R, Gabrilovich D, Heller R, Coppola D, et al. 2004. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med 10: 48–54 [DOI] [PubMed] [Google Scholar]
- Wansbury O, Panchal H, James M, Parry S, Ashworth A, Howard B 2008. Dynamic expression of Erbb pathway members during early mammary gland morphogenesis. J Invest Dermatol 128: 1009–1021 [DOI] [PubMed] [Google Scholar]
- Watson CJ 2006. Involution: Apoptosis and tissue remodelling that convert the mammary gland from milk factory to a quiescent organ. Breast Cancer Res 8: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CJ, Miller WR 1995. Elevated levels of members of the STAT family of transcription factors in breast carcinoma nuclear extracts. Br J Cancer 71: 840–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White DE, Kurpios NA, Zuo D, Hassell JA, Blaess S, Mueller U, Muller WJ 2004. Targeted disruption of β1-integrin in a transgenic mouse model of human breast cancer reveals an essential role in mammary tumor induction. Cancer Cell 6: 159–170 [DOI] [PubMed] [Google Scholar]
- Wiesen JF, Young P, Werb Z, Cunha GR 1999. Signaling through the stromal epidermal growth factor receptor is necessary for mammary ductal development. Development 126: 335–344 [DOI] [PubMed] [Google Scholar]
- Wood TL, Richert MM, Stull MA, Allar MA 2000. The insulin-like growth factors (IGFs) and IGF binding proteins in postnatal development of murine mammary glands. J Mammary Gland Biol Neoplasia 5: 31–42 [DOI] [PubMed] [Google Scholar]
- Xian W, Pappas L, Pandya D, Selfors LM, Derksen PW, drBruin M, Gray NS, Jonkers J, Rosen JM, Brugge JS 2009. Fibroblast growth factor receptor 1-transformed mammary epithelial cells are dependent on RSK activity for growth and survival. Cancer Res 69: 2244–2251 [DOI] [PubMed] [Google Scholar]
- Yamaji D, Na R, Feuermann Y, Pechhold S, Chen W, Robinson GW, Hennighausen L 2009. Development of mammary luminal progenitor cells is controlled by the transcription factor STAT5A. Genes Dev 23: 2382–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Cui Y, Watford WT, Bream JH, Yamaoka K, Hissong BD, Li D, Durum SK, Jiang Q, Bhandoola A, et al. 2006. Stat5a/b are essential for normal lymphoid development and differentiation. Proc Natl Acad Sci 103: 1000–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu WH, Woessner JF Jr, McNeish JD, Stamenkovic I 2002. CD44 anchors the assembly of matrilysin/MMP-7 with heparin-binding epidermal growth factor precursor and ErbB4 and regulates female reproductive organ remodeling. Genes Dev 16: 307–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit C, Coope R, Gomm JJ, Shousha S, Johnston CL, Coombes RC 2002. Fibroblast growth factor 8 is expressed at higher levels in lactating human breast and in breast cancer. Br J Cancer 86: 1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM 2006. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem 281: 15694–15700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Melenhorst JJ, Hennighausen L 2002. Loss of interleukin 6 results in delayed mammary gland involution: A possible role for mitogen-activated protein kinase and not signal transducer and activator of transcription 3. Mol Endocrinol 16: 2902–2912 [DOI] [PubMed] [Google Scholar]