Summary
Cellular quiescence is a state characterized by decreased cell size and metabolic activity. Quiescence acts to reduce the resources, energy and space. Quiescence might also protect cells from accumulating metabolic damage that could result in malignancy. Recent studies have shown that cell quiescence is an actively maintained rather than a default state in the absence of signals. Quiescence factors represent potential tumor suppressor genes because alterations in their expression or function contribute to progression of malignancies. There is growing evidence that quiescence is under active transcriptional control. The regulation of cell proliferation involves dozens of extracellular signals and intracellular factors of various types. In the present review we will focus on the role of Tob, a member of the APRO family members in regulating cellular quiescence and inhibition of cellular proliferation.
Introduction
Cellular quiescence is a state characterized by decreased cell size and metabolic activity. Quiescence in naive lymphocytes acts to reduce the resources, energy and space, required to maintain a vast repertoire of T and B cells, only a small fraction of which will be clonally selected by antigen during the lifetime of the host. Quiescence might also protect cells from accumulating metabolic damage that could result in malignancy. Recent studies have shown that quiescence in lymphocytes is an actively maintained rather than a default state in the absence of signals. Quiescence factors represent potential tumor suppressor genes because alterations in their expression or function contribute to progression of lymphoid malignancies 1. There is growing evidence that quiescence is under active transcriptional control. DNA subtraction and DNA microarray experiments have shown that activation of T and B cells involves not only increased expression of genes promoting growth but suppression of a ‘quiescent’ gene expression program 2, 3. The significance of active regulation of the quiescent state is further supported by the fact that transcriptional control of cellular and organismal quiescence appears to be an evolutionarily conserved phenomenon. A single transcription factor [DAF-16 (Dauer-formation mutant 16)] is required for the nematode Caenorhabditis elegans to enter a quiescent developmental state known as dauer arrest 4. Cell quiescence is a state distinct from cell senescence, which defines a condition of cell cycle arrest beyond the restriction point. Arrest beyond the restriction point is characterized by expression of high levels of cyclins and cdk inhibitors, activation of mitogenic pathways, cell hypertrophy and eventually cell senescence 5, 6.
The regulation of cell proliferation involves dozens of extracellular signals and intracellular factors of various types. Here we will focus on the role of Tob, a member of the APRO family of proteins in regulating cellular quiescence and inhibition of cellular proliferation. Tob is a member of the Tob/BTG antiproliferative (APRO) protein family (Matsuda 2001-FEBS let). The APRO family includes TOB1, TOB2, BTG1, PC3/TIS21/BTG2, ANA/BTG3 and PC3/PC3K 7, 8. The members of this family have been isolated from various cell types, including malignant cells, Swiss 3T3 cells and PC12 cells. Specifically, the first member of the family PC3/TIS21/BTG2, was identified as induced by nerve growth factor during neuronal differentiation of rat PC12 cells 9. TIS21 is induced by the tumor promoter tetradecanoyl phorbol acetate in Swiss murine 3T3 cells 10. BTG1 was identified and cloned from B-cell chronic lymphocytic leukemia cells, near the breakpoint of a t(8;12) chromosomal translocation 11. Because of the sequence homology and the antiproliferative effects of PC3 and BTG1, it was hypothesized and proposed that these factors are members of a novel family of antiproliferative proteins 9, 11. Tob (transducer of ErbB2; later termed Tob1) was identified next in a breast cancer cell line 12. BTG3/ANA 13, Tob2 14 and BTG4 15 were identified later in the human genome. Orthologs of human APRO family of genes have been identified in several vertebrate and invertebrate genomes, suggesting a high degree of evolutionary conservation.
Structure and expression of APRO genes
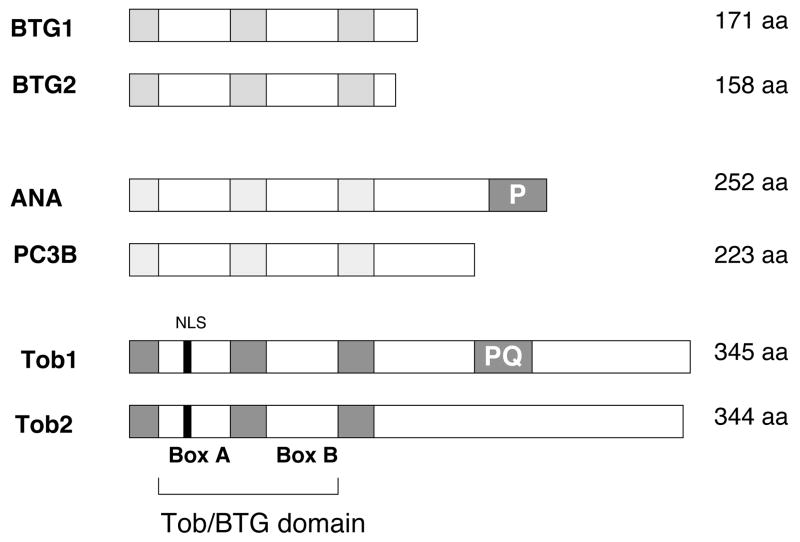
More than 20 members of the APRO family have been now isolated from several species from nematodes to humans with phylogenetically well-conserved homology 8. The APRO proteins have no homology to known functional motifs. However, a homologous domain at their N-terminal region, which has been termed the Tob/BTG domain, is conserved among all the members of this family. Based on the presence of this structural element it was determined that APRO proteins constitute a distinct gene family. Six members of the Tob/BTG family have been identified in humans. Based on their aminoacid sequence homology the six human Tob/BTG family proteins can be subgrouped into three classes (Figure 1). The Tob/BTG homology domain can be divided in two short, relatively more conserved elements, box A and box B, separated by a spacer sequence of 27 non-conserved amino acids 16. Among vertebrate proteins of the Tob subfamily, box A has a consensus sequence of KYEGHWYP(E/D)KP(Y/L)KGSG(F/Y)RC(I/V) and box B has a consensus sequence of (L/V)P(Q/E)(D/E)LSVWIDPFEVSYQIGE. Tob1 and Tob2 proteins have a highly conserved N-terminal region. In vertebrates, the sequence identity for the Tob N-terminal 117 amino acid region, which includes box A and box B domains, ranges from 70.1 to 100%. Amphioxus and Drosophila Tob proteins share a sequence identity of greater than 57.3% with vertebrate Tob1 and Tob2 in this region 17. Fog-3, a Tob orthologue identified in C. elegans 18, shares a sequence identity with other Tob proteins of only 22.2–29.9% in the N-terminal region, whereas its sequence homology with other APRO family members is even lower, suggesting that, during evolution, Fog-3 might have diverged earlier than the other APRO proteins from their common ancestor.
Figure 1.
Human Tob/BTG family members. Based on the presence of a conserved domain, the Tob/BTG domain, it was determined that Tob/BTG proteins constitute a distinct gene family. The Tob/BTG homology domain can be divided in two short, relatively more conserved elements, box A and box B, separated by a spacer sequence of non-conserved amino acids. Based on their aminoacid similarities the six human Tob/BTG family proteins can be sub-grouped into three classes. Tob and Tob2 also contain a predicted nuclear localization signal (NLS) within this region. Six human Tob/BTG family members have been identified.
Tob proteins are distributed throughout the cytoplasm and nucleus. Notably, subcellular distribution varies during cell cycle phases so that higher levels are detected in the cytoplasm during late S phase than during other phases of the cell cycle 19. The nuclear localization of TOB1 is mediated by a bipartite nuclear localization signal (NLS) that is conserved among all vertebrate proteins of the APRO family. NLS contains a consensus sequence of RRR-X12-KKK at amino acid positions 22–39 19. In human TOB1, in addition to NLS four nuclear export signals (NES)19 play significant role in subellular protein distribution 19, 20. Mutations in the TOB1 NLS result in preferential distribution of TOB1 protein to the cytoplasm, and significant reduction of its antiproliferative activity in NIH3T3 cells 19. Contrasting these observations, another other report indicated that addition of the potent SV40 NLS to the N-terminus of human TOB1 leads to exclusive nuclear accumulation of TOB1 and reduced antiproliferative activity 20. Although these data are obviously inconclusive, it seems that mechanisms of nuclear versus cytoplasmic localization of TOB proteins will have significant regulatory effects on their functions.
Post-transcriptional modifications and interactions of Tob proteins
There is compelling evidence that the APRO family proteins are involved in inhibition of cell growth in various cells including neuronal cells, fibroblasts, epithelial cells and T lymphocytes 7, 8. Consistent with such function, the levels of these proteins change dynamically during the cell cycle. In fibroblasts, TOB1 protein is detectable during the G0 phase, its level declines dramatically during the G1 and S phase and returns to base line during the G2 phase 21. In lymphocytes, TOB1 protein must be degraded rapidly during early G1 to ensure progression through late G1 phase. Forced, sustained expression of TOB1 prevents cell cycle progression mediated by antigenic stimulation in T cells. In contrast, elimination of endogenous TOB1 reduces the threshold of T cell activation and abrogates the requirement of costimulation for optimal cellular proliferation and clonal expansion 22. It seems that downregulation of TOB1 expression during cell cycle entry is mediated both at the mRNA and protein level 21-24. Resting, unstimulated T cells express high levels of Tob transcript, which is eliminated rapidly after activation. Consistent with downregulation of Tob at the protein level, treatment with proteasome inhibitors prevents the degradation of endogenous Tob1 protein in HeLa cells and inhibits the degradation of transiently expressed TOB1, TOB2, BTG1, and BTG2 in HEK293 cells 23.
TOB1 is subject to phosphorylation at multiple sites, specifically serine/threonine residues. Receptor tyrosine kinase-activated p90rsk1 can mediate phosphorylation of human TOB1 21. Ser152, Ser154, and Ser164 of human TOB1 and mouse Tob1 can be rapidly phosphorylated by ERK1, ERK2 and JNK2 upon growth-factor stimulation 20. Phosphorylation mediated by any of these kinases negatively regulates the antiproliferative activity of Tob proteins. Importantly, Ser152, Ser154, and Ser164 and neighboring residues of TOB1 are conserved in other vertebrate Tob1 and Tob2 orthologs and in amphioxus Tob, suggesting the importance of these conserved phosphorylation sites in regulating Tob function 20, 21, 24.
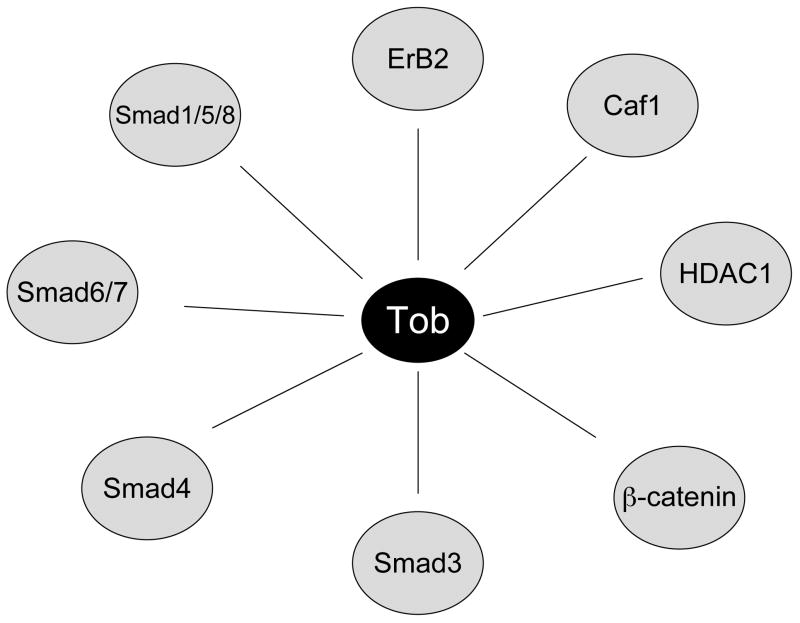
The N-terminal region of APRO family proteins is important for forming complexes with target proteins, and for exerting biological effects (Figure 2) 25-29. Specifically, the box B domain plays a particularly critical role in forming protein complexes and mediating biological activity 14. BTG1 and BTG2 associate with PRMT1, a protein arginine methyltransferase and it has been proposed that they may mediate anti-proliferative function via this interaction 30. TOB proteins can associate with Caf1 a homologue of the yeast Caf1/Pop2, a component of the CCR4 complex, the yeast homologue of which regulates cell growth 28, 29, 31, 32. In breast cancer cells, TOB1 can associate with ErbB-2, a kinase linked to epidermal growth factor receptor, which has been implicated in malignant cell growth and clinical trials with anti-ErbB-2 monoclonal antibody have shown efficiency in the treatment of metastatic breast cancer. The association of TOB with ErbB-2 suggests that it may potentially modify growth signals 12. Tob1 also interacts with Smads, which have DNA binding activity and regulate transcription of various genes resulting in either induction or inhibition of cell growth depended on the cell type and context 33, 34.
Figure 2.
Identified partners of Tob. Tob protein can interact with several components of various signaling pathways, thereby regulating their function.
A common feature among the APRO family proteins involves their ability to function as transcriptional regulators. Caf1 is a component of the CCR4 transcriptional regulatory complex. Among the members of this complex, a few like CCR4, Caf1 and DBF2 promote transcription whereas others, like the Not proteins, promote transcription but also function as transcriptional repressors 8, 14, 31, 35. Therefore, Tob proteins may regulate gene transcription both positively and negatively via their interaction with the CCR4 transcriptional complex. Caf1 has also been shown to belong to a deadenylase complex and regulate RNA stability 31. This effect of Caf1 may also have a functional role in the regulation of gene expression. BTG1 and BTG2 associate with HoxB9 and enhance HoxB9 DNA binding and HoxB9-dependent transcription 36. In an impressively similar fashion, in T lymphocytes Tob associates with Smad2 and Smad4 and enhances Smad4 DNA binding and Smad-dependent transcription 33. In contrast, in osteoblasts, Tob associates with Smads and enhances Smad DNA binding but inhibits Smad-mediated transcription 34. These data link Tob to the TGF-b family mediated signaling and regulation of transcription, which has a role in morphogenesis, as well as in cell survival, proliferation and differentiation.
TOB proteins are poly(A)-binding proteins and function as positive regulators of cytoplasmic mRNA deadenylation 37. Due to this property, in addition to regulating cytokine transcription in T cells, TOB proteins can act as positive regulators of cytoplasmic mRNA deadenylation. In mammalian cells, mRNA decay begins with deadenylation, which involves two consecutive phases mediated by the PAN2-PAN3 and the CCR4-CAF1 complexes. The regulation of the critical deadenylation step occurs at the RNA-processing bodies (P-bodies), which are thought to be a site where poly(A)-shortened mRNAs get degraded 38. These studies showed that TOB can simultaneously interact with the poly(A) nuclease complex CCR4-CAF1 and the cytoplasmic poly(A)-binding protein PABPC1 and colocalizes with P-bodies, suggesting a role of TOB in linking deadenylation to the P-bodies. Although the functional outcome and the biological significance of this observation currently remain unclear, this observation is a major advancement of our understanding on active mechanisms by which TOB proteins may exert their function.
Expression of Tob genes in developing and mature tissues
Although the APRO proteins are members of the same gene family, the regulation of their expression appears to be distinct. Murine TIS21 and TIS7, two of the first characterized members of the family, are induced by PMA in Swiss 3T3 cells and their rat homologues PC3 and PC4 are induced by nerve growth factor in PC12 pheochromocytoma cells 36, 39, 40. Similarly, in osteoblasts, Tob mRNA is not expressed in unstimulated cells but is induced only after BMP stimulation 34. In contrast, BTG1 is expressed in unstimulated, peripheral blood mononuclear cells and is downregulated by mitogenic stimulation with PHA. Similarly, Tob is expressed in high levels in resting T lymphocytes and is downregulated following mitogenic stimulation 11. Thus, although inhibition of cell cycle progression is a common functional feature of the APRO family members, these proteins appear to undergo distinct regulation of their expression in various cell types.
Expression patterns of Tob genes during early embryonic development have been studied in several species (reviewed in ref. 5). Transcripts of zebrafish tob1a 25 and tob1b 41 and Xenopus xTob2 27 are ubiquitously distributed during blastula and gastrula stages of development, suggesting a role in early embryogenesis in these species. At later stages of embryonic development, the expression of Tob genes occurs in distinct regions, such as the notochord, hatching gland, blood islands, and gut, depending on the species. During segmentation, Tob genes are expressed in somites, which give rise to axial skeleton, skeletal muscle, and dermis. In developing limbs of mouse embryos, Tob1 transcript displays significant expression in osteoblasts, in hypertrophic chondrocytes and low level expression in osteoclasts 34. Consistent with the hypothesis that Tob genes play roles in development of the somites and their derivatives, TOB1 protein is stably expressed in the proliferating basal layer of the epidermis and in primary human keratinocytes 42. In adults, mouse and human Tob1 and Tob2 and human TOB1and TOB2 are expressed in skeletal muscle 12, 14, 43, 44. Tob is expressed in the embryonic CNS of Drosophila and amphioxus Tob is expressed in the nerve cord during the late neurula and larva stages 17, 45. Notably, Tob transcripts are also present in adult brain tissues 12, 14, 43, 44, where they seem to serve important functions in learning and memory 46.
Tob family members are transcriptional regulators. As such it is expected that they may have a significant function during embryogenesis. Consistent with this hypothesis overexpression of tob1a in zebrafish embryos can cause embryonic ventralization, including loss of the head and the notochord, whereas translational inhibition of endogenous tob1a mRNA by an antisense morpholino results in dorsalized phenotypes, such as loss of the ventral fin and a shorter twisted tail 25. Further studies revealed that Tob1a can inhibit transcription mediated by b-catenin, a factor essential for dorsal development of amphibian and fish embryos 47. Tob1a acts upstream of b-catenin and competes with the Lef1/Tcf cofactors for binding to b-catenin, effectively blocking formation of a Lef1/Tcf–b-catenin protein complex that can stimulate the transcription of several genes 48. Tob1 interacts with Smad3 and prevents its binding to p300, which functions as a transcriptional cofactor, thereby inhibiting transcriptional activity of Smad3 downstream of TGF-b/Nodal signals 49. Xenopus Tob2 (xTob2) physically interacts with Smad6 and Smad7 and enhances their inhibitory activity on BMP-mediated transcription, thereby enhancing the ability of Smad6 to inhibit BMP signaling on the ventral side of embryos, and inhibiting ventral development 27. Thus Tob proteins have the ability to antagonize both BMP and catenin-mediated signaling.
Functions of the Tob members of the APRO family
a. Tob1 is involved in bone formation and metabolism
The role of Tob genes in bone formation and resorption has been well established. Tob1-deficient adult mice have a higher amount of bone mass than wild type control animals. This effect is due to increased numbers of osteoblasts and an accelerated rate of bone formation in the Tob1-deficient mice. Tob1 acts as a negative regulator of BMP2 signal during bone formation and mediates its effect by interacting with the BMP2 downstream regulators Smasd1, 5, 8 and 4 thereby promoting their nuclear translocation 34. The exact mechanism by which enhancement of nuclear translocation of Smads by Tob suppresses their transcriptional activity remains unclear. An additional mechanisms by which Tob1 regulates bone formation may involve inhibition of ligand dependent transcriptional activation induced by androgen or estrogen receptor due to the direct binding of Tob1 to the steroid hormone receptors 26. The regulatory role of Tob in bone formation and mineralization may provide new potentials for therapeutic interventions in metabolic diseases of the bone.
b. Tob1 is a regulator of T-cell activation and has a potential role as a biomarker for the clinical activity of autoimmune diseases in vivo
We identified Tob, unexpectedly, in a search for genes that are specifically expressed in quiescent or anergic CD4+ T lymphocytes. In contrast, T cell receptor engagement in the presence of either CD28 costimulation or IL-2 rapidly and dramatically downregulated the expression of Tob 33. These observations prompted us to investigate the role of Tob in regulating the activation status of T cells. When forced expression of Tob was introduced into peripheral blood T cells it was found that Tob, but not a control protein, inhibited anti-CD3+anti-CD28 induced proliferation. Tob also suppressed transcription of cytokines such as interleukin 2 (IL-2), IL-4 and interferon g (IFN-g) and positive regulators of the cell cycle such as cyclin E and cyclin A, thereby blocking T cell proliferation. In contrast, elimination of endogenous Tob reduced the threshold of T cell activation and rendered the cells capable of responding with maximal proliferation and cytokine production after TCR engagement in the absence of costimulation. These results indicate that Tob is an important player in actively maintaining the quiescent state of T lymphocytes and sets a threshold for activation.
In osteoblasts Tob associates Smad1, Smad5 and Smad8 all of which are positive signal transducers of bone morphogenetic protein (BMP), a member of transforming growth factor (TGF)-b superfamily 34. TGF-b has a critical role in inhibiting T cell proliferation and maintaining of the T cell quiescent state 50. This effect requires Smad expression and activation. TGF-b signal transduction is initiated by receptor phosphorylation of transcription factors Smad2 and Smad3. We observed that Tob interacted with Smads, preferentially Smad2 and Smad4 in T lymphocytes in vivo. In addition, Tob enhanced the ability of Smad4 to bind DNA-bearing Smad-binding sites in vitro and enhanced SMAD-mediated transcription. Thus Tob functionally interacts with Smads in regulating gene transcription. The promoter of IL-2, a cytokine that plays a critical role in T-cell activation, contains SMAD binding sites at the negative regulatory element suggesting that TOB1 may interfere with T cell activation and IL-2 production by enhancing a SMAD-mediated inhibitory effect on IL-2 transcription 33.
Because Tob is a transcriptional regulator we investigated gene expression pattern regulated by Tob. We used suppression subtractive hybridization and we identified Twisted Gastrulation (Tsg) as one of the genes suppressed by Tob expression 51. Tsg is an evolutionarily conserved, secreted, morphogenetic protein that interacts with Drosophila Decapentaplegic (Dpp) and its vertebrate orthologs BMP2/4. As a result, Tsg affects the binding of Dpp/BMP2/4 to their cellular receptors and subsequent downstream signaling and modulates Dpp- and BMP2/4-mediated morphogenetic effects in Drosophila and vertebrate embryos, respectively. Tsg is expressed by thymocytes and plays a role in regulating early thymocyte differentiation. We examined Tsg expression and function in peripheral blood human T cells and observed that Tsg mRNA was almost undetectable in unstimulated T cells and was highly upregulated after activation by TCR/CD3 and either CD28, IL-2 or PMA. Recombinant Tsg had no effect on proliferation of primary T cells during stimulation by TCR/CD3. In contrast, Tsg had a potent inhibitory effect on proliferation and cytokine production of primed alloreactive CD4+ cells stimulated by TCR/CD3. Surprisingly, Tsg did not affect phosphorylation of the BMP-specific Smad1, but induced phosphorylation of the TGF-b-specific Smad2 and mediated DNA binding on Smad3/4 consensus binding sites, suggesting that acted downstream of TGF-b. Consistently, TGF-b but not BMP inhibited proliferation of pre-activated alloreactive CD4+ T cells and Tsg enhanced this inhibitory effect. In vitro association assays using recombinant Tsg and TGF-b revealed a direct interaction of these proteins. These results indicate that Tsg functions as an agonist of TGF-b downstream signaling in pre-activated mature human CD4+ T cells. Thus, similarly to the positive regulatory role of Tob in TGF-b mediated signaling in resting cells, Tsg appears to enhance TGF-b signaling in activated cells, when Tob expression is downregulated.
TOB1 also interacts with PABP and iPABP, an inducible poly(A) binding protein (iPABP/PABPC4) which binds to the poly(A) tail of eukaryotic mRNAs and promotes mRNA translation 52. Expression of iPABP is low in resting human T cells and is rapidly elevated upon T-cell activation 53. When iPABP, Tob and IL-2 genes were artificially co-expressed in mouse fibroblasts, binding of TOB1 to iPABP inhibited promotion of IL-2 mRNA translation by iPABP 52. These data suggest that in addition to suppressing IL-2 gene transcription, Tob may also diminish IL-2 production at the translation level.
The above studies provide molecular explanations and identify mechanisms that might underlie the functional outcome of Tob mediated interactions in T cell immunity, using experimental systems. A recent report provided a significant clinical insight to the role of Tob in regulating autoimmune responses in vivo 54. By using material from a group of patients with multiple sclerosis, this study provided evidence that Tob might represent a clinically useful biomarker for the identification of patients at high risk for progression to clinically definitive sclerosis after the initial clinically isolated neurological syndrome. Using microarrays to study gene expression in naïve CD4+ T cells from 37 patients with clinically isolated neurological syndrome at the time of diagnosis and after one year, these researchers determined that 92% of patients who displayed suppressed TOB1 expression converted to clinically definitive sclerosis within one year. Moreover, a genetic association was observed between Tob1 variation and MS progression in an independent cohort of patients. This report provided the most striking evidence for the clinical significance regarding the role of Tob in regulating T cell activation and immune quiescence in vivo.
c. Tob1 regulates malignant transformation and tumor development
Tob proteins have antiproliferative potential, suggesting that they play important roles in suppressing tumor development. Yoshida et al. found that, at 18 months of age, 77% of Tob1−/− mice had spontaneously developed a variety of tumors, including hemangiosarcomas, lung carcinomas, and hepatocellular adenomas, whereas only 16% of wild-type mice at the same age had developed tumors: primarily malignant lymphomas and lung adenomas 55. After treatment with diethylnitrosamine, a liver-specific carcinogen, the rate of liver tumor formation was significantly higher in Tob1−/− mice than in wild-type mice. These results indicate that mice lacking Tob1 are much more susceptible to cancer than wild-type mice are. In humans, TOB1 is expressed at lower levels in lung cancer tissue than in adjacent lung tissues 55, 56. In lung cancer tissue TOB was present in the phosphorylated form, which is inactive, whereas this inactive form of TOB1 is rare in the adjacent lung tissues. Similar results have been obtained for thyroid cancer: high levels of phosphorylated TOB1 occur in papillary carcinoma, and TOB1 protein is not detected in anaplastic carcinomas 57. In squamous cell carcinoma, an invasive carcinoma of the surface epidermis, TOB1 protein levels are lower than in normal epidermis 42. The association of certain types of human cancers with decreased expression or inactivation of TOB1 suggests that it may be, an important tumor suppressor. However, the mechanism of this effect remains elusive.
TOB1 was first identified as a binding partner of the c-erbB-2 gene product p185erbB2; kinase-active p185erbB2 alleviates TOB1-mediated suppression of cell proliferation 12. That observation suggested that TOB1 might inhibit cell proliferation through intervention in oncogenic pathways. Similarly to BTG proteins 28, 29, TOB1 and TOB2 can also interact with human Caf1 14. The yeast Caf1 homolog (CCR4), with other cofactors, can form transcriptional complexes that activate or suppress target gene transcription. CAF1 directly interacts with cyclin dependent kinases (CDKs) cyclin E, cyclin A, cyclin B, and cyclin D1 14, 58. Thus, it is possible that Tob proteins regulate cell proliferation by modulating the activities of certain CDKs through their interaction with CAF1 14. Interestingly, Tob-1–deficient cells have increased levels of cyclin D1 expression. Moreover, Tob1 inhibits cyclin D1 transcription promoter activity by interacting with and recruiting histone deacetylase 1 (HDAC1) to the cyclin D1 promoter 27. Notably, in zebrafish embryos Tob controls dorsal development by interacting with beta-catenin and antagonizing beta-catenin nuclear translocation and transcriptional activity 25. Thus, suppression of beta-catenin-regulated transcription that induces expression of proliferative genes including Myc and Cyclin D1, may be a mechanism mediating the anti-proliferative function of Tob in other cell types. Consistent with this hypothesis, the Wnt signaling pathway that prevents beta-catenin degradation and promotes beta-catenin-mediated transcription has important roles in lymphogenesis, hematopoiesis and regulation of transformation events, known to contribute to the pathogenesis of various malignancies 59.
Conclusions and perspectives
Over the past ten years, compelling research work from various laboratories using distinct experimental systems has provided evidence that Tob proteins function as important negative regulators of the cell cycle. Despite the multiple interacting partners of Tob/BTG (APRO) family members that have been identified, currently, the precise mechanism regulating this anti-proliferative effect has not been fully elucidated. Tob proteins associate with positive regulators of the cell cycle and this interaction leads to blockade of cell cycle progression. They also interact with transcription factors altering the ultimate outcome of DNA binding as they function either as transcriptional repressors or transcriptional enhancers. The Tob members of the APRO family regulate development and replication of various cell types, including lymphocytes, bone, neuronal cells and epithelial cells. They are also involved in regulating malignant transformation and replicating properties of various cancers. Thus, identification of mechanisms that control expression of the APRO/Tob genes may help identify which extracellular factors maintain lymphocyte quiescent and homeostasis but may also provide understanding of mechanisms regulating malignant transformation and tumor development.
Acknowledgments
Supported by NIH grants: AI43552, CA104596, CA123855, HL087870
References
- 1.Yusuf I, Fruman DA. Regulation of quiescence in lymphocytes. Trends Immunol. 2003;24:380–6. doi: 10.1016/s1471-4906(03)00141-8. [DOI] [PubMed] [Google Scholar]
- 2.Glynne R, Ghandour G, Rayner J, Mack DH, Goodnow CC. B-lymphocyte quiescence, tolerance and activation as viewed by global gene expression profiling on microarrays. Immunol Rev. 2000;176:216–46. doi: 10.1034/j.1600-065x.2000.00614.x. [DOI] [PubMed] [Google Scholar]
- 3.Feske S, Giltnane J, Dolmetsch R, Staudt LM, Rao A. Gene regulation mediated by calcium signals in T lymphocytes. Nat Immunol. 2001;2:316–24. doi: 10.1038/86318. [DOI] [PubMed] [Google Scholar]
- 4.Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans Nature. 1997;389:994–9. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 5.Demidenko ZN, Blagosklonny MV. Growth stimulation leads to cellular senescence when the cell cycle is blocked. Cell Cycle. 2008;7:3355–61. doi: 10.4161/cc.7.21.6919. [DOI] [PubMed] [Google Scholar]
- 6.Blagosklonny MV. Cell senescence: hypertrophic arrest beyond the restriction point. J Cell Physiol. 2006;209:592–7. doi: 10.1002/jcp.20750. [DOI] [PubMed] [Google Scholar]
- 7.Jia S, Meng A. Tob genes in development and homeostasis. Dev Dyn. 2007;236:913–21. doi: 10.1002/dvdy.21092. [DOI] [PubMed] [Google Scholar]
- 8.Matsuda S, Rouault J-P, Magaurd J-P, Berther C. In search of a function for the TIS21/PC3/BTG1/TOB family. FEBS lett. 2001;497:67–72. doi: 10.1016/s0014-5793(01)02436-x. [DOI] [PubMed] [Google Scholar]
- 9.Bradbury A, Possenti R, Shooter R, Tirone F. Molecular cloning of PC3, a putatively secreted protein whose mRNA is induced by nerve growth factor and depolarization. Proc Natl Acad Sci. 1991;88:3353–7. doi: 10.1073/pnas.88.8.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fletcher BS, Lim RW, Varum BC, Kujubu DA, Koski RA, Herschman HR. Structure and expression of TIS21, a primary response gene induced by growth factors and tumor promoters. J Biol Chem. 1991;266:14511–8. [PubMed] [Google Scholar]
- 11.Rouault J-P, Rimokh R, Tessa C, Paranhos G, Ffrench M, Duret L, et al. BTG1, a member of a new family of antiproliferative genes. EMBO J. 1992;11:1663–70. doi: 10.1002/j.1460-2075.1992.tb05213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuda S, Kawamura-Tsuzuku J, Ohsugi M, Yoshida M, Emi M, Nakamura Y, et al. Tob, a novel protein that interacts with p185ebrB2, is associated with anti-proliferative activity. Oncogene. 1996;12:705–13. [PubMed] [Google Scholar]
- 13.Yoshida Y, Matsuda S, Ikematsu N, Kawamura-Tsuzuku J, Inazawa J, Umemori H, et al. ANA, a novel member of Tob/BTG1 family, is expressed in the ventricular zone of the developing central nervous system. Oncogene. 1998;16:2687–93. doi: 10.1038/sj.onc.1201805. [DOI] [PubMed] [Google Scholar]
- 14.Ikematsu N, Yoshida Y, Kawamura-Tsuzuku J, Ohsugi M, Onda M, Hirai M, et al. Tob2, a novel anti-proliferative Tob/BTG1 family member, associates with a component of the CCR4 transcriptional regulatory complex capable of binding cyclin-dependent kinases. Oncogene. 1999;18:7432–41. doi: 10.1038/sj.onc.1203193. [DOI] [PubMed] [Google Scholar]
- 15.Buanne P, Corrente G, Micheli L, Palena A, lavia, Spadafora C, et al. Cloning of PC3B, a novel member of the PC3/BTG/Tob family of growth inhibitory genes, highly expressed in the olfractory epithlium. Genomics. 2000;68:253–63. doi: 10.1006/geno.2000.6288. [DOI] [PubMed] [Google Scholar]
- 16.Guehenneux F, Duret L, Callanan MB, Bouhas R, Hayette S, Berthet C, et al. Cloning of the mouse BTG3 gene and definition of a new gene family (the BTG family) involved in the negative control of the cell cycle. Leukemia. 1997;11:370–5. doi: 10.1038/sj.leu.2400599. [DOI] [PubMed] [Google Scholar]
- 17.Bourbon HM, Gonzy-Treboul G, Peronnet F, Alin MF, Ardourel C, Benassayag C, et al. A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech Dev. 2002;110:71–83. doi: 10.1016/s0925-4773(01)00566-4. [DOI] [PubMed] [Google Scholar]
- 18.Chen PJ, Singal A, Kimble J, Ellis RE. A novel member of the tob family of proteins controls sexual fate in Caenorhabditis elegans germ cells. Dev Biol. 2000;217:77–90. doi: 10.1006/dbio.1999.9521. [DOI] [PubMed] [Google Scholar]
- 19.Kawamura-Tsuzuku J, Suzuki T, Yoshida Y, Yamamoto T. Nuclear localization of Tob is important for regulation of its antiproliferative activity. Oncogene. 2004;23:6630–8. doi: 10.1038/sj.onc.1207890. [DOI] [PubMed] [Google Scholar]
- 20.Maekawa M, Yamamoto T, Nishida E. Regulation of subcellular localization of the antiproliferative protein Tob by its nuclear export signal and bipartite nuclear localization signal sequences. Exp Cell Res. 2004;295:59–65. doi: 10.1016/j.yexcr.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki T, Tsuzuku J-K, Ajima R, Nakamura T, Yoshida Y, Yamamoto T. Phosphorylation of three regulatory serines of Tob by Erk1 and Erk2 is required for Ras-mediated cell proliferation and transformation. Genes & Dev. 2002;16:1357–70. doi: 10.1101/gad.962802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tzachanis D, Lafuente EM, Boussiotis VA. Tob is a negative regulator of T cell activation and cytokine transcription. Mod Asp Immunol. 2002;2:117–20. [Google Scholar]
- 23.Sasajima H, Nakagawa K, Yokosawa H. Antiproliferative proteins of the BTG/Tob family are degraded by the ubiquitin-proteasome system. Eur J Biochem. 2002;269:3596–604. doi: 10.1046/j.1432-1033.2002.03052.x. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki T, Matsuda S, Tsuzuku JK, Yoshida Y, Yamamoto T. A serine/threonine kinase p90rsk1 phosphorylates the anti-proliferative protein Tob. Genes Cells. 2001;6:131–8. doi: 10.1046/j.1365-2443.2001.00406.x. [DOI] [PubMed] [Google Scholar]
- 25.Xiong B, Rui Y, Zhang M, Shi K, Jia S, Tian T, et al. Tob1 controls dorsal development of zebrafish embryos by antagonizing maternal beta-catenin transcriptional activity. Dev Cell. 2006;11:225–38. doi: 10.1016/j.devcel.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 26.Kawate H, Wu Y, Ohnaka K, Nawata H, Takayanagi R. Tob proteins suppress steroid hormone receptor-mediated transcriptional activation. Mol Cell Endocrinol. 2005;230:77–86. doi: 10.1016/j.mce.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida Y, von Bubnoff A, Ikematsu N, Blitz IL, Tsuzuku JK, Yoshida EH, et al. Tob proteins enhance inhibitory Smad-receptor interactions to repress BMP signaling. Mech Dev. 2003;120:629–37. doi: 10.1016/s0925-4773(03)00020-0. [DOI] [PubMed] [Google Scholar]
- 28.Prevot D, Morel A-P, Voeltzel T, Rostan M-C, Rimokh R, Magaud J-P, et al. Relationships of the antiproliferative proteins BTG1 and BTG2 with CAF1, the human homolog of a component of the yeast CCR4 transcripitonal complex. J Biol Chem. 2001;276:9640–8. doi: 10.1074/jbc.M008201200. [DOI] [PubMed] [Google Scholar]
- 29.Rouault J-P, Prevot D, Berthet C, Birot AM, Billaud M, Magaud JP, et al. Interaction of BTG1 and p53-regulated BTG2 gene products with mCaf1, the murine homolog of a component of the yeast CCR4 transcriptional regulatory complex. J Biol Chem. 1998;273:22563–9. doi: 10.1074/jbc.273.35.22563. [DOI] [PubMed] [Google Scholar]
- 30.Tang J, Fankel A, Cook RJ, Kim S, Paik WK, Williams KR, et al. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J Biol Chem. 2000;275:7723–30. doi: 10.1074/jbc.275.11.7723. [DOI] [PubMed] [Google Scholar]
- 31.Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Sacharomyces cerevisiae. Cell. 2001;104:377–86. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 32.Yoshida Y, Hosoda E, Nakamura T, Yamamoto T. Association of ANA, a member of the antiproliferative Tob family proteins, with a Caf1 component of the CCD4 transcriptional regulatory complex. Jpn J Cancer Res. 2001;92:592–6. doi: 10.1111/j.1349-7006.2001.tb01135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tzachanis D, Freeman GJ, Hirano N, van Puijenbroek AAFL, Delfs MW, Berezovskaya A, et al. Tob is a negative regulator of activation that is expressed in anergic and quiescent T cells. Nature Immunol. 2001;2:1174–82. doi: 10.1038/ni730. [DOI] [PubMed] [Google Scholar]
- 34.Yoshida Y, Tanaka S, Umemori H, Minowa O, Usui M, Ikematsu N, et al. Negative regulation of BMP/Smad signaling by Tob in osteoblasts. Cell. 2000;103:1085–97. doi: 10.1016/s0092-8674(00)00211-7. [DOI] [PubMed] [Google Scholar]
- 35.Hy L, JHT, Chiang YC, Draper MP, Jonstoin LH, Denis CL. DBF2, a cell cycle-regulated protein kinase, is physically and functionally associated with the CCR4 transcriptional regulatory complex. EMBO J. 1997;16:5289–98. doi: 10.1093/emboj/16.17.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prevot D, Voeltzel T, Bitor A-M, Morel A-P, Rostan M-C, Magaud J-P, et al. The leukemia-associated protein Btg1 and the p53-regulated protein Btg2 interact with the homeoprotein Hoxb9 and enhance its transcriptional activation. J Biol Chem. 2000;275:147–53. doi: 10.1074/jbc.275.1.147. [DOI] [PubMed] [Google Scholar]
- 37.Ezzeddine N, Chang TC, Zhu W, Yamashita A, Chen CY, Zhong Z, et al. Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol Cell Biol. 2007;27:7791–801. doi: 10.1128/MCB.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng D, Ezzeddine N, Chen CY, Zhu W, He X, Shyu AB. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol. 2008;182:89–101. doi: 10.1083/jcb.200801196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varnum BC, Reddy ST, Koski RA, Herschman HR. Synthesis, degradation and subcellular localization of protein encoded by the primary response genes TIS7/PC4 and TIS21/PC3. J Cell Physiol. 1994;158:205–13. doi: 10.1002/jcp.1041580125. [DOI] [PubMed] [Google Scholar]
- 40.Rouault J-P, Falette N, Guehenneux F, Guillot C, Rimokh R, Wang Q, et al. Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nature Gen. 1996;14:482–6. doi: 10.1038/ng1296-482. [DOI] [PubMed] [Google Scholar]
- 41.Shi K, Zhang L, Meng A. Cloning and expression analysis of zebrafish tob1 gene. Dev Genes Evol. 2004;214:309–11. doi: 10.1007/s00427-004-0405-5. [DOI] [PubMed] [Google Scholar]
- 42.Park GT, Seo EY, Lee KM, Lee DY, Yang JM. Tob is a potential marker gene for the basal layer of the epidermis and is stably expressed in human primary keratinocytes. Br J Dermatol. 2006;154:411–8. doi: 10.1111/j.1365-2133.2005.07037.x. [DOI] [PubMed] [Google Scholar]
- 43.Ajima R, Ikematsu N, Ohsugi M, Yoshida Y, Yamamoto T. Cloning and characterization of the mouse tob2 gene. Gene. 2000;253:215–20. doi: 10.1016/s0378-1119(00)00270-5. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida Y, Matsuda S, Yamamoto T. Cloning and characterization of the mouse tob gene. Gene. 1997;191:109–13. doi: 10.1016/s0378-1119(97)00049-8. [DOI] [PubMed] [Google Scholar]
- 45.Holland ND, Zhang SC, Clark M, Panopoulou G, Lehrach H, Holland LZ. Sequence and developmental expression of AmphiTob, an amphioxus homolog of vertebrate Tob in the PC3/BTG1/Tob family of tumor suppressor genes. Dev Dyn. 1997;210:11–8. doi: 10.1002/(SICI)1097-0177(199709)210:1<11::AID-AJA2>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 46.Jin M, Wang XM, Tu Y, Zhang XH, Gao X, Guo N, et al. The negative cell cycle regulator, Tob (transducer of ErbB-2), is a multifunctional protein involved in hippocampus-dependent learning and memory. Neuroscience. 2005;131:647–59. doi: 10.1016/j.neuroscience.2004.10.044. [DOI] [PubMed] [Google Scholar]
- 47.De Robertis EM, Kuroda H. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu Rev Cell Dev Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bellipanni G, Varga M, Maegawa S, Imai Y, Kelly C, Myers AP, et al. Essential and opposing roles of zebrafish beta-catenins in the formation of dorsal axial structures and neurectoderm. Development. 2006;133:1299–309. doi: 10.1242/dev.02295. [DOI] [PubMed] [Google Scholar]
- 49.Tian T, Meng AM. Nodal signals pattern vertebrate embryos. Cell Mol Life Sci. 2006;63:672–85. doi: 10.1007/s00018-005-5503-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 51.Tzachanis D, Li L, Lafuente EM, Berezovskaya A, Freeman GJ, Boussiotis VA. Twisted gastrulation (Tsg) is regulated by Tob and enhances TGF-beta signaling in activated T lymphocytes. Blood. 2007;109:2944–52. doi: 10.1182/blood-2006-03-006510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okochi K, Suzuki T, Inoue J, Matsuda S, Yamamoto T. Interaction of anti-proliferative protein Tob with poly(A)-binding protein and inducible poly(A)-binding protein: implication of Tob in translational control. Genes Cells. 2005;10:151–63. doi: 10.1111/j.1365-2443.2005.00826.x. [DOI] [PubMed] [Google Scholar]
- 53.Yang H, Duckett CS, Lindsten T. iPABP, an inducible poly(A)-binding protein detected in activated human T cells. Mol Cell Biol. 1995;15:6770–6. doi: 10.1128/mcb.15.12.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corvol JC, Pelletier D, Henry RG, Caillier SJ, Wang J, Pappas D, et al. Abrogation of T cell quiescence characterizes patients at high risk for multiple sclerosis after the initial neurological event. Proc Natl Acad Sci U S A. 2008;105:11839–44. doi: 10.1073/pnas.0805065105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshida Y, Nakamura T, Komoda M, Satoh H, Suzuki T, Tsuzuku JK, et al. Mice lacking a transcriptional corepressor Tob are predisposed to cancer. Genes Dev. 2003;17:1201–6. doi: 10.1101/gad.1088003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iwanaga K, Sueoka N, Sato A, Sakuragi T, Sakao Y, Tominaga M, et al. Alteration of expression or phosphorylation status of tob, a novel tumor suppressor gene product, is an early event in lung cancer. Cancer Lett. 2003;202:71–9. doi: 10.1016/j.canlet.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 57.Ito Y, Suzuki T, Yoshida H, Tomoda C, Uruno T, Takamura Y, et al. Phosphorylation and inactivation of Tob contributes to the progression of papillary carcinoma of the thyroid. Cancer Lett. 2005;220:237–42. doi: 10.1016/j.canlet.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 58.Bogdan JA, Adams-Burton C, Pedicord DL, Sukovich DA, Benfield PA, Corjay MH, et al. Human carbon catabolite repressor protein (CCR4)-associative factor 1: cloning, expression and characterization of its interactions with B-cell tranlocation protein BTG1. Biochem J. 1998;336:471–81. doi: 10.1042/bj3360471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–98. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]