Abstract
Approximately 10 to 15% of patients with AIDS but without ocular opportunistic infections will have a presumed neuroretinal disorder (HIV-NRD), manifested by reduced contrast sensitivity and abnormal visual fields. The loss of contrast sensitivity often is sufficient to impair reading speed. To evaluate the effect of host genetics on HIV-NRD, we explored validated AIDS restriction gene variants CCR5Δ32, CCR2-64I, CCR5 P1, SDF-3`A, IL-10-5`A, RANTES -403A, RANTES -28G, RANTES-In1.1C, CX3CR1-249I, CX3CR1-280M, IFNG-179T, MDR1-3435T, and MCP-1364G, each of which has been implicated previously to influence HIV-1 infection, AIDS progression, therapy response, and antiviral drug metabolism, and an IL-10 receptor gene, IL-10R1, in the Longitudinal Study of the Ocular Complications of AIDS (LSOCA) cohort. In European Americans (cases=55, controls=290), IL-10-5`A variant and its promoter haplotype (HR=2.09, CI: 1.19–3.67, P = 0.01); in African Americans (cases=54, controls=180) RANTES-In1.1C and the associated haplotype (HR=2.72, CI: 1.48–5.00, P = 0.001), showed increased HIV-NRD susceptibility. While sample sizes are small and P values do not pass a strict Bonferroni correction, our results suggest that, in European Americans, an IL-10-related pathway, and, in African Americans, chemokine receptor ligand polymorphisms in RANTES are risk factors for HIV- NRD development. Clearly, further studies are warrented.
Keywords: AIDS, HIV-1, host genetics, HIV-neuroretinal disorder
INTRODUCTION
Prior to the introduction of highly active antiretroviral therapy (HAART), ocular complications, particularly ocular opportunistic infections (OIs), were common among patients with the acquired immunodeficiency syndrome (AIDS).1 Although the incidence of ocular OIs has been reduced ~80% by HAART,2, 3 the impact of other non-infectious problems is more evident, particularly an HIV-related presumed neuroretinal disorder (HIV-NRD), manifested by abnormal contrast sensitivity, color vision, and visual fields. 4–7 The decrease in contrast sensitivity often is sufficiently severe to impair reading speed. The pathogenesis of HIV-NRD is not well understood currently, but hypotheses include direct infection of neural tissue, indirect damage due to immune reaction against HIV infection, and HIV-microangiopathy-related cumulative damage to optic nerve and retina.
Host genetics have been shown to affect the acquisition of HIV infection, progression to AIDS, and the efficacy of antiretroviral therapy. 8–12 HIV-NRD may be an outcome of worse AIDS prognosis. 4, 6 Therefore, it is also possible that host genetics that affect progression to AIDS, and the efficacy of antiretroviral therapy may also affect the development of HIV-NRD. In this study, we evaluated host genetic factors that may influence HIV-NRD development. We evaluated the effects of variants in the genes CCR5Δ32, CCR2-64I, CCR5 P1, SDF-3`A, IL-10-5`A, RANTES -403A, RANTES -28G, RANTES-In1.1C, CX3CR1-249I, CX3CR1-280M, IFNG-179T, MDR1-3435T, and MCP-1364G each of which has been shown to influence HIV-1 infection, AIDS progression, therapy response, and antiviral drug metabolism. Previous studies showed that tumor necrosis factor (TNF) leads to damage in optic nerves. 13–16 IL-10 is a major regulator/suppressor of TNF and other inflammatory cytokines. 17, 18 Moreover, genetic polymorphisms in IL-10R1 have been shown to diminish IL-10 signalling through the IL-10 receptor complex. 19, 20 As our initial screen identified as IL-10-5`A a genetic risk factor for HIV-NRD, we extended our analyses to polymorphisms in the primary IL-10 receptor gene, IL-10R1, that have a crucial role in the IL-10 signaling pathway. Our study participants were HIV-infected European American and African American patients enrolled in the Longitudinal Study of the Ocular Complications of AIDS (LSOCA) cohort.
PATIENTS and METHODS
Study population and clinical assessment of HIV-NRD
Study patients included 345 European American and 234 African American individuals enrolled in the LSOCA cohort, who did not have ocular OIs. All patients in this study were enrolled beginning in September 1998 and diagnosed with AIDS according to the 1993 Centers for Disease Control and Prevention surveillance case definition for AIDS. Details of the study design and implementation have been published previously.2, 3 Eighty seven percent of the European and 86% of African American patients were receiving HAART. The date of HIV-NRD diagnosis was defined as the first date when a patient had log unit contrast sensitivity less than 1.5, in at least one eye. Clinical methods for assessing HIV-NRD in LSOCA have been described previously. 4 The LSOCA program, including a specimen bank for immunologic and genetic testing, was reviewed and approved by the institutional review boards at the participating clinical centers and at the resource centers, and written consent was obtained from each participant.
Genotyping and Haplotype construction
Previously identified functional polymorphisms rs333, rs1799864, rs1799988, rs1801157, rs1800872, rs2107538, rs2280788, rs2280789, rs3732379, rs3732378, rs2069709, rs1045642, and rs2857657, were genotyped for CCR5Δ32, CCR2-64I, CCR5 P1, SDF-3`A, IL-10-5`A, RANTES -403A, RANTES -28G, RANTES-In1.1C, CX3CR1-249I, CX3CR1-280M, IFNG-179T, MDR1-3435T, and MCP-1364G(intronic 767G, representative of haplotype 7) mutations, respectively. Additionally, 11 haplotype tagging single nucleotide polymorphisms (SNPs) (promoter region: rs17351243, rs4072227, rs6667202, rs1800890, rs1800896 and rs1800894; intronic: rs3021094, rs3024508; 3’ UTR: rs3024496, rs3024498, and rs3024500) covering the IL-10 region were also selected. rs1800896 (−1082) and rs1800872 (−592) were used to construct the proximal promoter IL-10 haplotypes of ATA, ACC and GCC that were reported to be associated with differential IL-10 production. 21–24 rs1800871 (−819) was not genotyped due to complete linkage with rs1800872. Functional and haplotype tagging SNPs for IL-10R1 region were rs3135932 (replacement), rs2228055 (replacement), rs4252279 (Intron), rs4252314 (Intron), rs4252286 (Intron), rs2229113 (replacement), and rs2229114 (replacement). All SNPs were genotyped with the ABI-TaqMan method (Applied Biosystems, Foster City, CA, USA). We couldn’t get clear genotyping results for a few individuals for the IL-10 and IL-10R1 SNPs and they were omitted from SNP based association analyses.
All haplotypes are inferred by the expectation maximization algorithm using SAS Genetics (SAS Institute, Cary, NC, USA) and the HaploView software. 25 The presence of CCR5_59353C (rs1799988) in the absence of CCR2-64I and CCR5Δ32 defines the CCR5 P1 promoter haplotype +.P1.+.26 The RANTES -403A, RANTES -28G, and RANTES-In1.1C genotypes define the RANTES haplotypes. RANTES -H1=G-C-T, RANTES -H2=A-C-T and RANTES -H3=A-C-C (low producer haplotype).8, 27
Statistical Analyses
Each SNP and haplotype found at ≥ 1% frequency in the study population were evaluated for NRD development with three different models of inheritance: allelic, dominant and codominant. Allelic analyses were used to examine individual allele effects. Genotypes were coded as 0, 1, and 2 copies of the rare allele for the codominant model. The dominant model analyzed genotypes as absence or presence of the rare alleles. Odds Ratios (OR) for the codominant model were calculated by logistic regression and for the allelic and dominant models by 2×2 tables. Nominal P values are reported. As the patients were diagnosed with AIDS prior to study entry, a staggered entry 28 approach was adapted, and time to HIV-NRD was analyzed using the Cox proportional hazards model. The method of staggered entry allows inclusion of prevalent cases into a survival analysis of time from diagnosis to event. The main assumption is that prevalent cases without the event of interest at baseline have the same risk over time as incident cases without the event of interest at the same time as entry into the study of the prevalent cases. The method of staggered entry creates risk sets (i.e., number at risk) to compare incident and prevalent AIDS patients at similar times since AIDS diagnosis. Unlike standard survival methods in which the number at risk can only decrease over time, the number at risk can increase or decrease over time. We checked the main assumption using only incident cases and did not see a significant loss in statistical power. As the main assumption was not violated, this method can control for varying lengths of followup. Cox models were adjusted for square root of nadir CD4+ T-cell cell count, highest log10HIV-1 load, age, and gender. To increase sample size and statistical power, analyses were extended to include each eye separately by a sandwich estimate of covariates in the Wald tests for the global null hypothesis and null hypotheses of individual parameters (by PROC PHREG covsandwich procedure in SAS). Only patients with visual acuity 20/20 Snellen equivalents (Standard ETDRS letters=85) or better were included in the Cox analyses to avoid cases of decreased contrast sensitivity attributable to other major ocular complications, cataracts, or glaucoma. All analyses were performed with SAS version 9.1 (SAS Institute, Cary, NC, USA). Throughout the manuscript cases represent study patients who developed HIV-NRD and controls represent study patients who did not develop HIV-NRD.
RESULTS
There were significant differences in terms of male gender percentage (P = 0.001), age (P = 0.01), CD4+T cell counts (P = 0.01), and time since AIDS diagnosis (P = 0.01) at study enrollment between European American (n = 345) and African American study patients (n = 234; Table 1). However, the HIV-NRD cases and controls did not differ significantly from each other for the clinical variables considered within European American patients, except more of the control patients were on HAART compared to HIV-NRD cases (89% vs. 78%; P = 0.01). African American patients with HIV-NRD were slightly older compared to the patients that did not develop HIV-NRD (44.7±7.5 vs. 40.5±7.9; P = 0.003).
TABLE 1.
Clinical aspects of European American and African American LSOCA patients used in this study
| European Americans |
African Americans |
|||||||
|---|---|---|---|---|---|---|---|---|
| Cases (n=55) |
Controls (n=290) |
Cases (n=54) |
Controls (n=180) |
|||||
| Mean±SD |
Median |
Mean±SD |
Median |
Mean±SD |
Median |
Mean±SD |
Median |
|
| Variable | (25th%,75th%-tile) | (25th%,75th%-tile) | (25th%,75th%-tile) | (25th%,75th%-tile) | ||||
| Male gender (%)a | 92 | 88 | 73 | 61 | ||||
| Age at study entry (yrs)a |
44.6±9.6 | 44.0 (38,50) |
43.0±7.7 | 42.0 (38,47) |
44.7±7.5 | 46.0 (41,49) |
40.5±7.9 | 43.0 (35,46) |
| Nadir CD4+ T-cell count (cells/mL) |
74.5±63.5 | 55.0 (17,127) |
77.7±99.4 | 48.0 (17,99) |
60.4±63.7 | 36.0 (8,98) |
59.9±73.6 | 30.0 (10,93) |
| CD4+ T-cell count (cells/mL) |
301.7±201.7 | 284.0 (147,387) |
255.0±205.6 | 220.0 (93,347) |
209.4±186.7 | 162.0 (68,284) |
207.2±202.9 | 146.0 (76,283) |
| Peak HIV viral load (log10 copies/mL) |
5.5±2.4 | 5.4 (4.9,5.7) |
5.6±2.4 | 5.3 (4.5,5.7) |
5.5±2.4 | 5.2 (4.4,5.7) |
5.5±2.3 | 5.3 (4.7,5.7) |
| Baseline HIV viral load (log10 copies/mL) |
4.8±2.3 | 2.9 (2.2,4.4) |
4.8±2.2 | 2.7 (2.2,4.6) |
5.1±2.3 | 3.7 (2.6,4.9) |
4.9±2.3 | 3.2 (2.3,4.7) |
| Time since AIDS diagnosis (yrs)b |
5.1±3.7 | 4.9 (2.0,7.0) |
5.0±3.6 | 4.7 (2.1,7.2) |
4.6±4.0 | 3.6 (1.0,7.5) |
4.2±3.6 | 3.5 (1.0,6.5) |
| HAART use (%) c | 78 | 89 | 84 | 87 | ||||
Significantly different between NRD cases and controls in African Americans (P = 0.003)
Years before study entry (see Methods for staggered entry study design)
Significantly different between NRD cases and controls in European Americans (P = 0.01)
European American Analyses
In SNP based analyses, we observed an increased frequency of IL-10-5A` (rs1800872) variant in the European American patients with HIV-NRD. Persons carrying this allele were more likely to develop HIV-NRD in all three models (allelic, codominant and dominant) of association tests (Table 2). Further analysis of IL-10 region haplotype tagging SNPs identified two other variants with reduced risk of HIV-NRD development (Supplemental Table 1). We also observed a trend towards HIV-NRD susceptibility associated with an intronic variant in IL-10R1 (Supplemental Table 1).
TABLE 2.
Allelic distribution and association tests of genetic polymorphisms in HIV-NRD cases and controls
| European Americans |
African Americans |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele Frequency (%) |
Allelic |
Codominant |
Dominant |
Allele Frequency (%) |
Allelic |
Codominant |
Dominant |
||||||||||
| Gene-variant | SNP | cases (n=55) |
controls (n=290) |
OR | P | OR | P | OR | P | cases (n=54) |
controls (n=180) |
OR | P | OR | P | OR | P |
| CCR5-Δ32 | rs333 | 7 | 9 | 0.72 | 0.32 | 0.70 | 0.38 | 0.71 | 0.40 | 2 | 2 | 1.11 | 0.90 | 1.12 | 0.90 | 1.11 | 0.90 |
| CCR2-64I | rs1799864 | 8 | 10 | 0.72 | 0.43 | 0.71 | 0.42 | 0.73 | 0.47 | 20 | 13 | 1.79 | 0.04 | 1.75 | 0.05 | 2.24 | 0.01 |
|
CCR5- 59353C |
rs1799988 | 56 | 51 | 1.23 | 0.32 | 1.25 | 0.30 | 1.69 | 0.20 | 46 | 41 | 1.24 | 0.49 | 1.21 | 0.51 | 1.26 | 0.62 |
| SDF-3A` | rs1801157 | 19 | 21 | 0.89 | 0.67 | 0.89 | 0.67 | 0.87 | 0.66 | 7 | 4 | 1.72 | 0.22 | 1.78 | 0.21 | 1.78 | 0.21 |
| IL-10-5A` | rs1800872 | 31 | 21 | 1.70 | 0.02 | 1.74 | 0.02 | 2.06 | 0.01 | 38 | 40 | 1.15 | 0.55 | 1.07 | 0.75 | 1.06 | 0.84 |
|
RANTES- 403A |
rs2107538 | 19 | 22 | 0.86 | 0.56 | 0.85 | 0.55 | 0.74 | 0.34 | 57 | 56 | 1.02 | 0.90 | 1.03 | 0.90 | 1.15 | 0.74 |
|
RANTES- 28G |
rs2280788 | 0 | 3 | -a | - | -a | - | -a | - | 0.9 | 0.6 | 1.64 | 0.68 | 1.65 | 0.69 | 1.65 | 0.68 |
|
RANTES- In1.1C |
rs2280789 | 9 | 14 | 0.59 | 0.13 | 0.59 | 0.13 | 0.60 | 0.17 | 27 | 17 | 1.76 | 0.03 | 1.72 | 0.03 | 2.17 | 0.01 |
|
CX3CR1- 249I |
rs3732379 | 28 | 27 | 1.04 | 0.87 | 1.04 | 0.88 | 1.03 | 0.93 | 10 | 12 | 0.80 | 0.55 | 0.79 | 0.55 | 0.79 | 0.55 |
|
CX3CR1- 280M |
rs3732378 | 16 | 15 | 1.00 | 0.91 | 1.03 | 0.92 | 1.14 | 0.74 | 3 | 3 | 1.04 | 0.96 | 0.97 | 0.96 | 1.08 | 0.91 |
| IFNG-179T | rs2069709 | 0 | 0 | -a | - | -a | - | -a | - | 0.9 | 2 | 0.40 | 0.37 | 0.39 | 0.38 | 0.39 | 0.36 |
|
MDR1- 3435T |
rs1045642 | 56 | 46 | 1.48 | 0.11 | 1.41 | 0.14 | 1.26 | 0.55 | 18 | 22 | 1.33 | 0.30 | 1.33 | 0.31 | 0.71 | 0.30 |
|
MCP1-767G (H7) |
rs2857657 | 17 | 19 | 0.85 | 0.60 | 0.85 | 0.61 | 0.66 | 0.28 | 5 | 5 | 1.08 | 0.86 | 0.92 | 0.87 | 0.92 | 0.97 |
Adjusting the models for square root of nadir CD4+ T-cell cell count, highest log10HIV-1 load, age, and gender does not significantly change the results.
OR not calculated in European Americans due to small sample size and lack of HIV-NRD cases.
Following individual SNP analyses, haplotypes were constructed and analyzed. There were 19 and 8 haplotypes with ≥ 1% frequency inferred for IL-10 and IL-10R1, respectively (Figure 1, supplemental figure 1). A strong linkage disequilibrium (LD) pattern around IL-10 driving the non-independent nominally significant SNP based HIV-NRD associations was evident (i.e. high D′ values observed between several SNPs, Figure 1).
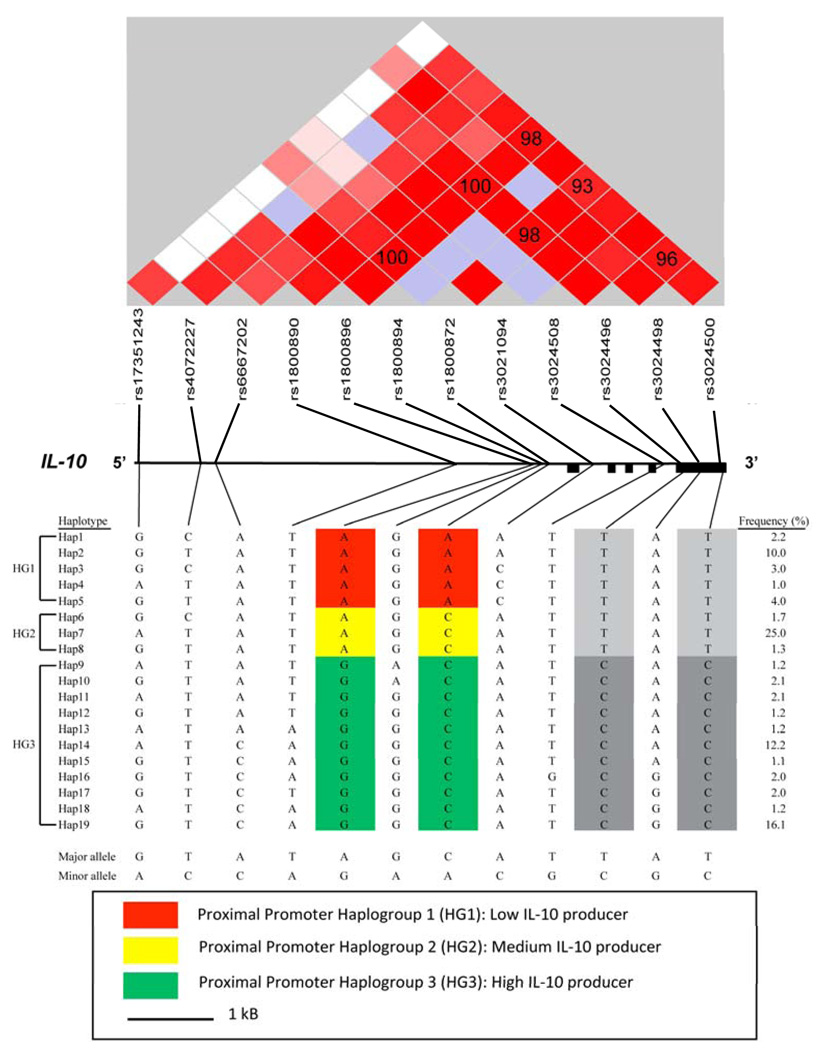
FIGURE 1.
Inferred haplotypes, their frequencies and LD structure across the IL-10 region in European American samples. Black filled rectangles show IL-10 exons. Lines indicate the actual physical position of SNPs with respect to each other. Brightness of red color represents the pairwise D′ (%) values. High D′ values (D′ > 80) are shown with bright red, low D′ values are shown in light red and blue squares. The P values associated with D′ estimates ranged from 10−22 for the rs1800872 and rs1800896 pair to 10−73 for the rs3024500 and rs3024496 pair. Pairwise D′ values (%) for these SNPs are shown in the squares. Previously described proximal promoter haplogroups assocaited with low, medium and high IL-10 production 21–24 are combined to form HG1 (low IL-10 producer), HG2 (medium IL-10 producer) and HG3 (high IL-10 producer), respectively. The minor alleles of rs3024496 and rs3024500, highlighted in dark grey color, show high LD with HG3 (P = 10−20) (see Supplementary Table 1 for association of these two SNPs with HIV-NRD development).
Three well studied IL-10 proximal promoter haplotypes ATA, ACC, and GCC, including rs1800896 (A/G), rs1800871 (C/T) and rs1800872 (C/A), were easily discerned using rs1800896 (A/G) and rs1800872 (C/A) as AA, AC and GC by red, yellow and green highlights respectively in Figure 1. Three IL-10 haplotypes showed increased HIV-NRD susceptibility, each of which had AA (rs1800896-A and rs1800872-A) haplotype (highlighted red in Figure 1; Supplemental Table 2). The effect of the fifth haplotype (Hap5) was more evident compared to the other haplotypes (Supplemental Table 2), which, in part, may be due to its larger sample size. Individual IL-10 proximal promoter AA, AC, and GC haplotypes were pooled to form combined haplotype groups HG1, HG2 and HG3 (Figure 1). Similar to individual AA haplotypes, the combined HG1 haplotypes showed increased HIV-NRD susceptibility in all models of association (Table 3). The combined HG3 haplotypes showed decreased susceptibility, although the results were not always significant (Table 3). Moreover, IL-10R1-Hap5 was enriched in the HIV-NRD cases (Supplemental Table 3). Finally, patients with HIV-NRD had more CCR5 P1 promoter allele defining haplotype +.P1.+ than expected by chance (Table 3).
TABLE 3.
Haplotype analyses for HIV-NRD development in European American and African American patients
| European Americans |
African Americans |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype Frequency (%) |
Allelic |
Codominant |
Dominant |
Haplotype Frequency (%) |
Allelic |
Codominant |
Dominant |
|||||||||
| Gene Haplotypes | cases (n=55) |
controls (n=290) |
OR | P | OR | P | OR | P | cases (n=54) |
controls (n=180) |
OR | P | OR | P | OR | P |
| CCR5 | ||||||||||||||||
| +.P1.+ | 43 | 34 | 1.45 | 0.07 | 1.90 | 0.04 | 1.89 | 0.04 | 22 | 25 | 0.88 | 0.63 | 0.87 | 0.57 | 0.79 | 0.46 |
| RANTES | ||||||||||||||||
| H1(G-C-T) | 81 | 79 | 1.13 | 0.65 | 1.13 | 0.65 | 1.01 | 0.67 | 36 | 40 | 0.86 | 0.50 | 0.83 | 0.46 | 1.00 | 1.00 |
| H2(A-C-T) | 10 | 7 | 1.49 | 0.26 | 1.35 | 0.33 | 1.27 | 0.57 | 39 | 44 | 0.82 | 0.38 | 0.87 | 0.46 | 0.70 | 0.25 |
| H3(A-C-C) | 9 | 14 | 0.61 | 0.16 | 0.61 | 0.16 | 0.63 | 0.22 | 25 | 17 | 1.67 | 0.05 | 1.63 | 0.06 | 2.12 | 0.02 |
| IL-10a | (n=55) | (n=290) | (n=50) | (n=172) | ||||||||||||
| HG1 | 32 | 21 | 1.74 | 0.01 | 1.70 | 0.02 | 2.05 | 0.01 | 38 | 39 | 0.96 | 0.86 | 0.96 | 0.87 | 0.95 | 0.65 |
| HG2 | 30 | 30 | 0.99 | 0.97 | 0.99 | 0.97 | 0.95 | 0.86 | 28 | 28 | 0.99 | 0.97 | 0.99 | 0.97 | 1.01 | 0.97 |
| HG3 | 38 | 49 | 0.66 | 0.05 | 0.67 | 0.06 | 0.71 | 0.26 | 33 | 32 | 1.10 | 0.72 | 1.09 | 0.72 | 1.32 | 0.39 |
Adjusting the models for square root of nadir CD4+ T-cell cell count, highest log10HIV-1 load, age, and gender does not significantly change the results.
HG1, HG2 and HG3 correspond to the combined proximal promoter IL-10 haplotypes (see Figure 1 for details).
The effects of individual SNPs and haplotypes on HIV-NRD development were also evaluated by the Cox proportional hazards models adjusted for age, gender, CD4+ T cell count, and HIV-1 viral load of patients. The increased HIV-NRD risk associated with IL-10-5A’ (rs1800872) and HG1 (Table 4), and IL-10R1 rs4252314 (Supplemental Table 4), were still evident. When each eye was assessed individually, all the aforementioned susceptible and protective variant effects were stronger (HR: 2.02 – 2.46, P = 0.02 – 0.0002; Table 4 and Supplemental Table 4 footnotes).
TABLE 4.
Cox proportional hazards analyses for HIV NRD development in European American and African American patients
| European Americans |
African Americans |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene/haplotype | SNP | events/n | HR | (95% CI) | P | events/n | HR | (95% CI) | P |
| RANTES-403A | rs2107538 | 48/299 | 0.67 | (0.37–1.24) | 0.21 | 49/198 | 1.09 | (0.48–2.46) | 0.84 |
| RANTES-28G | rs2280788 | 0/239 | - | - | - | 52/219 | 1.78 | (0.24–13.19) | 0.57 |
| RANTES-In1.1C | rs2280789 | 50/329 | 0.53 | (0.26–1.10) | 0.09 | 52/218 | 2.56 | (1.40–4.68) | 0.002 |
| H1(G-C-T) | 50/328 | 0.69 | (0.21–2.30) | 0.55 | 52/218 | 1.08 | (0.59–1.96) | 0.81 | |
| H2(A-C-T) | 50/328 | 1.28 | (0.57–2.91) | 0.55 | 52/218 | 0.55 | (0.31–0.99) | 0.04 | |
| H3(A-C-C) | 50/328 | 0.56 | (0.27–1.16) | 0.11 | 52/218 | 2.72 | (1.48–5.00) | 0.001 | |
| IL-10-5A` | rs1800872 | 49/330 | 2.07 | (1.18–3.63) | 0.01a | 52/219 | 1.28 | (0.72–2.27) | 0.40 |
| HG1 | 49/329 | 2.09 | (1.19–3.67) | 0.01a | 48/207 | 1.31 | (0.72–2.37) | 0.38 | |
| HG2 | 49/329 | 0.71 | (0.51–1.58) | 0.90 | 48/207 | 0.99 | (0.55–1.77) | 0.97 | |
| HG3 | 49/329 | 0.76 | (0.42–1.36) | 0.35 | 48/207 | 1.33 | (0.73–2.40) | 0.35 | |
When each eye is considered separately, IL-10-5A` HR=2.46, P = 0.0002; IL-10 HG1 haplotype HR=2.44, P = 0.0002 in European Americans.
African American Analyses
African Americans patients with the CCR2-64I (rs1799864) and RANTES-In1.1C (rs2280789) variants showed increased risk of HIV-NRD development in allelic, dominant, and codominant models of association (Table 2). Haplotype analyses confirmed the RANTES association where the H3 (A-C-C) haplotype carrying the In1.1C variation showed higher odds of developing HIV-NRD (Table 3). Similar to European American analyses, there were 18 and 7 haplotypes with ≥ 1% frequency constructed for IL-10 (Supplemental Figure 2, Supplemental Table 5) and IL-10R1, respectively (Supplemental Figure 1, Supplemental Table 3). Whereas only six IL-10 haplotypes were common between European American and African American individuals, the IL-10R1 haplotypes were nearly identical in both groups. The IL-10R1 Hap5, with increased frequency in European American HIV-NRD cases, also suggested an increased risk of HIV-NRD for African American patients (Supplemental Table 3).
The Cox analyses strengthened the observation of increased HIV-NRD risk associated with RANTES-In1.1C and H3 (A-C-C), and also indicated a protective role for H2 (A-C-T) (Table 4). Although CCR2-64I still trended for HIV-NRD susceptibility (Supplemental Table 4) and IL-10R1 Hap3 for protection (HR = 0.49; P = 0.06; Supplemental Table 3), the results were less significant after clinical covariates were adjusted. When each eye was analyzed independently, similar HIV-NRD association trends were observed, with a possible increased HIV-NRD risk for patients with SDF-3A` variant (HR = 2.24, CI: 1.24 – 4.03, P = 0.007; see Supplemental Table 4 footnotes).
DISCUSSION
We investigated the role of host genetics in HIV-NRD development and explored the influence of variants of several genes known to influence other aspects of HIV infection. Our analyses suggest that European American patients with the IL-10-5A` variant and with the associated haplotype (proximal promoter HG1) are more likely to progress to HIV-NRD. Moreover, IL-10R1 receptor variants may also influence this complication in European Americans. On the other hand, RANTES polymorphisms (RANTES-In1.1C ) and associated haplotypes (H2 and H3) are the main effectors on HIV-NRD development in African American patients.
In this study, we focused on 11 different genes. Using a strict Bonferroni correction, each gene necessitates a P value ≤ 4.5 × 10−3 to be considered statistically significant. Moreover, some genes were analyzed for more than one SNP (variant) and inferred haplotypes. When all these individual tests considered (>100), the expected Bonferroni significance cut off goes down to roughly P < 10−5. None of the P values observed in this study will meet this conservative cutt-off value. However, given the linkage disequilibrium pattern around these genes, it is clear that neither the individual SNPs nor the inferred haplotype association tests are independent comparisons. In other words, if we assume all individual SNP and haplotype comparisons as independent and we correct for multiple tests, type 1 error will inflate. Using a gene based P value cut off of 4.5 × 10−3, only the RANTES associations in African Americans would be considered statistically significant. Given the size of the available sample (especially the number of HIV-NRD cases), our limited statistical power is not surprising. When each eye was considered independently, our sample size (nearly) doubled and the observed association list expanded to include variants +.P1.+ (CCR5 promoter) haplotype and SDF-3A` in European and African Americans, respectively. Overall, IL-10 (and possibly its receptor), RANTES, and SDF associations suggest a potential biological basis for our results.
IL-10 is a key regulatory cytokine involved in a wide spectrum of immune responses, particularly the suppression of T helper type-1 (Th1) immune responses involved in cellular immunity. 18 Variations in the promoter region of IL-10 affect IL-10 production. 21, 22, 24, 29, 30 Moreover, one of these variants, the low producer IL-10-5A` and its associated ATA haplotype (represented by HG1 in this study), has been shown to influence HIV-1 infection and accelerate progression to AIDS 23, 31, 32 in European Americans. RANTES, a CC chemokine receptor 5 (CCR5) ligand, is a potent inhibitor of HIV-1 cell entry and replication. 33 RANTES variants and haplotypes influence RANTES production and have been shown to affect HIV-1 infection, progression to AIDS, and HAART outcome. 8, 10, 34–37 SDF-1 is a natural ligand for CXCR4 receptor and a potent inhibitor of HIV-1 cell entry and replication. 33 The SDF-3A` variant is associated with increased SDF production 38, 39 and has also been show to affect AIDS progression and response to HAART. 10, 40–42 Finally, the significant effects of CCR5 promoter haplotype variant (+.P1.+) on AIDS progression and response to HAART are well documented. 10, 12, 26 Most of the genetic risk factors for HIV-NRD observed in this study are ones that make a patient more susceptible to AIDS. In other words, the patients who are more susceptible to HIV-NRD development in this study are genetically similar to patients from earlier studies who have increased susceptibility to both faster AIDS progression and HAART failure. However, the CCR2-64I and SDF-3A` variants have been previously reported to have protective effects against AIDS progression, 11, 39, 42 although their effect on AIDS patients’ prognosis who received HAART were inconclusive 12, 41, 43–45 if not suggesting a negative effect on some reports. 10, 46
Clearly, interpretations of gene-disease interactions are difficult because of the complexity of these relationships. HIV-NRD is a rare disease with no known etiology; it occurs only in patients with advanced AIDS. Therefore, it is impossible to completely untangle AIDS effects from NRD-specific effects. In addition, there is an inherent survival bias in any study of HIV-NRD; all affected individuals have survived to advanced stages of AIDS and have probably experienced other AIDS-related illnesses. A possible and simple explanation for the similar genetic associations of HIV-NRD and AIDS progression is that HIV-NRD is present in patients with a worse HIV prognosis. The host genetics may be affecting the severity of the AIDS progression rather than exerting a direct effect on neuroretinal tissue and the development of HIV-NRD.
There are, however, several reasons to suspect that the presence of HIV-NRD may be more complex than a simple indicator of more severe HIV infection. The first comes from comparisons of AIDS-related clinical parameters. CD4+ T-cell counts, HIV viral load, and age are the major clinical parameters that influence HIV infection, progression, and response to therapy. Patients with lower CD4+ T-cell counts and higher HIV viral loads (or rebounds after therapy) are expected to be more prone to faster HIV progression to AIDS and associated complications. If the sole explanation was that HIV-NRD was a marker for more severe HIV infection, one might expect significant differences between HIV-NRD cases and controls in these clinical parameters even though CD4+ T-cell count and HIV viral load comparisons may not be a comprehensive representation of disease severity in a seroprevalent cohort. We did not see significant difference in either of these parameters between HIV-NRD cases and AIDS controls in either European Americans or in African Americans. Moreover, the statistical models were adjusted for these four major HIV-related clinical variables, and the genetic associations with HIV-NRD were still evident. However, we do acknowledge that CD4+ T-cell count and HIV viral load comparisons may not be a true representation of disease severity in a seroprevalent cohort. Still, the presence of HIV-NRD may be more complex than a simple indicator of more severe HIV infection.
Second, not all of the variants associated with susceptibility and rapid progression to AIDS influenced HIV-NRD significantly, and two protective variants (CCR2-64I and SDF-3A`) that are associated with slower AIDS progression actually increased the risk of HIV-NRD. One can speculate that increased proinflammatory signaling could be beneficial from a standpoint of AIDS progression, but that long-term activation of these pathways resulting from a longer chronic stage of HIV-infection could have other detrimental effects including damage to neuronal tissues in the eyes. However, for the moment this must remain speculative as genetic associations are complicated and need replication and functional follow up not possible in this particular rare-incidence effect cohort.
Finally, neurotoxic effects of HIV infection itself in neural tissues are well documented.47 For example, neuropathologies affect up to 40% of adult patients with AIDS, 48 and autopsy studies have shown a substantial (up to 50%) loss of optic nerve fibers in patients with AIDS. 49 In the cohort in this genetic study, 18% of the European American patients with HIV-NRD also had an HIV-related neurological disorder, whereas only 8% of the patients without HIV-NRD developed a similar neurological disorder. The fractions of HIV-related neurological complications in African Americans were 12% and 8% for patients with and without HIV-NRD, respectively. Given that only a small fraction of patients with neurological damage from HIV can be diagnosed reliably in patients with AIDS, these fractions could represent an underestimate of ongoing neuronal damage in patients with AIDS in this cohort.
Neuroimmunological studies of patients with AIDS provide information suggesting potential mechanisms of neurodegeneration associated with HIV-1 infection. 50 Previously, both in vivo and in vitro examinations showed that cytokine expression, especially the tumor necrosis factor (TNF) leads to myelin and/or membrane damage in optic nerves. 13–16 IL-10 is a major regulator/suppressor of TNF and other inflammatory cytokines. 17, 18 The association of low IL-10 producer variant in European Americans and increased HIV-NRD risk may be explained in part by an increased immune activity (i.e. higher TNF production) leading to an increased damage to the optic neurons. In addition to the damaging cytokines, prostaglandins, proteases, arachidonic acid and other metabolites, viral gp120, gp41, tat, vpr, and nef proteins can be directly neurotoxic. 51 These neurotoxic viral proteins are produced irrespective of productive HIV infection and can be transported along the neural pathways causing damage at remote sites. 52 In other words, the cascade of reactions leading to neurotoxicity may be started by relatively small amounts of viral proteins and need not depend on high viral loads or viral reproduction in a cell. 53 HIV-1 infection of nervous system, and therefore the potential start of destructive lesions in neural tissues, occurs at an early stage, well before HAART typically is begun. Hence, the presence of these neuronal complications in the HAART era, despite substantially more effective therapies, improved CD4+ T cell counts, and decreased HIV loads, may not be surprising.
Another crucial observation is the presence of HIV coreceptors CCR5 and CXCR4 in neurons. 54, 55 Studies suggest an important role for gp120 activated CXCR4 and CCR5 in HIV-associated neuronal damage. 56, 57 It has been shown that the CCR5 ligand, RANTES, can protect neurons against gp-120-induced toxicity, 51, 56, 57 whereas SDF-1 can induce toxicity and trigger neuronal death in a CXCR4-dependent manner. 51, 56, 57 These reports suggest a biological basis for the increased HIV-NRD risk in African Americans associated with low RANTES and high SDF producing variants, and may also explain the opposite effect of SDF-3A′ on AIDS (protective) vs. HIV-NRD (susceptible).We observed different gene variants to be associated with HIV-NRD in African Americans and European Americans. This may be due to allele frequency differences between the two ethnic groups, genetic heterogenety in African Americans or other clinical and/or social factors that we cannot account for in this study.
In conclusion, some host genetic risk factors that influence AIDS progression, response to HAART, and overall immune health, appear to affect ocular health in HIV-infected patients. Our results suggest a role for the IL-10 pathway in European Americans and for the chemokine ligands, RANTES and SDF-1, in African Americans leading to damage to retina and/or optic nerve. It will be intersting to study a cohort of patients who develop HIV-NRD independent of AIDS to test if the observed assocaitions are specific for HIV-NRD.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the patients and staff of all the participating centers in the study. We are also grateful to Michael Malasky, Mary Thompson, Bailey Kessing, Christiana Martin, Nick Edler, Nicole Shifflett, Katy Limpert, Natalie Baggett, Kelly Subramanian and Alyssa Drosdak for their assistance. Finally, we acknowledge the constructive criticism from two reviewers that helped in the final version of the manuscript. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does any mention of trade names, commercial products, or organizations imply endorsement by the U.S. government. See supplemental Acknowledgements for LSOCA Clinical Centers Credit Roster.
LSOCA Grant Support:
Supported by cooperative agreements from the National Eye Institute to The Johns Hopkins University School of Medicine (U10 EY 08052), The Johns Hopkins University Bloomberg School of Public Health (U10 EY 08057), and the University of Wisconsin, Madison School of Medicine (U10 EY 08067). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Eye Institute or the National Institutes of Health.
Additional support provided by National Center for Research Resources through General Clinical Research Center grants:
5M01 RR 00350 (Baylor College of Medicine)
5M01 RR 05096 (LSU/Tulane/Charity Hospital)
5M01 RR00096 (New York University Medical Center, New York)
5M01 RR 00865 (University of California, Los Angeles)
5M01 RR00046 (University of North Carolina)
5M01 RR00043 (University of Southern California)
ULI RR024996 (Weill Medical College of Cornell University)
Support also provided through cooperative agreements:
U01 AI 27674 (Louisiana State University/Tulane)
U01 AI 27660 (University of California, Los Angeles)
U01 AI 27670 (University of California, San Diego)
U01 AI 27663 (University of California, San Francisco)
U01 AI25868 (University of North Carolina)
U01 AI32783 (University of Pennsylvania)
Sources of support: Supported in part by contract N01-CO-12400 from the National Cancer Institute to the Laboratory of Genomic Diversity, Frederick, MD, with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN26120080001E and cooperative agreements from the National Eye Institute, National Institutes of Health, U10-EY-08052 to the Mount Sinai School of Medicine, New York, NY, U10-EY-08057 to the Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, and U10-EY-08067 to the University of Wisconsin, Madison, Madison, WI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Participating Clinical Centers
Baylor College of Medicine, Cullen Eye Institute, Houston, TX: Richard Alan Lewis, MD, MS (Director); John Michael Bourg; Victor Fainstein, MD; Zbigniew Krason, CRA; Joseph F. Morales, CRA; Silvia Orengo-Nania, MD; Tobias C. Samo, MD; Steven Spencer, BA, COMT; Mitchell P. Weikert, MD. Former Members: Richard C. Allen, MD; Pamela Frady, COMT; Ronald Gross, MD; Allison Schmidt, CRA; Laura Shawver, COT/CCRP; James Shigley, CRA; Benita Slight, COT; Rachel Sotuyo, COT; Stephen Travers, CRA.
Emory University Eye Center, Atlanta, GA: Sunil K. Srivastava, MD (Director); Allison Gibbs, BS; Deborah Gibbs, COMT; Debora Jordan, CRA; Bob Myles, CRA; Janna Rutter, CRA. Former Members: Antonio Capone, Jr. MD; David Furukuwa, PA; Baker Hubbard, MD; Daniel F. Martin, MD.
Indiana University, Indianapolis, IN: Former Members: Mitchell Goldman, MD (Director); Janice Brown; Thomas Ciulla, MD; Jean Craft, RN, CS; Ronald Danis, MD; Paul Fry; Hua Gao,MD; Samir Gupta, MD; Janet Hernandez, RN; Debra Poe; Linda Pratt, RN; James D. Richardson, MD; Tim Steffens, CRA; L. Joseph Wheat, MD; Beth Zwickl, RN, CS, MSN.
Johns Hopkins University School of Medicine, Baltimore, MD: J.P. Dunn, MD (Director); Diane M. Brown, RN; Dennis Cain; David Emmert; Mark Herring; Adam Jacobowitz, MD; Henry A. Leder, MD; Alison G. Livingston, RN, BSN; Yavette Morton; Kisten D. Nolan, RN, BSN, MPH; Richard D. Semba, MD, MPH; Priscilla Soto; Jennifer E. Thorne, MD, PhD. Former Members: Patricia Barditch-Crovo, MD; Marie-Lyne Bélair, MD; Stephen G. Bolton, CRNP; Joseph B. Brodine; Lisa M. Brune, RN, BSN; Anat Galor, MD; Douglas A. Jabs, MD, MBA; Meera Kapoor; Sanjay R. Kedhar, MD; John H. Kempen, MD, PhD; Stephen J. Kim, MD; Armando L. Oliver, MD; George B. Peters, III, MD; Ricardo Stevenson, MD; Michelle Tarver-Carr, MD, PhD; Susan Wittenberg, MD; Michelle Yue Wang, MD.
Louisiana State University Health Sciences Center, New Orleans, LA: Donald Bergsma, MD (Director); Rebecca Clark, MD; Robin Cooper, COMT; Jasmine Elison, MD; Butler Fuller, MD; Christine Jarrott, RN, ACRN; Lynn Otillio, COT; Maria Reinoso, MD; Christine Romero, COT, ROUB. Former Members: Bruce Barron, MD; Robin Bye, RN; Mandi Conway, MD; Larry Dillon, COT/CRA; Audrey Lombard, RN; Gholman Peyman, MD.
New Jersey Medical School, Newark, NJ: Former Members: Ronald Rescigno, MD (Director); Neelakshi Bhagat, MD; Rosa Paez-Boham, COMT; Marta Paez-Quinde.
New York Hospital - Cornell Medical Center, New York, NY: Murk-Hein Heinemann, MD (Director); Susana Coleman; Sara Daniel; Roberta Janis, RN, BSN; Aziz Khanifer, MD; Andrzej Kozbial; Diane Iglesias Rivera, COA; Kent Sepkowitz, MD. Former Members: Kenneth Boyd; Robinson V.P. Chan, MD; Cynthia Chiu, MD; Charles Cole, MD; Charles Doering, MD; Jasmine Elison, MD; Sangwoo Lee, MD; Fang Lu; Joseph Murphy; Sophia Pachydaki, MD; Christina Peroni, MD; Firas M. Rahhal, MD; Ashok Reddy, MD; Scott Warden, MD.
New York University Medical Center, New York, NY: Dorothy N. Friedberg, MD, PhD (Director); Adrienne Addessi, MA, RN; Douglas Dieterich, MD; Monica Lorenzo-Latkany, MD; Maria Pei, COA. Former Member: Alex McMeeking, MD.
Northwestern University, Chicago, IL: Alice T. Lyon, MD (Director); Lori Ackatz, RN, MPH; Manjot Gill, MD; Lori Kaminski, RN, MS; Rukshana Mirza, MD; Robert Murphy, MD; Frank Palella, MD; Carmen Ramirez; Zuzanna Rozenbajgier; Dawn Ryan; Evica Simjanoski; Former Members: Alexander Habib; Jill Koecher; Jeevan Mathura, MD; Annmarie Muñana, RN; Jonathan Shankle; David V. Weinberg, MD; James Yuhr.
Rush University, Chicago, IL: Former Members: Mathew W. MacCumber, MD, PhD (Director); Bruce Gaynes, OD, PharmD; Christina Giannoulis; Pamela Hulvey; Harold Kessler, MD; Heena S. Khan; Andrea Kopp; Pauline Merrill, MD; Frank Morini; Nada Smith; Allen Tenorio, MD; Denise Voskuil-Marre; Kisung Woo.
University of California, Irvine: Former Members: Baruch D. Kuppermann, MD, PhD (Director); Bogdan Alexandiescu, MD; Donald N. Forthal, MD; Jeff Grijalva, COT; Faisal Jehan, MD; Karen Lopez; Rosie Magallon, BA; Nader Moinfar, MD; Bret Trump; Melody Vega, COA; Randy Williams.
University of California, Los Angeles: Gary N. Holland, MD (Director); Robert D. Almanzor, COA; Margrit E. Carlson, MD; Jose T. Castellanos, COT; Jeffrey A. Craddock, COT; Serina Gonzales; Ann K. Johiro, MN, RN,BC, FNP-C, AACRN, AAHIVS; Partho S. Kalyani, MD; Michael A. Kapamajian, MD; David L. LeBeck; Kristin M. Lipka; Susan S. Ransome, MD. Former Members: Suzette A. Chafey, RN, NP; Alexander C. Charonis, MD; Peter J. Kappel, MD; Ardis A. Moe, MD; Germán Piñón; Angela Sanderson; Kayur H. Shah, MD; Robert Stalling, COA; Dennis Thayer, CRA; Jean D. Vaudaux, MD.
University of California, San Diego: William R. Freeman, MD (Director); Denise Cochran; Igor Kozak, MD; Megan Loughran; Luzandra Magana; Victoria Morrison, MD; Vivian Nguyen; Stephen Oster, MD. Former Members: Sunan Chaidhawanqual, MD; Lingyun Cheng, MD; Tom Clark; Mark Cleveland; Randall L. Gannon; Claudio Garcia, MD; Daniel Goldberg, MD; Joshua Hedaya, MD; Marietta Karavellas, MD; Tiara Kemper; Brian Kosobucki; Alona Mask; Nicole Reagan MD; Mi-Kyoung Song, MD; Francesca Torriani, MD; Dorothy Wong; Tekeena Young.
University of California, San Francisco: Jacque Duncan, MD (Director); Fermin Ballesteros, Jr.; Robert Bhisitkul, MD, PhD; Debra Brown; David Clay; Michael Deiner; Donald Eubank; Mark Jacobson, MD; Mary Lew, COT; Todd Margolis, MD, PhD. Former Members: Judith Aberg, MD; Jacqueline Hoffman; Alexander Irvine, MD; James Larson; Jody Lawrence, MD; Michael Narahara; Monique Trinidad.
University of North Carolina, Chapel Hill: Travis A. Meredith, MD (Director); Sandy Barnhart; Debra Cantrell; Seema Garg, MD, PhD; Elizabeth Hartnett, MD; Maurice B. Landers, MD; Sarah Moyer; David Wohl, MD;. Former Members: Stephanie Betran; Kelly DeBoer; David Eifrig, MD; John Foley, MD; Angela Jeffries; Jan Kylstra, MD; Barbara Longmire; Sharon Myers; Fatima N’Dure, COA; Kean T. Oh, MD; Jeremy Pantell; Susan Pedersen, RN; Cadmus Rich, MD; Cecilia A. Sotelo, RN; Charles van der Horst, MD; Samir Wadhvania.
University of Pennsylvania Medical Center, Philadelphia, PA: Charles W. Nichols, MD (Director); Mark Bardsley, BSN; Cheryl C. Devine; Jay Kostman, MD; Albert Maguire, MD; William Nyberg; Leslie Smith, RN. Former Members: Chris Helker, RN; RobRoy MacGregor, MD; Karen McGibney, RN; Keith Mickelberg, RN.
University of Southern California, Los Angeles, CA: Former Members: Jennifer I. Lim, MD (Director); Rizwan Bhatti, MD; John Canzano, MD; Thomas S. Chang, MD; Alexander Charonis, MD; Lawrence Chong, MD; Robert Equi, MD; Amani Fawzi, MD; Christina Flaxel, MD; Jesus Garcia; Todd Klesert, MD; Francoise Kramer, MD; Lori Levin, MPH; Tracy Nichols, COA, CRA; Christopher Pelzek, MD; Margaret Podilla, BS; Len Richine; Danny Romo, COA; Srinivas Sadda, MD; Richard Scartozzi, MD; Robert See, MD; Kevin Shiramizu, MD; Mark Thomas; A. Frances Walonker, CO, MPH; Alexander Walsh, MD; Ziquiang Wu, MD.
University of South Florida, Tampa, FL: Peter Reed Pavan, MD (Director); JoAnn Leto, COT; Brian Madow, MD; Richard Oehler, MD; Nandesh Patel, MD; Wyatt Saxon; Susan Sherouse, COT. Former Members: Andrew Burrows, MD; Steve Carlton; Burton Goldstein, MD; Sandra Gompf, MD; Bonnie Hernandez, COT; Mohan Iyer, MD; Patrick Kelty, MD; Amy Kramer, COT; Sharon Millard, RN, COT; Jeffrey Nadler, MD; Scott E. Paulter, MD; Jennifer Tordilla-Wadia, MD; Nancy Walker, COA.
University of Texas Medical Branch, Galveston, TX: Former Members: Garvin Davis, MD (Director); Robert Blem, MD; J. Mike Bourg, VA; John Horna, BS; Craig Kelso; Zbigniew Krason, BS; Helen K. Li, MD; Lan-Chi Nguyen, COMT; Rhonda Nolen, BS, CRC; Michelle Onarato, MD; David Paar, MD; Steven Rivas; Vicky Seitz, COT; Happy Spillar; Sami Uwaydat, MD.
Chairman’s Office, Mount Sinai School of Medicine, New York, New York: Douglas A. Jabs, MD, MBA (Study Chairman); Yasmin Hilal, MHS; Melissa Nieves, BA; Karen Pascual, BBA; Jill Slutsky, MPA; Maria Stevens, CM. Former member: Judith C. Southall.
Coordinating Center, The Johns Hopkins University Bloomberg School and Public Health: Curtis L. Meinert, PhD (Director); Alka Ahuja, MS; Debra A. Amend-Libercci; Karen L. Collins; Betty J. Collison; Ryan Colvin; John Dodge; Michele Donithan, MHS; Cathleen Ewing; Kevin Frick, PhD; Janet T. Holbrook, MS, MPH, PhD; Milana R. Isaacson, BS; Rosetta M. Jackson; Hope Livingston; Lee McCaffrey, MA; Milo Puhan, PhD; Girlie Reyes; Jacki Smith; Michael Smith; Elizabeth Sugar, PhD; Jennifer E. Thorne, MD, PhD; James A. Tonascia, PhD; Mark L. Van Natta, MHS; Annette Wagoner. Former members: Carley Benham; Gregory Foster; Judith Harle; Adele M. Kaplan Gilpin, JD, PhD; John H. Kempen, MD, PhD; Barbara K. Martin, PhD; Nancy Min, MPH, PhD; Laurel Murrow, MS; Maria J. Oziemkowska, MS, MPH; Wai Ping Ng, BS; Pamela E. Scott, MA; Erica Smothers; Emily West; Claudine Woo, MPH; Albert Wu, MD, MPH; Alice Zong.
Fundus Photograph Reading Center, University of Wisconsin: Ronald Danis, MD (Director); Charles Chandler; Sapna Gangaputra, MD, MPH; Gregory Guilfoil; Larry Hubbard, MAT; Jeffrey Joyce; Thomas Pauli; Nancy Robinson; Dennis Thayer; Jeong Won Pak; Grace Zhang. Former members: Michael Altaweel, MD; Jane Armstrong; Matthew D. Davis, MD; Sheri Glaeser; Katrina Hughes; Dolores Hurlburt; Linda Kastorff; Michael Neider, BA; Therese Traut; Marilyn Vanderhoof-Young; Hugh Wabers;.
National Eye Institute, Bethesda, MD: Natalie Kurinij, PhD.
Officers of the Study: Douglas A. Jabs, MD, MBA (Chair); Ronald Danis, MD; Natalie Kurinij, PhD; Curtis L. Meinert, PhD; Jennifer E. Thorne, MD, PhD. Former Members: Matthew D. Davis, MD; Janet T. Holbrook, MS, MPH, PhD.
Steering Committee: Douglas A. Jabs, MD, MBA (Chair); Ronald Danis, MD; James P. Dunn, MD; Gary N. Holland, MD; Milana R. Isaccson, BS; Mark Jacobson, MD; Natalie Kurinij, PhD; Richard Lewis, MD, MS; Kisten D. Nolan, RN, BSN, MPH; Curtis L. Meinert, PhD; William Nyberg; Frank Palella, MD; Jennifer E. Thorne, MD, PhD. Former Members: Adrienne Addessi, MA, RN; Lisa Brune, RN, BSN; Rebecca Clark, MD; Tom Clark, CRA; Janet Davis, MD; Matthew D. Davis, MD; William R. Freeman, MD; Dorothy Friedberg, MD; James Gilman; Janet T. Holbrook, MS, MPH, PhD; John Horna; Larry Hubbard, MAT; Mark Jacobson, MD; Daniel F. Martin, MD; Travis A. Meredith, MD; Annmarie Muñana, RN; Robert Murphy, MD; P. Reed Pavan, MD; Steven Spencer, BA, COMT; Tim Steffens, CRA; Dennis Thayer; Charles van der Horst, MD; Fran Wallach.
Policy and Data Monitoring Board: John P. Phair, MD (Chair); Brian P. Conway, MD; Barry R. Davis, MD, PhD; Douglas A. Jabs, MD, MBA; Natalie Kurinij, PhD; Curtis L. Meinert, PhD; David Musch, PhD; Robert B. Nussenblatt, MD; Jennifer E. Thorne, MD, PhD; Richard Whitley, MD. Former Members: B. William Brown, Jr., PhD; Matthew D. Davis, MD; James Grizzle, PhD; Argye Hillis, PhD; Janet T. Holbrook, MS, MPH, PhD; Harmon Smith, PhD; James A. Tonascia, PhD.
Visual Function Quality Assurance Committee: Steven Spencer, BA, COMT (Chair); Robert D. Almanzor; Deborah Gibbs, COMT; Milana Isaacson, BS; Mary Lew, COT ;Richard Alan Lewis, MD, MS (Advisor);. Former Members: Ferman Ballesteros; Jeff Grijalva, COT; Karen Lopez; Laura G. Neisser, COT; Rosa Paez-Boham, COST.
REFERENCES
- 1.Jabs DA. Ocular manifestations of HIV infection. Trans Am Ophthalmol Soc. 1995;93:623–683. [PMC free article] [PubMed] [Google Scholar]
- 2.Jabs DA, Van Natta ML, Holbrook JT, et al. Longitudinal study of the ocular complications of AIDS: 1. Ocular diagnoses at enrollment. Ophthalmology. 2007;114:780–786. doi: 10.1016/j.ophtha.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 3.Jabs DA, Van Natta ML, Holbrook JT, et al. Longitudinal study of the ocular complications of AIDS: 2. Ocular examination results at enrollment. Ophthalmology. 2007;114:787–793. doi: 10.1016/j.ophtha.2006.07.065. [DOI] [PubMed] [Google Scholar]
- 4.Freeman WR, Van Natta ML, Jabs D, et al. Vision function in HIV-infected individuals without retinitis: report of the Studies of Ocular Complications of AIDS Research Group. Am J Ophthalmol. 2008;145:453–462. doi: 10.1016/j.ajo.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller AJ, Plummer DJ, Dua R, et al. Analysis of visual dysfunctions in HIV-positive patients without retinitis. Am J Ophthalmol. 1997;124:158–167. doi: 10.1016/s0002-9394(14)70780-9. [DOI] [PubMed] [Google Scholar]
- 6.Plummer DJ, Marcotte TD, Sample PA, et al. Neuropsychological impairment-associated visual field deficits in HIV infection. HNRC Group. HIV Neurobehavioral Research Center. Invest Ophthalmol Vis Sci. 1999;40:435–442. [PubMed] [Google Scholar]
- 7.Shah KH, Holland GN, Yu F, et al. Contrast sensitivity and color vision in HIV-infected individuals without infectious retinopathy. Am J Ophthalmol. 2006;142:284–292. doi: 10.1016/j.ajo.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 8.An P, Nelson GW, Wang L, et al. Modulating influence on HIV/AIDS by interacting RANTES gene variants. Proc Natl Acad Sci U S A. 2002;99:10002–10007. doi: 10.1073/pnas.142313799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrington M, O'Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 10.Hendrickson SL, Jacobson LP, Nelson GW, et al. Host genetic influences on highly active antiretroviral therapy efficacy and AIDS-free survival. J Acquir Immune Defic Syndr. 2008;48:263–271. doi: 10.1097/QAI.0b013e31816fdc5f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Brien SJ, Nelson GW. Human genes that limit AIDS. Nat Genet. 2004;36:565–574. doi: 10.1038/ng1369. [DOI] [PubMed] [Google Scholar]
- 12.O'Brien TR, McDermott DH, Ioannidis JP, et al. Effect of chemokine receptor gene polymorphisms on the response to potent antiretroviral therapy. AIDS. 2000;14:821–826. doi: 10.1097/00002030-200005050-00008. [DOI] [PubMed] [Google Scholar]
- 13.Lin XH, Kashima Y, Khan M, et al. An immunohistochemical study of TNF-alpha in optic nerves from AIDS patients. Curr Eye Res. 1997;16:1064–1068. doi: 10.1076/ceyr.16.10.1064.9017. [DOI] [PubMed] [Google Scholar]
- 14.Madigan MC, Sadun AA, Rao NS, et al. Tumor necrosis factor-alpha (TNF-alpha)-induced optic neuropathy in rabbits. Neurol Res. 1996;18:176–184. doi: 10.1080/01616412.1996.11740399. [DOI] [PubMed] [Google Scholar]
- 15.Petrovich MS, Hsu HY, Gu X, et al. Pentoxifylline suppression of TNF-alpha mediated axonal degeneration in the rabbit optic nerve. Neurol Res. 1997;19:551–554. doi: 10.1080/01616412.1997.11740856. [DOI] [PubMed] [Google Scholar]
- 16.Saadati HG, Khan IA, Lin XH, et al. Immunolocalization of IL-1beta and IL-6 in optic nerves of patients with AIDS. Curr Eye Res. 1999;19:264–268. doi: 10.1076/ceyr.19.3.264.5319. [DOI] [PubMed] [Google Scholar]
- 17.Ho AS, Moore KW. Interleukin-10 and its receptor. Ther Immunol. 1994;1:173–185. [PubMed] [Google Scholar]
- 18.Moore KW, de Waal Malefyt R, Coffman RL, et al. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 19.Gasche C, Grundtner P, Zwirn P, et al. Novel variants of the IL-10 receptor 1 affect inhibition of monocyte TNF-alpha production. J Immunol. 2003;170:5578–5582. doi: 10.4049/jimmunol.170.11.5578. [DOI] [PubMed] [Google Scholar]
- 20.Grundtner P, Gruber S, Murray SS, et al. The IL-10R1 S138G loss-of-function allele and ulcerative colitis. Genes Immun. 2009;10:84–92. doi: 10.1038/gene.2008.72. [DOI] [PubMed] [Google Scholar]
- 21.Crawley E, Kay R, Sillibourne J, et al. Polymorphic haplotypes of the interleukin-10 5' flanking region determine variable interleukin-10 transcription and are associated with particular phenotypes of juvenile rheumatoid arthritis. Arthritis Rheum. 1999;42:1101–1108. doi: 10.1002/1529-0131(199906)42:6<1101::AID-ANR6>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 22.Mormann M, Rieth H, Hua TD, et al. Mosaics of gene variations in the Interleukin-10 gene promoter affect interleukin-10 production depending on the stimulation used. Genes Immun. 2004;5:246–255. doi: 10.1038/sj.gene.6364073. [DOI] [PubMed] [Google Scholar]
- 23.Shin HD, Winkler C, Stephens JC, et al. Genetic restriction of HIV-1 pathogenesis to AIDS by promoter alleles of IL10. Proc Natl Acad Sci U S A. 2000;97:14467–14472. doi: 10.1073/pnas.97.26.14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turner DM, Williams DM, Sankaran D, et al. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24:1–8. doi: 10.1111/j.1365-2370.1997.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 25.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 26.Martin MP, Dean M, Smith MW, et al. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science. 1998;282:1907–1911. doi: 10.1126/science.282.5395.1907. [DOI] [PubMed] [Google Scholar]
- 27.Liu H, Chao D, Nakayama EE, et al. Polymorphism in RANTES chemokine promoter affects HIV-1 disease progression. Proc Natl Acad Sci U S A. 1999;96:4581–4585. doi: 10.1073/pnas.96.8.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarwater PM, Mellors J, Gore ME, et al. Methods to assess population effectiveness of therapies in human immunodeficiency virus incident and prevalent cohorts. Am J Epidemiol. 2001;154:675–681. doi: 10.1093/aje/154.7.675. [DOI] [PubMed] [Google Scholar]
- 29.Gibson AW, Edberg JC, Wu J, et al. Novel single nucleotide polymorphisms in the distal IL-10 promoter affect IL-10 production and enhance the risk of systemic lupus erythematosus. J Immunol. 2001;166:3915–3922. doi: 10.4049/jimmunol.166.6.3915. [DOI] [PubMed] [Google Scholar]
- 30.Dumont FJ. Therapeutic potential of IL-10 and its viral homologues: an update. Expert Opinion on Therapeutic Patents. 2003;13:1551–1577. [Google Scholar]
- 31.Vasilescu A, Heath SC, Ivanova R, et al. Genomic analysis of Th1-Th2 cytokine genes in an AIDS cohort: identification of IL4 and IL10 haplotypes associated with the disease progression. Genes Immun. 2003;4:441–449. doi: 10.1038/sj.gene.6363983. [DOI] [PubMed] [Google Scholar]
- 32.Oleksyk TK, Shrestha S, Truelove AL, et al. Extended IL10 haplotypes and their association with HIV progression to AIDS. Genes Immun. 2009;10:309–322. doi: 10.1038/gene.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 34.Cocchi F, DeVico AL, Garzino-Demo A, et al. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 35.Gonzalez E, Dhanda R, Bamshad M, et al. Global survey of genetic variation in CCR5, RANTES, and MIP-1alpha: impact on the epidemiology of the HIV-1 pandemic. Proc Natl Acad Sci U S A. 2001;98:5199–5204. doi: 10.1073/pnas.091056898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDermott DH, Beecroft MJ, Kleeberger CA, et al. Chemokine RANTES promoter polymorphism affects risk of both HIV infection and disease progression in the Multicenter AIDS Cohort Study. AIDS. 2000;14:2671–2678. doi: 10.1097/00002030-200012010-00006. [DOI] [PubMed] [Google Scholar]
- 37.Trkola A, Gordon C, Matthews J, et al. The CC-chemokine RANTES increases the attachment of human immunodeficiency virus type 1 to target cells via glycosaminoglycans and also activates a signal transduction pathway that enhances viral infectivity. J Virol. 1999;73:6370–6379. doi: 10.1128/jvi.73.8.6370-6379.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia-Moruja C, Rueda P, Torres C, et al. Molecular phenotype of CXCL12beta 3'UTR G801A polymorphism (rs1801157) associated to HIV-1 disease progression. Curr HIV Res. 2009;7:384–389. doi: 10.2174/157016209788680543. [DOI] [PubMed] [Google Scholar]
- 39.Tiensiwakul P. Stromal cell-derived factor (SDF) 1-3'A polymorphism may play a role in resistance to HIV-1 infection in seronegative high-risk Thais. Intervirology. 2004;47:87–92. doi: 10.1159/000077831. [DOI] [PubMed] [Google Scholar]
- 40.Brambilla A, Villa C, Rizzardi G, et al. Shorter survival of SDF1-3'A/3'A homozygotes linked to CD4+ T cell decrease in advanced human immunodeficiency virus type 1 infection. J Infect Dis. 2000;182:311–315. doi: 10.1086/315650. [DOI] [PubMed] [Google Scholar]
- 41.Puissant B, Roubinet F, Massip P, et al. Analysis of CCR5, CCR2, CX3CR1, and SDF1 polymorphisms in HIV-positive treated patients: impact on response to HAART and on peripheral T lymphocyte counts. AIDS Res Hum Retroviruses. 2006;22:153–162. doi: 10.1089/aid.2006.22.153. [DOI] [PubMed] [Google Scholar]
- 42.Winkler C, Modi W, Smith MW, et al. Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. ALIVE Study, Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC) Science. 1998;279:389–393. doi: 10.1126/science.279.5349.389. [DOI] [PubMed] [Google Scholar]
- 43.Bogner JR, Lutz B, Klein HG, et al. Association of highly active antiretroviral therapy failure with chemokine receptor 5 wild type. HIV Med. 2004;5:264–272. doi: 10.1111/j.1468-1293.2004.00219.x. [DOI] [PubMed] [Google Scholar]
- 44.Wit FW, van Rij RP, Weverling GJ, et al. CC chemokine receptor 5 delta32 and CC chemokine receptor 2 64I polymorphisms do not influence the virologic and immunologic response to antiretroviral combination therapy in human immunodeficiency virus type 1-infected patients. J Infect Dis. 2002;186:1726–1732. doi: 10.1086/345677. [DOI] [PubMed] [Google Scholar]
- 45.Yamashita TE, Phair JP, Munoz A, et al. Immunologic and virologic response to highly active antiretroviral therapy in the Multicenter AIDS Cohort Study. AIDS. 2001;15:735–746. doi: 10.1097/00002030-200104130-00009. [DOI] [PubMed] [Google Scholar]
- 46.Lathey JL, Tierney C, Chang SY, et al. Associations of CCR5, CCR2, and stromal cell-derived factor 1 genotypes with human immunodeficiency virus disease progression in patients receiving nucleoside therapy. J Infect Dis. 2001;184:1402–1411. doi: 10.1086/324427. [DOI] [PubMed] [Google Scholar]
- 47.Scaravilli F, Bazille C, Gray F. Neuropathologic contributions to understanding AIDS and the central nervous system. Brain Pathol. 2007;17:197–208. doi: 10.1111/j.1750-3639.2007.00047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levy RM, Bredesen DE, Rosenblum ML. Neurological manifestations of the acquired immunodeficiency syndrome (AIDS): experience at UCSF and review of the literature. J Neurosurg. 1985;62:475–495. doi: 10.3171/jns.1985.62.4.0475. [DOI] [PubMed] [Google Scholar]
- 49.Tenhula WN, Xu SZ, Madigan MC, et al. Morphometric comparisons of optic nerve axon loss in acquired immunodeficiency syndrome. Am J Ophthalmol. 1992;113:14–20. doi: 10.1016/s0002-9394(14)75746-0. [DOI] [PubMed] [Google Scholar]
- 50.Kaul M, Lipton SA. Mechanisms of neuroimmunity and neurodegeneration associated with HIV-1 infection and AIDS. J Neuroimmune Pharmacol. 2006;1:138–151. doi: 10.1007/s11481-006-9011-9. [DOI] [PubMed] [Google Scholar]
- 51.Kaul M, Garden GA, Lipton SA. Pathways to neuronal injury and apoptosis in HIV-associated dementia. Nature. 2001;410:988–994. doi: 10.1038/35073667. [DOI] [PubMed] [Google Scholar]
- 52.Bruce-Keller AJ, Chauhan A, Dimayuga FO, et al. Synaptic transport of human immunodeficiency virus-Tat protein causes neurotoxicity and gliosis in rat brain. J Neurosci. 2003;23:8417–8422. doi: 10.1523/JNEUROSCI.23-23-08417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li W, Galey D, Mattson MP, et al. Molecular and cellular mechanisms of neuronal cell death in HIV dementia. Neurotox Res. 2005;8:119–134. doi: 10.1007/BF03033824. [DOI] [PubMed] [Google Scholar]
- 54.Asensio VC, Campbell IL. Chemokines in the CNS: plurifunctional mediators in diverse states. Trends Neurosci. 1999;22:504–512. doi: 10.1016/s0166-2236(99)01453-8. [DOI] [PubMed] [Google Scholar]
- 55.Miller RJ, Meucci O. AIDS and the brain: is there a chemokine connection? Trends Neurosci. 1999;22:471–479. doi: 10.1016/s0166-2236(99)01408-3. [DOI] [PubMed] [Google Scholar]
- 56.Hesselgesser J, Taub D, Baskar P, et al. Neuronal apoptosis induced by HIV-1 gp120 and the chemokine SDF-1 alpha is mediated by the chemokine receptor CXCR4. Curr Biol. 1998;8:595–598. doi: 10.1016/s0960-9822(98)70230-1. [DOI] [PubMed] [Google Scholar]
- 57.Kaul M, Lipton SA. Chemokines and activated macrophages in HIV gp120-induced neuronal apoptosis. Proc Natl Acad Sci U S A. 1999;96:8212–8216. doi: 10.1073/pnas.96.14.8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.