Abstract
Advances in microscopy automation and image analysis have given biologists the tools to attempt large scale systems-level experiments on biological systems using microscope image readout. Fluorescence microscopy has become a standard tool for assaying gene function in RNAi knockdown screens and protein localization studies in eukaryotic systems. Similar high throughput studies can be attempted in prokaryotes, though the difficulties surrounding work at the diffraction limit pose challenges, and targeting essential genes in a high throughput way can be difficult. Here we will discuss efforts to make live-cell fluorescent microscopy based experiments using genetically encoded fluorescent reporters an automated, high throughput, and quantitative endeavor amenable to systems-level experiments in bacteria. We emphasize a quantitative data reduction approach, using simulation to help develop biologically relevant cell measurements that completely characterize the cell image. We give an example of how this type of data can be directly exploited by statistical learning algorithms to discover functional pathways.
The diffraction limit makes high-throughput fluorescence microscopy more challenging in prokaryotes, but approaches such as quantitative data reduction now allow systems-level analysis of bacteria by this technique.
Over the last decade, advances in microscope automation, fluorescent labeling, sample preparation, image processing, pattern recognition, and statistical learning have ushered in a new era of cell biology experiments based on unbiased genome-wide cell-based assays. In eukaryotic cell biology, these advances have been exploited to investigate the distribution and patterning of labeled subcellular structures and the “location proteome” defined by the distribution and intensity of localized protein reporters (Chen et al. 2006). In the prokaryotic world, the challenges surrounding fluorescence microscopy at the diffraction limit has meant a somewhat slower adoption of highly automated methods and analysis designed for large-scale genetic screens or assays. Nevertheless, large-scale approaches are now being used more routinely during investigations of bacterial systems, from the identification of entire location proteomes (Werner et al. 2009) to the detection of modular transcription and signaling pathways (Christen and Fero 2009). Because of the size and complexity of the eukaryotic cell, generalized pattern recognition and classification approaches are often crucial. In these approaches, conditioned, parameterized, preclassified image data are used by statistical learning algorithms to build models that capture the differences between classes of images. A properly constructed model can then be used to automatically analyze a previously unclassified experimental data set (Boland et al. 1998; Boland and Murphy 1999; Boland and Murphy 2001; Huang and Murphy 2004; Chen and Murphy 2005; Jones et al. 2009). In prokaryotic cell biology a similar situation exists, with one major difference; the simplicity of the bacterial cell allows us to entertain the idea of a cell parameterization based not only on generalized image properties, but also on specific cell measurements of high biological relevance. For example, instead of measuring hard to interpret properties based on generalized image parameters and a global image transform, such as a Zernike or Fourier transform, one can conceive of measuring a more relevant set of parameters related directly to gene function or to the biology of interest, such the position and quantity of both localized and delocalized fluorescent signals, the length and width of the cell, the degree of pinch at the division plane or sporulation septum, the precise location of the poles or other points of inflection, as well as the location of membrane structures such as pili or stalks and the membrane outline itself. In this way, albeit with a certain degree of additional work, the image can be reduced to a set of high information content parameters. This type of data can be used more efficiently in any type of subsequent analysis, whether direct or via a model-building statistical learning exercise. In addition, reducing the cell to a small set of highly informative parameters allows the option of combining those single-cell measurements into quantitative ensemble-based phenotypes where the entire distribution of measured values is recorded rather than just means and standard deviations. With the addition of genotype information, this data can then be used directly in unsupervised statistical learning approaches such as hierarchical clustering to do quantitative phenotypic profiling (Ohya et al. 2005). In this article we outline the process of doing such an automated analysis on a bacterial system. To keep our discussion firmly rooted in a practical example, we will use a case-study approach based on work performed in the α-proteobacteria Caulobacter crescentus. However, most of what we discuss can be applied to other bacteria and single-cell eukaryotes. It should also be noted that the efficacy of a quantitative analysis depends on the constructs used to express fluorescently tagged reporter proteins. Even modest overproduction of some proteins, particularly those involved in signal transduction and cell division, can have deleterious effects on cell viability and on cell shape (Gregory et al. 2008). We will assume that for the purposes of quantitative analyses, tagged protein reporters are fully functional proteins produced from genes in their native chromosomal context.
AUTOMATED LIVE CELL FLUORESCENT MICROSCOPY
Live cell, 100×, oil immersion, fluorescence microscopy presents some problems for a high-throughput system not found in lower resolution systems. First, the geometry is tightly constrained, with a ∼2 mm gap between the focal plane and the condenser, and ∼0.24 mm spacing between the 100× objective and the focal plane. The cells must be kept alive, which usually means imaging on a 1% agarose + nutrient gel pad and limiting the amount of time the samples are under observation. Fluorescence microscopy usually entails imaging in multiple optical channels; the multiplexed mutant screen example described later used one phase contrast plus three fluorescent channels. This means the optical system has to be stable for several seconds while cubes or filter wheels are automatically switched. The need for optical coupling oil also drives the design of the microscopy plate and choice of inverted versus upright scope geometry.
Data Acquisition and Autofocusing
Acquisition timescales range from tens of milliseconds for optical channel exposure times to seconds required for some mechanical system motions, all the way to the hundreds of minutes to needed to push through thousands of samples. Several issues arise when trying to automatically image multiple live samples. Image drift, the availability of nutrients and oxygen over long time periods, the cells doubling time, and drying out of agarose pads are among the issues that determine how long a sample plate can sit on the microscope and thus how long a data-taking run can last. Experience shows that data-taking runs lasting approximately 1 h work well. If imaging in a 96-well microplate format this implies a throughput of 96 sample-wells per hour. Thus, we have roughly 30 s to: move to the well, coarse focus, fine focus, image capture, and write the data in phase contrast plus several fluorescent channels. Focusing is a large contributor to the time it takes to image a sample well. Because of fluorophore bleaching, using any fluorescent channel to optimize focus is usually unwise. This leads to using an alternative white-light channel such as phase contrast or DIC to set the focus for subsequent fluorescent images. There are several schemes for calculating z-focus quality scores (Groen et al. 1985; Firestone et al. 1991; Price and Gough 1994; Geusebroek et al. 2000) but in our experience we have had good results with a simple autocorrelation function. Images are acquired in a fast loop while changing the z-focus depth. The z-focus quality score is calculated for each z-focus depth. A fast parabolic fit to the points surrounding the maximum of the graph of all z-focus quality scores yields the optimal z-focus depth. This is a very fast way of finding the best focus and works well for multiple closely spaced image fields. However, when the automated imaging system has to move over a macroscopic distance, for example to the next well in a 96- or 384-well microscopy plate, the fine focusing described earlier will not be sufficient for finding a badly out-of-focus image field. Even very small tilts in the plane of the microplate can put an image very far out of focus after translating the objective by several millimeters. To help the autofocusing heuristic an overall calibration or offset is needed to account for any small misalignments between the perpendicular to the sample plane and the optical axis. One method is to use an offset calculated by manually taking focused image data at 3–4 points on the microscopy plate and then fitting a plane to the calibration points. This is performed before automated data taking begins. The z-coordinate of the of the fitted calibration plane at the location of the center of each microscopy-plate-well becomes the zero offset for subsequent autofocusing. Alternatively, if available on the microscope setup, one can use laser range finding to calculate the distance to the cover slip. This distance plus the cover slip thickness gives the starting point for subsequent fine focusing. The fine focusing step is implemented with a precision piezo electric objective insert or peizo electric actuated stage insert. An approach using the image data is used because the best focus is only found using the full image as observed through the optical system and CCD camera. Even though the procedure outlined earlier will yield satisfactory results in the fluorescent channels, the correct focal plane in the white-light channel used for autofocusing will not necessarily match the correct focal plane for different fluorescent channels. However, this is a repeatable physical effect and can be compensated for by measuring the focal plane offsets for each fluorescent channel versus each other and versus the optimal phase contrast focal plane. Once measured, this becomes a constant offset when setting the z-focus for that particular fluorescent channel.
Image Stability
Two important considerations for automating data taking and analysis are that one can take a well-focused fluorescent image without resorting to a z-stack, and that repeated frames, such as those found in a time series, are in good alignment. During the analysis, it is possible to correct for out-of-alignment frames, as it is possible to automatically search for the best-focused image out of a z-stack. However, correcting for issues resulting from poor data takes time and is likely to result in an increase in the overall systematic error of a quantitative measurement. An environmentally controlled microscope setup will minimize drift in x–y or z-focus coordinates regardless of vibration, temperature, or humidity changes in the surrounding environment. Image drift over some relevant time scale should be less than the maximum resolution of the optical system. For 100× fluorescent microscopy this means stability against drift at less than a ten nano meters per second. Systems that achieve this level of stability can image in phase contrast plus several fluorescent channels while keeping the optical drift at the subpixel level. Note that it can take several minutes for the microscopy plate to come into thermal equilibrium after being introduced to a controlled environment. Acquiring images too early can lead to significant drift and result in misregistered and out-of-focus images.
Data Model and Storage
For flexibly analyzing large amounts of data the images must be indexed, for example by the optical channel imaged (c), the z-depth of focus in a multiple z-stack set of images (z), a time index for the purposes of time-lapse (t), the selected image field for a particular sample (f), and finally an index for the unique sample (w). Once image data is acquired it needs to be associated with its corresponding metadata and stored. Getting complete metadata associated with the correct image data can be a vexing problem. Image information from the microscope and camera that can be acquired automatically can be easily saved. Ensuring that information provided by the experimenter is properly saved is more difficult. One scheme requires the upload of an experimental metadata file at the start of a data-taking run. However, user friendly tools for properly annotating image data with user-generated experimental metadata after the fact are also important. Regarding computer power and data storage, the capability of software allowing individual investigators to organize, manage, and analyze a large amount of image data is improving so rapidly that any comment made here will be quickly out of date. In our experience, data analysis ambitions tend to scale with increasing computer and data storage capabilities, to the point where dedicated image servers are used for image libraries and a multiple node compute clusters are used for image analysis.
AUTOMATED QUANTITATIVE IMAGE ANALYSIS
Our example of an automated analysis work flow starts with a high-quality phase contrast or DIC image and one or more fluorescent-channel images for each image field as seen in Figure 1. In the first analysis stage, cell finding is followed by subcellular feature detection and parameterization at the single-cell level as shown in Figure 2. The data reduction factor at this stage is 10–20-fold depending on the complexity of the image. In the second stage of the analysis, the individual cell measurements are combined into high-statistics ensemble averages and standard deviations. A typical ensemble might consist of all of the cells from all of the images taken for a particular sample well. At this stage ensemble averages are translated in to biologically relevant phenotypes and the results can be tabulated, plotted, or subjected to more advanced classification algorithms. Note that by making individual cell measurements, we have access to the distribution of each measured parameter on an ensemble basis and not just the average and standard deviation. There are observable phenotypes that do not significantly change the average or standard deviation, but do noticeably change the distribution of measured values. For example, high intensity measurements in the tail of a distribution of localized fluorescence signals may disappear in a particular mutant background, while the core distribution remains intact (Christen and Fero 2009). The reason for separating the cell detection and parameterization from the rest of the analysis is that the processing time for the two stages differs markedly. Because cell parameterization can require 100 times more CPU time than ensemble characterization, it is not wise to directly couple the two parts of the analysis. Once the images have been processed and the cells reduced to their data summary form, it is not necessary to repeat this processing step. Instead, the investigator can concentrate on refining the second stage of the analysis, which is where most of the biological content lies.
Figure 1.
Automated image analysis. Stored images are segmented by individual cell and parameterized using all available image information. Parameterized data is stored to disk for second-pass analysis where cell parameters are converted to biologically relevant metrics that can be used for quantitative phenotyping.
Figure 2.
Measurements made on an individual bacterial cell. (A) The membrane profile from a subpixel resolution spline fit to a phase-contrast image is combined with, (B) subpixel resolution measurements of fluorescent foci and delocalized fluorescent signal to provide, (C) a parameterized view of the cell including localized and delocalized fluorescent contributions.
STAGE ONE: CELL LEVEL MEASUREMENTS
Cell Finding and Image Segmentation
Every automated analysis starts with finding cells. Under the controlled experimental conditions of a bacterial fluorescence microscopy experiment, the cell density can be tuned to provide a distribution of cells with a minimal number of touching or overlapping cells. Unlike many eukaryotic cell types, where cell finding and segmentation can be a challenging task (Kasson et al. 2005), cell finding in bacterial systems is a fairly straight forward, usually involving either a thresholding or edge-detection, region-filling approach performed on the phase contrast channel. To compensate for uneven illumination of the phase-contrast channel a simple block-filtering algorithm can be used for background flattening. This step followed by an automatically adjusted thresholding and region-finding algorithm works well to identify cells. It is not crucial at this stage to perfectly identify the cell boundary. Rather, the aim is to properly tag the potential cell locations. After tagging the location of each cell, the original image can be segmented into a number of subimages, one for each cell, in preparation for detailed examination.
Membrane Profiling
When analyzing the smaller subimages the cell-finding algorithm is repeated, in a more refined way, to get a good measure of the cell profile in two dimensions. The goal, in our example, is a membrane profile representing a section halfway through the cell in the z-direction. This can be a difficult step, if care is not taken delicate structures that are marginally visible are lost. In some cases it is possible to assume that the membrane boundary has no sharp discontinuities and a spline-fit (De Boor 1978) can be fit to a threshold boundary yielding a correct subpixel representation of the membrane profile as shown in Figure 2A. Once a membrane profile is available, rod shaped cells can be defined by an internal coordinate scheme defined by the longitudinal midcell line, the poles, and several measures of cell width. Note that here we have taken care to define an internal coordinate system defined by an midcell line that runs from one cell pole to the other, with orthogonal width measures at right angles to the midcell line.
Cell Image Simulation
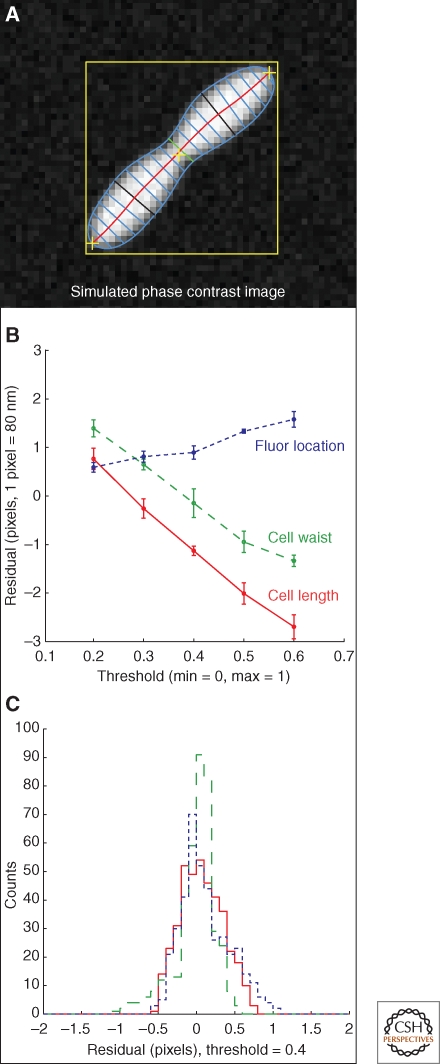
Vital to the process of developing algorithms that yield results with believable error estimates is the implementation of a simple cell-image simulation, as shown in Figure 3A. With a simulation we know, at the outset, what the “true” (expected) values are for the parameters that will eventually be measured by the analysis algorithm. Starting with an idealized cell profile, one can apply the effect of the optical system (in this case the optical point-spread-function plus the finite pixel size of the imaging system), plus an additional dose of simulated noise that emulates the noise seen in the actual images, to create a pseudo-cell image that for our purposes cannot be distinguished from a real cell image. When an analysis algorithm is applied to this image, its output can be compared with the input to the simulation to yield a residual plot of the expected value minus the measured value as a function of the measured parameter, as shown in Figure 3B. As seen in the figure, this analysis has quantified the degree of the negative correlation between a threshold parameter used in the membrane finding algorithm and two measures of cell size. This immediately tells the investigator that, depending on the threshold parameter used, the measurement may have a systematic error of up to four pixels. The measurement only falls into line with the input value to the simulation for fairly low values of the threshold. If these threshold values are too low for reliable cell finding, one option is to use a higher threshold, and then correct the length and width measurements with a factor determined from the simulation. The next step is to run a simple Monte Carlo simulation where numerous simulated cells are generated with varying sizes and amounts of noise. A histogram of the residual values determined from the difference between expected and measured values (with corrections) will produce a Gaussian-like distribution, the width of which can be taken to be a lower bound on the systematic error because of the optical properties of the system and the measurement algorithm itself. Figure 3C shows the result of such a Monte Carlo simulation for 400 randomly produced cells. The residual histograms show that subpixel resolution can be achieved if parameter-dependent algorithms are properly tuned. Without the adjustments determined from cell-image simulation, systematic errors larger than the inherent resolution of the optical system can be unwittingly introduced. Finally, the algorithm can be checked for accuracy with a standard of known range in diameters such as those given by commercially available polystyrene beads (Werner et al. 2009).
Figure 3.
Using cell image simulation when developing algorithms. (A) An image simulation of a bacterial cell has been used to test a membrane-fitting algorithm. (B) The difference between the predicted and measured length and width of the cell as a function of threshold shows a systematic bias. (C) The effect on measuring a fluorescent spot relative to the measured pole of the cell. After correcting for the threshold effect a simulation run of 400 cells shows the resolution of the algorithms, given by the width of the residual histograms as shown.
Localized and Delocalized Florescent Signals
Properly imaged fluorescent signals from GFP-protein fusions have excellent signal-to-noise characteristics. It is often a simple matter to find the fluorescent maxima and, with criteria based on proximity of the peaks to the structures determined by the membrane profile, assign the fluorescence to the correct cell as shown in Figure 2B. If point-like in origin, the fluorescent signal will look like a well defined two-dimensional Gaussian peak with a width defined by the point spread function of the optical system, as shown in Figure 2C. Knowledge of the underlying source of the fluorescent signal puts powerful constraints on the signal and helps to eliminate spurious pixel noise (with widths much smaller than the PSF) (Szelezniak 2007) and out-of-focus channels or fluorescent contamination or aggregates (with widths much larger than the PSF). Slightly less simple to determine is the level of the localized fluorescent signal, and substantially more difficult to determine is the level or gradient across the cell of nonlocalized signal. When one signal underlies another it is often hard to disentangle them because one may be a background contribution to the other. To make the analysis rigorous we recommend renormalizing the image data, scaling it between zero and one, where a baseline of zero is set to the value of the minimum-intensity pixel and a normalized intensity of one was assigned to the brightest pixel. The scale factors and offsets should be preserved so that after analysis the image can be translated back into the original fluorescence units. Cell image simulation is again useful here for developing a well understood algorithm that will yield a measurement with reasonable systematic errors. To characterize the amplitude of the localized signal one can fit a two-dimensional Gaussian to the data with starting parameters for a nonlinear search algorithm given by (1) starting XY coordinates given by the peak centroid, (2) a starting width determined by the PSF, and (3) a starting amplitude given by the amplitude of the brightest pixel in the fluorescent peak. The output from the fit can be compared with a simulated point like light source and compared as a method for tuning the peak finding algorithm. In this way it is possible to obtain subpixel resolution on the XY coordinate of the center of the peak. The absolute amplitude of the localized signal is more difficult and remains dependent on the amount of background attributed to delocalized, cytoplasmic signal and whether it contributes to the localized signal in an additive way. We have tested various techniques via cell image simulation to account for the delocalized signal and have found one that works reasonably well. The technique involves counting the number of discrete “found” regions inside the cell boundary in the (unfiltered) fluorescent channel of interest, as a continually varying threshold is applied. It is observed that the threshold yielding the maximum number of found regions corresponds well with the median of the cytoplasmic contribution to cell fluorescence.
STAGE TWO: ENSEMBLE LEVEL MEASUREMENTS
Cell Cycle Example
We have divided image analysis into an image processing stage where the individual cells are parameterized, and a second stage where ensemble averages are made and the cells are characterized biologically. We have mentioned the advantages of doing this from a processing point of view, the second stage being much less CPU intensive than the first. Another advantage is procedural; we have left the difficult technical task of reducing the cell image to a set of numbers and can now concentrate on the biology. This stage of the analysis is often repeated and tuned numerous times during the course of an analysis, with input from various collaborators, so it is useful not to have to reanalyze all of the images every time someone has a good idea about what to measure. What is measured at the ensemble level depends on the experiment being performed and is thus often unique. For example, if one were performing a cell cycle time-course experiment on polar protein abundance and localization with synchronized cells, it would be reasonable to plot the peak polar signal averaged over an ensemble of high quality cells for each time point in the cell cycle. An example of this type of analysis is shown in Figure 4 for a Caulobacter crescentus cell cycle experiment. The values shown are from ensemble averages over many cells and the error bars reflect the error in those measurements. Although it took several hundred seconds to parameterize all of the imaged cells during the first stage of this analysis, the second stage, involving selection of high quality data, determining the mean of the peak fluorescent signals in the three channels and plotting the data, takes only seconds.
Figure 4.
Polar localized protein concentration as a function of cell cycle. (A) An example of cell cycle dependent protein localization in Caulobacter. (B) An example single-cell image shows the correspondence with the visible changes in localized protein concentration (C) Automated analysis detects time variation in polar fluorescence intensities of the CpaE-CFP, PleC-YFP, and DivJ-RFP reporter fusions.
Mutant Screening Example
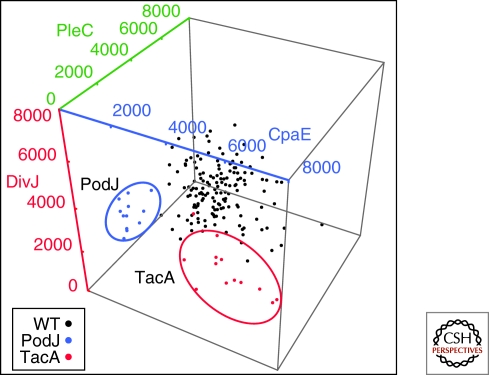
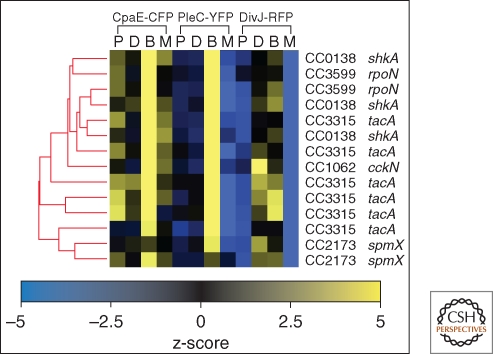
A large-scale mutant screen provides a more challenging example. In this case it is necessary to process images from thousands of mutants, and then classify them on the basis of several criteria such as polar-localized signal amplitude and location, cytoplasmic-delocalized signal amplitude, localized protein polarity, as well as a host-cell shape parameters. As an example we show in Figure 5 the localized fluorescence amplitude signals from a triply labeled strain of Caulobacter crescentus used in a high throughput screen for mutants that mislocalized one or more of the reporter proteins. Each data point is an ensemble average over a number of cells whose peak signal is in the high intensity tail of the peak signal distribution. The data shows the distinct difference between reporter signals resulting from transposon disruptions in the PodJ and TacA genes and the unmutagenized control strains. In Figure 5 we can simultaneously observe three-dimensional phenotypic data, labeled by genotype. However, incorporating more fluorescence based phenotypic information requires analysis tools capable of dealing with higher-dimensional data. Because we know the genotype of every mutant analyzed for each fluorescent phenotype, we can use hierarchical clustering as shown in Figure 6, taking advantage of all the fluorescent data available in the screen. The cluster shown in Figure 6 represents a robust cluster of mutants that emerged from a much larger dendrogram of over 800 mutants. The genes identified in the cluster correspond to the elements of a pathway controlling the relocalization of the sensor histidine kinase DivJ to the stalked pole at the swarmer to stalked transition (Biondi et al. 2006; Radhakrishnan et al. 2008; Christen and Fero 2009). Thus, it is possible to completely automate a fluorescence microscopy based screen that leads to functional gene module identification.
Figure 5.
Ensemble measurement of polar localized protein based on individual cell measurements. The polar fluorescence intensities of the CpaE-CFP, PleC-YFP, and DivJ-RFP reporter fusions are plotted for the nonmutagenized control strain (black) as well as gene disruptions in two separate open reading frames coding for the PodJ (blue) and TacA (red) proteins.
Figure 6.
Example of Genotype-Phenotype microscopy based analysis. A collection of automatically determined cell metrics is used to identify a functional cluster indicating a gene module coordinating temporal and spatial protein localization. Heat map representation of the calculated z-score values from localized protein abundance (L), delocalized protein (D), the fraction of cells identified as being biopolar (B), and the fraction similarly identified as monopolar (M) are shown for a highly significant gene cluster designated by the dendrogram.
Shape Characterization
The shape of a mutant or non–wild-type cell is often a critical part of a fluorescence microscopy experiment. Although one of the easiest things to pick out by eye, it is surprisingly difficult to parameterize mutant shapes, because it is hard to anticipate all the shapes one might encounter. The membrane parameterization techniques we have described do a reasonably good job following a complex membrane profile but the chances of missing an interesting phenotype increase as the cells begin to differ markedly from what was expected by the algorithm developer. Thus, mutant shape characterization can benefit from more global analysis techniques. In these approaches, the cell is not assumed to have a particular form that yields to parameterization in the way that we have described above. Rather, the cells are interpreted as observations that can be compared either to each other or to a control set and tested for similarity, or to a set of alternative models for classification. This process lends itself to the use of pattern recognition and supervised or unsupervised statistical learning techniques. Supervised techniques require a training data set and produce a model that can then be applied to a test set for purposes of classification. The canonical examples of these approaches are classification and regression trees and various types of neural nets (Hastie et al. 2009). In an unsupervised approach, no assumption about what is expected is made and the algorithm is expected to bootstrap its way to finding unique classes based on the data itself (Heidmann 2005). Image data is not often directly amenable to these approaches and is usually first reduced in complexity by the computation of numerical representations of the image. After a cell has been thus re-interpreted it can be classified via one of the supervised or unsupervised approaches. Good examples of morphology analysis are found in Pincus and Jones (Pincus and Theriot 2007; Jones et al. 2009).
CONCLUDING REMARKS
Well-defined prokaryotic fluorescence microscopy experiments can now be performed in an almost completely automated way. Once a process has been developed, image data acquisition, first and second stage analysis and the presentation of final results can all be done without ever requiring the viewing, converting, or measuring of individual images by the biologist. In making this statement we do not which to trivialize the effort involved in developing automated acquisition and analysis tools. A brief reflection on cost vs. benefit will make it clear to the experimenter whether the upfront investment in automated data taking and analysis is required. We have illustrated a seamless pipeline approach, which requires a minimum amount of user intervention but a maximum of software developer effort. Unless the experiment is very high throughput, most experiments today can be performed quite successfully using nonautomated image acquisition and semiautomated quantitative analysis like those available using stand-alone tools such as ImageJ (Girish and Vijayalakshmi 2004; Collins 2007). However, as the interest in performing high-throughput assays and screens at the genomic or meta-genomic level in prokaryotic systems grows, the value of integrated, automated systems for data acquisition and analysis will become evident.
ACKNOWLEDGMENTS
This work was funded by National Institutes of Health Grant K25 GM070972-01A2 (M.F.).
Footnotes
Editors: Lucy Shapiro and Richard Losick
Additional Perspectives on Cell Biology of Bacteria available at www.cshperspectives.org
REFERENCES
- Biondi EG, Skerker JM, Arif M, Prasol MS, Perchuk BS, Laub MT 2006. A phosphorelay system controls stalk biogenesis during cell cycle progression in Caulobacter crescentus. Mol Microbiol 59: 386–401 [DOI] [PubMed] [Google Scholar]
- Boland MV, Murphy RF 1999. Automated analysis of patterns in fluorescence-microscope images. Trends Cell Biol 9: 201–202 [DOI] [PubMed] [Google Scholar]
- Boland MV, Murphy RF 2001. A neural network classifier capable of recognizing the patterns of all major subcellular structures in fluorescence microscope images of HeLa cells. Bioinformatics 17: 1213–1223 [DOI] [PubMed] [Google Scholar]
- Boland MV, Markey MK, Murphy RF 1998. Automated recognition of patterns characteristic of subcellular structures in fluorescence microscopy images. Cytometry 33: 366–375 [PubMed] [Google Scholar]
- Chen X, Murphy RF 2005. Objective clustering of proteins based on subcellular location patterns. J Biomed Biotechnol 2005: 87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Velliste M, Murphy RF 2006. Automated interpretation of subcellular patterns in fluorescence microscope images for location proteomics. Cytometry 69: 631–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen B, Fero M, Hillson N, Bowman G, Hong S-H, Shapiro L, McAdams H 2010. High-throughput identification of protein localization dependency networks. Proc Natl Acad Sci 107: 4681–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins TJ 2007. ImageJ for microscopy. BioTechniques 43: 25–30 [DOI] [PubMed] [Google Scholar]
- De Boor C 1978. A practical guide to splines Springer-Verlag, New York [Google Scholar]
- Firestone L, Cook K, Culp K, Talsania N, Preston K Jr 1991. Comparison of autofocus methods for automated microscopy. Cytometry 12: 195–206 [DOI] [PubMed] [Google Scholar]
- Geusebroek JM, Cornelissen F, Smeulders AW, Geerts H 2000. Robust autofocusing in microscopy. Cytometry 39: 1–9 [PubMed] [Google Scholar]
- Girish V, Vijayalakshmi A 2004. Affordable image analysis using NIH Image/ImageJ. Ind J Cancer 41: 47. [PubMed] [Google Scholar]
- Gregory JA, Becker EC, Pogliano K 2008. Bacillus subtilis MinC destabilizes FtsZ-rings at new cell poles and contributes to the timing of cell division. Genes Develop 22: 3475–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen FC, Young IT, Ligthart G 1985. A comparison of different focus functions for use in autofocus algorithms. Cytometry 6: 81–91 [DOI] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman JH 2009. The elements of statistical learning : Data mining, inference, and prediction Springer, New York [Google Scholar]
- Heidmann G 2005. Unsupervised image categorization. Image and Vision Computing 23: 861–876 [Google Scholar]
- Huang K, Murphy RF 2004. From quantitative microscopy to automated image understanding. J Biomed Optics 9: 893–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TR, Carpenter AE, Lamprecht MR, Moffat J, Silver SJ, Grenier JK, Castoreno AB, Eggert US, Root DE, Golland P, et al. 2009. Scoring diverse cellular morphologies in image-based screens with iterative feedback and machine learning. Proc Natl Acad Sci 106: 1826–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasson PM, Huppa JB, Davis MM, Brunger AT 2005. A hybrid machine-learning approach for segmentation of protein localization data. Bioinformatics 21: 3778–3786 [DOI] [PubMed] [Google Scholar]
- Ohya Y, Sese J, Yukawa M, Sano F, Nakatani Y, Saito TL, Saka A, Fukuda T, Ishihara S, Oka S, et al. 2005. High-dimensional and large-scale phenotyping of yeast mutants. Proc Natl Acad Sci 102: 19015–19020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus Z, Theriot JA 2007. Comparison of quantitative methods for cell-shape analysis. J Micros 227: 140–156 [DOI] [PubMed] [Google Scholar]
- Price JH, Gough DA 1994. Comparison of phase-contrast and fluorescence digital autofocus for scanning microscopy. Cytometry 16: 283–297 [DOI] [PubMed] [Google Scholar]
- Radhakrishnan SK, Thanbichler M, Viollier PH 2008. The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus. Genes Dev 22: 212–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szelezniak M 2007. Cluster finder in the HFT readout.
- Werner JN, Chen EY, Guberman JM, Zippilli AR, Irgon JJ, Gitai Z 2009. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc Natl Acad Sci 106: 7858–7863 [DOI] [PMC free article] [PubMed] [Google Scholar]