SUMMARY
RNA interference (RNAi) provides a powerful reverse genetics approach to analyze gene functions both in tissue culture and in vivo. Because of its widespread applicability and effectiveness it has become an essential part of the tool box kits of model organisms such as Caenorhabditis elegans, Drosophila, and the mouse. In addition, the use of RNAi in animals in which genetic tools are either poorly developed or nonexistent enables a myriad of fundamental questions to be asked. Here, we review the methods and applications of in vivo RNAi to characterize gene functions in model organisms and discuss their impact to the study of developmental as well as evolutionary questions. Further, we discuss the applications of RNAi technologies to crop improvement, pest control and RNAi therapeutics, thus providing an appreciation of the potential for phenomenal applications of RNAi to agriculture and medicine.
In vivo RNAi technology is replacing classical genetics in screens and conditional targeting of gene function. It also has applications in crop improvement, pest control, and medicine.
1. INTRODUCTION
Genetic screening is one of the most powerful methods available for gaining insights into complex biological processes. Indeed, much of what we have learned from model organisms such as Saccharomyces cerevisiae, Caenorhabditis elegans, and Drosophila melanogaster can be traced back to genetic screens designed to identify sets of mutations that perturb specific processes. For example, screens in yeast have led to the identification of key regulators of the cell cycle (Hartwell et al. 1974); screens in C. elegans have identified the genetic regulation of programmed cell death (Horvitz et al. 1999); and screens for mutations that cause embryonic lethality in Drosophila have elucidated the logic of body patterning in a multicellular organism (Nusslein-Volhard and Wieschaus 1980; St Johnston and Nusslein-Volhard, 1992).
Over the years many improvements and tools for genetic manipulation have become available, and as a result there now exist powerful “tool-boxes” for each model organism (Nagy et al. 2003; Venken and Bellen 2005; Kaletta and Hengartner 2006). Sophistication in approaches and tools facilitates the ease of genetic screening as well as the identification of genetic alteration(s) and requisite follow-up analyses of mutant phenotypes. For example, innovations such as mosaic analysis and tissue-specific expression of transgenes have allowed researchers to study gene function in a wider variety of tissues, stages, and contexts.
Soon after the initial discovery by Fire and Mello (Fire et al. 1998) that double-stranded RNAs (dsRNAs) can be used to knockdown the activity of individual genes, many RNA interference (RNAi)-based methods were (and continue to be) added to the tool-boxes of various organisms. These methods have truly revolutionized the field of functional genomics because of their relative ease, and most significantly, because RNAi, unlike more traditional genetic screening methods, provides a powerful reverse genetic approach, especially for organisms in which genetics is difficult, as is the case with mammalian systems. Importantly, the power of RNAi-based methods for genetic analyses became fully realized when the genome sequences of various organisms were completed (C. elegans Sequencing Consortium, 1998; Adams et al. 2000; Venter et al. 2001; Waterston et al. 2002; The Rat Genome Sequencing Project Consortium, 2004). Thus, the identification of all genes in the C. elegans, Drosophila, mouse, rat, and human genomes has led to the construction of numerous genome-wide RNAi resources, allowing reverse genetic screens either in tissue culture or in vivo. Today, genome-wide RNAi screening is possible in vivo in C. elegans, in tissue culture cells and in vivo in Drosophila, and in cell lines from mice, rats, and humans.
RNAi is a well-established tool for studies in tissue culture and, following the first genome-wide RNAi screen performed in Drosophila cells (Boutros et al. 2004), RNAi high-throughput screening (HTS) has become routine both in Drosophila and mammalian cells. Cell-based screening has been extensively reviewed in the past (Echeverri and Perrimon 2006; Perrimon and Mathey–Prevot, 2007; Boutros and Ahringer 2008; Mohr et al. 2011). In this review, we focus on in vivo methods and applications of RNAi. In most organisms, methods for in vivo RNAi are still in development and we discuss the state of the field, what has been learned so far, and future development. In particular, we describe the application of in vivo RNAi to characterize the function of pleiotropic genes and discuss its impact for the study of organisms for which genetic tools are either nonexistent or poorly developed.
2. RNAi REAGENTS FOR IN VIVO SCREENING
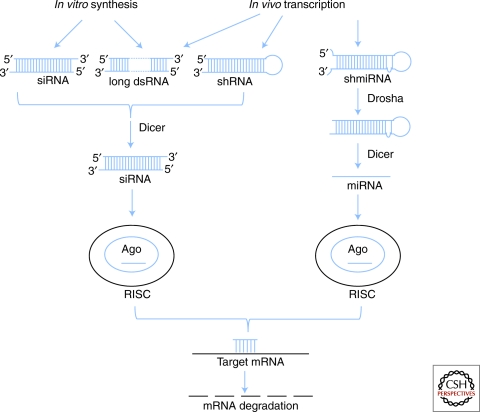
Four different types of RNAi reagents are used for in vivo studies: synthetic siRNAs, small hairpin RNAs (shRNAs), small hairpin microRNAs (shmiRNAs), and long dsRNAs (reviews by Echeverri and Perrimon 2006; Lee and Kumar 2009) (Fig. 1).
Figure 1.
SiRNAs, shRNAs, shmiRNAs, and long dsRNAs pathways.
Synthetic siRNAs are small RNA duplexes composed of 19 complementary base pairs (bps) and 2-nucleotide 3′ overhangs. They are transfected into cells or injected into animals. On entering cells one strand of the siRNA duplex is incorporated into the multi-subunit ribonucleoprotein complex (RISC) and directs RISC to the target mRNA by complementary base-pairing, resulting in mRNA degradation. The effects of the siRNAs are transient, especially in actively dividing cells.
In contrast, shRNA and shmiRNA-synthesizing vectors allow for controlled or continuous expression of small transcripts in the cell that contain both the sense and antisense strand complementary to the selected mRNA target. They are either transfected into cells as plasmid DNAs or delivered using viral particles, and are maintained as extra-chromosomal copies or stably integrated in the genome as transgenes. The 50–70 bps single-stranded RNA transcripts fold back to form a stem-loop structure. ShRNAs are processed in the cytoplasm by the ribonuclease Dicer to generate siRNAs. ShmiRNAs are a variation of shRNAs in that sequences for the silencing trigger are embedded in an endogenous miRNA expression cassette. ShmiRNAs therefore exploit the endogenous microRNA pathway for the biogenesis and subsequent loading of siRNAs into RISC, and are usually more effective in knocking down target mRNAs than shRNAs.
For use as RNAi reagents, long dsRNAs are usually 200–500 nucleotides (nts) in length. They can be injected into animals and in many cases, into eggs; delivered via bacteria (see sections on C. elegans and Planaria later); expressed as transgenes (see sections on C. elegans, Drosophila and mouse); or delivered into cultured cells by transfection or bathing (Drosophila). With the exception of the esiRNA (endoribonuclease-prepared siRNAs) method (Yang et al. 2002), whereby long dsRNAs are used to produce a pool of small, diced siRNAs that is then transfected into cells, long dsRNAs are not used in mammalian systems as they trigger an unwanted interferon response that can mask gene-specific effects.
3. IN VIVO RNAi SCREENING IN C. ELEGANS
The discovery that a dsRNA introduced into the nematode C. elegans is able to degrade a specific mRNA (Fire et al. 1998) marked the beginning of the revolution in in vivo RNAi. Importantly, RNAi in C. elegans is both systemic and transitive. First, injection, or expression, of a dsRNA into one tissue can lead to gene silencing in other tissues (Fire et al. 1998; Winston et al. 2002). Genetic analysis of this systemic effect has identified a number of genes involved in the phenomenon, including the multispan transmembrane protein SID-1, which is sufficient to confer cellular uptake of dsRNA to cells (Feinberg and Hunter 2003). Second, RNAi in C. elegans is transitive, whereby an RNA-dependent RNA polymerase (RdRP) is involved in an amplification step of RNAi and as a result, siRNAs that are derived from regions upstream of the original dsRNA sequences are produced (Alder et al. 2003).
The relative ease of methods required to perform RNAi experiments in C. elegans makes this genetically amenable model organism a logical choice for the development of technologies to study gene function on a genome-wide scale (review by Sugimoto et al. 2004). DsRNAs can be introduced into the nematode by simply soaking the animal in a solution of dsRNA, by feeding the worms bacteria that express long dsRNAs, by injection of dsRNA, or by generating transgenic hairpin-expressing animals.
Many genome-wide RNAi screens have been performed in the past 10 yr in C. elegans to interrogate a large variety of biological questions in developmental biology, cell signaling, aging, metabolic regulation, and neurodegenerative diseases, to name a few. These screens have been performed either in a wild-type strain or specific mutant backgrounds and either by injection or by feeding (see reviews by Kaletta and Hengartner 2006; Boutros and Ahringer 2008). In a landmark study based on injection into eggs, Sönnichsen et al. (2005) performed a genome-wide screen to identify all genes required for the first two rounds of cell division by examining embryonic phenotypes using time-lapse microscopy. However, the method of choice is large-scale RNAi screening by feeding worms bacteria that produce dsRNAs (Timmons and Fire 1998) because first, the method is less tedious by far, and second, RNAi libraries in bacteria that cover most of the 20,000 C. elegans genes are available (Fraser et al. 2000; Kim et al. 2005; Boutros and Ahringer 2008). For example, Ashrafi et al. (2003) screened 16,757 genes for their roles in fat storage in living worms using Nile Red staining of tissue lipids. They isolated 305 genes that when knocked down, lead to reduced body fat and 112 genes that lead to increased fat storage, representing a core set of fat regulatory genes as well as pathway-specific fat regulators.
Importantly, RNAi screening in C. elegans can easily be performed in various combinations, either in mutant backgrounds or by using multiple RNAs, to identify synthetic phenotypes. Such screens are a powerful means to gain an understanding of the structure of signaling networks, disease susceptibility, and identification of new drug targets. (Lehner et al. 2006), for example, systematically tested approximately 65,000 pairs of genes for their abilities to interact genetically and identified 350 genetic interactions between components of the EGF/Ras, Notch, and Wnt pathways.
Finally, an important issue with large-scale RNAi screening (also discussed later) is the rate of false positive and negative results associated with the method. False positives that occur when novel unexpected phenotypes are associated with RNAi lines appear to be a minor contributor to false discovery in C. elegans (Sönnichsen et al. 2005). False negatives on the other hand, because of the variability of knockdown associated with the feeding techniques, can be more of an issue (depending on the screen) and may account for the 10%–30% variability observed between screens even if they are performed in the same laboratory (Simmer et al. 2003).
4. IN VIVO RNAi SCREENING IN DROSOPHILA
In Drosophila, feeding methods for RNAi delivery, as in C. elegans, do not appear to work; however, RNAi reagents can be delivered either by injection into precellular blastoderm embryos or as transgenes. Importantly, although there have been reports that systemic and transitive RNAi may occur in Drosophila (Saleh et al. 2009; Lipardi and Patterson 2009), this does not appear to occur when the dsRNA is produced from a transgene (Roignant et al. 2003).
Injection of dsRNAs as short as 200 bps and as long as 2000 bps, as well as short 21–22 nts siRNAs injected into embryos, have been shown to have potent interfering activities (Kennerdell and Carthew 1998; Misquitta and Paterson 1999; Williams and Rubin 2002; Misquitta et al. 2008). This approach has been used, for example, to clarify the role of the MyoD-related gene nautilus in embryonic somatic muscle formation (Misquitta and Paterson, 1999), and the roles of both the Frizzled1 and Frizzled2 receptors in Wingless signaling (Kennerdell and Carthew 1998). RNAi injection has been used systematically to screen more than 5000 genes for cardiogenic and embryonic nervous system phenotypes. For the heart screen, dsRNA-injected embryos that carry the D-mef2-lacZ transgene to detect cardiac cells were examined. For the nervous system screen, embryos were stained using the 22C10 antibody that detects the entire peripheral nervous system and a subset of central nervous system neurons. This approach led to the identification of many new genes involved in either heart or neural development (Kim et al. 2004) (http://flyembryo.nhlbi.nih.gov/).
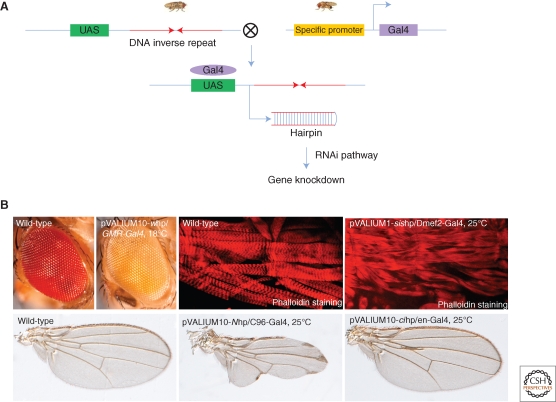
RNAi by injection has somewhat limited applications as this approach is restricted to studies of gene function during embryonic development and maternally loaded proteins may mask embryonic phenotypes. Transgenic RNAi, on the other hand, has been widely used to study gene function in somatic tissues. Importantly, and unlike in C. elegans, in Drosophila RNAi is cell-autonomous, and because of this, targeted expression of RNAi constructs using the Gal4/UAS system (Brand and Perrimon 1993) can be used for cell- or tissue-specific interrogation of gene function. Indeed, this approach has been used extensively (Fig. 2). To date, transgenic RNAi lines have been shown to be potent in all somatic tissues, including neurons and muscles. However, for unknown reasons they do not appear to be effective in the female germ line.
Figure 2.
Transgenic RNAi in Drosophila. (A) Tissue expression of the transgenic RNAi construct is achieved following a cross between a UAS-hairpin and a Gal4 driver line. The main advantage of this method, in addition to its relatively simple design and fast execution time, is that it allows spatial and temporal control of the knockdown construct, which is essential for characterizing genes with pleiotropic functions. As thousands of Gal4 lines are available, appropriate Gal4 drivers are basically available for most questions to be addressed in the intact animal. (B) Examples of tissue specific RNAi phenotypes generated in the eye (knockdown of the white gene in the eye using the GMR-Gal4 driver), muscle (knockdown of the sallimus (sis) gene in the eye using the Dmef2-Gal4 driver), and wings (knockdown of the Notch (N) gene and cubitus interruptus (ci) genes in the wing using the C96-Gal4 and en-Gal4 drivers, respectively).
Several groups, working independently, have developed vectors that have been used to generate transgenic RNAi fly strains. The initial vectors were based on transgenes having an inverted–repeat configuration, driven from either a single promoter or symmetrically transcribed from opposing promoters (Lam and Thummel 2000; Fortier and Belote 2000; Martinek and Young 2000; Kennerdell and Carthew 2000; Giordano et al. 2002). Because these vectors generated variable RNAi silencing effects, a number of modifications were introduced based on the observation in plants that intron-spliced hairpin RNAs are more efficient at gene silencing than the hairpin loop RNA (Smith et al. 2000). Thus, a number of groups designed vectors that include intron sequences from genes such as mub (Reichhart et al. 2002), white (Lee and Carthew 2003), Ret (Pili-Floury et al. 2004), or fushi-tarazu (ftz) (Kondo et al. 2006), as well as genomic/cDNA hybrids (Kalidas and Smith 2002). Additionally, the position of the ftz intron within the construct, e.g., located to the end of the hairpin structure, was tested (Fig. 3) (Dietzl et al. 2007). Altogether, these intron-containing vectors gave more robust RNAi phenotypes than the inverted–repeat configuration, most likely because of the enhanced formation of duplex dsRNAs following the splicing event and/or enhanced export of the processed mRNAs from the nucleus. Finally, in addition to the RNAi vectors that generate long dsRNAs, small hairpin microRNA-based (shmiRNA) RNAi constructs that generate a single siRNA have been shown to work as Drosophila transgenes (Chen et al. 2007; Haley et al. 2008). Although these vectors appear to work effectively in the soma, their overall effectiveness, especially compared with long dsRNAs, has not been tested systematically.
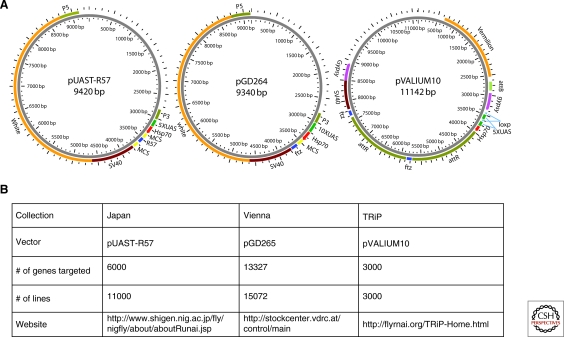
Figure 3.
Structure of the vectors (A) and transgenic RNAi resources (B) available in Drosophila.
A major source of variability between these first generation vectors is caused by the method of transgenesis used, in which the constructs are integrated into the genome at random positions using P-element based transformation. Indeed, Dietzl et al. (2007) estimated that only 60% of their RNAi lines are effective with this most likely because of position effects associated with a large number of the random insertions. To solve this source of variability, a series of vectors, the “VALIUM” series, were generated. These rely on the phiC31-mediated site-specific integration approach (Groth et al. 2004) and the RNAi constructs were strategically integrated into sites in the genome that had been preselected for optimal expression (Ni et al. 2008; Ni et al. 2009). Specifically, a number of genomic sites were identified for which high levels of induced, Gal4-driven gene expression is observed, and importantly, low basal levels are seen in the absence of the Gal4 driver (Markstein et al. 2008). Furthermore, a series of related VALIUM vectors were built and tested for their ability to produce optimal RNAi effects. From these analyses, one optimal vector, VALIUM10, proved excellent for somatic RNAi (Figs. 2, 3) (Ni et al. 2009).
In flies, transgenic RNAi is particularly applicable to studies relevant to human biology such as cancer and metastasis, inflammation and wound healing, metabolic disorders, immunity, aging, and central nervous system disorders. This is exemplified by the hundreds of fly lines generated by individual laboratories, which can be identified either from the published literature or in the Drosophila database Flybase (http://flybase.bio.indiana.edu/). Building on the proven strength of transgenic RNAi, three independent efforts have already generated large-scale resources, such that RNAi lines that cover most of the Drosophila 13,929 protein-encoding genes (Tweedie et al. 2009) are now available (Fig. 3).
Two recently published large-scale screens, both using the Dietzl et al. (2007) library, illustrate that in flies complex developmental processes can be dissected on a genome-wide level using transgenic RNAi. First, Mummery-Widmer et al. (2009) screened for novel components of the Notch pathway by examining the effect of RNAi lines on external sensory organ development. In particular, they identified six new genes involved in asymmetric cell division and 23 novel genes regulating Notch signaling. Among the many interesting genes identified as Notch regulators were genes involved in nuclear import and the COP9 signallosome. In the second whole-genome study, Cronin et al. (2009) screened the RNAi lines for their ability to be resistant or susceptible to the ingestion of pathogenic Gram-negative bacteria Serratia marcescens. The initial screen identified 95 resistant and 790 susceptible candidates that were subsequently analyzed using different Gal4 drivers to determine the site of action of the identified genes. A number of genes involved in intracellular processes, the immune system, the stress response, as well as genes associated with stem cell proliferation, growth, and cell death were shown to be required in the gut. Others, involved in phagocytosis and the stress response, were required in macrophages. Building on these observations, the authors characterized a requirement for the JAK/STAT pathway in response to intestinal Serratia infection.
An important issue with regard to Drosophila RNAi screens in tissue culture concerns false positives that occur from sequence specific off-target effects (OTEs) (Kulkarni et al. 2006; Ma et al. 2006; review by Perrimon and Mathey-Prevot 2007). OTEs can be avoided by selecting sequences that do not contain 19 nts or longer cross-hybridizing stretches to other genes or tri-nucleotide CAN (CA[AGCT]) repeats. In this regard, a number of software tools are available for identifying the most common off-target sequences so that they can be excluded from RNAi constructs. These include E-RNAi from the German Cancer Research Center (http://www.dkfz.de/signaling2/ernai/; Arziman et al. 2005) and SnapDragon from the Drosophila RNAi Screening Center (http://www.flyrnai.org/cgi-bin/RNAi_find_primers.pl; Flockhart et al. 2006). In vivo, although it is difficult to fully evaluate the rates of false positives and negatives in general, as it depends on the reagents used, the specific Gal4 driver used, and the temperature at which the flies are screened, the consensus is that OTEs appear negligible if sequences that avoid potentially problematic sequences are used (Dietzl et al. 2007; Ni et al. 2008; Ni et al. 2009). The rate of false negatives, however, in the fly screens, as discussed in the case of C. elegans, may be more of an issue. For example, Dietzl et al. (2007) estimate that 40% of their RNAi lines may not generate phenotypes because of low transgene expression. In some cases the effectiveness of individual RNAi lines can be improved by co-expressing Dicer2 (Dietzl et al. 2007) but in general, the newer generation of RNAi lines based on targeted insertion (Ni et al. 2009) are more likely to significantly decrease the overall rate of false negatives in RNAi screens.
Although false positives associated with transgenic RNAi appear to be less of an issue than RNAi in tissue culture, results derived from a single fly line still have to be taken with caution. To validate a transgenic RNAi phenotype, two simple follow-up experiments can be performed. First, the result can be confirmed with a second independent line, which becomes easier as more transgenic RNAi lines are generated. Second, and most conclusive, the RNAi induced phenotype can be rescued via expression of a transcript that can confer gene activity but evades the RNAi treatment, such as by having a divergent nucleotide sequence or exogenous 3′UTR (Stielow et al. 2008). Using genomic DNA of closely related species, Kondo et al. (2009) have established a cross-species RNAi rescue method useful to rescue RNAi phenotypes. This straightforward and reliable method, based on genomic DNA fragments contained in fosmids, can be used to quickly build the construct needed to generate the transgenic flies harboring genomic DNA of a sibling Drosophila species that can confer activity but avoid knockdown.
5. APPLICATIONS OF IN VIVO RNAi TO THE DEVELOPMENTAL BIOLOGY OF EMERGING SYSTEMS AND EVO-DEVO
The application of RNAi to non-model and/or emerging model organisms in which few, if any, genetic tools are available is providing biologists a powerful means to characterize the roles of specific genes throughout development and evolution. As transformation methods are available in only a handful of organisms, RNAi is being delivered to emerging models either by feeding or following injection of RNAi reagents.
A large-scale RNAi screen for gene function has been performed in the planarian, Schmidtea mediterranea, an organism not previously accessible to extensive genetic manipulation. As in C. elegans, RNAi is delivered to a Planarian by feeding. In a landmark study, Reddien et al. (2005) screened 1065 genes, 5%–7% of the total, and described phenotypes associated with 240 of them. Many of these showed specific defects in regeneration, and in particular, defects were observed during stem cell/neoblast proliferation in amputated animals. Many biological insights are emerging from this work; for example, a recent study implicates the Wnt/beta-catenin pathway in antero-posterior polarity of the blastema during regeneration (Petersen and Reddien 2008).
RNAi is being used to address evo-devo questions in jellyfish, wasps, beetles, crickets, spiders, etc. For example, in the long germ band parasitic wasp Nasonia vitripennis, injection of pupae with dsRNAs (Lynch and Desplan 2006) has been used to examine the logic of antero-posterior patterning, and in particular mechanisms that differ from the short germ band patterning of Drosophila. Further, these in vivo RNAi studies have clarified the ancestral roles of the bicoid and caudal genes as patterning organizers, and helped to elucidate how these functions have evolved in higher dipterans such as Drosophila (Olesnicky et al. 2006; Brent et al. 2007).
6. IN VIVO RNAi IN VERTEBRATE MODELS
RNAi-based methods are now a common tool for gene perturbation in mammalian tissue culture cells (mouse, rat, monkey, and human). Beyond cell culture screens (see recent review by Mohr et al. 2011), RNAi is being used in a number of in vivo studies in which the RNAi reagents are delivered topically and directed to specific tissues or organs, such as the retina, brain, or muscles; using ex vivo, for example in hematopoietic cells; or delivered as transgenes (Sandy et al. 2005).
RNAi can be achieved locally by delivering synthetic siRNAs or using shRNAs delivered via viral particles or following transfection of plasmid DNAs. Many vectors have been built, based on either shRNA or shmiRNA designs, to optimize the level of expression of the RNAi reagent. Furthermore, much effort has been devoted to the development of methods for conditional RNAi that include irreversible and reversible approaches (Sandy et al. 2005; Lee and Kumar 2009). For example, a number of vectors based on the Cre/loxP and FLP-FRT systems have been used to induce RNAi in an irreversible way. In addition, Tet-, Ecdysone-, LacR, HIV-1 tat-, and HIV-1 LTR-, based systems have been explored for reversible conditional RNAi systems. In addition, vectors have been built for generating transgenic animals that can be either inserted into the genome at random or at targeted sites to ensure expression. To date, most of the published studies are still at the proof of principle stage (Sandy et al. 2005; Lee and Kumar 2009). Importantly, more studies are needed to evaluate technical aspects of RNAi effects, such as the level of knockdown in various cell types, variability because of the insertion site, potential epigenetic silencing of the construct, etc. Regardless of these limitations, from intense ongoing efforts will emerge many exciting applications for RNAi-based methods in the coming years (see review by Lee and Kumar 2009).
To date, and unlike the mouse, RNAi in Xenopus and zebra fish has not had a great impact, due in part to mixed results on the efficacy of some of the RNAi reagents, the prevalent use of the well-established method of antisense oligonucleotide morpholinos, and the lack of effective methods for controlled gene expression. In Xenopus, injection of siRNAs or long dsRNAs into oocytes and early blastomeres appears to work well (Zhou et al. 2002; Nakano et al. 2002), and gene silencing via transgenesis has been shown, although some difficulties have been observed in the silencing of genes at later stages of development (Li and Rohrer 2006). Similarly, in zebra fish, although a few studies have shown that dsRNAs, shRNAs and siRNAs can be effective for gene knockdown, a number of studies report that unexplained morphological abnormalities can be associated with RNAi-injected embryos (Wargelius et al. 1999; Skromne and Prince 2008). Altogether, it is not clear to what extent, at least in the absence of major technical advances, RNAi-based methods will become mainstream in fish or Xenopus.
Finally, in chick embryos, electroporation of siRNAs, as well as delivery using Replication Competent Avian Splice (RCAS) retroviruses to introduce hairpins intotissues, have been used successfully (Harpavat and Cepko, 2006). The RCAS approach is of particular interest as it is long lasting and transmissible because infected cells release more virus that spread to neighboring cells.
7. IN VIVO RNAi IN PLANTS: APPLICATIONS TOWARD CROP IMPROVEMENT
In plants, as in C. elegans, RNAi is both systemic and heritable. The siRNAs move between cells through channels in cell walls, thus enabling communication and transport throughout the plant. In addition, methylation of promoters targeted by RNAi confers heritability, as the new methylation pattern is copied in each new generation of the cell (Jones et al. 2001). Interestingly, in plants, endogenously encoded miRNAs rather than inhibiting translation are nearly or perfectly complementary to their target genes and induce mRNA cleavage by interaction with RISC.
The focus of in vivo RNAi applications in plants is directed toward the improvement of plant productivity and/or nutritional value (see reviews by Kusaba, 2004; Tang et al. 2007; Hebert et al. 2008). Among the exciting applications in which RNAi could have a major impact in agriculture is the improvement of essential food crops such as corn and rice. In addition, RNAi could be used to engineer food plants rendering them rich in dietary protein; for example, lowering the levels of natural plant toxins in cotton seeds could make this abundant plant appropriate for human consumption. Although we are still far from seeing RNAi-modified plant products in agriculture, especially considering the controversies and concerns surrounding growing genetically modified plants for human consumption, a number of successful applications have already emerged, particularly the ability of RNAi to confer resistance to common plant viruses (Zadeh and Foster 2004) and fortification of plants such as tomatoes with dietary antioxidants (Niggeweg et al. 2004).
8. IN VIVO RNAi APPLICATIONS TOWARD VIRUS AND PEST CONTROL
RNAi may have important agricultural applications as illustrated by ongoing attempts to use RNAi approaches to remedy the colony collapse disorder (CCD) in European honeybees. In recent years, millions of beehives have disappeared, most likely because of the spread through bee colonies of a lethal virus, the Israeli acute paralysis virus (IAPV). The current working hypothesis is that IAPV infection, together with poor nutrition and exposure to pesticides, weakens bee colonies to the extent that they simply disappear. One RNAi-based strategy being explored to fight IAPV infection is to feed bees siRNAs targeting specific IAPV sequences such that, following viral entry into bee cells, translation of viral proteins is blocked (Cox-Foster and vanEngelsdorp 2009).
RNAi is also becoming an important tool to combat insect pests, in particular Anopheles gambia, the vector for Plasmodium, the protozoan responsible for malaria. RNAi reagents are being used to dissect host-pathogen interactions and have already provided fundamental insight into the insect defense mechanisms to control the protozoan, such as the identification of the pattern-recognition receptor TEP1 in host defense (Blandin et al. 2004).
Methods to disseminate RNAi expressing transgenes that may confer resistance to a pathogen within a population are also being explored. One of the strategies being considered for the control of pathogen-laden pests is to rapidly convert a pathogen-bearing insect population to a genetically modified population that is resistant to the pathogen. For example, if wild mosquito populations could be replaced with malaria or dengue-resistant ones, this alone may provide an effective means to control these devastating diseases. The challenge with such an approach is to develop a method for rapid replacement of the wild population. In one clever demonstration, Chen et al. (2007) reported an RNAi-based method in Drosophila that achieves the selfish drive of a genetic element into a population. Although the approach is still at an early stage in development, the results of this study show the feasibility of RNAi-based population replacement.
9. IN VIVO RNAi APPLICATIONS IN MEDICINE
RNAi has the potential to offer more specificity and flexibility than traditional drugs to silence gene expression. In addition, because any protein that causes or contributes to a disease is susceptible to RNAi, previous disease targets considered “undruggable” are now accessible. Not surprisingly, RNAi has become a major focus for biotechnology and pharmaceutical companies, which are now in the early stages of developing RNAi therapeutics, mostly based on siRNAs, to target viral infection, cancer, hypercholesterolemia, cardiovascular disease, macular degeneration, and neurodegenerative diseases (Sah et al. 2006).
Critical issues with RNAi as a therapeutic are delivery, specificity and stability of the RNAi reagents. Delivery is currently considered the biggest hurdle as the introduction of siRNAs systemically into body fluids can result in their degradation, off-target effects, and immune detection and subsequent reactions (see for example Zimmermann et al. 2006). Thus, many efforts are focused on developing ways to modify an RNAi or attach them to delivery agents that will protect them until they reach their therapeutic destinations. These include delivery as particles or complexes using lipid nanoparticles that encapsulate the siRNA or combining siRNA molecules with peptide-based polymers. Additional advances in therapeutic applications are likely to come from chemical modifications or other approaches to improving the specificity and potency of RNAi reagents.
10. CONCLUDING REMARKS
RNAi-based methods are providing unprecedented tools useful to address fundamental questions in the biology of living organisms. As exemplified by in vivo screens in C. elegans and Drosophila as reviewed here, these tools are enhancing and/or replacing more classical genetic approaches and manipulations. Further, as most organisms possess the cellular machinery for RNAi, this near-universal approach makes loss-of-function studies approachable in organisms in which genetic tools do not exist. Finally, with the growing appreciation for the fundamental potential of RNAi and a burgeoning collection of RNAi technologies and reagents, the diversity in in vivo applications to biology, medicine, and agriculture is seemingly limitless.
ACKNOWLEDGMENTS
We thank Rui Zhou and Stephanie Mohr for comments on the manuscript and Luping Liu, Donghui Yang-Zhou and Martha Reed assistance on the figures. This work is supported by GM084947 to N.P. and the HHMI.
Footnotes
Editors: John F. Atkins, Raymond F. Gesteland, and Thomas R. Cech
Additional Perspectives on RNA Worlds available at www.cshperspectives.org
REFERENCES
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG et al. 2000. The Genome sequence of Drosophila melanogaster. Science 287: 2185–2195 [DOI] [PubMed] [Google Scholar]
- Alder MN, Dames S, Gaudet J, Mango SE 2003. Gene silencing in Caenorhabditis elegans by transitive RNA interference. RNA 9: 25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arziman Z, Horn T, Boutros M 2005. E-RNAi: A web application to design optimized RNAi constructs. Nucleic Acids Res 33: W582–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Rukun G 2003. Genome-wide RNAi analysis in Caenohabitis elegans fat regulatory genes. Nature 421: 268–272 [DOI] [PubMed] [Google Scholar]
- Blandin S, Shiao SH, Moita LF, Janse CJ, Waters AP, Kafatos FC, Levashina EA 2004. Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell 116: 661–670 [DOI] [PubMed] [Google Scholar]
- Boutros M, Ahringer J 2008. The art and design of genetic screens: RNA interference. Nat Rev Genet 9: 554–566 [DOI] [PubMed] [Google Scholar]
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Paro R, Perrimon N 2004. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303: 832–835 [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Brent AE, Yucel G, Small S, Desplan C 2007. Permissive and instructive anterior patterning rely on mRNA localization in the wasp embryo. Science 315: 1841–1843 [DOI] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282: 2012–2018 [DOI] [PubMed] [Google Scholar]
- Chen CH, Huang H, Ward CM, Su JT, Schaeffer LV, Guo M, Hay BH 2007. A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science 316: 597–600 [DOI] [PubMed] [Google Scholar]
- Cox-Foster D, vanEngelsdorp D 2009. Saving the honeybee. Sci Am 300: 40–47 [DOI] [PubMed] [Google Scholar]
- Cronin SJ, Nehme NT, Limmer S, Liegeois S, Pospisilik JA, Schramek D, Leibbrandt A, Simoes Rde M, Gruber S, Puc U, et al. 2009. Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science 325: 340–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S et al. 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156 [DOI] [PubMed] [Google Scholar]
- Echeverri CJ, Perrimon N 2006. High-throughput RNAi screening in cultured cells: A user's guide. Nat Rev Genet 7: 373–384 [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408: 325–330 [DOI] [PubMed] [Google Scholar]
- Feinberg EH, Hunter CP 2003. Transport of dsRNA into cells by the transmembrane protein SID-1. Science 301: 1545–1547 [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811 [DOI] [PubMed] [Google Scholar]
- Flockhart I, Booker M, Kiger A, Boutros M, Armknecht S, Ramadan N, Richardson K, Xu A, Perrimon N, Mathey-Prevot B 2006. FlyRNAi: The Drosophila RNAi screening center database. Nucleic Acids Res 34: D489–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortier E, Belote JM 2000. Temperature-dependent gene silencing by an expressed inverted repeat in Drosophila. Genesis 26: 240–244 [DOI] [PubMed] [Google Scholar]
- Giordano E, Rendina R, Peluso I, Furia M 2002. RNAi triggered by symmetrically transcribed transgenes in Drosophila melanogaster. Genetics 160: 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166: 1775–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B, Hendrix D, Trang V, Levine M 2008. A simplified miRNA-based gene silencing method for Drosophila melanogaster. Dev Biol 321: 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpavat S, Cepko CL 2006. RCAS-RNAi: A loss-of-function method for the developing chick retina. BMC Developmental Biol 6: doi: 10.1186/1471-213X-6-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Culotti J, Pringle JR, Reid BJ 1974. Genetic control of the cell division cycle in yeast. Science 183: 46–51 [DOI] [PubMed] [Google Scholar]
- Hebert CG, Valdes JJ, Bentley WE 2008. Beyond silencing–engineering applications of RNA interference and antisense technology for altering cellular phenotype. Curr Opin Biotechnol 19: 500–505 [DOI] [PubMed] [Google Scholar]
- Horvitz HR 1999. Genetic control of programmed cell death in the nematode Caenorhabditis elegans. Cancer Res 59: 1701s–1706s [PubMed] [Google Scholar]
- Jones L, Ratcliff F, Baulcombe DC 2001. RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr Biol 11: 747–757 [DOI] [PubMed] [Google Scholar]
- Kaletta T, Hengartner MO 2006. Finding function in novel targets: C. elegans as a model organism. Nat Rev Drug Discov 5: 387–398 [DOI] [PubMed] [Google Scholar]
- Kalidas S, Smith DP 2002. Novel genomic cDNA hybrids produce effective RNA interference in adult Drosophila. Neuron 33: 177–184 [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW 1998. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95: 10171026. [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW 2000. Heritable gene silencing in Drosophila using double-stranded RNA. Nat Biotechnol 18: 896–898 [DOI] [PubMed] [Google Scholar]
- Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, Rual JF, Kennedy S, Dybbs M, Bertin N, Kaplan JM et al. 2005. Functional genomic analysis of RNA interference in C. elegans. Science 308: 1164–1167 [DOI] [PubMed] [Google Scholar]
- Kim YO, Park SJ, Balaban RS, Nirenberg M, Kim Y 2004. A functional genomic screen for cardiogenic genes using RNA interference in developing Drosophila embryos. Proc Natl Acad Sci 101: 159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Booker M, Perrimon N 2009. Cross-species RNAi rescue platform in Drosophila melanogaster. Genetics Aug 31. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Inagaki S, Yasuda K, Kageyama Y 2006. Rapid construction of Drosophila RNAi transgenes using pRISE, a P-element-mediated transformation vector exploiting an in vitro recombination system. Genes Genet Syst 81: 129–134 [DOI] [PubMed] [Google Scholar]
- Kulkarni MM, Booker M, Silver SJ, Friedman A, Hong P, Perrimon N, Mathey-Prevot B 2006. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat Methods 3: 833–838 [DOI] [PubMed] [Google Scholar]
- Kusaba M 2004. RNA interference in crop plants. Curr Opin Biotechnol 15: 139–143 [DOI] [PubMed] [Google Scholar]
- Lam G, Thummel CS 2000. Inducible expression of double-stranded RNA directs specific genetic interference in Drosophila. Curr Biol 10: 957–963 [DOI] [PubMed] [Google Scholar]
- Lee YS, Carthew RW 2003. Making a better RNAi vector for Drosophila: Use of intron spacers. Methods 30: 322–329 [DOI] [PubMed] [Google Scholar]
- Lee SK, Kumar P 2009. Conditional RNAi: Towards a silent gene therapy. Adv DrugDeliv Rev 61: 650–664 [DOI] [PubMed] [Google Scholar]
- Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG 2006. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet 38: 896–903 [DOI] [PubMed] [Google Scholar]
- Lipardi C, Paterson BM 2009. Identification of an RNA-dependent RNA polymerase in Drosophila involved in RNAi and transposon suppression. Proc Natl Acad Sci 106: 15645–15650 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lipardi C, Baek HJ, Wei Q, Paterson BM 2005. Analysis of short interfering RNA function in RNA interference by using Drosophila embryo extracts and schneider cells. Methods Enzymol 392: 351–371 [DOI] [PubMed] [Google Scholar]
- Li M, Rohrer B 2006. Gene silencing in Xenopus laevis by DNA vector-based RNA interference and transgenesis. Cell Res 16: 99–105 [DOI] [PubMed] [Google Scholar]
- Lynch JA, Desplan C 2006. A method for parental RNA interference in the wasp Nasonia vitripennis. Nat Protoc 1: 486–494 [DOI] [PubMed] [Google Scholar]
- Ma Y, Creanga A, Lum L, Beachy PA 2006. Prevalence of off-target effects in Drosophila RNA interference screens. Nature 443: 359–363 [DOI] [PubMed] [Google Scholar]
- Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N 2008. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet 40: 476–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinek S, Young MW 2000. Specific genetic interference with behavioral rhythms in Drosophila by expression of inverted repeats. Genetics 156: 1717–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misquitta L, Paterson BM 1999. Targeted disruption of gene function in Drosophila by RNA interference (RNA-i): A role for nautilus in embryonic somatic muscle formation. Proc Natl Acad Sci 96: 1451–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misquitta L, Wei Q, Paterson BM 2008. Collection of Drosophila Embryos for RNA Interference (RNAi). Cold Spring Harb Protoc 10.1101/pdb.prot4917 [DOI] [PubMed] [Google Scholar]
- Mohr S, Bakal C, Perrimon N 2011. RNAi: Results and Challenges. Ann Rev Biochem (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummery-Widmer JL, Yamazaki M, Stoeger T, Novatchkova M, Bhalerao S, Chen D, Dietzl G, Dickson BJ, Knoblich JA 2009. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature 458: 987–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Perrimon N, Sandmeyer S, Plasterk R 2003. Tailoring the genome: The power of genetic approaches. Nat Genet 33: 276–284 [DOI] [PubMed] [Google Scholar]
- Nakano H, Amemiya S, Shiokawa K, Taira M 2002. RNA interference for theorganizer-specific gene Xlim-1 in Xenopus embryos. Biochem Biophys Res Commun 274: 434–439 [DOI] [PubMed] [Google Scholar]
- Ni JQ, Liu LP, Binari R, Hardy R, Shim HS, Cavallaro A, Booker M, Pfeiffer BD, Markstein M, Wang H, et al. 2009. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics 182: 10891100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Markstein M, Binari R, Pfeiffer B, Liu LP, Villalta C, Booker M, Perkins L, Perrimon N 2008. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat Methods 5: 49–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niggeweg R, Michael AJ, Martin C 2004. Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat Biotechnol 22: 746–754 [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287: 795–801 [DOI] [PubMed] [Google Scholar]
- Olesnicky EC, Brent AE, Tonnes L, Walker M, Pultz MA, Leaf D, Desplan C 2006. A caudal mRNA gradient controls posterior development in the wasp Nasonia. Development 133: 3973–3982 [DOI] [PubMed] [Google Scholar]
- Perrimon N, Mathey-Prevot B 2007. Applications of high-throughput RNA interference screens to problems in cell and developmental biology. Genetics 175: 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW 2008. Smed-betacatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science 319: 327–330 [DOI] [PubMed] [Google Scholar]
- Pili-Floury S, Leulier F, Takahashi K, Saigo K, Samain E, Ueda R, Lemaitre B 2004. In vivo RNA interference analysis reveals an unexpected role for GNBP1 in the defense against Gram-positive bacterial infection in Drosophila adults. J Biol Chem 279: 12848–12853 [DOI] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428: 493–521 [DOI] [PubMed] [Google Scholar]
- Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sanchez Alvarado A 2005. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell 8: 635–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichhart JM, Ligoxygakis P, Naitza S, Woerfel G, Imler JL, Gubb D 2002. Splice-activated UAS hairpin vector gives complete RNAi knockout of single or double target transcripts in Drosophila melanogaster. Genesis 34: 160–164 [DOI] [PubMed] [Google Scholar]
- Roignant JY, Carre C, Mugat B, Szymczak D, Lepesant JA, Antoniewski C 2003. Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. RNA 9: 299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah DW 2006. Therapeutic potential of RNA interference for neurological disorders. Life Sci 79: 1773–1780 [DOI] [PubMed] [Google Scholar]
- Saleh MC, Tassetto M, van Rij RP, Goic B, Gausson V, Berry B, Jacquier C, Antoniewski C, Andino R 2009. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature 458: 346–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandy P, Ventura A, Jacks T 2005. Mammalian RNAi: A practical guide. Biotechniques 39: 215–224 [DOI] [PubMed] [Google Scholar]
- Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, Kamath RS, Fraser AG, Ahringer J, Plasterk RH 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol 1: E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skromne I, Prince VE 2008. Current perspectives in zebrafish reverse genetics: Moving forward. Dev Dyn 237: 861–882 [DOI] [PubMed] [Google Scholar]
- Smith NA, Singh SP, Wang MB, Stoutjesdijk PA, Green AG, Waterhouse PM 2000. Total silencing by intron-spliced hairpin RNAs. Nature 407: 319–320 [DOI] [PubMed] [Google Scholar]
- Sönnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E et al. 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434: 462–469 [DOI] [PubMed] [Google Scholar]
- St Johnston D, Nüsslein-Volhard C 1992. The origin of pattern and polarity in the Drosophila embryo. Cell 68: 201–219 [DOI] [PubMed] [Google Scholar]
- Stielow B, Sapetsching A, Kruger I, Kunert N, Brehm M, Suske G 2008. Indentification of SUMO-dependant chromatin-associated transcriptional repression components by genome-wide RNAi screen. Mol Cell 29: 742–754 [DOI] [PubMed] [Google Scholar]
- Sugimoto A 2004. High-throughput RNAi in Caenorhabditis elegans: Genome-wide screens and functional genomics. Differentiation 72: 81–91 [DOI] [PubMed] [Google Scholar]
- Tang G, Galili G, Zhuang X 2007. RNAi and microRNA: Breakthrough technologies for the improvement of plant nutritional value and metabolic engineering. Metabolomics 3: 357–369 [Google Scholar]
- Timmons L, Fire A 1998. Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R et al. 2009. FlyBase: Enhancing Drosophila Gene ontology annotations. Nucleic Acids Res 37: D555559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, Bellen HJ 2005. Emerging technologies for gene manipulation in Drosophila melanogaster. Nat Rev Genet 6: 167–178 [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. 2001. The sequence of the human genome. Science 291: 1304–1351 [DOI] [PubMed] [Google Scholar]
- Wargelius A, Ellingsen S, Fjose A 1999. Double-stranded RNA induces specific developmental defects in zebrafish embryos. Biochem Biophys Res Commun 263: 156–161 [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Williams RW, Rubin GM 2002. ARGONAUTE1 is required for efficient RNA interference in Drosophila embryos. Proc Natl Acad Sci 99: 6889–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston WM, Molodowitch C, Hunter CP 2002. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295: 2456–2459 [DOI] [PubMed] [Google Scholar]
- Yang D, Buchholz F, Huang Z, Goga A, Chen CY, Brodsky FM, Bishop JM 2002. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc Natl Acad Sci 99: 9942–9947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadeh AH, Foster GD 2004. Transgenic resistance to tobacco ringspot virus. Acta Virol 48: 145–152 [PubMed] [Google Scholar]
- Zhou Y, Ching YP, Kok KH, Kung HF, Jin DY 2002. Post-transcriptional suppression of gene expression in Xenopus embryos by small interfering RNA. Nucleic Acids Res 30: 1664–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M et al. 2006. RNAi-mediated gene silencing in nonhuman primates. Nature 441: 111–114 [DOI] [PubMed] [Google Scholar]