Abstract
The budding yeast nucleus, like those of other eukaryotic species, is highly organized with respect to both chromosomal sequences and enzymatic activities. At the nuclear periphery interactions of nuclear pores with chromatin, mRNA, and transport factors promote efficient gene expression, whereas centromeres, telomeres, and silent chromatin are clustered and anchored away from pores. Internal nuclear organization appears to be function-dependent, reflecting localized sites for tRNA transcription, rDNA transcription, ribosome assembly, and DNA repair. Recent advances have identified new proteins involved in the positioning of chromatin and have allowed testing of the functional role of higher-order chromatin organization. The unequal distribution of silent information regulatory factors and histone modifying enzymes, which arises in part from the juxtaposition of telomeric repeats, has been shown to influence chromatin-mediated transcriptional repression. Other localization events suppress unwanted recombination. These findings highlight the contribution budding yeast genetics and cytology have made to dissecting the functional role of nuclear structure.
Yeast is an ideal model system for studies of nuclear function. Recent work has identified new proteins that control chromatin positioning and define specific “zones” for DNA repair and transcription.
With 16 chromosomes and a closed mitosis, the budding yeast nucleus provides an ideal model system for understanding structure–function relationships within one of the cell's most complex compartments. Eukaryotic chromatin has an intrinsic higher-order structure based on the folding of the nucleosomal fiber. Yet beyond the intrinsic folding of the 10- and 30-nm fibers, sequence-determined subchromosomal domains and entire chromosomes assume non-random positions in the nucleus. Here we discuss a few general principles of nuclear organization. The first is an order imposed by enzymatic function. Examples include replication foci, splicing centers, or sites of DNA repair by homologous recombination (Lamond and Spector 2003). The second is a global territorial organization that stems from a nonoverlapping positioning of entire chromosomes within the nucleus (Cremer et al. 2006). The third principle reflects binding characteristics of particular sequence elements, such as simple repeats, and the factors they recruit to assemble unique higher-order levels of chromatin folding (Gasser et al. 2004).
The most straightforward way to show the functional importance of nuclear and chromosomal structure is to identify the proteins and sequences involved and to mutate those elements. Any resulting alteration in nuclear function might stem from the change in organization. These loss-of-function approaches then need to be coupled with gain-of-function assays that restore or promote a particular function or spatial organization. For these manipulations budding yeast is an ideal organism. Yeast has extremely efficient homologous recombination machinery that allows for targeted gene disruption, mutagenesis, chromosomal modification, and in vivo tagging of both DNA and proteins for live imaging. These techniques can be coupled with powerful biochemical and molecular genetic techniques, many of which give genome-wide read-outs. Such approaches have provided basic insights into the relationship of nuclear structure and function (e.g., Andrulis et al. 1998; Taddei et al. 2004; Taddei et al. 2006).
Unfortunately, some elements of nuclear organization present in multicellular organisms are missing in yeast. Having a closed mitosis means that disassembly and reassembly of the yeast nuclear envelope does not occur during the cell cycle, although yeast has less pronounced changes in its nuclear membrane during cell division. Coupled with the yeast cell's closed mitosis is a lack of nuclear intermediate filament proteins or lamins, which form a rigid network underlying the inner nuclear membrane (INM). The nuclearlamina stabilizes nuclear shape and help anchor interphase chromatin (Gruenbaum et al. 2005). Yeast cells also lack many of the well-characterized subnuclear “bodies” with the exception of a nucleolar domain that functions in snoRNA maturation (Verheggen et al. 2001) and which appears to be equivalent to Cajal bodies (Gall, 2000). Despite lacking detectable splicing compartments (Misteli, 2000) or PML (promyelocytic leukemia) (Zhong et al. 2000) bodies, yeast supports many of the activities coordinated by these compartments, and recent evidence suggests that the yeast nuclear pore provides a platform for events related to gene expression and double strand break processing (Akhtar and Gasser 2007; Taddei 2007; Nagai et al. 2008). Other aspects of subnuclear organization, such as the distribution of chromatin within the nucleus, are conserved from yeast to man.
NUCLEAR ORGANIZATION
Nuclear Envelope Associated Proteins and the Nuclear Pore Complex
The nucleus is delimited by a double membrane called the nuclear envelope (NE) that is contiguous with the endoplasmic reticulum. The NE separates chromatin from the cytoplasm and provides anchorage for various nuclear and chromosomal structures, including the spindle pole body, and separate clusters of centromeres and telomeres. Trafficking between the nucleoplasm and the cytoplasm occurs through approximately 200 nuclear pore complexes (NPCs), which allow the free diffusion of small molecules, whereas regulating the transport of macromolecules. NPCs also provide a platform for mRNA transcription and quality control, as well as its export. These proteinaceous assemblies of approximately 50 MDa contain 456 nucleoporins of 30 different types (D'Angelo and Hetzer 2008). Each pore is a doughnut-shaped structure with eightfold symmetry around a central channel. Flexible protein filaments emanate from the core into both the cytoplasm and nucleoplasm, providing binding sites for both transport proteins and chromatin. A detailed map for the relative position of each nucleoporin was calculated based on multiple molecular, biochemical, and structural data revealing a strongly modular structure (Alber et al. 2007).
Beside nucleoporins, many yeast proteins were shown by imaging techniques to be associated with the nuclear envelope (Huh et al. 2003). Of particular interest are the integral proteins of the inner nuclear membrane (INM) (Lusk et al. 2007) including: Doa10, a RING domain containing protein that targets nuclear proteins for degradation, Mps3, a member of the SUN (Sad1, UNC-84) family that is a shared component of the INM and of the spindle pole body (SPB; Jaspersen et al. 2002), and Helix–extension–helix-1 and -2 (Src1 or Heh1 and Heh2) (King et al. 2006), which are orthologs of the mammalian lamin associated protein MAN1. These proteins interact with the nuclear lamina in mammals, as well as with chromatin through a small protein called BAF1. Although yeast lack the lamins, the roles of SUN-domain and Man1 family proteins in chromosome anchorage seem to be conserved (Grund et al. 2008). In addition, an INM-associated protein called Esc1 (Enhancer of silent chromatin 1) shares features with lamina components: Esc1 binds but does not span the inner nuclear membrane, and anchors silent chromatin through a component of the silencer complex, Sir4 (Taddei et al. 2004). Intriguingly, overexpression of Esc1 induces INM expansion (Hattier et al. 2007). Given their intimate proximity, it is not surprising that functional cross-talk among INM proteins such as Esc1 and the nuclear pore basket proteins Nup60, Mlp1 and Mlp2 (Therizols et al. 2006; Lewis et al. 2007; Palancade et al. 2007) has been detected.
Chromosome Folding
Observations of chromosome folding were originally recorded by one of the founders of the field of cytology, Carl Rabl (Rabl 1885), who described the fold-back conformation of anaphase chromosomes in spotted salamander larvae. In the so-called Rabl conformation a chromosome folds back at its centromere, such that telomeres are juxtaposed (Fig. 1). Today we know that this conformation results from the attachment of microtubules to kinetochores, which assemble at centromeric chromatin, and which lead the movement of chromosomes as chromosomes are pulled by microtubules into daughter cells. The telomeres, on the other hand, lag passively behind.
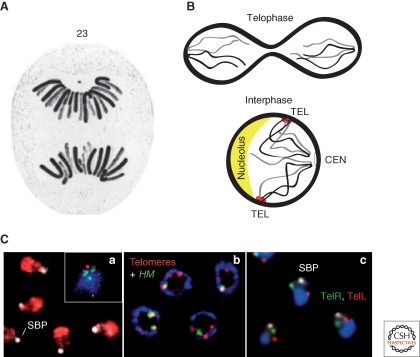
Figure 1.
Chromosomes have distinct orientation within the yeast interphase nucleus, which results from their Rabl configuration at anaphase: centromeres cluster and lead the way into daughter nuclei, with telomeres following behind. Telomeres of equal-length chromosome arms tend to cluster together in yeast interphase nuclei. (A) In 1885, Carl Rabl drew the folded conformation of anaphase chromosomes in spotted salamander larvae, in which anaphase chromosomes fold back on themselves because of attachment of the centromere to microtubules through the kinetochore. Centromeres lead the way as chromosomes are actively pulled into daughter cells. (B) The Rabl organization persists in the interphase yeast nucleus. (C) Confocal fluorescence images show (a) the clustering of yeast centromeres (green by FISH) near the spindle pole body (SPB, red) or in the larger image the nucleolus (red) opposite the SPB (anti-Spc42, white); (b) yeast telomeric foci are labeled with anti-Sir4 protein and FISH for the silent HM loci in yeast; (c) visualization of the centromere and right and left telomeres of Chr VI through Tel6R-CFP-lacO, Tel6L-YFP-TetR, and centromere staining (white).
Evidence that yeast chromosomes assume a Rabl conformation came initially from the work of Loidl and coworkers, who used fluorescent in situ hybridization (FISH) to identify chromosomal landmarks such as centromeres and telomeres (Jin et al. 2000). Using chromosome conformation capture (3C) to model chromosome conformation, the Kleckner lab predicted that yeast Chr III folds as a contorted ring, with a strong bend near the centromere and the telomeres in close proximity to each other (Dekker et al. 2002). Gasser and co-workers confirmed and extended these observations showing that not only Chr III, but other small metacentric chromosomes, such as Chr VI, form small loops in vivo with closely juxtaposed right and left telomeres. In contrast, telomeres on chromosome arms of vastly different lengths did not interact (Bystricky et al. 2005; Schober et al. 2008). Importantly, these studies showed that the Rabl configuration is not unique to anaphase in yeast, but persists throughout the cell cycle.
Using a genome-wide conformation capture approach, in which spatial chromosome interactions can be mapped, O'Sullivan and co-workers have recently shown that yeast chromosome placement is not random, i.e., some chromosomes were found never to interact, whereas others showed favored interactions (Rodley et al. 2009). Furthermore, this method detected intra-chromosomal loops, much like those described by microscopic analysis for chromosomes 3 and 6 (Bystricky et al. 2005; Schober et al. 2008). Consistently, a new imaging and data analysis approach revealed a strong confinement of several loci into “gene territories” reflecting the architectural constraints imposed on chromosomes by nuclear structures (i.e., centromere attachments to the SPB and telomere anchoring to the NE). Importantly, this organization can be significantly remodeled upon transcriptional activation (Berger et al. 2008).
Chromatin Dynamics
Chromatin in living cells is subjected to constant motion, which can be best described as a constrained random walk (Marshall et al. 1997) (Fig. 2). Rapid time-lapse imaging led to the distinction of at least two types of motion in yeast: small random movements (<0.2 µm within 1.5 s) that occur constantly, as well as larger, more directional movements (i.e., >0.5 µm in a 10.5 s interval) (Heun et al. 2001). The smaller movements are observed for internal loci as well as for peripheral silent domains, although the active loci in the nuclear lumen make large movements more frequently (i.e., a large movement is >0.5µm within 10.5 s) (Heun et al. 2001). Intriguingly, changes in cellular energy levels because of depletion of glucose or the addition of protonophores to deplete membrane potential, abolish large movements (Marshall et al. 1997; Heun et al. 2001). This suggests that interphase chromatin movement at least responds to ATP levels. Recent work suggests that these most likely stem from the action of chromatin remodelers, rather than RNA or DNA polymerases. Indeed, highly transcribed galactose-inducible genes that become tethered at nuclear pores show less mobility, despite their high level of transcription (Taddei et al. 2006; Cabal et al. 2006). Moreover, chromatin movement decreases, rather than increases, as cells enter S phase and DNA polymerases are active (Heun et al. 2001). Given that the mobility of a genomic locus is constrained by its continuity with the rest of the chromosome (Gartenberg et al. 2004), it is likely that the S-phase associated drop in mobility reflects the association of DNA in replication factories (Kitamura et al. 2006).
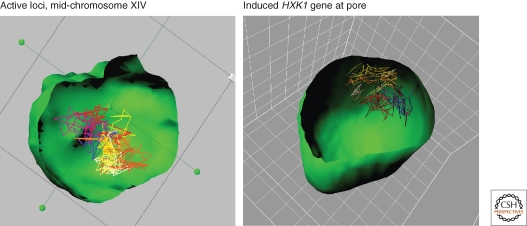
Figure 2.
Rapid time-lapse imaging of GFP-lacI tagged chromosomal loci in yeast cells bearing GFP-Nup49, allows 3D tracking of an internal locus (Gartenberg et al. 2004). 3D stacks are taken at 1.5 s intervals and the spatial rendering and alignment is performed with Imaris (Bitplane, Zürich). Tracks show movement over 7.5 min (300 stacks). To the left is a region of Chr IVX that shows low level transcription upon activation by glucose. To the right is shown the movement of the induced HXK1, a subtelomeric gene that is strongly induced in the absence of glucose (Taddei et al. 2006). Reductions in radius of constraint and diffusion coefficients are monitored for the pore-associated gene.
DNA BASED COMPARTMENTS: NUCLEOLUS, TELOMERES, tRNA
The Nucleolus
The most evident subnuclear compartment is the nucleolus, a crescent-shaped structure abutting the nuclear envelope and occupying roughly one third of the nucleus opposite the spindle pole body (Yang et al. 1989; Bystricky et al. 2005). This subcompartment is the site of RNA pol I-mediated rDNA transcription and ribosome subunit assembly and can be seen as a factory dedicated to ribosome biogenesis. Its morphology is strongly influenced by the cell growth rate, probably as a result of adapting the rate of ribosome production to the needs of the cell (Oakes et al. 1993; Powers and Walter, 1999). The nucleolus is a prime example of a functional organization principle, for it compartmentalizes different steps of ribosome biogenesis although the nucleolar machineries are in permanent exchange with the nucleoplasm and other nuclear bodies. The nucleolus further generates a domain of retention/sequestration of molecules that are normally active only outside the nucleolus (Sirri et al. 2008).
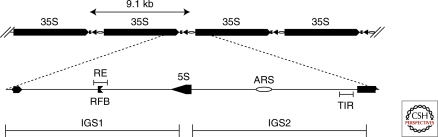
In budding yeast, rRNA is encoded in 100–200 tandem repeats. Each repeat unit is 9.1 kb in size and yields a 35S precursor rRNA, transcribed by RNA polymerase I and a 5S rRNA, transcribed by RNA polymerase III. The 5S unit is surrounded by two intergenic spacers, IGS1 and IGS2 (Fig. 3). Within this spacer there is a so-called polar replication fork barrier (RFB), a recombination enhancer (RE), a RNA polymerase I transcription initiation region (TIR) and an origin of replication (ARS, from autonomously replicating sequence).
Figure 3.
Organization of the rDNA repeats. The 9.1 kb unit encodes the 35S precursor for all the 25S, 18S, and 5.8S rRNA and the 5S rRNA. The 35S is transcribed by RNA polI whereas the 5S is transcribed by RNA pol III. The 9.1 kb unit is repeated up to 200 times on chr XII. Other abbreviations are as follows: IGS, intergenic spacer; RE recombination enhancer; RFB, Replication fork block, TIR, PolI transcription initiation region, open circle—origin of DNA replication.
This extended array of tandem repeats serves as an ideal template for homologous recombination, which is highly efficient in budding yeast. However, suppression of this recombination and the ensuing stability of the yeast rDNA array, are absolutely critical for cell growth and survival. By stabilizing the rDNA repeats the cell avoids or delays replicative senescence (Sinclair and Guarente 1997). The mechanisms that suppress recombination within the rDNA array involve both local nucleosomal organization mediated by Sir2 (Gottlieb and Esposito 1989; Bryk et al. 1997; Fritze et al. 1997; Smith and Boeke 1997) and long-range chromatin structure that involves tethering at the nuclear envelope.
tRNA genes
Although the 274 Pol III-transcribed tRNA genes are scattered throughout the yeast genome, many of these are clustered close to the nucleolus (Thompson et al. 2003; Wang et al. 2005). This association could create an environment that fosters the coregulation of transcription by RNA pol III and RNA pol I, given that the other major RNA pol III transcript is the 5S rRNA, which is part of the rDNA repeat (Fig. 3). Intriguingly, some tRNA genes remain associated with the nucleolus throughout the cell cycle, although they are not present on Chr XII, which contains the rDNA. This unusual positioning depends both on microtubules and on the action of condensin (Haeusler et al. 2008). Given the high number of RNA pol III-transcribed genes, their positioning is likely to affect the spatial organization of a large portion of the genome. Intriguingly, RNA pol II genes in the vicinity of tRNA genes become silenced through a phenomenon known as tRNA-gene mediated gene silencing (tgm) (Wang et al. 2005). Consistent with a link between nucleolar tethering and tRNA-associated silencing, it was shown that conditional ablation of condensin subunits leads to a coordinate loss of both nucleolar tRNA clustering and tgm (Haeusler et al. 2008). We note, however, that not all active tRNA genes relocate to the nucleolus. It remains to be determined what characterizes those that do show nucleolar association.
Telomere Foci: Assemblies of Repetitive DNA and Silencing Factors
Another type of subnuclear compartment stems from the clustering of the 32 yeast telomeres into three to eight foci, which are mainly found at the nuclear periphery. These foci are unfavorable for RNA polII-driven transcription. The budding yeast telomeric repeat consist of 250–300 base pairs of irregular tandem repeats (called TG1-3) (Shampay et al. 1984). A crucial feature of telomeric DNA is the 3′ overhang of the G-rich strand, which is 10–15bp in length (Larrivee et al. 2004). Toward the end of S phase after completion of replication, an extended G-rich overhang is generated, providing a template for the action of telomerase, a conserved ribonucleoprotein complex with reverse transcriptase activity (reviewed in Zakian 1996; Hug and Lingner 2006). The protein subunits of this complex were identified in a screen for “ever shorter telomeres” and are called Est1, Est2, and Est3 (Lundblad and Szostak 1989; Lendvay et al. 1996), whereas the RNA moiety that templates the extension of TG repeats is called Tlc1 (Singer and Gottschling 1994). Tlc1 and Est2 form the catalytic core of telomerase, whereas Est1 and Est3 are auxiliary subunits that are dispensable for in vitro activity (Lingner et al. 1997). The end-binding factor yKu interacts with telomerase and helps recruit it to telomeres (Stellwagen et al. 2003), as does the ssDNA binding factor Cdc13 (reviewed in Fisher et al. 2004).
Unlike this association with ssDNA, the Repressor Activator protein 1 binds the double-stranded telomeric repeat (Rap1) (Shore and Nasmyth 1987). Rap1 is a conserved factor with a double Myb-like domain that mediates high affinity binding to its consensus within the TG1-3 repeat. The Rap1 carboxyl terminus is a binding site for silencing factors Sir3 and Sir4 (Moretti et al. 1994; Marcand et al. 1997; Wotton and Shore 1997), which form a stochiometric complex with the NAD-dependent histone deacetylase Sir2, to mediate transcriptional repression in subtelomeric zones (Aparicio et al. 1991; Martino et al. 2009). The same site binds Rif1, which together with Rif2, antagonizes Sir4 interaction, and feeds back to limit telomerase activity (Wotton and Shore 1997).
To prevent spreading of silent chromatin, subtelomeric regions contain boundaries, which are characterized by the presence of acetylated histone variant Htz1 (or H2A.Z) (Meneghini et al. 2003; Babiarz et al. 2006). In addition to the immediate zone of SIR-mediated repression, characterized by nucleosomes that lack detectable acetylation or methylation, there is a subtelomeric region called HAST for (Hda1-affected subtelomeric) that is characterized by a continuous stretch of Hda1-deacetylated chromatin that extends 10–25 kb inwards from the telomeric repeat (Robyr et al. 2002). Thus telomeres are not only designed to ensure end replication, but they nucleate special domains with respect to SIR protein spreading and histone modifications.
MECHANISMS UNDERLYING NUCLEAR COMPARTMENTATION
Anchoring of DNA at the Nuclear Envelope
The positioning of chromosomes within the nucleus depends on reversible interactions of chromosomal landmarks with structural features of the nucleus, such as the spindle pole body (SPB) and the inner nuclear envelope. Centromeres are held in a cluster near the SPB through short microtubules that persist through interphase (Jin et al. 2000; Bystricky et al. 2004). The tethering of telomeres in perinuclear foci is achieved by two redundant pathways that require Sir4, a silencing factor, and the yKu70/yKu80 heterodimer (Hediger et al. 2002; Taddei et al. 2004). Sir4 anchors repressed chromatin to the NE through its partitioning and anchoring domain (PAD, aa 950 to 1262). PAD specifically binds Esc1, a low-abundance acidic protein associated exclusively with the inner face of the NE (Andrulis et al. 2002; Gartenberg et al. 2004; Taddei et al. 2004). By electron microscopy it was shown that Esc1 localizes in patches along the nuclear membrane independently of Sir4 and is excluded from nuclear pores (Taddei et al. 2004).
Although yKu80 and Sir4 interact, a mutant allele of yKu80 that loses interaction with Sir4 (yKu80-4) (Taddei et al. 2004; Roy et al. 2004) can still tether chromatin to the NE (Taddei et al. 2004). Given that this occurs in strains deleted for the genes encoding Sir4 and Esc1, it was proposed that yKu should bind at least one additional membrane-bound factor. This is achieved indirectly through the ability of yKu to bind telomerase (Schober et al. 2009).
Intriguingly, the requirements of the Sir4-independent yKu tethering pathway varied with phases of the cell cycle: yKu80 tethering was dependent on yKu70 in G1-phase but not in S-phase cells (Taddei et al. 2004). Conversely, the domain of yKu80 that binds a stem-loop in the telomerase RNA Tlc1, was found to be necessary for telomere anchoring in S-phase, but not in G1-phase cells (Schober et al. 2009). The S-phase specific anchoring by yKu not only required Tlc1, but Est1 as well, which forms a complex with the catalytic subunit of telomerase, Est2. It was shown that a targeted Est2 fusion protein can anchor DNA to the nuclear perimeter in an Est1-dependent manner, and that Est1 interacts with an integral nuclear membrane protein, Mps3 (Fig. 4) (Schober et al. 2009; Jaspersen et al. 2002; Antoniacci et al. 2007). An acidic amino-terminal domain of this SUN-domain family member extends into the nucleoplasm where it contacts Est1 to tether telomerase-bound telomeres. Yet Mps3 also appears to contribute to an alternative telomere anchoring pathway mediated by Sir4 (Bupp et al. 2007). In both cases Mps3-dependent anchoring is specific to S-phase cells.
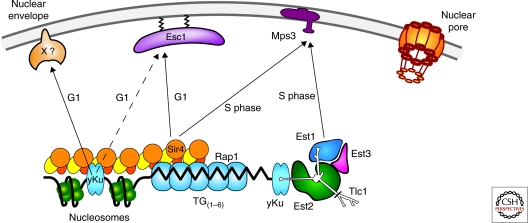
Figure 4.
Parallel mechanisms lead to yeast telomere attachment at the nuclear envelope. At different stages of the cell cycle the telomere associated proteins mediate different contacts with inner nuclear membrane components. The Sir4-PAD domain binds the Esc1 C terminus, as well as yKu80 and Mps3. yKu80 binds telomerase, which also associates with Mps3 in S phase through Est1. There is an unidentified anchor for yKu in G1 phase that is neither Esc1- nor Mps3-dependent.
The budding yeast Mps3 also organizes the SPB (Jaspersen et al. 2002; Nishikawa et al. 2003), yet by deletion of its N-terminal domain, the role of Mps3 in chromatin tethering could be separated from its essential role in spindle pole body organization (Bupp et al. 2007). Importantly, antagonism of the yKu-telomerase-Mps3 interaction led to hyper-recombination among telomeres, suggesting that this mechanism protects replicating ends from unequal strand invasion (Schober et al. 2009). This may be particularly relevant in S phase. Finally, it has been shown that the yKu-mediated peripheral positioning is switched off after DNA replication, leading to the dislodgment of telomeres from the nuclear envelope after replication (Ebrahimi and Donaldson 2008). Such release may facilitate mitotic separation of replicated telomeres.
Besides these pathways other proteins such as Asf1, Rtt109, Esc2, and Ctf18 have been implicated yeast telomers anchoring, although it could not be determined whether their effects were direct or indirect (Hiraga et al. 2006, 2008). Indeed, mutations in proteins of the nuclear pore such as Mlp1, Mlp2, or Nup133 were shown to interfere with mRNA export and cell cycle progression, which could indirectly affect perinuclear organization.
Trans-Association of Chromatin Loci
The high concentration of macromolecules within the nucleoplasm (between 100 and 400 mg/ml) produces volume exclusion effects that enhance attractive interactions among macromolecules, a phenomenon known as macromolecular crowding. This effect has been proposed to favor the formation of compartments by driving bulky components into structurally compact organizations (Richter et al. 2007). Combination of these effects with specific intermolecular interactions and attachment to nuclear structures such as the NE could account for the formation of subnuclear compartments.
One example of a functional compartment is provided by telomere clusters onto which silencing factors concentrate. The bifunctional role of Sir4 as a mediator of repression and an anchor for silent chromatin provides a mechanism for the self-organization of repressive compartments. In vivo, a nonsilent telomere can be localized at the NE through interaction with yKu, and thus be brought into proximity of other telomeres which generate a zone enriched for Sir proteins. Recruitment to this compartment results in a higher probability that a subtelomeric gene gets repressed. Once Sir-dependent repression is established, the Sir4 anchoring pathway ensures that the silent chromatin stays perinuclear. In this way, the formation of silent chromatin is self-reinforcing, thanks to its ability to bind to the same sites as telomeres.
Intriguingly, telomeres are not evenly distributed at the nuclear periphery but rather form discrete foci. We cannot rule out that telomere clustering arises in part from volume exclusion effects and molecular crowding (Iborra 2007), but it has also been shown that Sir proteins interact with themselves and with each other, establishing trans-interactions between telomeres. Recent work has shown that interactions between silent domains depends not only on silencing proteins Sir2, Sir3, and Sir4, but also on Sir1 and Esc2, two proteins involved in establishment of silencing at HM loci (Miele et al. 2009). Trans-interactions were not dependent on yKu or Esc1, suggesting that the mechanisms of NE-tethering and telomere clustering can be at least partially separated. Nonetheless, the self-organization of heterochromatin into perinuclear foci illustrates a self-perpetuating mechanism that applies to other chromatin-based domains of epigenetic character.
FUNCTIONAL CONSEQUENCES OF NUCLEAR ORGANIZATION ON GENE TRANSCRIPTION
Nuclear Organization and Gene Silencing
Chromatin-based compartments like those discussed earlier not only concentrate factors where they are needed, but also keep factors away from sites where they would interfere with other types of regulation (Taddei et al. 2009). Specifically, the clustering of telomeres leads to the sequestration of SIRs, which was shown both to favor subtelomeric repression and to prevent promiscuous effects on a distinct subset of promoters (Taddei et al. 2009). Essential to such a phenomenon is the fact that the Sir3 protein level, estimated at 1400 copies per cell (Ghaemmaghami et al. 2003), is both tightly regulated and limiting for the spread of silent chromatin (Renauld et al. 1993). Sir4 abundance is also tightly regulated, and low level overexpression of Sir4 actually disrupts telomeric repression.
It has been shown that each Rap1 tail and each nucleosome in the silent subtelomeric domain binds one SIR complex comprising Sir2, Sir3 and Sir4 in precisely equal molar ratio (Martino et al. 2009). This means that each telomere of ∼16 Rap1 binding sites (Gilson et al. 1993) and ∼18 subtelomeric nucleosomes (Renauld et al. 1993; Strahl-Bolsinger et al. 1997), provides binding sites for 30–40 SIR complexes. Because each haploid yeast cell has 32 telomeres clustered in three to eight telomeric foci, a focus contains several hundred potential SIR binding sites. The clustering of telomeric repeats could thus create a “sink” for limiting amounts of Sir proteins, and could account for the strikingly unequal distribution of this repressive complex within the nucleoplasm (Gotta et al. 1996).
Confirming the functional consequences of this unequal distribution of Sir proteins, it was found that silencer-nucleated repression is highly sensitive to the distance of the reporter from a telomere (Renauld et al. 1993; Stavenhagen and Zakian 1994; Thompson et al. 1994; Maillet et al. 1996; Marcand et al. 1996). Reciprocally, the tethering of a weakened HMR silencer to the NE favored the repression of a reporter gene (Andrulis et al. 1998). Importantly, this effect depends on the ability of telomere clusters to concentrate SIR factors at the nuclear envelope (Mondoux et al. 2007; Taddei et al. 2009). Thus, it appears that transcriptional silencing is not inherent to position, but rather requires facilitated access to a local high concentration of SIR proteins. Confirming this, when telomere anchoring is impaired, by deletion of YKU70 and ESC1, transcription is affected differently at different loci: genes at internal loci flanked by silencers show enhanced repression, whereas telomere-proximal repression is lost (Maillet et al. 2001; Taddei et al. 2009). In conclusion, the efficiency with which silent chromatin is formed depends on spatial concern, such as the local concentration of SIR proteins, and the strength of cis-acting nucleation elements.
In apparent contradiction with this interpretation is the absence of a correlation between the colocalization of a subtelomeric gene in telomere clusters (visualized as Rap1-GFP foci) and the efficiency of TPE (Mondoux et al. 2007). This may be because of the dynamic behavior of individual telomeres, which move into and out of Rap1 and SIR foci rapidly, presumably without loss of their epigenetic status, because they retain SIR factors (Schober et al. 2008). Indeed, a repressed ring of chromatin maintains a stable state of repression, even when it is excised from its subtelomeric chromosomal context, suggesting that a high local concentration of SIR factors is dispensable once silencing is established (Gartenberg et al. 2004).
The promiscuous repression of nontelomeric genes by released SIR factors, was analyzed genome-wide using a computational program that maps putative transcriptional factor sites. The affected promoters carry specific transcription factor motifs for either Abf1 or the PAC factors, Pdf1 and Pdf2, which are part of the Rpd3L histone deacetylase complex (RNA Polymerase A and C promoters; Dequard-Chablat et al. 1991; Zhu et al. 2009). A related motif, RRPE (ribosomal RNA processing element; Hughes et al. 2000) also correlated weakly with promiscuous SIR regulation. Intriguingly, these elements are primarily bound to promoters of genes expressing enzymes and proteins involved in ribosome biogenesis. Down-regulation may be seen as a global attempt to suppress growth, and perhaps favor a stress survival pathway.
These results make a forceful argument that changes in the spatial distribution of repetitive sequences that bind silencing factors can regulate patterns of gene expression genome-wide. The cell may exploit such a mechanism by controlling SIR complex release, an event that can respond rapidly to environmental insult (Martin et al. 1999; Ai et al. 2002). Consistently, SIR dispersion, or modulation of TPE, have been observed in the presence of various forms of stress (Stone and Pillus 1996; Martin et al. 1999; McAinsh et al. 1999; Mills et al. 1999; Ray and Runge 1999; Bi et al. 2004; Mercier et al. 2005), stress-induced redistribution of SIR proteins may derepress subtelomeric genes required for use of alternative carbon sources, and simultaneously contribute to the down-regulation of genes involved in ribosome biogenesis.
Nuclear Organization and Inducible Gene Expression
Over the last years nuclear pore complexes (NPCs) emerged as a major player in organizing gene activity. First, the nucleoporin Nup2 was shown to exert a strong boundary activity that can block the spread of heterochromatin when targeted on both sides of a reporter gene (Ishii et al. 2002). In this case, NPC tethering was proposed to establish a protected chromatin domain by creating a small DNA loop. Subsequently, a series of genome-wide studies identified nuclear pore components to be associated with highly active genes (Casolari et al. 2004, 2005; Schmid et al. 2006). Furthermore, specific inducible genes (INO1, HXK1, GAL1, GAL2, HSP104) were shown to associate with the nuclear periphery upon activation (Akhtar and Gasser 2007; Taddei 2007) (Fig. 5). Although many active genes seem to be associated with the NE, a stable interaction is not an obligate feature of gene activity (Casolari et al. 2004) and activation of the same promoter by different pathways or with different 3′UTR can alter a gene's position within the nucleus (Abruzzi et al. 2006; Taddei et al. 2006).
Figure 5.
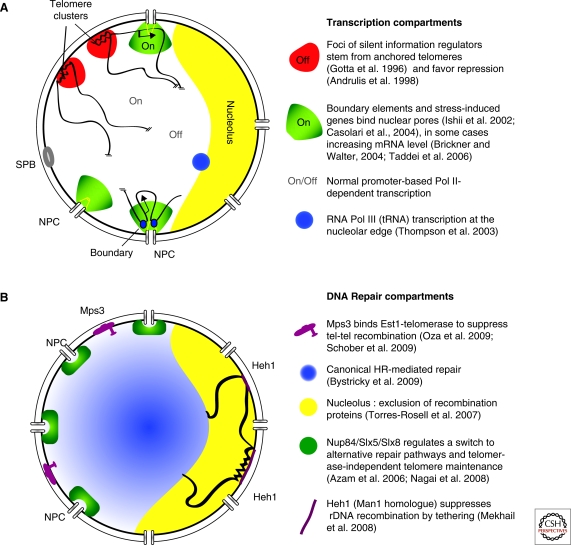
Nuclear subcompartments of yeast. (A) Transcription subcompartments within the yeast nucleus. Telomere clustering favors repression through SIR factors whereas pore association correlates with induced expression of certain genes and boundary function. (B) DNA repair compartments within the yeast nucleus. The nucleoplasm is the site of Rad52-mediated recombination whereas sequestration at Mps3 or nuclear pores either suppresses recombination between telomeres or processes a DSB for alternative repair pathways. For the rDNA, binding to the INM protein Heh1 (Src1) prevents rDNA recombination.
How specific genes associate with the NPC is still unclear and each step of mRNA production, maturation and export has been implicated at some level (Taddei 2007 for review). If multiple steps contribute to the stable association of active genes to NPCs, the importance of each individual step to NPC anchoring is probably specific for each gene. One possible scenario is that promoter-bound factors such as SAGA and Sus1 promote an initial, transient contact of the gene with pore proteins. This may be later stabilized by factors recruited for transcript termination, processing, quality control and export. These multiple anchors could favor the formation of active gene loops connecting promoters and 3′ sequences, thus improving the recycling of polymerases and elongation efficiency (O'Sullivan et al. 2004).
Gene-NPC association might be particularly important for inducible genes such as galactose- and heat-shock controlled promoters for which NPC association involves the histone acetyltransferase SAGA complex. This complex is connected to the mRNA export machinery by one of its components, Sus1, which is also part of the Sac3-Thp1-Cdc31 complex that binds the NPC through Nup1. The SAGA complex is generally required for the induction of stress-responsive genes, suggesting a rationale for gene relocalization: Stress-induced genes are likely to require rapid and high level expression and export, which could be facilitated by their positioning at pores. Although, mutations affecting gene-NPC association show normal activation level for GAL1 (Cabal et al. 2006) it is still possible that the kinetics of induction are affected in these mutants. Importantly, association with the nuclear periphery has been shown to enhance the activation of at least some genes (Brickner and Walter 2004; Menon et al. 2005; Taddei et al. 2006) and could provide an additional layer of regulation for fine-tuning expression levels.
Gene-NPC association has also been proposed to reflect an epigenetic mark that enables past events to be “remembered” (Brickner et al. 2007). It was reported that both the GAL1 and INO1 genes remain associated with the NPC for several generations after the inducing agent is removed. These genes appeared to be more rapidly re-activated in a pore-bound state than after a long-term repression. However, this mechanism has been called into question because other mechanisms involving cytoplasmic factors also can account for the rapid reactivation of these genes (Zacharioudakis et al. 2007).
The mechanism through which NPC-association can influence expression is still unclear. In 1985, Blobel proposed the “gene gating” hypothesis according to which a “circumscribed space subjacent to the NPC and extending into the interior of the nucleus was envisioned to serve as the locale where transcription and much of the co- and posttranscriptional processing would occur” (Blobel 1985). Thus, the NPC was proposed to be a scaffold to build an assembly line favoring the coordination of the different processes that occur at an active gene. Because of their eightfold symmetry, each pore may accommodate multiple genes, forming a factory for highly efficiency transcription, in which factors that promote initiation or elongation can concentrate. The existence of internal transcription factories has been proposed in differentiation mammalian nuclei or in cultured cells to coordinate expression of coregulated genes (Sexton et al. 2007). Such internal transcription factories might also exist in yeast although it remains unclear what provides the scaffolding for promoter recruitment.
The Dual Role of the Nuclear Envelope
Given its role in the creation of repressive telomeric compartments and its ability to support, if not promote, high-level gene induction, one can conclude that the nuclear periphery has a dual role in regulating gene expression. This dual role is corroborated by the structural organization of the nuclear envelope. Indeed, microscopy studies show that repressive and activating compartments at the nuclear periphery coexist but do not overlap, as visualized by the positioning of subtelomeric domains in-between nuclear pores (Taddei et al. 2004). This proximity between repressive (telomere clusters) and activating compartments (NPCs) could favor the efficiency and reversibility of gene induction. This might be especially relevant for subtelomeric genes, which are mainly involved in the usage of alternative carbon sources and have to be induced only under specific growth conditions (Fabre et al. 2005). Consistently, increasing the association of the HXK1 subtelomeric gene with the nuclear periphery improves both its repression on glucose medium and its activation in the absence of glucose (Taddei et al. 2006).
FUNCTIONAL CONSEQUENCES OF NUCLEAR ORGANIZATION ON GENOME STABILITY
Although these studies indicate important functions for yeast nuclear structure in the regulation of gene expression, recent work suggests that DNA repair may also respond to spatial cues in the nucleus. The most obvious elements that organize DNA repair are foci that form at double strand breaks (DSBs) as they are processed for homologous recombination (HR). This can be followed through the binding of fluorescently tagged Rad52, a protein that facilitate Rad51-dependent strand invasion, preceding the formation of a Holliday junction (New et al. 1998). A Rad52-containing HR center is able to recruit more than one DSB (Lisby et al. 2003), suggesting that there is a self-recognition event that allows breaks to congregate. Foci of Rad52, whether spontaneous or induced by DNA damage, are strongly enriched in the nuclear interior (Bystricky et al. 2009). Given that sister chromatid exchange is the dominant pathway for homologous recombination in mitotically dividing cells (Kadyk and Hartwell 1992), the internal enrichment of Rad52 foci argues that most sister chromatid exchange occurs in the nuclear interior. Indeed, when the HMR or HML sequences are available as donors for the cleaved MATa locus, MAT stays internal and recruits the appropriate donor, e.g., HMLα, from a more peripheral position for gene conversion (Nagai et al. 2008; Bystricky et al. 2009). The inward shift is only seen for the appropriate donor locus, and thus is mating-type dependent.
Pore Association of Persistent DSBs, Collapsed Forks and Critically Short Telomeres
If a donor sequence is not available, and the HO endonuclease is continuously expressed, the cut at the MAT locus persists. Such irreparable breaks are seen to shift to the nuclear periphery in a process dependent on Mec1 kinase and the histone variant Htz1 (Nagai et al. 2008; Kalocsay et al. 2009; Oza et al. 2009). Several lines of evidence implicate a nuclear pore subcomplex as the binding site, although the INM protein Mps3 has also been implicated transiently in the relocation process (Nagai et al. 2008; Kalocsay et al. 2009; Oza et al. 2009). Alternatively one might argue that the breaks move nonspecifically to the NE because crosslinking to both nuclear pores and Mps3, was reported (Nagai et al. 2008; Kalocsay et al. 2009; Oza et al. 2009).
Links between DNA damage and nuclear pores was supported by an epistasis-miniarray profile (E-MAP) analysis (Collins et al. 2007b), in which the Nup84 subcomplex of the nuclear pore, containing Nup133, Nup120 and Nup84, was clustered with a conserved SUMO-dependent ubiquitin ligase complex, Slx5/Slx8. The Slx5/Slx8 complex plays an important role in the maintenance of genome integrity particularly during DNA replication (Perry et al. 2008). In addition, Slx5, which bears two SUMO interacting motifs, interacts physically with components of the proteasome (Collins et al. 2007a). Tellingly, there is an accumulation of SUMOylated proteins in cells lacking either Slx5 or Slx8 (Wang et al. 2006; Xie et al. 2007), suggesting that the Slx5/Slx8 complex normally helps the proteasome degrade SUMOylated proteins through its ubiquitin ligase activity (Wang et al. 2006; Xie et al. 2007). Indeed, an accumulation of SUMO conjugates has been shown to be responsible for the DNA damage sensitivity of the slx8 mutant in fission yeast (Prudden et al. 2007).
Mutation of any component of the Nup84 complex, i.e., Nup84, Nup120, or Nup133 nucleoporins, renders cells hypersensitive to DNA damage, such as irradiation, MMS, bleomycin and HU (Bennett et al. 2001; Loeillet et al. 2005; Therizols et al. 2006). The same is true for mutation of Slx5 or Slx8 (Nagai et al. 2008). Furthermore, mutants of the Nup84 complex have been shown to be synthetically lethal with mutants of DNA repair, recombination and replication genes (Loeillet et al. 2005; Pan et al. 2006; Collins et al. 2007b; Nagai et al. 2008). A direct involvement of nuclear pores in DNA repair was supported by the fact that Nup84 and Slx5/Slx8 colocalize and coimmunoprecipitate (Nagai et al. 2008). ChIP assays have shown that irreparable DSBs and collapsed, but not stalled, replication forks, becomes associated with the Nup84–Slx5/Slx8 complex. Moreover, the rate of spontaneous gene conversion could be enhanced by tethering the recipient site of recombination to the nuclear periphery. The enhancement of DNA repair was dependent on Slx8 and Nup84 proteins, suggesting that Slx5/Slx8 - Nup84 complex facilitates at least one kind of strand invasion event (Nagai et al. 2008). Short telomeres have also been reported to associate with the nuclear pore complex (Khadaroo et al. 2009).
Importantly, pore binding is not detected if DSBs can be readily repaired by HR (Nagai et al. 2008). This suggests that only lesions that are refractive to DNA repair by canonical HR pathways associate stably with pores. At the pore a repair intermediate may be shunted to alternative recombination and repair pathways through steps that are controlled by SUMO recognition, ubiquitylation, and degradation, in other words through Slx5/Slx8 and the proteasome. The yeast proteasome was also shown by ChIP to be recruited to irreparable DSBs, and was genetically shown to be required for recombination-mediated modes of DNA repair (Krogan et al. 2004). In mammals, the proteasome has been implicated in the differential choice of recombination pathways; namely the up-regulation of gene-conversion at the expense of single-strand annealing (Gudmundsdottir et al. 2007). Consistently, Slx5/Slx8 has been shown to counteract Rad51-independent recombination, such as single-strand annealing and the break induced replication (Burgess et al. 2007).
An increased dependence on Rad51-independent recombination pathways favors deleterious genomic rearrangements, such as gene amplifications, deletions and translocations (Haber and Debatisse 2006). Thus, we suggest the possibility that the pore-associated DNA repair pathway mediated by Slx5/Slx8 complex helps counteract error-prone Rad51-independent recombination, whereas up-regulating alternative pathways for recovery. Alternative pathways may be particularly important at collapsed replication forks and at telomeres. Which pathways are used, will depend on the targets of Slx5/Slx8 action. Based on E-MAP data likely pathways will include Pol32, Rad27, Est1, and Srs2 (Nagai et al. 2008). Intriguingly, the outgrowth of type II survivors of telomerase inhibition, which requires TG repeat amplification by recombination, requires not only Rad52 and Sgs1, but Slx5 and Slx8 (Azam et al. 2006; McEachern and Haber 2006). Given the involvement of the Slx5/8 E3 ligase and proteasome, it seems likely that shifting the mode of DNA repair at these loci may require the degradation of an inhibitor, one that could be the target of SUMO-directed ubiquitination by Slx5/Slx8 (Collins et al. 2007a; Wang et al. 2006; Xie et al. 2007). Finally, there may be crosstalk between Mps3 and these pathways, because the mps3Δ75-150 mutant, which fails to bind DSBs and telomeres, delayed recombinational repair of DSBs and decreased gross chromosomal rearrangements in either a slx5 deletion or in the pif1-m2 mutant, which abrogates the negative regulation of telomerase activity by the Pif1 helicase (Oza et al. 2009). Consistently, overexpression of the Mps3N domain increased telomere exchange (Schober et al. 2009).
Regulation of Recombination at the rDNA Locus
In budding yeast, the maintenance of rDNA repeat number is important for cell growth as well as for preventing premature senescence. It has been proposed that HR helps to maintain rDNA copy number (Smith 1974). Indeed, expansion or contraction of rDNA repeats appears to stem from HR, which follows on Fob1-induced replication fork blocking (Kobayashi et al. 1998). Although several sequence elements that stimulate recombination are found within the rDNA (Keil and Roeder 1984; Kobayashi et al. 1998), the frequency of HR is nonetheless significantly lower than what one would expect for a repetitive array in yeast. This argues that recombination is actively suppressed at the rDNA locus to maintain rDNA repeat homeostasis.
Interestingly, components of the HR machinery, namely Rad52, Rad51, Rad55, and Rad59, are excluded from the nucleolus, although sensors of DSBs, such as Mre11 and Rfa1 are present (Torres-Rosell et al. 2007). Furthermore, a single DSB in the rDNA induced by the I-SceI endonuclease shifts away from the nucleolus when Rad52 was recruited, suggesting repair of rDNA by HR can only take place outside of the nucleolus. Nucleolar exclusion of Rad52 foci requires the Smc5/Smc6 complex, which harbors E3 SUMO ligase activity, and requires SUMOylation of Rad52. Given that mutants that impair nucleolar exclusion of HR, i.e., mutants of Smc5 or Smc6, show rDNA hyper-recombination and unequal sister chromatid exchange, one can conclude that the extrusion of HR proteins from the nucleolus is functionally important for genome stability (Burgess et al. 2007; Torres-Rosell et al. 2007).
Although it is well-established that the yeast nucleolus abuts the nuclear periphery (Oakes et al. 1998), the functional significance of this localization has been poorly understood. Recent work shows that the perinuclear anchoring of rDNA repeats by the inner nuclear membrane proteins Heh1(also called Src1) and Nur1, contributes to the maintenance of rDNA repeat stability (Mekhail et al. 2008). Heh1 shares homology with human Man1 (King et al. 2006), an INM protein containing a conserved LEM (LAP-Emerin-Man1) domain. Deletion of Heh1 or Nur1 releases rDNA from the nuclear periphery, and like loss of Smc5/Smc6, this release correlates with increase unequal sister chromatid exchange. Importantly, unlike Sir2, which also functions to reduce unequal sister chromatid exchange (Gottlieb and Esposito 1989), neither Heh1 nor Nur1 is required for rDNA silencing. The artificial tethering of repeats to the INM through a Sir2-Heh1 fusion suppresses rDNA instability in the absence of Lrs4, a protein required for peripheral tethering of the rDNA. Thus, the peripheral tethering appears to limit access of the repetitive arrays to the machinery mediating recombination and sister chromatid exchange (Mekhail et al. 2008).
CONCLUDING REMARKS
The yeast nucleus provides a readily manipulated genetic system for the analysis of nuclear organization. Its main advantage is that yeast provides ample means to test both gain and loss of function mutations on nuclear organization and genetic function. Data show clearly that there are zones that regulate DNA repair (Fig. 5B) just as there are zones that influence transcription (Fig. 5A). The one drawback of yeast is that it does not differentiate into highly differentiated cell types. It does, however, show changes in nuclear organization as the culture adapts to changes in carbon source or encounters other conditions of metabolic stress. These changes reinforce the notion of “cell type-” or “condition-” specific levels of nuclear organization. Most importantly, yeast cells allow us to test for subtle changes in the efficiency of replication, DNA repair and transcription, as a result of changes in nuclear structure. Although the field is still at an early stage, the number of genes recognized as regulating aspects of nuclear structure increases constantly, and both microscopy and genomic assays improve at staggering rates. These are forceful arguments for pursuing yeast as a model in which to test the relationship between nuclear structure and function.
ACKNOWLEDGMENTS
We would like to acknowledge critical reading by V. Dion and H. Ferreira, and many fruitful discussions with the Gasser laboratory. We thank S. Nagai for the DNA repair figure and K. Bystricky images in Figure 1. S.M.G. acknowledges support of the Novartis Research Foundation, the National Center for Competence in Research, “Frontiers-in-Genetics” and the EU NOE Epigenome. A.T. is supported by the French “Agence Nationale pour la Recherche” (ANR), and received funding from the European Research Council under the European Community's Seventh Framework Programme (FP7/2007-2013 Grant Agreement no. [210508]).
Footnotes
Editors: David L. Spector and Tom Misteli
Additional Perspectives on The Nucleus available at www.cshperspectives.org
REFERENCES
- Abruzzi KC, Belostotsky DA, Chekanova JA, Dower K, Rosbash M 2006. 3′-end formation signals modulate the association of genes with the nuclear periphery as well as mRNP dot formation. EMBO J 25: 4253–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai W, Bertram PG, Tsang CK, Chan TF, Zheng XF 2002. Regulation of subtelomeric silencing during stress response. Mol Cell 10: 1295–1305 [DOI] [PubMed] [Google Scholar]
- Akhtar A, Gasser SM 2007. The nuclear envelope and transcriptional control. Nat Rev Genet 8: 507–17 [DOI] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. 2007. The molecular architecture of the nuclear pore complex. Nature 450: 695–701 [DOI] [PubMed] [Google Scholar]
- Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R 1998. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 394: 592–595 [DOI] [PubMed] [Google Scholar]
- Andrulis ED, Zappulla DC, Ansari A, Perrod S, Laiosa CV, Gartenberg MR, Sternglanz R 2002. Esc1, a nuclear periphery protein required for Sir4-based plasmid anchoring and partitioning. Mol Cell Biol 22: 8292–8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniacci LM, Kenna MA, Skibbens RV 2007. The nuclear envelope and spindle pole body-associated Mps3 protein bind telomere regulators and function in telomere clustering. Cell Cycle 6: 75–79 [DOI] [PubMed] [Google Scholar]
- Aparicio OM, Billington BL, Gottschling DE 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66: 1279–1287 [DOI] [PubMed] [Google Scholar]
- Azam M, Lee JY, Abraham V, Chanoux R, Schoenly KA, Johnson FB 2006. Evidence that the S.cerevisiae Sgs1 protein facilitates recombinational repair of telomeres during senescence. Nucleic Acids Res 34: 506–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Halley JE, Rine J 2006. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev 20: 700–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CB, Lewis LK, Karthikeyan G, Lobachev KS, Jin YH, Sterling JF, Snipe JR, Resnick MA 2001. Genes required for ionizing radiation resistance in yeast. Nat Genet 29: 426–434 [DOI] [PubMed] [Google Scholar]
- Berger AB, Cabal GG, Fabre E, Duong T, Buc H, Nehrbass U, Olivo-Marin JC, Gadal O, Zimmer C 2008. High-resolution statistical mapping reveals gene territories in live yeast. Nature Methods 5: 1031–1037 [DOI] [PubMed] [Google Scholar]
- Bi X, Yu Q, Sandmeier JJ, Elizondo S 2004. Regulation of transcriptional silencing in yeast by growth temperature. J Mol Biol 344: 893–905 [DOI] [PubMed] [Google Scholar]
- Blobel G 1985. Gene gating: A hypothesis. Proc Natl Acad Sci 82: 8527–8529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner JH, Walter P 2004. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol 2: e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH 2007. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol 5: e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ, Curcio MJ 1997. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev 11: 255–269 [DOI] [PubMed] [Google Scholar]
- Bupp JM, Martin AE, Stensrud ES, Jaspersen SL 2007. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J Cell Biol 179: 845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess RC, Rahman S, Lisby M, Rothstein R, Zhao X 2007. The Slx5-Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol Cell Biol 27: 6153–6162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K, Heun P, Gehlen L, Langowski J, Gasser SM 2004. Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proc Natl Acad Sci U S A 101: 16495–16500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K, Laroche T, van Houwe G, Blaszczyk M, Gasser SM 2005. Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J Cell Biol 168: 375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K, Van Attikum H, Montiel MD, Dion V, Gehlen L, Gasser SM 2009. Regulation of nuclear positioning and dynamics of the silent mating type loci by the yeast Ku70/80 complex. Mol Cell Biol 29: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, et al. 2006. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441: 770–773 [DOI] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Drubin DA, Rando OJ, Silver PA 2005. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes Genes Dev 19: 1188–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117: 427–439 [DOI] [PubMed] [Google Scholar]
- Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ 2007a. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics 6: 439–450 [DOI] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M et al. 2007b. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810 [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer M, Dietzel S, Müller S, Solovei I, Fakan S 2006. Chromosome territories – a functional nuclear landscape. Curr Opin Cell Biol 18: 307–316 [DOI] [PubMed] [Google Scholar]
- D'Angelo MA, Hetzer MW 2008. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol 18: 456–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dequard-Chablat M, Riva M, Carles C, Sentenac A 1991. RPC19, the gene for a subunit common to yeast RNA polymerases A (I) and C (III). J Biol Chem 266: 15300–15307 [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N 2002. Capturing chromosome conformation. Science 295: 1306–1311 [DOI] [PubMed] [Google Scholar]
- Ebrahimi H, Donaldson AD 2008. Release of yeast telomeres from the nuclear periphery is triggered by replication and maintained by suppression of Ku-mediated anchoring. Genes Dev 22: 3363–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre E, Muller H, Therizols P, Lafontaine I, Dujon B, Fairhead C 2005. Comparative genomics in hemiascomycete yeasts: Evolution of sex, silencing, and subtelomeres. Mol Biol Evol 22: 856–873 [DOI] [PubMed] [Google Scholar]
- Fisher TS, Taggart AK, Zakian VA 2004. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat Struct Mol Biol 11: 1198–1205 [DOI] [PubMed] [Google Scholar]
- Fritze CE, Verschueren K, Strich R, Easton Esposito R 1997. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J 16: 6495–6509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG 2000. Cajal bodies: The first 100 years. Annu Rev Cell Dev Biol 16: 273–300 [DOI] [PubMed] [Google Scholar]
- Gasser SM, Hediger F, Taddei A, Neumann FR, Gartenberg MR 2004. The function of telomere clustering in yeast: The Circe effect. Cold Spring Harb Symp Quant Biol 69: 327–337 [DOI] [PubMed] [Google Scholar]
- Gartenberg MR, Neumann FR, Laroche T, Blaszczyk M, Gasser SM 2004. Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell 119: 955–967 [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS 2003. Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Gilson E, Roberge M, Giraldo R, Rhodes D, Gasser SM 1993. Distortion of the DNA double helix by RAP1 at silencers and multiple telomeric binding sites. J Mol Biol 231: 293–310 [DOI] [PubMed] [Google Scholar]
- Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM 1996. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol 134: 1349–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S, Esposito RE 1989. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56: 771–776 [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL 2005. The nuclear lamina comes of age. Nat Rev Mol Cell Biol 6: 21–31 [DOI] [PubMed] [Google Scholar]
- Grund SE, Fischer T, Cabal GG, Antunez O, Perez-Ortin JE, Hurt E 2008. The inner nuclear membrane protein Src1 associates with subtelomeric genes and alters their regulated gene expression. J Cell Biol 182: 897–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsdottir K, Lord CJ, Ashworth A 2007. The proteasome is involved in determining differential utilization of double-strand break repair pathways. Oncogene 26: 7601–7606 [DOI] [PubMed] [Google Scholar]
- Haber JE, Debatisse M 2006. Gene amplification: Yeast takes a turn. Cell 125: 1237–1240 [DOI] [PubMed] [Google Scholar]
- Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR 2008. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev 22: 2204–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattier T, Andrulis ED, Tartakoff AM 2007. Immobility, inheritance and plasticity of shape of the yeast nucleus. BMC Cell Biol 8: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger F, Neumann FR, Van Houwe G, Dubrana K, Gasser SM 2002. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr Biol 12: 2076–2089 [DOI] [PubMed] [Google Scholar]
- Heun P, Laroche T, Shimada K, Furrer P, Gasser SM 2001. Chromosome dynamics in the yeast interphase nucleus. Science 294: 2181–2186 [DOI] [PubMed] [Google Scholar]
- Hiraga S, Botsios S, Donaldson AD 2008. Histone H3 lysine 56 acetylation by Rtt109 is crucial for chromosome positioning. J Cell Biol 183: 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S, Robertson ED, Donaldson AD 2006. The Ctf18 RFC-like complex positions yeast telomeres but does not specify their replication time. EMBO J 25: 1505–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug N, Lingner J 2006. Telomere length homeostasis. Chromosoma 115: 413–25 [DOI] [PubMed] [Google Scholar]
- Hughes JD, Estep PW, Tavazoie S, Church GM 2000. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J Mol Biol 296: 1205–1214 [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Iborra FJ 2007. Can visco-elastic phase separation, macromolecular crowding and colloidal physics explain nuclear organisation? Theor Biol Med Model 4: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK 2002. Chromatin boundaries in budding yeast: The nuclear pore connection. Cell 109: 551–562 [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Giddings TH Jr, Winey M 2002. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J Cell Biol 159: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin QW, Fuchs J, Loidl J 2000. Centromere clustering is a major determinant of yeast interphase nuclear organization. J Cell Sci 113: 1903–1912 [DOI] [PubMed] [Google Scholar]
- Kadyk LC, Hartwell LH 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132: 387–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalocsay M, Hiller NJ, Jentsch S 2009. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell 33: 335–343 [DOI] [PubMed] [Google Scholar]
- Keil RL, Roeder GS 1984. Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell 39: 377–386 [DOI] [PubMed] [Google Scholar]
- Khadaroo B, Teixeira MT, Luciano P, Eckert-Boulet N, Germann SM, Simon MN, Gallina I, Abdallah P, Gilson E, Géli V, et al. 2009. The DNA damage response at eroded telomeres and tethering to the nuclear pore complex. Nat Cell Biol Jul 13. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- King MC, Lusk CP, Blobel G 2006. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature 442: 1003–1007 [DOI] [PubMed] [Google Scholar]
- Kitamura E, Blow JJ, Tanaka TU 2006. Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell, 125: 1297–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Heck DJ, Nomura M, Horiuchi T 1998. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: Requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev 12: 3821–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N, Lam MHY, Fillingham J, Keogh MC, Gebbia M, Li J, Datta N, Cagney G, Buratowski S, Emili A, et al. 2004. Proteasome involvement in the repair of DNA double-strand breaks. Molecular Cell 16: 1027–1034 [DOI] [PubMed] [Google Scholar]
- Lamond AI, Spector DL 2003. Nuclear speckles: A model for nuclear organelles. Nat Mol Cell Biol 4: 605–612 [DOI] [PubMed] [Google Scholar]
- Larrivee M, LeBel C, Wellinger RJ 2004. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev 18: 1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144: 1399–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A, Felberbaum R, Hochstrasser M 2007. A nuclear envelope protein linking nuclear pore basket assembly, SUMO protease regulation, and mRNA surveillance. J Cell Biol 178: 813–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cech TR, Hughes TR, Lundblad V 1997. Three ever shorter telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci 94: 11190–11195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Mortensen UH, Rothstein R 2003. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol 5: 572–577 [DOI] [PubMed] [Google Scholar]
- Loeillet S, Palancade B, Cartron M, Thierry A, Richard GF, Dujon B, Doye V, Nicolas A 2005. Genetic network interactions among replication, repair and nuclear pore deficiencies in yeast. DNA Repair (Amst) 4: 459–468 [DOI] [PubMed] [Google Scholar]
- Lundblad V, Szostak JW 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57: 633–643 [DOI] [PubMed] [Google Scholar]
- Lusk CP, Blobel G, King MC 2007. Highway to the inner nuclear membrane: rules for the road. Nat Rev Mol Cell Biol 8: 414–420 [DOI] [PubMed] [Google Scholar]
- Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser SM 1996. Evidence for silencing compartments within the yeast nucleus: A role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev 10: 1796–1811 [DOI] [PubMed] [Google Scholar]
- Maillet L, Gaden F, Brevet V, Fourel G, Martin SG, Dubrana K, Gasser SM, Gilson E 2001. Ku-deficient yeast strains exhibit alternative states of silencing competence. EMBO Rep 2: 203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S, Gilson E, Shore D 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275: 986–990 [DOI] [PubMed] [Google Scholar]
- Marcand S, Buck SW, Moretti P, Gilson E, Shore D 1996. Silencing of genes at nontelomeric sites in yeast is controlled by sequestration of silencing factors at telomeres by Rap 1 protein. Genes Dev 10: 1297–1309 [DOI] [PubMed] [Google Scholar]
- Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, Murray AW, Agard DA, Sedat JW 1997. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr Biol 7: 930–939 [DOI] [PubMed] [Google Scholar]
- Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM 1999. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 97: 621–633 [DOI] [PubMed] [Google Scholar]
- Martino F, Kueng S, Robinson P, Tsai-Pflugfelder M, van Leeuwen F, Ziegler M, Cubizolles F, Cockell MM, Rhodes D, Gasser SM 2009. Reconstitution of yeast silent chromatin: Multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol Cell 33: 323–334 [DOI] [PubMed] [Google Scholar]
- McAinsh AD, Scott-Drew S, Murray JA, Jackson SP 1999. DNA damage triggers disruption of telomeric silencing and Mec1p-dependent relocation of Sir3p. Curr Biol 9: 963–966 [DOI] [PubMed] [Google Scholar]
- McEachern MJ, Haber JE 2006. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem 75: 111–135 [DOI] [PubMed] [Google Scholar]
- Mekhail K, Seebacher J, Gygi SP, Moazed D 2008. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature 456: 667–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini MD, Wu M, Madhani HD 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112: 725–736 [DOI] [PubMed] [Google Scholar]
- Menon BB, Sarma NJ, Pasula S, Deminoff SJ, Willis KA, Barbara KE, Andrews B, Santangelo GM 2005. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc Natl Acad Sci 102: 5749–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier G, Berthault N, Touleimat N, Kepes F, Fourel G, Gilson E, Dutreix M 2005. A haploid-specific transcriptional response to irradiation in Saccharomyces cerevisiae. Nucleic Acids Res 33: 6635–6643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele A, Bystricky K, Dekker J 2009. Yeast silent mating type loci form heterochromatic clusters through silencer protein-dependent long-range interactions. PLoS Genet 5: e1000478 Epub 2009 May 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills K, Sinclair D, Guarente L 1999. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell 97: 609–620 [DOI] [PubMed] [Google Scholar]
- Misteli T 2000. Cell biology of transcription and pre-mRNA splicing: Nuclear architecture meets nuclear function. J Cell Sci 113: 1841–1849 [DOI] [PubMed] [Google Scholar]
- Mondoux MA, Scaife JG, Zakian VA 2007. Differential nuclear localization does not determine the silencing status of Saccharomyces cerevisiae telomeres. Genetics 177: 2019–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D 1994. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev 8: 2257–2269 [DOI] [PubMed] [Google Scholar]
- Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, Varela E, Hediger F, Gasser SM, Krogan NJ 2008. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322: 597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC 1998. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391: 407–410 [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Terazawa Y, Nakayama T, Hirata A, Makio T, Endo T 2003. Nep98p is a component of the yeast spindle pole body and essential for nuclear division and fusion. J Biol Chem 278: 9938–9943 [DOI] [PubMed] [Google Scholar]
- Oakes M, Aris JP, Brockenbrough JS, Wai H, Vu L, Nomura M 1998. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J Cell Biology 143: 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes M, Nogi Y, Clark MW, Nomura M 1993. Structural alterations of the nucleolus in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Mol Cell Biol 13: 2441–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ 2004. Gene loops juxtapose promoters and terminators in yeast. Nat Genet 36: 1014–1018 [DOI] [PubMed] [Google Scholar]
- Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL 2009. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev 23: 912–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B, Liu X, Garcia-Rubio M, Aguilera A, Zhao X, Doye V 2007. Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Mol Biol Cell 18: 2912–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD 2006. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124: 1069–1081 [DOI] [PubMed] [Google Scholar]
- Perry JJ, Tainer JA, Boddy MN 2008. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci 33: 201–208 [DOI] [PubMed] [Google Scholar]
- Powers T, Walter P 1999. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol Biol Cell 10: 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, McGowan CH, Boddy MN 2007. SUMO-targeted ubiquitin ligases in genome stability. EMBO J 26: 4089–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl C 1885. Über Zelltheilung. Morphologisches Jahrbuch 10: 214–330 [Google Scholar]
- Ray A, Runge KW 1999. Varying the number of telomere-bound proteins does not alter telomere length in tel1Δ cells. Proc Natl Acad Sci U S A 96: 15044–15049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renauld H, Aparicio OM, Zierath PD, Billington BL, Chhablani SK, Gottschling DE 1993. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev 7: 1133–1145 [DOI] [PubMed] [Google Scholar]
- Richter K, Nessling M, Lichter P 2007. Experimental evidence for the influence of molecular crowding on nuclear architecture. J Cell Sci 120: 1673–1680 [DOI] [PubMed] [Google Scholar]
- Robyr D, Suka Y, Xenarios I, Kurdistani SK, Wang A, Suka N, Grunstein M 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109: 437–446 [DOI] [PubMed] [Google Scholar]
- Rodley CD, Bertels F, Jones B, O'Sullivan JM 2009. Global identification of yeast chromosome interactions using Genome conformation capture. Fungal Genet Biol 46: 879–886 [DOI] [PubMed] [Google Scholar]
- Roy R, Meier B, McAinsh AD, Feldmann HM, Jackson SP 2004. Separation-of-function mutants of yeast Ku80 reveal a Yku80p-Sir4p interaction involved in telomeric silencing. J Biol Chem 279: 86–94 [DOI] [PubMed] [Google Scholar]
- Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK 2006. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol Cell 21: 379–91 [DOI] [PubMed] [Google Scholar]
- Schober H, Ferreira H, Kalck V, Gehlen LR, Gasser SM 2009. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev 23: 928–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober H, Kalck V, Vega-Palas MA, Van Houwe G, Sage D, Unser M, Gartenberg MR, Gasser SM 2008. Controlled exchange of chromosomal arms reveals principles driving telomere interactions in yeast. Genome Res 18: 261–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Umlauf D, Kurukuti S, Fraser P 2007. The role of transcription factories in large-scale structure and dynamics of interphase chromatin. Semin Cell Dev Biol 18: 691–697 [DOI] [PubMed] [Google Scholar]
- Shampay J, Szostak JW, Blackburn EH 1984. DNA sequences of telomeres maintained in yeast. Nature 310: 154–157 [DOI] [PubMed] [Google Scholar]
- Shore D, Nasmyth K 1987. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell 51: 721–732 [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L 1997. Extrachromosomal rDNA circles–a cause of aging in yeast. Cell 91: 1033–1042 [DOI] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266: 404–409 [DOI] [PubMed] [Google Scholar]
- Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D 2008. Nucleolus: The fascinating nuclear body. Histochem Cell Biol 129: 13–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GP 1974. Unequal crossover and the evolution of multigene families. Cold Spring Harb Symp Quant Biol 38: 507–513 [DOI] [PubMed] [Google Scholar]
- Smith JS, Boeke JD 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev 11: 241–254 [DOI] [PubMed] [Google Scholar]
- Stavenhagen JB, Zakian VA 1994. Internal tracts of telomeric DNA act as silencers in Saccharomyces cerevisiae. Genes Dev 8: 1411–1422 [DOI] [PubMed] [Google Scholar]
- Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE 2003. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev 17: 2384–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EM, Pillus L 1996. Activation of an MAP kinase cascade leads to Sir3p hyperphosphorylation and strengthens transcriptional silencing. J Cell Biol 135: 571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev 11: 83–93 [DOI] [PubMed] [Google Scholar]
- Taddei A 2007. Active genes at the nuclear pore complex. Curr Opin Cell Biol 19: 305–310 [DOI] [PubMed] [Google Scholar]
- Taddei A, Hediger F, Neumann FR, Bauer C, Gasser SM 2004. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J 23: 1301–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM 2006. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441: 774–778 [DOI] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Nagai S, Erb I, van Nimwegen E, Gasser SM 2009. The functional importance of telomere clustering: global changes in gene expression result from SIR factor dispersion. Genome Res 19: 611–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therizols P, Fairhead C, Cabal GG, Genovesio A, Olivo-Marin JC, Dujon B, Fabre E 2006. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J Cell Biol 172: 189–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JS, Johnson LM, Grunstein M 1994. Specific repression of the yeast silent mating locus HMR by an adjacent telomere. Mol Cell Biol 14: 446–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M, Haeusler RA, Good PD, Engelke DR 2003. Nucleolar clustering of dispersed tRNA genes. Science 302: 1399–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, Jentsch S, Rothstein R, Aragon L, Lisby M 2007. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol 9: 923–931 [DOI] [PubMed] [Google Scholar]
- Verheggen C, Mouaikel J, Thiry M, Blanchard JM, Tollervey D, Bordonne R, Lafontaine DL, Bertrand E 2001. Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors and a novel nuclear domain. EMBO J 20: 5480–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Jones GM, Prelich G 2006. Genetic analysis connects SLX5 and SLX8 to the SUMO pathway in Saccharomyces cerevisiae. Genetics 172: 1499–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Haeusler RA, Good PD, Thompson M, Nagar S, Engelke DR 2005. Silencing near tRNA genes requires nucleolar localization. J Biol Chem 280: 8637–8639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton D, Shore D 1997. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev 11: 748–760 [DOI] [PubMed] [Google Scholar]
- Xie Y, Kerscher O, Kroetz MB, McConchie HF, Sung P, Hochstrasser M 2007. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J Biol Chem 282: 34176–34184 [DOI] [PubMed] [Google Scholar]
- Yang CH, Lambie EJ, Hardin J, Craft J, Snyder M 1989. Higher order structure is present in the yeast nucleus: autoantibody probes demonstrate that the nucleolus lies opposite the spindle pole body. Chromosoma 98: 123–128 [DOI] [PubMed] [Google Scholar]
- Zacharioudakis I, Gligoris T, Tzamarias D 2007. A yeast catabolic enzyme controls transcriptional memory. Curr Biol 17: 2041–2046 [DOI] [PubMed] [Google Scholar]
- Zakian VA 1996. Structure, function, and replication of Saccharomyces cerevisiae telomeres. Annu Rev Genet 30: 141–72 [DOI] [PubMed] [Google Scholar]
- Zhong S, Salomoni P, Pandolfi PP 2000. The transcriptional role of PML and the nuclear body. Nat Cell Biol 2:, E85–90 [DOI] [PubMed] [Google Scholar]
- Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, Saulrieta K, Smith Z, Shah MV, Radhakrishnan M et al. 2009. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res 19: 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]