Abstract
Promoter CpG island hypermethylation has become recognized as an important mechanism for inactivating tumor suppressor genes or tumor-related genes in human cancers of various tissues. Gene inactivation in association with promoter CpG island hypermethylation has been reported to be four times more frequent than genetic changes in human colorectal cancers. Hepatocellular carcinoma is also one of the human cancer types in which aberrant promoter CpG island hypermethylation is frequently found. However, the number of genes identified to date as hypermethylated for hepatocellular carcinoma (HCC) is fewer than that for colorectal cancer or gastric cancer, which can be attributed to fewer attempts to perform genome-wide methylation profiling for HCC. In the present study, we used bead-array technology and coupled methylation-specific PCR to identify new genes showing cancer-specific methylation in HCC. Twenty-four new genes have been identified as hypermethylated at their promoter CpG island loci in a cancer-specific manner. Of these, TNFRSF10C, HOXA9, NPY, and IRF5 were frequently hypermethylated in hepatocellular carcinoma tissue samples and their methylation was found to be closely associated with inactivation of gene expression. Further study will be required to elucidate the clinicopathological implications of these newly found DNA methylation markers in hepatocellular carcinoma.
Keywords: Bead Array; CpG Islands; DNA Methylation; Carcinoma, Hepatocellular
INTRODUCTION
A CpG island is an approximately 1-kb DNA sequence with a high density of CpG dinucleotides. About 70% of human genes are known to harbor CpG islands in their promoter sequences (1, 2). In normal cells, promoter CpG islands are usually protected from aberrant hypermethylation, except for those on imprinting genes or genes of inactivated X chromosomes (3, 4). However, in association with carcinogenesis, hundreds of promoter CpG islands undergo aberrant hypermethylation, which represses gene transcription of active genes or enforces suppression of already inactive genes (5). Promoter CpG island hypermethylation has become recognized as an important mechanism for inactivation of tumor suppressor genes or tumor-related genes in human cancers, and occurs in virtually all types of human cancers (6). In addition to gene inactivation, promoter CpG island hypermethylation has received attention for its potential utility as a biomarker for tumor detection or prediction of prognosis or response of tumor cells to chemotherapeutic agents (7). Hepatocellular carcinoma (HCC) is one of the human cancers in which researchers are actively studying epigenetic changes. More than 113 genes have been found to be hypermethylated in HCC in a cancer-specific manner and some of them have been demonstrated to have a close association between hypermethylation and poor prognosis for HCC patients, supporting the potential usefulness of DNA methylation markers as prognostic markers (8-10).
However, the number of genes that have been identified to date as harboring hypermethylation in their promoter CpG island loci is far fewer for HCC than for colon cancer or gastric cancer, which can be attributed to fewer attempts to perform genome-wide methylation analysis compared with colon cancer or gastric cancer. Considering that about 400 genes may be inactivated through promoter CpG island hypermethylation in colorectal cancers (11), there remain many genes that may be hypermethylated in HCC.
Among the various technologies available for large-scale analysis of DNA methylation, the GoldenGate methylation solution is a powerful and accurate screening tool for analyzing methylation status in 807 genes (12). With the advent of this technique, we were able to analyze the methylation status of 687 promoter CpG island loci in liver cancer. The aim of the present study was to identify additional genes that are hypermethylated in HCC in a cancer-specific manner that have not yet been reported as such in the literature thus far.
MATERIALS AND METHODS
Cell lines and 5-aza-dC treatment
We used eight different human HCC cell lines (SNU398, SNU475, SNU739, SNU761, SNU878, SNU886, HepG2, and Huh7) obtained from the Korean Cell Line Bank (Seoul, Korea). All of the cell lines except Huh7 were grown in RPMI-1640 medium supplemented with 10% fetal bovine serum. Huh7 was grown in DMEM medium supplemented with 10% fetal bovine serum. All cell lines were cultured in a humidified 37℃, 5% CO2 incubator. The cell lines were seeded at 3×105/mL in their respective culture media and treated with 1 µM and 5 µM 5-aza-dC (Sigma Chemical Co., St. Louis, MO, USA) for 96 hr; media and drugs were replaced every 24 hr. As a control, cell lines were mock-treated in parallel with the addition of an equal volume of PBS without the drug. We prepared total RNA using the RNeasy Mini kit (Qiagen, Valencia, CA, USA).
Tissue samples
Fresh-frozen samples of HCC and paired non-neoplastic liver tissues were obtained from patients (n=5) who underwent curative resection for HCC at the Seoul National University Hospital in 2008. In addition, after microscopic examination of HCC samples from patients who underwent curative resection at the Seoul National University Hospital from 2001 to 2002, we selected 50 cases of HCC that had sufficient amounts of archival neoplastic and non-neoplastic tissues for the DNA methylation study. For the bead array study requiring fresh tissue samples, we obtained informed consent from the patients. The Institutional Review Board approved this study.
Methylation microarray
DNA methylation analysis of individual genes was performed using GoldenGate Methylation Solution utilizing the current Cancer Panel I platform (Illumina, San Diego, CA, USA), which probes 1,505 CpG loci selected from 807 genes. Using this method, oligonucleotide primers query putative methylation sites in bisulfite-converted genomic DNA. Bisulfite modification of genomic DNA was performed using the EZ DNA methylation kit (Zymo Research, Orange, CA, USA). Similar to methylation-specific PCR, two primer sets are designed for each CpG target; one primer set corresponds to the unmethylated, bisulfite-converted sequence (uracil) while the other corresponds to the unconverted sequence (5-methyl cytosine). The methylation level (beta value) is determined by the ratio of the fluorescent signals from methylated and unmethylated alleles. The beta value identifies the level of DNA methylation at a CpG site, ranging from 0 in the case of almost all unmethylated sites to 1 for completely methylated sites (12). Of 1,505 CpG sites (selected from 807 genes) included in GoldenGate Methylation Cancer Panel I, 1,044 of the CpG sites are located within CpG islands, and 461 are located outside of CpG islands. Comparing the average beta values for the 1,044 CpG sites between HCC and non-neoplastic liver tissues, cancer-specific hypermethylation was determined to be present when the average beta value for a CpG site was significantly greater (P<0.05) for the HCC samples (n=5) than for the non-neoplastic liver tissues (n=5).
Methylation-specific polymerase (MSP) chain reaction
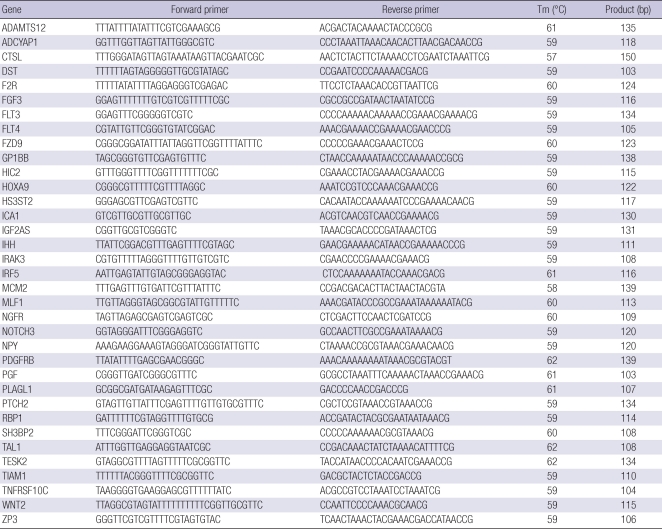
For MSP analysis, DNA was extracted following a standard phenol-chloroform extraction method. Bisulfite modification of genomic DNA was carried out using the EZ DNA methylation Kit (Zymo Research). Primers were designed using two web sites, MSPPrimer (http://www.mspprimer.org) and MethPrimer (http://www.urogene.org/methprimer). Primer sequences and PCR conditions are shown in Table 1. MSP was performed as previously described (13).
Table 1.
Primers for methylation-specific polymerase chain reaction
Quantitative RT-PCR
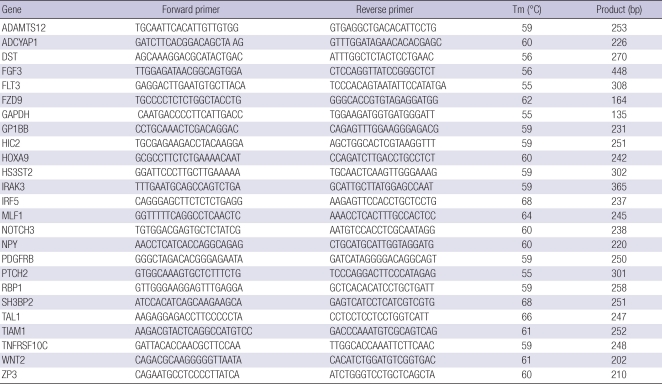
Total RNA was prepared using the RNeasy (Qiagen) kit according to the manufacturer's protocols. A total of 5 µg of RNA was reverse transcribed using Oligo dT and Superscript III (Invitrogen, Carlsbad, CA, USA). Quantitative RT-PCR amplification reactions were performed using the SYBR® Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) with a 7300 Real-Time PCR System (Applied Biosystems). The expression levels of the genes were normalized to the expression of GAPDH. Primer sequences and PCR conditions are shown in Table 2.
Table 2.
Primers for quantitative RT-PCR
RESULTS
Microarray analysis of methylated genes
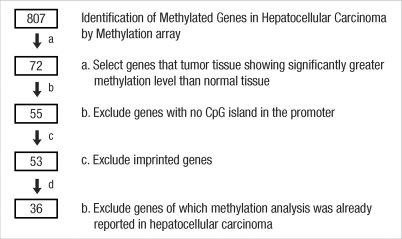
The methylation profiles of five pairs of HCC and non-neoplastic liver tissues were analyzed for 807 genes using the Illumina GoldenGate Methylation Solution. In total, 72 annotated genes (81 CpG sites) were found to be hypermethylated in tumor tissues. After excluding genes without a CpG island in the promoter (18 genes), imprinted genes (2 genes), and genes for which methylation of their promoter CpG island loci has already been reported for HCC (17 genes), we explored DNA hypermethylation for the remaining 36 genes using MSP. These 36 genes were considered to be potential novel methylation markers for HCC. This gene filtering approach is depicted in Fig. 1.
Fig. 1.
Flow chart for selection of candidate methylation markers. We used 5 paired hepatocellular carcinoma/normal tissues to screen for candidate methylation markers using a methylation array. We obtained 72 candidates that showed significant hypermethylation in hepatocellular carcinoma tissues. We removed genes with no CpG island loci in their promoters, imprinted genes, and genes for which methylation status was already known for hepatocellular carcinoma. Thus, we selected 36 genes to further examine for methylation analysis using methylation-specific PCR.
Validation of the methylation status of candidate genes in the HCC cell lines using MSP
For MSP analysis, we attempted to design primers for the 36 genes using both MSPPrimer (http://www.mspprimer.org) and MethPrimer (http://www.urogene.org/methprimer). However, for one gene (HOXB2), neither tool was able to design primers, because it has a short CpG island in the promoter. Therefore, we examined the methylation status of the remaining 35 genes in the 8 HCC cell lines. It was found that 26 of the genes (HOXA9, TNFRSF10C, NPY, TIAM1, PDGFRB, IRAK3, SH3BP2, IRF5, HIC2, TAL1, HS3ST2, MLF1, IGF2AS, ADCYAP1, FGF3, WNT2, ADAMTS12, FLT3, PTCH2, GP1BB, RBP1, FZD9, DST, NOTCH3, PLAGL1, and ZP3) were methylated in at least one HCC cell line (26 of 35; 74.3%). The remaining 9 genes (TESK2, IHH, MCM2, CTSL, F2R, PGF, NGFR, FLT4, and ICA1) were not found to be methylated in the HCC cell lines using the MSP assay (Figs. 2, 3).
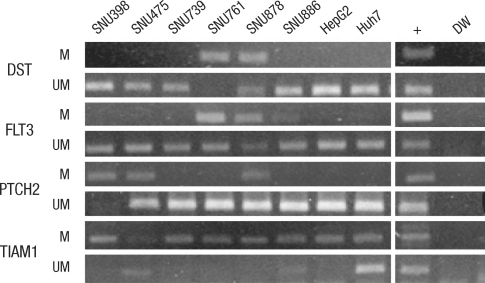
Fig. 2.
Representative examples of MSP analysis of DST, FLT3, PTCH2 and TIAM1 in 8 HCC cell lines. DNA extracted from 8 HCC cell lines were amplified with primers specific to the methylated (M) or unmethylated (UM) CpG islands of each gene after modification with sodium bisulfite. +, positive control; DW, distilled water. Positive controls for methylated MSP and unmethylated DNA are M.SssI-treated placental DNA and whole-genome amplified DNA, respectively.
Fig. 3.
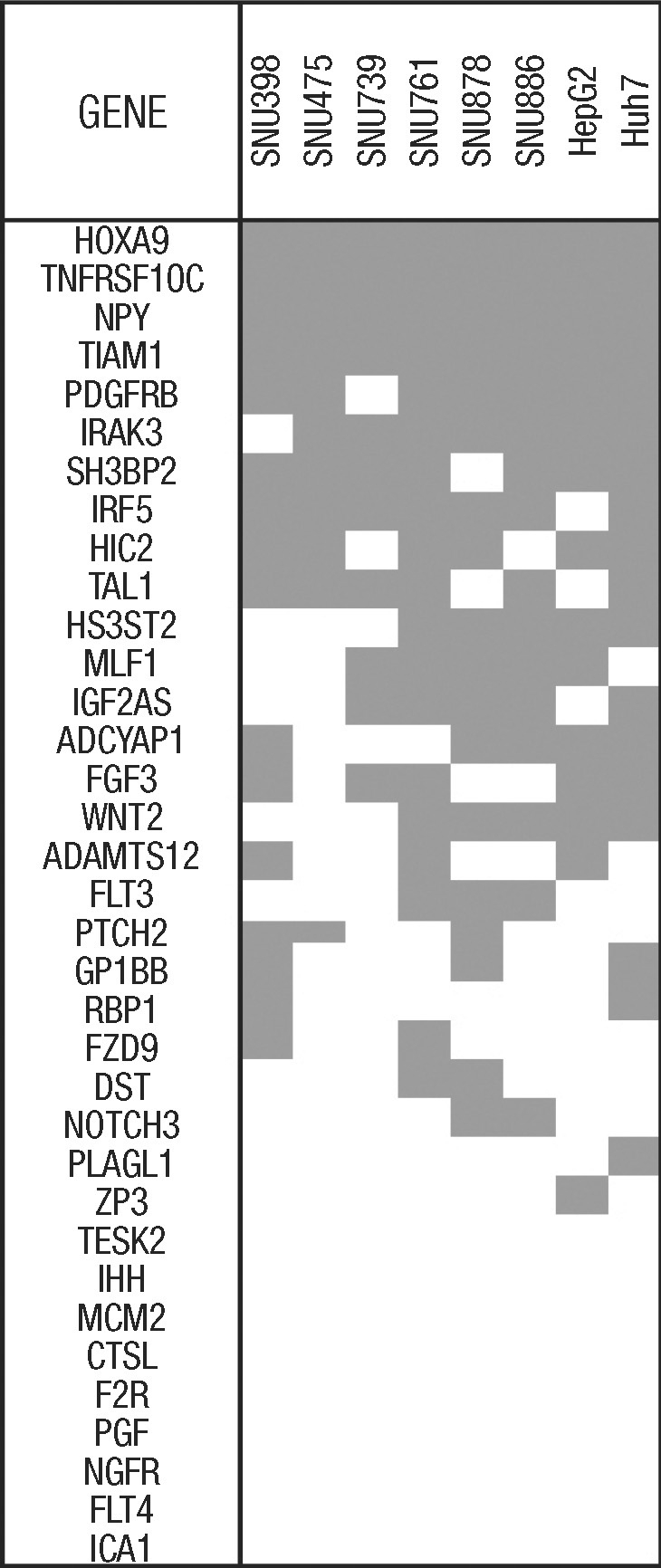
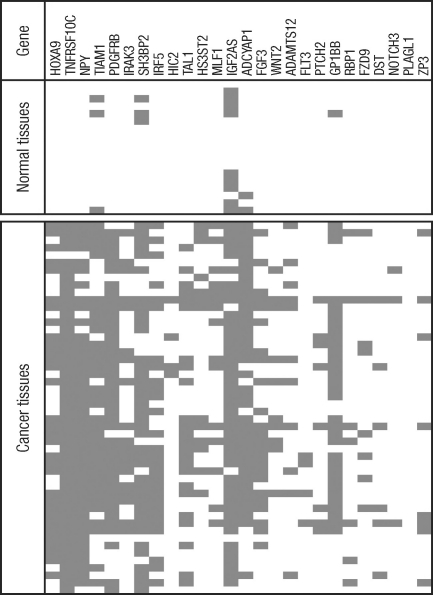
Summary of methylation-specific PCR for hepatocellular carcinoma (HCC) cell lines. We examined eight HCC cell lines for methylation status of the 35 genes and found that 26 of the genes (HOXA9, TNFRSF10C, NPY, TIAM1, PDGFRB, IRAK3, SH3BP2, IRF5, HIC2, TAL1, HS3ST2, MLF1, IGF2AS, ADCYAP1, FGF3, WNT2, ADAMTS12, FLT3, PTCH2, GP1BB, RBP1, FZD9, DST, NOTCH3, PLAGL1, and ZP3) were methylated in at least one HCC cell line. Data are color-coded as follows: gray fill indicates the presence of methylation, whereas white fill indicates the absence of methylation.
Confirmation of hypermethylation of newly developed candidate genes in liver tissues
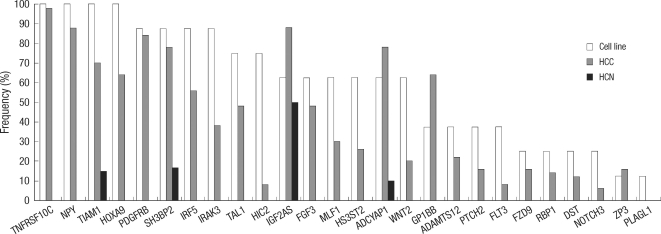
To determine whether the genes that were hypermethylated in the HCC cell lines were hypermethylated in a cancer-specific manner, we analyzed the methylation status of the 26 genes in 18 normal liver samples and 50 primary HCC samples using methylation-specific PCR. Of the candidate 26 genes that showed methylation in the cell lines, promoter methylation was detected in 25 of the genes in primary tumor tissues. In contrast, only five genes (5 of 26; 19.2%) were methylated in normal tissues. Moreover, all but one of these genes (IGF2AS) were methylated at low frequencies in the non-neoplastic liver tissues (Fig. 4). Thus, the vast majority of the genes were found to show cancer-specific methylation at frequencies of 6%-98% (Fig. 5). Thus, 24 new cancer-specific CpG island loci were identified through our approach (Table 3).
Fig. 4.
Summary of methylation-specific PCR results in hepatocellular carcinoma (n=50) and non-neoplastic liver tissue samples (n=18). Out of 26 genes showing methylation in HCC cell lines, promoter methylation was detected in 25 genes in primary tumor tissues. Data are color-coded as follows: gray fill indicates the presence of methylation, while white fill indicates absence of methylation.
Fig. 5.
Comparison of hypermethylation frequencies for 26 genes. Hepatocellular carcinoma (HCC) cell lines (white column), HCC tissue samples (gray column), and non-neoplastic liver tissue samples (HCN, black column).
Table 3.
New cancer-specific methylated genes
Gene expression and induction after 5-aza-dC treatment in HCC cell lines
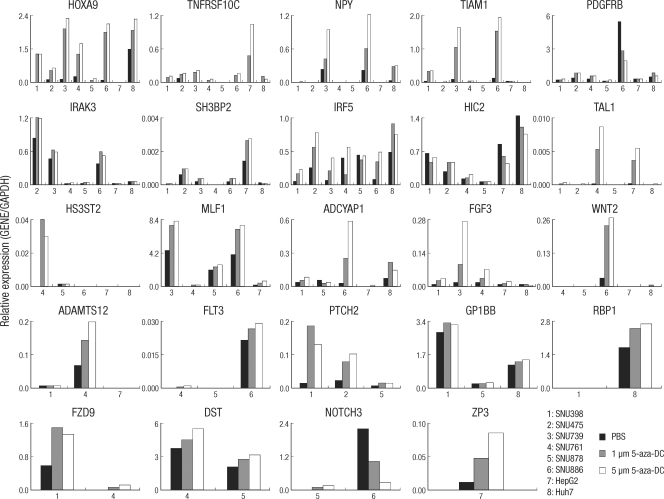
For hypermethylated and transcriptionally silenced genes, the DNA demethylating agent 5-aza-dC is known to induce gene re-expression (14, 15). To identify whether promoter CpG island hypermethylation was closely associated with gene expression in the 24 newly identified cancer-specific methylated genes, we treated the 8 HCC cell lines with 5-aza-dC (1 and 5 µM for 96 hr) and performed quantitative RT-PCR of the mock-treated and 5-aza-dC treated cell lines. TNFRSF10C, NPY, TIAM1, and HOXA9 were methylated in all cell lines and showed up-regulation of mRNA expression in all cell lines with 5-aza-dC, although TIAM1 was not re-expressed in two cell lines, SNU-878 and Huh7. Most of the methylated genes exhibited re-expression after 5-aza-dC treatment; in contrast, SNU-886 in PDGFRB; SNU-878 in IRF5; SNU-398, HepG2, and Huh7 in HIC2; HepG2 and Huh7 in HS3ST2; SNU-878 in ADCYAP1; and SNU-886 in NOTCH3 were down-regulated by 5-aza-dC. In addition, HepG2 and SNU-761 in WNT2, HepG2 in ADAMTS12, and SNU-878 in FLT3 were not re-expressed (Fig. 6). In such cases, histone modification may be related to transcription suppression (16, 17).
Fig. 6.
Analysis of mRNA expression of candidate genes in hepatocellular carcinoma cell lines by quantitative RT-PCR. Cells were either mock-treated (PBS) or treated with 5-aza-dC (1AZA [1 µM] or 5AZA [5 µM] for 96 hr) as indicated. The expression patterns and re-expression patterns for each gene after 5-aza-dC treatment are shown for the cell lines that were methylated. Numbers along the horizontal axis indicate the cell lines. Gene expression levels were determined by quantitative RT-PCR and normalized to GAPDH levels.
DISCUSSION
Although many studies have reported aberrant hypermethylation of genes in HCC, for example, identifying E-cadherin, RASSF1A, GSTP, SOCS1, SFRP1, and PTEN as tumor suppressor gene silenced by hypermethylation, most of these studies were limited to analysis of a single or a few specific genes (8-10). Therefore, the true extent of promoter CpG island hypermethylation in HCC remains largely unknown. With advancements in microarray technology, the number of genes found to be hypermethylated in HCC in a cancer-specific manner is expected to increase exponentially, leading to a better understanding of epigenetic modulation of tumor-related genes in hepatocarcinogenesis. The GoldenGate bead-array technology incorporates a strong analytical methylation platform and provides reliable and highly reproducible data (12). Here, we present identification of new genes demonstrating cancer-specific methylation in HCC through the use of bead-array technology. The fidelity of the bead-array results was confirmed using methylation-specific PCR for genes selected according to our criteria (Fig. 1). As a result of the present approach, 24 genes were newly identified as cancer-specific methylation loci for HCC.
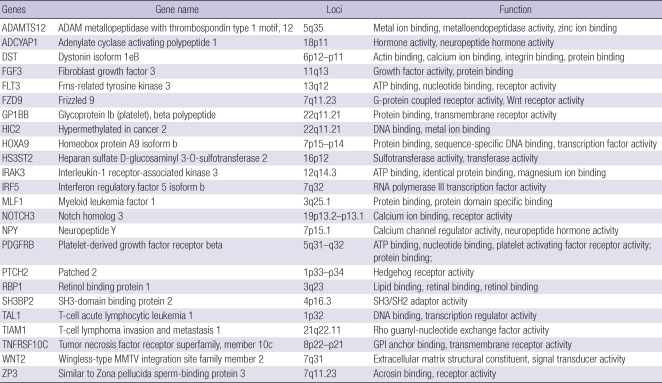
Table 3 summarizes the functions of the genes that were newly identified to be hypermethylated in a cancer-specific manner in HCC. TNFRSF10C, HOXA9, NPY, and IRF5 were frequently methylated in our panel of cell lines and HCC tissues and their methylation was correlated with low expression levels or silencing. These genes were re-expressed in the majority of the cell lines after 5-aza-dC treatment. Therefore, these genes are considered to be repressed by DNA hypermethylation in their promoter CpG islands. Although it is unclear whether TNFRSF10C plays a pro-apoptotic role or an anti-apoptotic role in tumors, hypermethylation of TNFRSF10C has been reported in various human cancers, including breast, lung, prostate, and neuroblastomas (18). HOXA9 encodes a DNA-binding transcription factor that may regulate gene expression, morphogenesis, and differentiation. Expression of HOXA9 proteins is spatially and temporally regulated during embryonic development (19). HOXA9 promoter CpG island methylation has been reported in ovary and lung cancer (20, 21). NPY is involved in cell motion and cell proliferation as well as neuropeptide hormone activity (22, 23). In prostate cancer, low NPY expression is closely associated with aggressive clinical behavior (24). In another recent study, NPY was shown to be frequently methylated in neuroblastomas (25). IRF5, a member of the interferon regulatory factor family, is known to be related to innate immune system activity and is also a critical regulator of DNA damage-induced apoptosis (26). Because IRF5 is critical for induction of apoptosis, IRF5 deficiency predisposes cells to tumorigenic transformation (27). However, methylation of IRF5 in cancer has rarely been reported.
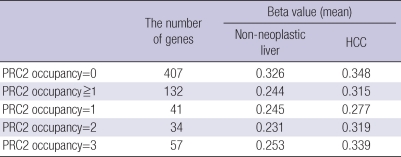
Recently, the role of chromatin structure and epigenetic alterations in controlling gene transcriptional activity during embryonic stem (ES) cell self-renewal and differentiation has been intensively investigated (28, 29). Recent studies have attempted to identify the relationships between DNA methylation, histone modifications, and promoter occupancy of pluripotent regulators such as PcG in regulation gene expression in ES cells (30). Gene promoter chromatin patterns, including PcG-mediated repressive histone modifications, may render certain genes vulnerable to DNA hypermethylation. These changes may enhance the likelihood of tumor initiation and progression from cell transformation (30). To identify whether polycomb repressive complex 2 (PRC2) occupancy information obtained from embryonic stem cells may predict the vulnerability of individual CpG island loci to hypermethylation, we compared the average methylation level (beta value) of each CpG island locus between normal tissues and tumor tissues and correlated it with PRC2 occupancy. In PRC2-positive CpG islands, the methylation level of the tumor group was higher than the methylation level of the normal group. In addition, the differences between the two groups were larger when the CpG island loci were occupied by two or more components of the PRC2 complex. However, there were no differences among PRC2-negative CpG islands (Supplementary Table 1). Thus, PRC2 may indicate that these genes are preferred substrates for targeted methylation.
Supplementary Table 1.
The average methylation level (beta value) of each CpG island locus between non-neoplastic tissues and HCC and correlated it with polycomb repressive complex 2 (PRC2) occupancy. The value of PRC2 occupancy represents the number of occupied components of PRC2 in the gene promoter
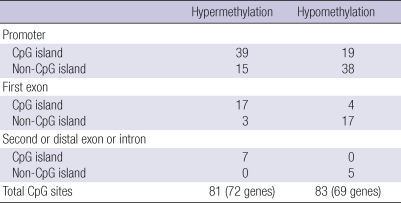
In addition to hypermethylation of promoter CpG island loci, some hypomethylated CpG sites were also identified through comparison of methylation levels (bead array-determined beta values) of CpG sites in HCC. CpG sites showing hypomethylation in a cancer-related manner appear to be different from those that were hypermethylated with respect to their relationships with CpG island loci. In the present study, we found that 69 genes (83 CpG sites) were significantly hypomethylated in tumor tissues. Of these 83 CpG sites, only 23 sites (27.7%) were located in promoter CpG island loci, in contrast with the 63 of 81 (77.8%) hypermethylated CpG sites that were located in CpG island loci (Supplementary Table 2). The identified hypomethylated genes tend to be involved in biological processes of immunity, neuronal activity, lipid metabolism, and transport, whereas the hypermethylated genes tend to be involved in nucleic acid metabolism, cell proliferation, differentiation, and the cell cycle. For example, IL16, involved in chemokine-mediated immunity, was significantly hypomethylated in HCC compared to non-neoplastic liver tissues (P=0.003). GABRA5, with neurotransmitter receptor activity, was hypomethylated (P<0.001). APOC1, which processes lipid metabolism and transport, was hypomethylated (P=0.001). In addition, certain oncogenes, including HGF, BLK, and PGR were hypomethylated in HCC. Interestingly, hypomethylation of histone deacetylase 1 (HDAC1), which plays a key role in the regulation of eukaryotic gene repression, was prominent and had the largest significant difference in methylation levels between HCC and non-neoplastic liver tissues (P<0.001).
Supplementary Table 2.
The summarization of the number of CpG sites undergoing hypermethylation or hypomethylation in association with cancerization according to their genomic location. CpG sites located in CpG island loci tend to undergo hypermethylation in association with cancerization whereas CpG sites located in non-CpG island tend to become hypomethylated
In summary, we have identified 24 novel cancer-specific methylation markers for HCC using array-based methylation profiling and coupled MSP. Of these, TNFRSF10C, HOXA9, NPY, and IRF5 were frequently hypermethylated in HCC tissues and their promoter hypermethylation was correlated with inactivation of gene expression in cell lines. The clinicopathologic implications of these newly identified DNA methylation markers will need to be further investigated in large-scale studies of HCC samples.
Footnotes
This study was supported by a grant of the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare & Family Affairs, Republic of Korea (A090126).
References
- 1.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schübeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 3.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 4.Panning B, Jaenisch R. RNA and the epigenetic regulation of X chromosome inactivation. Cell. 1998;93:305–308. doi: 10.1016/s0092-8674(00)81155-1. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet. 2007;16 Spec No 1:R50–R59. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- 6.Baylin SB, Chen WY. Aberrant gene silencing in tumor progression: implications for control of cancer. Cold Spring Harb Symp Quant Biol. 2005;70:427–433. doi: 10.1101/sqb.2005.70.010. [DOI] [PubMed] [Google Scholar]
- 7.Teodoridis JM, Strathdee G, Brown R. Epigenetic silencing mediated by CpG island methylation: potential as a therapeutic target and as a biomarker. Drug Resist Updat. 2004;7:267–278. doi: 10.1016/j.drup.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Calvisi DF, Ladu S, Gorden A, Farina M, Lee JS, Conner EA, Schroeder I, Factor VM, Thorgeirsson SS. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest. 2007;117:2713–2722. doi: 10.1172/JCI31457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tischoff I, Tannapfe A. DNA methylation in hepatocellular carcinoma. World J Gastroenterol. 2008;14:1741–1748. doi: 10.3748/wjg.14.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lou C, Yang B, Gao YT, Wang YJ, Nie FH, Yuan Q, Zhang CL, Du Z. Aberrant methylation of multiple genes and its clinical implication in hepatocellular carcinoma. Zhonghua Zhong Liu Za Zhi. 2008;30:831–836. [PubMed] [Google Scholar]
- 11.Schuebel KE, Chen W, Cope L, Glockner SC, Suzuki H, Yi JM, Chan TA, Van Neste L, Van Criekinge W, van den Bosch S, van Engeland M, Ting AH, Jair K, Yu W, Toyota M, Imai K, Ahuja N, Herman JG, Baylin SB. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–1723. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, Doucet D, Thomas NJ, Wang Y, Vollmer E, Goldmann T, Seifart C, Jiang W, Barker DL, Chee MS, Floros J, Fan JB. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho NY, Kim BH, Choi M, Yoo EJ, Moon KC, Cho YM, Kim D, Kang GH. Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J Pathol. 2007;211:269–277. doi: 10.1002/path.2106. [DOI] [PubMed] [Google Scholar]
- 14.Christman JK. 5-Azacytidine and 5-aza-2'-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 15.Jones PA. Altering gene expression with 5-azacytidine. Cell. 1985;40:485–486. doi: 10.1016/0092-8674(85)90192-8. [DOI] [PubMed] [Google Scholar]
- 16.Yan C, Boyd DD. Histone H3 acetylation and H3 K4 methylation define distinct chromatin regions permissive for transgene expression. Mol Cell Biol. 2006;26:6357–6371. doi: 10.1128/MCB.00311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lund AH, van Lohuizen M. Polycomb complexes and silencing mechanisms. Curr Opin Cell Biol. 2004;16:239–246. doi: 10.1016/j.ceb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 18.Shivapurkar N, Toyooka S, Toyooka KO, Reddy J, Miyajima K, Suzuki M, Shigematsu H, Takahashi T, Parikh G, Pass HI, Chaudhary PM, Gazdar AF. Aberrant methylation of trail decoy receptor genes is frequent in multiple tumor types. Int J Cancer. 2004;109:786–792. doi: 10.1002/ijc.20041. [DOI] [PubMed] [Google Scholar]
- 19.Gehring WJ, Affolter M, Burglin T. Homeodomain proteins. Annu Rev Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- 20.Rauch T, Wang Z, Zhang X, Zhong X, Wu X, Lau SK, Kernstine KH, Riggs AD, Pfeifer GP. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci USA. 2007;104:5527–5532. doi: 10.1073/pnas.0701059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Q, Lothe RA, Ahlquist T, Silins I, Trope CG, Micci F, Nesland JM, Suo Z, Lind GE. DNA methylation profiling of ovarian carcinomas and their in vitro models identifies HOXA9, HOXB5, SCGB3A1, and CRABP1 as novel targets. Mol Cancer. 2007;6:45. doi: 10.1186/1476-4598-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hallden G, Hadi M, Hong HT, Aponte GW. Y receptor-mediated induction of CD63 transcripts, a tetraspanin determined to be necessary for differentiation of the intestinal epithelial cell line, hBRIE 380i cells. J Biol Chem. 1999;274:27914–27924. doi: 10.1074/jbc.274.39.27914. [DOI] [PubMed] [Google Scholar]
- 23.Minth CD, Andrews PC, Dixon JE. Characterization, sequence, and expression of the cloned human neuropeptide Y gene. J Biol Chem. 1986;261:11974–11979. [PubMed] [Google Scholar]
- 24.Liu AJ, Furusato B, Ravindranath L, Chen YM, Srikantan V, McLeod DG, Petrovics G, Srivastava S. Quantitative analysis of a panel of gene expression in prostate cancer--with emphasis on NPY expression analysis. J Zhejiang Univ Sci B. 2007;8:853–859. doi: 10.1631/jzus.2007.B0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abe M, Watanabe N, McDonell N, Takato T, Ohira M, Nakagawara A, Ushijima T. Identification of genes targeted by CpG island methylator phenotype in neuroblastomas, and their possible integrative involvement in poor prognosis. Oncology. 2008;74:50–60. doi: 10.1159/000139124. [DOI] [PubMed] [Google Scholar]
- 26.Pitha PM, Au WC, Lowther W, Juang YT, Schafer SL, Burysek L, Hiscott J, Moore PA. Role of the interferon regulatory factors (IRFs) in virus-mediated signaling and regulation of cell growth. Biochimie. 1998;80:651–658. doi: 10.1016/s0300-9084(99)80018-2. [DOI] [PubMed] [Google Scholar]
- 27.Yanai H, Chen HM, Inuzuka T, Kondo S, Mak TW, Takaoka A, Honda K, Taniguchi T. Role of IFN regulatory factor 5 transcription factor in antiviral immunity and tumor suppression. Proc Natl Acad Sci USA. 2007;104:3402–3407. doi: 10.1073/pnas.0611559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 30.Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, Berman DM, Jenuwein T, Pruitt K, Sharkis SJ, Watkins DN, Herman JG, Baylin SB. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]