Abstract
Pharmacologically-motivated marine natural product investigations have yielded a large variety of structurally unique compounds with interesting biomedical properties, but the natural roles of these molecules often remain unknown. While secondary metabolites may function as antimicrobial chemical defenses, few studies have examined this hypothesis. In the present investigation, chromatographic fractions from 69 collections of Fijian red macroalgae representing at least 43 species were evaluated for growth inhibition of three microbial pathogens and saprophytes of marine macrophytes. At least one microbe was suppressed by fraction(s) of all evaluated algae, suggesting that antimicrobial defenses are common among tropical seaweeds. From these leads, peyssonoic acids A–B (1–2), novel sesquiterpene hydroquinones, were isolated from the crustose red alga Peyssonnelia sp. At ecologically realistic concentrations, both compounds inhibited growth of Pseudoalteromonas bacteriolytica, a bacterial pathogen of marine algae, and Lindra thalassiae, a fungal pathogen of marine algae, and exhibited modest antineoplastic activity against ovarian cancer cells. The peyssonoic acids included one novel carbon skeleton and illustrated the utility of ecological studies in natural product discovery.
Keywords: chemical ecology, chemical defense, antimicrobial, sesquiterpene, marine natural product, macroalga, Peyssonnelia
1. Introduction
Marine organisms including invertebrates, microbes, and seaweeds are widely recognized sources of structurally novel secondary metabolites.1,2 These natural products have provided promising drug leads, offered targets for synthetic organic chemists, and afforded opportunities for elucidation of unusual biosynthetic pathways. Secondary metabolite pathways probably evolved as a result of complex interactions between organisms in their native habitats, but the role of natural products in mediating such interactions remains poorly understood in the majority of cases.
Secondary metabolites may play a particularly important role in mediating marine host-microbe interactions in the ocean.3,4 While most microbes may be innocuous or beneficial to hosts, reports of disease outbreaks in a variety of marine invertebrates and seaweeds suggest the negative impact of some microbes on coral reef health and thus the potential for selection to resist pathogenic microbes.5,6 Among marine plants, coralline lethal orange disease devastated susceptible South Pacific coralline algal populations during the 1990s,7 red spot disease has impacted commercially valuable kelp populations,8 a slime mold wasting epidemic destroyed nearly all Zosteria marina eelgrass in the North Atlantic during the 1930s,9 and the pathogenic fungus Lindra thalassiae has been reported to cause raisin disease in the brown algae Sargassum spp. as well as disease in seagrasses.10,11 Microbial pathogens not only affect susceptible populations, but can also disturb the structure and function of entire marine communities.5
Disease outbreaks in marine plants appear sporadic and pathogens may target specific hosts.5 One possible explanation for this limited disease prevalence is that secondary metabolites defend some species against microbial attack,4 but only a handful of previous studies investigated this possibility and even fewer identified specific defensive metabolites from marine plants. Two previous surveys provided evidence for antimicrobial chemical defenses among these organisms.12,13 Known antimicrobial defenses among marine plants include six secondary metabolite classes: (1) halogenated furanones from the red alga Delisea pulchra impede colonization by a variety of genera of marine bacteria,14 (2) a poly-brominated 2-heptanone from the red alga Bonnemaisonia hamifera inhibits growth of co-occurring marine bacterial strains,15 (3) a macrocyclic polyketide from the brown alga Lobophora variegata inhibits growth of a pathogenic fungus,16 (4) a flavone glycoside from the seagrass Thalassia testudinum is growth-inhibitory toward a zoosporic fungus,17 (5) sulfated triterpenes from Penicillus capitatus and Tydemania expeditionis are effective antifungal defenses at ecologically realistic concentrations,18,19 and (6) diterpene-shikimate natural products from the red alga Callophycus serratus found at heterogeneous sites on algal surfaces defend this alga against a pathogenic marine fungus.20
Herein, we evaluate antimicrobial chemical defenses for 69 collections of Fijian red macroalgae, providing evidence that chemical defenses span a wide range of polarities and suggesting these defenses are not broad-spectrum but instead active against specific microbes. Although not commonly investigated as sources of novel natural products, members of the algal genus Peyssonnelia exhibited particularly strong antimicrobial activities in ecological assays. Bioassay-guided fractionation of extracts from Peyssonnelia sp. resulted in the discovery of peyssonoic acids A–B (1–2), growth inhibitors of both a bacterial and fungal pathogen of marine algae. The peyssonoic acids include a novel carbon connectivity pattern and illustrate the potential of ecologically-motivated studies in the discovery of novel chemistry.
2. Results and Discussion
2.1 Survey of 69 Fijian red macroalgae reveals antimicrobial chemical defenses are common
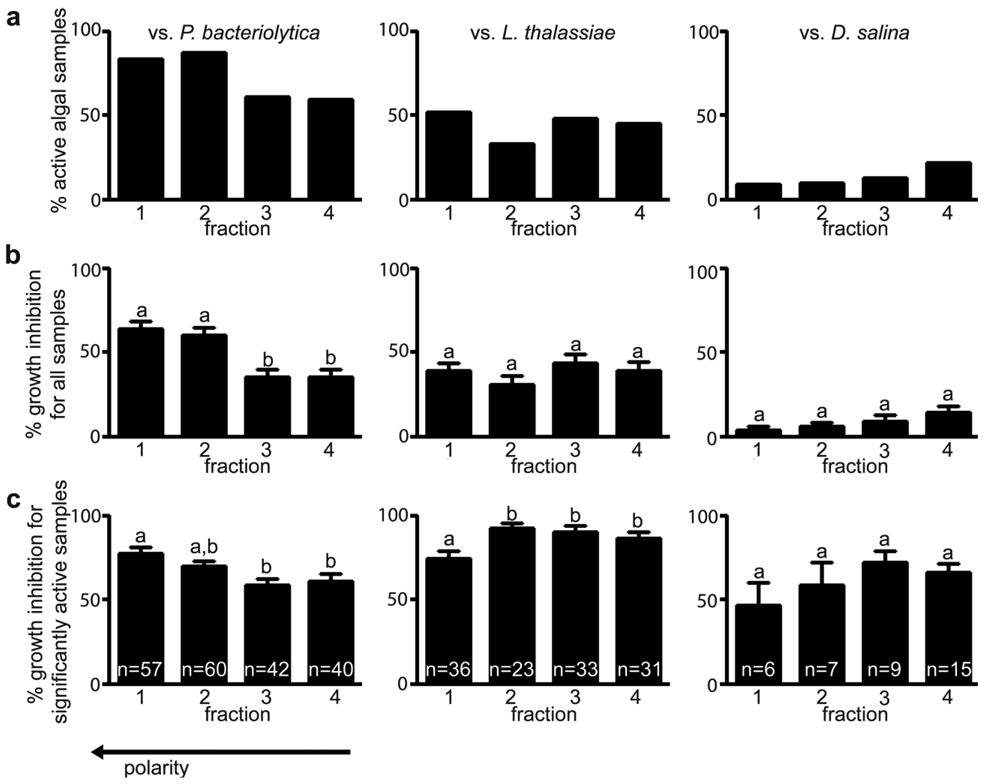
To determine the frequency of antimicrobial chemical defenses among tropical red macroalgae as well as better understand properties of these defenses, Fijian seaweeds were extracted and these extracts separated into four chromatographic fractions using reversed phase liquid chromatography. Among 69 collections of red macroalgae, including at least 43 distinct species (Supplemental Data), antimicrobial chemical defenses were prevalent against the bacterial pathogen Pseudoalteromonas bacteriolytica, known to cause red spot disease in kelp,8 and the fungal pathogen Lindra thalassiae, reported to infect phylogenetically distant hosts including Sargassum spp. brown algae and seagrasses.10 Both of these marine pathogens were inhibited by chromatographic fractions ranging from polar to lipophilic (Fig. 1a), suggesting that a variety of classes of secondary metabolites are employed as antimicrobial chemical defenses.
Fig. 1.
Antimicrobial activities of four chromatographic fractions prepared from extracts of 69 collections of Fijian red macroalgae against pathogenic bacterium Pseudoalteromonas bacteriolytica, pathogenic fungus Lindra thalassiae, and saprophytic fungus Dendryphiella salina. (a) Frequency of significant antimicrobial activity for fractions at natural whole tissue concentrations (n = 69 algal collections). (b) Comparison of inhibitory potency among all fractions evaluated at natural whole tissue concentrations. Different letters indicate fractions differing significantly in antimicrobial activity (p < 0.05, n = 69 algal collections; one-way ANOVA with Bonferroni multiple comparisons test; bars denote one standard error). (c) Comparison of inhibitory potency among significantly active algal collection fractions evaluated at natural whole tissue concentrations. Different letters indicate fractions differing significantly in antimicrobial activity (p < 0.05; one-way ANOVA with Bonferroni multiple comparisons test; bars denote one standard error); n represents the number of significantly active algal sample fractions compared.
At least one chromatographic fraction from all 69 seaweed samples significantly inhibited growth of one or more evaluated microbes, suggesting that red algae have been selected to deter deleterious microbes via chemical defenses. This high prevalence of defense corresponds with findings reported by Engel et al. 12 and Puglisi et al. 13 in surveys of antimicrobial defenses among tropical red, green, and brown algae. In the present study, for 99% of the 69 algal collections, at least one chromatographic fraction at natural whole tissue concentration was significantly inhibitory toward growth of pathogenic P. bacteriolytica; for 74% of collections, at least one fraction was active against pathogenic L. thalassiae. Dendryphiella salina, a saprophytic marine fungus, was more resistant to algal chemical defenses, with only 30% of collections exhibiting at least one fraction with significant activity against this saprophyte.
The most polar fractions were significantly more inhibitory toward P. bacteriolytica than less polar fractions (p < 0.05, n = 69; one-way ANOVA with Bonferroni’s multiple comparison test; Fig. 1b), when both active and inactive fractions were considered. Given this high frequency of antibacterial activity among polar chromatographic fractions, these samples should be the focus of future studies. As most known natural products from red macroalgae are lipophilic, it is likely these fractions contain novel antibacterial metabolites. In contrast, polar and nonpolar fractions did not differ significantly in their ability to suppress growth of the fungi L. thalassiae or D. salina (p > 0.05, n = 69; one-way ANOVA with Bonferroni’s multiple comparison test; Fig. 1b). However, when only significantly inhibitory fractions were evaluated, less polar fractions were significantly more potent than more polar fractions against L. thalassiae (p < 0.05, one-way ANOVA with Bonferroni’s multiple comparison test; Fig. 1c). The reverse was true for fractions inhibitory toward P. bacteriolytica (Fig. 1c), suggesting that different polar classes of compounds defend against this bacterium than against the fungus L. thalassiae. Further, no correlations were observed between antimicrobial potencies of individual fractions against L. thalassiae vs. D. salina, L. thalassiae vs. P. bacteriolytica, or D. salina vs. P. bacteriolytica (data not shown). This overall lack of broad-spectrum antibiotic activity suggests that algal chemical defense molecules are not multifunctional against a wide variety of microbial genera, but instead are more targeted in their effects.
The high prevalence of antimicrobial activities observed among chromatographic fractions from several orders of red macroalgae (Supplemental Data) suggests the potential of ecology-driven studies in the discovery of novel chemistry. A search of the MarinLit database revealed secondary metabolites have been previously reported for only 53% of these evaluated genera (n = 29 evaluated genera; Supplemental Data). Further, antimicrobial chemical defense compounds have been isolated and identified from only one of these genera, 20 indicating a wealth of ecologically active natural products remain to be discovered.
In addition to the abundance of chemical defense observed across different species, substantial variation in antimicrobial defense was observed among multiple collections identified as the same algal species by morphological- and/or molecular-based analyses (Supplemental Data). Despite the indication from 18S rRNA analyses that samples G-0011 and G-0163 were both Neogoniolithon frutescens, fractions from G-0011 were significantly more inhibitory toward both P. bacteriolytica and L. thalassiae than were fractions from G-0163. Analogously, among two collections identified as Corynocystis prostrata, collection G-0008 exhibited significantly greater antifungal activity than collection G-0026. Together, these results as well as additional comparisons of chemical defenses among members of the same species (Supplemental Data) suggest intraspecific variation in antimicrobial chemical defenses is common. The importance of intraspecific variation in secondary metabolite production has previously been recognized among macroalgal antiherbivore defenses, 21,22 and such intraspecific variation may result from a variety of biotic and abiotic factors.23
In contrast to variability observed for antimicrobial chemical defenses within several species and genera of red algae, some genera were consistently well-defended against microbial pathogens. For example, extracts from all nine collections of Peyssonnelia spp. were strongly inhibitory toward both P. bacteriolytica and L. thalassiae, suggesting Peyssonnelia represents a particularly well-defended algal genus. This prevalence of chemical defense, together with the paucity of known secondary metabolites from Peyssonnelia spp.,24,25 implicated these algae as a promising source of novel chemistry and insights into ecological function.
2.2 Bioassay-guided fractionation yields novel antimicrobial sequiterpene hydroquinones from Peyssonnelia sp
Peyssonnelia sp. collection G-0109, exhibiting particularly potent antimicrobial activity, was selected as a candidate for ecologically-guided natural product isolation and identification. Guided by growth inhibition of the pathogenic bacterium Pseudoalteromonas bacteriolytica, peyssonoic acids A–B (1–2) were isolated by reversed-phase column chromatography and HPLC (see Experimental).
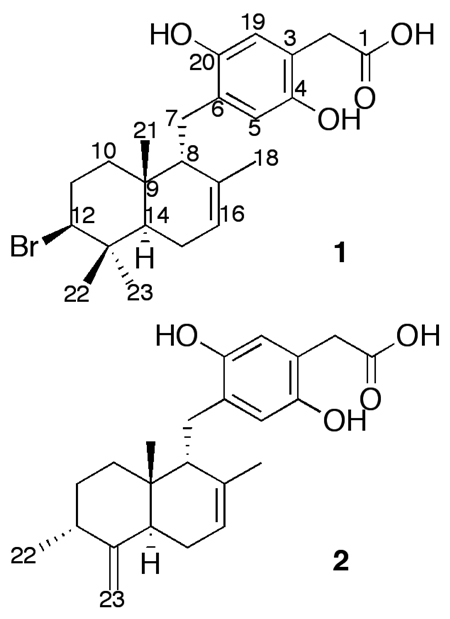 Structures of novel peyssonoic acids A and B (1–2).
Structures of novel peyssonoic acids A and B (1–2).
Peyssonoic acid A (1) displayed an [M – H]− molecular ion with m/z 449.1309 and a characteristic monobrominated isotopic pattern, supporting a molecular formula of C23H31O4Br. The structure of 1 was determined through examination of 1D and 2D NMR spectral data (Table 1, Supplemental Data). Assignments within the aromatic group of 1 were established by HMBC correlations from H-5 (δ 6.35) to C-3 (δ 122.6), C-4 (δ 150.1) and C-20 (δ 146.1), and from H-19 (δ 6.31) to aromatic C-4, C-6 (δ 126.6), and C-20. These assignments were confirmed and para dihydroxy substitution established by comparison of experimental 13C chemical shifts with empirical and literature values.25,26 HMBC correlations from H-19 to C-2 (δ 44.5), along with correlations from H-2 (δ 3.09) to C-1 (δ 175.3), C-3, C-4, and C-19 (δ 116.6) then established an acetic acid substituent attached at C-3. The connection to the sesquiterpene group was assigned at C-6 on the basis of HMBC correlations from H-5 to C-7 (δ 30.3) and from both H-7a (δ 2.13) and H-7b (δ 2.77) to C-5 (δ 117.8), C-6, and C-20.
Table 1.
13C and 1H NMR spectral data for 1-2 (125 MHz for 13C and 500 MHz for 1H; in DMSO).
| 1 | 2 | |||
|---|---|---|---|---|
| no. | δ 13C | δ 1H (JH,H) |
δ 13C | δ 1H (JH,H) |
| 1 | 175.3 | - | 175.3 | - |
| 2 | 44.5 | 3.09s | 44.5 | 3.10s |
| 3 | 122.6 | - | 122.5 | - |
| 4 | 150.1 | - | 150.1 | - |
| 5 | 117.8 | 6.35s | 117.5 | 6.40s |
| 6 | 126.6 | - | 126.7 | - |
| 7 | 30.3 | 2.13m, 2.77dd (6.2, 4.6) |
29.4 | 2.21dd (2.4, 12.2) 2.90dd (7.2, 15.0) |
| 8 | 53.7 | 1.78t (5.2) |
50.9 | 1.97m |
| 9 | 36.4 | - | 38.4 | - |
| 10 | 36.3 | 0.99m, 1.88m |
29.5 | 0.68m, 2.06td (2.0, 10.9) |
| 11 | 30.8 | 1.93m, 2.13m |
28.4 | 1.28brd (12.7), 1.63m |
| 12 | 70.9 | 4.20dd (3.0, 12.5) |
38.5 | 2.53m |
| 13 | 39.1 | - | 154.5 | - |
| 14 | 41.8 | 1.64m | 32.6 | 2.42brt (7.9) |
| 15 | 25.1 | 1.92m, 2.06m |
25.8 | 1.90m |
| 16 | 119.2 | 5.23s | 119.2 | 5.29s |
| 17 | 136.7 | - | 136.7 | - |
| 18 | 23.6 | 1.49s | 23.4 | 1.60s |
| 19 | 116.6 | 6.31s | 116.6 | 6.33s |
| 20 | 146.1 | - | 146.1 | - |
| 21 | 21.8 | 0.86s | 19.0 | 0.65s |
| 22 | 17.5 | 0.97s | 19.6 | 1.09d (7.2) |
| 23 | 29.9 | 1.00s | 106.8 | 4.50s, 4.81s |
| OH | - | 8.38brs | - | 8.51brs |
| OH | - | 13.39 brs | - | 13.37 brs |
The bromine-substituted drimane-type sesquiterpene group of 1 was elucidated primarily through analysis of HMBC and COSY data. HMBC correlations from singlet Me-18 (δ 1.49) to C-8 (δ 53.7), C-16 (δ 119.2), and C-17 (δ 136.7) prompted attachment of this methyl group to C-17 and established C-8—C-17—C-16 connectivity. HMBC correlations from singlet Me-21 (δ 0.86) to C-8, C-9 (δ 36.4), and C-10 (δ 36.3) next established connectivity between these carbons. COSY correlations between H-10a (δ 0.99) and H-11a (δ 1.93), between H-10b (δ 1.88) and H-11b (δ 2.13), and between both H-11 protons and H-12 (δ 4.20) prompted linkage of C-10—C-11—C-12, with these assignments confirmed by an HMBC correlation from H-11b to C-10. A bromine substituent was assigned at C-12 on the basis of downfield 13C and 1H chemical shifts (δ 70.9 and 4.20, respectively). HMBC correlations from both Me-22 (δ 0.97) and Me-23 (δ 1.00) to C-12 and quaternary C-13 (δ 39.1) supported C-12—C-13 connectivity, while HMBC correlations from Me-21, Me-22, and Me-23 to C-14 (δ 41.8) established connectivity between C-9 and C-14, thus sealing this ring. COSY correlations between H-14 (δ 1.64) and both H-15 protons (δ 1.92, 2.06) as well as an HMBC correlation between H-16 (δ 5.23) and C-15 (δ 25.1) then sealed the second ring, completing the drimane skeleton.
Relative stereochemical assignment for peyssonoic acid A (1) commenced with assignment of H-12 (δ 4.20) in an axial position based on a large J coupling constant (J = 12.5 Hz) observed for this proton (as well as a smaller 3.0 Hz coupling), which supported an axial-axial relationship and 180° dihedral angle between H-12 and a proton at C-11.26 Observation of a very intense COSY correlation between H-12 and H-11b (δ 2.13) prompted assignment of H-11b in an axial position on the opposite face of the ring from H-12. NOE correlations between H-12, H-14 (δ 1.64), and H-10b (δ 1.88) supported assignment of all of these atoms in axial positions on the same face of the drimane system. NOE correlations were not observed between any of these axial protons and Me-21 (δ 0.86) or H-11b. However, a strong correlation was noted between Me-21 and H-11b (δ 2.13), supporting assignment of Me-21 and H-11b in axial positions on the opposite face of the molecule. These assignments were reinforced by an NOE correlation observed between H-11b and Me-22 (δ 0.97), for which an axial position was supported by the upfield carbon chemical shift (δ 17.5) of this methyl relative to equatorial Me-23 (δ 29.9).27 Thus, with axial H-11b, Me-21, and Me-22 assigned on one face of the drimane system and axial H-10b, H-12, and H-14 established on the opposite face, a trans-fused drimane ring configuration was proposed for 1. The relative stereochemistry at C-8 was then assigned on the basis of NOE and J coupling constant arguments. A 6.2 Hz vicinal coupling observed for H-7b (δ 2.77) supported a dihedral angle of approximately 30° or 130° between H-7b and H-8 (δ 1.78). Further, the 5.2 Hz coupling observed for pseudotriplet H-8 suggested gauche relationships between this proton and both H-7a (δ 2.13) and H-7b. Observation of NOE correlations between H-7b and axial H-10b and between H-8 and axial Me-21 then established relative stereochemistry at C-8.
High-resolution mass spectral data established the molecular formula of peyssonoic acid B (2) as C23H30O4 (m/z 369.2080 [M – H]−). Comparison of 13C and 1H NMR spectral data as well as HMBC and COSY correlations between 1 and 2 indicated these molecules shared an acetic acid-substituted hydroquinone functionality and both possessed a sesquiterpene group attached at aromatic C-6 (Table 1, Supplemental Data). Comparison of molecular formulae for 1 and 2 indicated the loss of a bromine group and gain of an alkene moiety within the sesquitepene portion of 2. An exo-methylene substituent was assigned at C-13 on the basis of HMBC correlations from singlets H-23a (δ 4.50) and H-23b (δ 4.81) to C-12 (δ 38.5), C-13 (δ 154.5), and C-14 (δ 32.6). HMBC correlations from doublet Me-22 (δ 1.09) to C-11 (δ 28.4), C-12, and C-13 then established attachment of Me-22 to methine C-12. HMBC and COSY correlations supported assignment of the remainder of this decalin-type system identical to that of 1, and the sesquiterpene skeleton of 2 was verified by comparison with literature values. 25
For peyssonoic acid B (2), NOEs were observed between H-14 (δ 2.42) and Me-22 (δ 1.09), but not between H-12 (δ 2.53) and H-14, supporting assignment of H-14 and H-22 at axial positions on the same face of the drimane-type skeleton. NOEs between H-10b (δ 2.06) and both Me-22 and H-14 completed this series of 1,3-diaxial interactions. Me-21 was then assigned to an axial position on the opposite face based on NOEs observed between Me-21 (δ 0.65) and H-11b (δ 1.63), but not between Me-21 and H-14 or Me-22. This trans orientation of Me-21 and H-14 corresponded with the trans-fused bicyclic system proposed for 1. Finally, assignment of the C-8 stereocenter was established analogously to 1.
The most structurally similar known relatives of peyssonoic acids A and B (1–2) are peyssonols A–B, isolated from a Red Sea collection of Peyssonnelia sp. on the basis of activity against HIV-1 reverse transcriptase-associated RNA-dependent DNA polymerases.25 Peyssonol A differs from peyssonoic acid A (1) by substitution with a formyl group instead of an acetic acid group at C-3 on the hydroquinone ring and by a trans-fused drimane skeleton in 1 versus a cis-fused orientation in peyssonol A. Hence, peyssonoic acid A (1) represents a novel carbon skeleton with one additional carbon relative to peyssonol A. Peyssonoic acid B (2) shares a carbon skeleton with peyssonol B, and differs from this known metabolite in the presence of an acetic acid-substituted hydroquinone versus a methyl acetate group at the corresponding position in peyssonol B as well as in regioisomerization of one site of unsaturation in the drimane group. To our knowledge, 1 and 2 represent the first examples of terpene-hydroquinone natural products bearing an acetic acid-substituted hydroquinone.
2.3 Ecological and pharmacological activities of peyssonoic acids A–B (1–2)
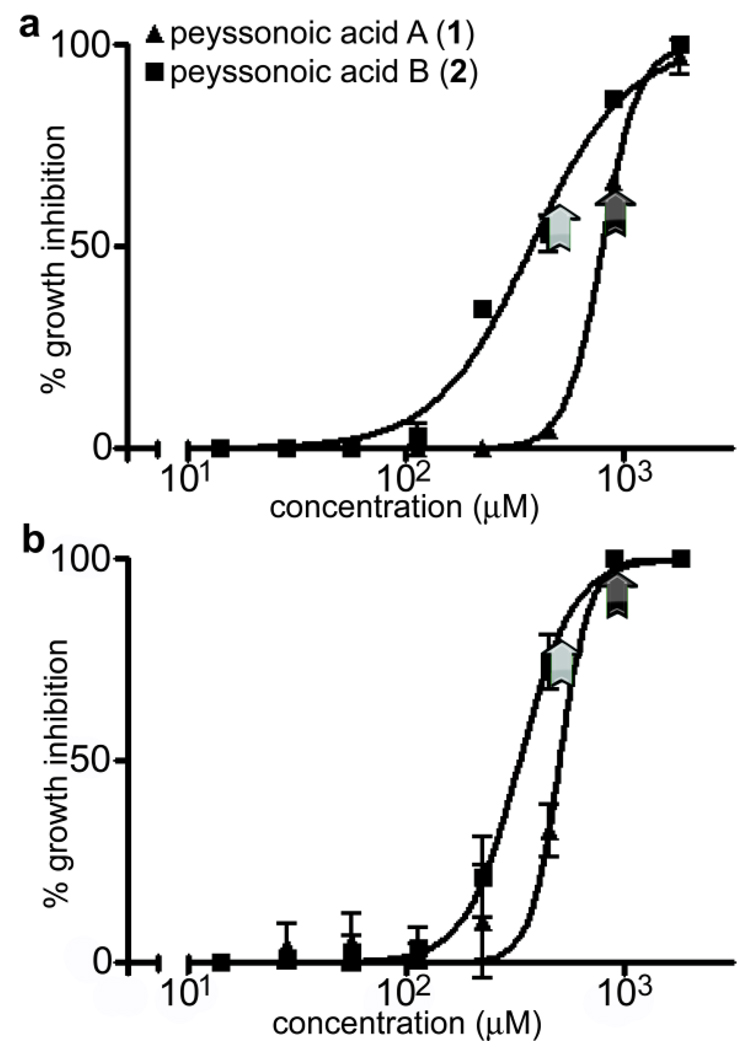
Both peyssonoic acids A–B (1–2) inhibited growth of the bacterial pathogen P. bacteriolytica and the fungal pathogen L. thalassiae (Fig. 2), suggesting their role as antimicrobial chemical defenses of Peyssonnelia sp. Against P. bacteriolytica, peyssonoic acid A (1) exhibited an average IC50 value of 799 µM, while peyssonoic acid B (2, IC50 377 µM) was significantly more inhibitory toward this pathogen (Fig. 2; p < 0.0001, F-test of logIC50 values). Against L. thalassiae, peyssonoic acid B (2, IC50 331 µM) was also significantly more inhibitory than peyssonoic acid A (1, IC50 506 µM; Fig. 2; p < 0.0001, F-test of logIC50 values). For both compounds, IC50 values observed against each marine pathogen were lower than natural isolated concentrations of these natural products. This suggests that, despite their weak potencies when compared to typical biomedical activity data, these secondary metabolites are present within organism tissues at concentrations sufficient for inhibition of these microbes in nature. Despite significant inhibition of D. salina observed with crude extracts, neither of these compounds inhibited growth of this marine saprophyte, suggesting Peyssonnelia sp. harbors other unidentified chemical defenses against this fungus.
Fig. 2.
Comparison of isolated concentrations (denoted by black arrow for 1 and gray arrow for 2) of peyssonoic acids A–B (1–2) and growth inhibition curves against ecologically relevant pathogens (a) P. bacterioltyica and (b) L. thalassiae (n = 3 subsample assays at 8 concentrations). In all cases, natural isolated concentrations were greater than experimental IC50 values, indicating these compounds represent effective antimicrobial chemical defenses of whole algal tissues. Further, peyssonoic acid B (2) was significantly more inhibitory toward both marine pathogens than peyssonoic acid A (1) (p < 0.0001; F-test of logIC50 values). Error bars denote one standard deviation.
While peyssonoic acids A–B (1–2) were strongly inhibitory toward a marine pathogenic bacterium and fungus at ecologically realistic concentrations, they were only modestly inhibitory toward human pathogenic bacteria and fungi (Table 2). The most notable pharmacological activity for both compounds was against human ovarian cancer cell lines, with 2 exhibiting stronger anticancer activity (IC50 13.5 µM) than 1 (IC50 34.5 µM).
Table 2.
Pharmacological activities of 1–2.
| antibacterial IC50 (µM) | anticancer IC50 (µM) | Antifungal MIC (µM) |
||||||
|---|---|---|---|---|---|---|---|---|
| cmpd | MRSAa | VREFb | antitubercular | meanc | A2780/ DDP-Sd |
cell line selectivity (IC50 max/IC50 min) |
WTCAe | ARCAf |
| 1 | >550 | >550 | >100 | 52.7 | 34.5 | 1.6 | >550 | >550 |
| 2 | 533 | 230 | >100 | 28.0 | 13.5 | 4.6 | >550 | >550 |
Methicillin-resistant Staphylococcus aureus;
Vancomycin-resistant Enterococcus faecalis;
Mean of 11 cancer cell lines (see Experimental section for details);
Ovarian tumor cell line;
Wild-type Candida albicans;
Amphotericin B-resistant C. albicans.
2.4 Conclusions
In the present study, all 69 evaluated tropical red algal samples harbored significant antimicrobial chemical defenses against at least one of three evaluated marine microbes. These chemical defenses spanned a full range of polarities, indicating a variety of chemical classes are involved in defending marine macroalgae against microbial attack. This abundance of defenses suggested the largely untapped potential of ecology-driven investigations in the discovery of novel chemistry. The potential of such approaches was illustrated by isolation and identification of peyssonoic acids A–B (1–2), novel antimicrobial defenses from the red alga Peyssonnelia sp. These sesquiterpene hydroquinones mark the first report of terpenoid natural products bearing an acetic acid-substituted hydroquinone and include one novel carbon framework.
3. Experimental
3.1 General
Semipreparative HPLC was performed with a Waters 1525 or 515 pump and a Waters 2996 diode-array UV detector or a Waters 2487 dual-wavelength absorbance detector. 1H, 13C, DEPT-135, HSQC, HMBC, COSY, NOESY, ROESY, and DPFGSE-NOE NMR experiments were conducted in DMSO with a Bruker DRX-500 instrument using a 5 mm broadband or inverse detection probe, and referenced to residual DMSO (δ 2.49 and δ 39.9 ppm for 1H and 13C, respectively). High resolution mass spectra were acquired using electrospray ionization with an Applied Biosystems QSTAR-XL hybrid quadrupole-time-of-flight tandem mass spectrometer and Analyst QS software. UV spectra were recorded in methanol with a Spectronic 21D spectromphotometer, and optical rotations were measured with a Jasco P-1010 spectropolarimeter. All statistical analyses were completed with either SYSTAT version 9 (Systat Software, Inc., Chicago, IL) or GraphPad version 4 (GraphPad Software, Inc., La Jolla, CA). HPLC grade solvents were used in semipreparative HPLC (Fisher Scientific Co.), and NMR solvents were obtained from Cambridge Isotope Laboratories.
3.2 Algal collection and identification
Algae were collected at depths of 2 – 20 m offshore from several islands in Fiji: Beqa (18° 23’ 56” S, 177° 57’ 59” E; 18° 22’ 47” S, 177° 59’ 37” E; 18° 20’ 86” S, 178° 2’ 1” E), Viti Levu (18° 12’ 9” S, 177° 39’ 65” E; 18° 14’ 51” S, 177° 47’ 27” E; 18° 9’ 61” S, 178° 25’ 57” E; 18° 14’ 41” S, 177° 46’ 85” E), Taveuni (16° 48’ 97” S, 179° 50’ 84” W; 16° 52’ 32” S, 179° 52’ 68” W; 16° 50’ 44” S, 179° 52’ 12” W; 16° 52’ 47” S, 179° 52’ 4” W; 16° 52’ 33” S, 179° 52’ 67” E), and Kadavu (18° 42’ 71” S, 178° 29’ 53” E; 18° 42’ 49” S, 178° 32’ 35” E). Peyssonoic acids A–B (1–2) were isolated from Peyssonnelia sp. (collection ID# G-0109), collected at Kadavu (18° 42’ 49” S, 178° 32’ 35” E).
Algal specimens were identified by comparison with previously described morphological traits28 and via 18S rRNA sequencing. Genomic DNA from ethanol-preserved red algal samples was extracted using the DNeasy Tissue Extraction Kit (Qiagen) and purified using polyethylene glycol. A region of the nuclear small subunit ribosomal RNA (18S rRNA) gene was amplified via PCR using established primers and reaction conditions. 29 18S rRNA fragments were sequenced in both directions. Sequences were manually edited in BioEdit version 7.0.5.330 and aligned using ClustalW. 31 Sequences were deposited in GenBank (accessions GQ227510-GQ227534). Phylogenetic relationships among sequences were determined in PAUP* (Sinauer Associates, Inc.) using parsimony criteria and 1000 bootstrap replicates, including the poriferan, Halichondria melanodocia, as an outgroup (GenBank accession AY737639). Vouchers are housed at the University of the South Pacific in Suva, Fiji, and at the Georgia Institute of Technology.
3.3 Algal extraction
Fresh algal material was extracted successively in MeOH (2×) and MeOH/DCM (2:1, 1:1). Extracts were reduced in vacuo and subjected to reversed-phase fractionation with HP20ss resin (Supelco). Fractions 1 and 2 were eluted with MeOH/H2O (1:1 and 4:1, respectively), fraction 3 with MeOH, and fraction 4 with acetone.
3.4 Ecological antimicrobial assays
Chromatographic fractions were evaluated for inhibition of three ecologically relevant marine microbes (see below). Assays were designed to approximate natural concentrations of metabolites experienced by microbes invading whole algal tissues. All fractions were tested at maximum concentrations volumetrically equivalent to those in the whole alga; fractions corresponding to a 1 mL wet volume of alga were incorporated into 1 mL of media, which was inoculated with an evaluated microbe.
3.4.1 Antifungal assays
Applying previously described methods, 16 antifungal assays were conducted with Lindra thalassiae (ATCC 56663), a fungal pathogen of a variety of marine macrophytes, and Dendryphiella salina, a fungal saprophyte of marine plants. Chromatographic fractions, corresponding to 1.2 mL of wet tissue volume, were each solubilized in a minimal volume of methanol or acetone and incorporated into of 1.2 mL molten YPM agar (16 g/L granulated agar, 2 g/L yeast extract, 2 g/L peptone, 4 g/L D-mannitol, 250 mg/L of both streptomycin sulfate and penicillin G in 1 L of natural seawater). For each fraction, three 400 µL subsamples of this mixture were dispensed into sterile 24-well microtiter plates, allowed to solidify, and an aliquot of L. thalassiae or D. salina suspension in sterile seawater added to each well. Control wells were prepared with YPM agar and solvent but no algal material. Plates were incubated at 28° C for three days and digital photographs collected for each well. The percent of each well covered in fungal hyphae was determined using the area calculator feature of ImageJ software (NIH), and treatments and controls compared using one-way ANOVA with Dunnett’s post test. Antifungal assays with pure 1–2 were completed at 1:1 serially diluted concentrations ranging from 1812 µM to 14 µM, and percent growth inhibition at each concentration was calculated relative to solvent-only controls. Growth inhibition data were fit to a sigmoidal dose-response curve, and mean log IC50 and standard error values computed. Reported IC50 values were calculated as the antilog of mean log IC50 values. 32 Antifungal activities of 1–2 were statistically compared with an F test of the log IC50 value for each compound.
3.4.2 Antibacterial assays
Assays using Pseudoalteromonas bacteriolytica, a known marine plant pathogen, were adapted from previous methods. 16 A 24 h shake culture of this bacterium was diluted 1:160 in Difco Marine Broth 2216 (BD Biosciences) and 195 µL of this mixture added to duplicate treatment and control wells of a 96-well plate. An equal amount of sterile broth was added to blank wells. Five microliters of 40× concentrated chromatographic fractions in DMSO were dispensed into treatment and blank wells, yielding a final concentration approximating that found in whole algal tissues; 5 µL of DMSO were added to corresponding control wells. Plates were incubated for 20 h at 30 °C, and turbidity measured at 600 nm. We corrected for the natural absorbance of chromatographic fractions by subtracting algal-containing sterile blank turbidities from values obtained for treatments. Corrected treatment turbidity values were compared with controls using one-way ANOVA with Dunnett’s post-test. For HPLC fractions and pure compounds from Peyssonnelia sp. (ID# G-0109), assays were completed at 1:1 serially diluted concentrations; log IC50 values computed and statistically compared analogously to antifungal assays.
3.5 Pharmacological assays
Pure peyssonoic acids A and B (1–2) were evaluated for activity against tumor cell lines BT-549, DU4475, MDA-MD-468, NCI-H446, PC-3, SHP-77, LNCaP-FGC, HCT116, MDA-MB-231, A2780/DDP-S, and Du145, representing breast, colon, lung, prostate and ovarian cancer cells. In vitro cytotoxicity was evaluated with the (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxylmethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium inner salt) MTS dye conversion assay as previously described.33 Antimalarial activity was determined with a previously reported SYBR Green based parasite proliferation assay.34,35 Antibacterial assays were performed against methicillin-resistant Staphylococcus aureus (ATCC 33591) and vancomycin-resistant Enterococcus faecium (ATCC 700221), and antifungal assays against both wild type and amphotericin B-resistant Candida albicans (ATCC 32354 and ATCC 90173, respectively), using previously reported methods.36 Antitubercular activity was assessed against Mycobacterium tuberculosis strain H37Rv (ATCC 27294) using the previously described alamar blue susceptibility test (MABA).37
3.6 Isolation of sesquiterpene hydroquinones A–B
Frozen Peyssonnelia sp. (ID# G-0109) was extracted exhaustively in methanol (2×) and methanol/dichloromethane (2:1 and 1:1) and fractionated with HP20ss resin (Supelco), following the same procedure applied in the algal survey (Experimental 3.3). Fraction 2, with the strongest activity against ecologically-relevant pathogen P. bacteriolytica, was subjected to multiple rounds of semipreparative reversed-phase HPLC with an Agilent Zorbax SB-C18 column (5 µm, 9.4 × 250 mm) using methanol:water and acetonitrile:water gradient mobile phases. Antibacterial properties of HPLC fractions were measured with the P. bacteriolytica assay described above, and final purification of active compounds 1–2 achieved using a Phenomenex Develosil C30 column (5 µm, 4.6 × 250 mm) with a methanol:water gradient mobile phase.
3.6.1 Peyssonoic acid A (1)
brown gum (8.2 mg; 0.13% plant dry mass); [α] 24 D 35.0 (c 0.343 g/100 mL, MeOH); UV (MeOH) λmax (log ε) 229 nm (4.24), 270 nm (3.89); 1H NMR (DMSO, 500 MHz) and 13C/DEPT NMR (DMSO, 125 MHz) data, Table 1; NOE, COSY, HMBC NMR data, Supplemental Data; HRESIMS [M – H]− m/z 449.1309 (calcd for C23H30O4Br, 449.1333).
3.6.2 Peyssonoic acid B (2)
brown gum (3.1 mg; 0.05% plant dry mass); [α] 24 D 200.0 (c 0.0800 g/100 mL, MeOH); UV (MeOH) λmax (log ε) 230 nm (4.35), 268 nm (3.81); 1H NMR (DMSO, 500 MHz) and 13C/DEPT NMR (DMSO, 125 MHz) data, Table 1; NOE, COSY, HMBC NMR data, Supplemental Data; HRESIMS [M – H]− m/z 369.2080 (calcd for C23H29O4, 369.2071).
Supplementary Material
Species identification, phylogenetic analysis, and antimicrobial chemical defense potencies for individual algal collections; 1H and 13C NMR spectra for 1–2; COSY, HSQC, HMBC, and NOE NMR spectral data for 1–2.
Acknowledgements
This work was supported by an NSF-IGERT predoctoral fellowship to A.L.L., by NIH ICBG grant # U01-TW007401 to M.E.H. and J.K., and by NSF grant OCE-0726689 to J.K. The authors thank A. Chequer, P.R. Jensen, C. Kauffman, C. Kicklighter, and G. Toth for collection assistance; K. Feussner and M. Sharma for extraction assistance; T. Davenport and S. Franzblau for antibacterial assays; D. Bostwick and C. Sullards for mass spectral analyses; and L. Gelbaum for NMR spectroscopy assistance. The authors thank the Government of Fiji for permission to perform research in their territorial waters, and the people of Serua, Kadavu, and Cakaudrove provinces for facilitating this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Blunt JW, Copp BR, Hu WP, Munro MHG, Northcote PT, Prinsep MR. Natural Product Reports. 2008;25:35–94. doi: 10.1039/b701534h. [DOI] [PubMed] [Google Scholar]
- 2.Blunt JW, Copp BR, Hu WP, Munro MHG, Northcote PT, Prinsep MR. Natural Product Reports. 2007;24:31–86. doi: 10.1039/b603047p. [DOI] [PubMed] [Google Scholar]
- 3.Lane AL, Kubanek J. In: Algal Chemical Ecology. Amsler CD, editor. Berlin: Springer; 2008. pp. 229–243. [Google Scholar]
- 4.Engel S, Jensen PR, Fenical W. Journal of Chemical Ecology. 2002;28:1971–1985. doi: 10.1023/a:1020793726898. [DOI] [PubMed] [Google Scholar]
- 5.Harvell CD, Kim K, Burkholder JM, Colwell RR, Epstein PR, Grimes DJ, Hofmann EE, Lipp EK, Osterhaus A, Overstreet RM, Porter JW, Smith GW, Vasta GR. Science. 1999;285:1505–1510. doi: 10.1126/science.285.5433.1505. [DOI] [PubMed] [Google Scholar]
- 6.Harvell D, Aronson R, Baron N, Connell J, Dobson A, Ellner S, Gerber L, Kim K, Kuris A, McCallum H, Lafferty K, McKay B, Porter JW, Pascual M, Smith G, Sutherland K, Ward J. Frontiers in Ecology and the Environment. 2004;2:375–382. [Google Scholar]
- 7.Littler MM, Littler DS. Science. 1995;267:1356–1360. doi: 10.1126/science.267.5202.1356. [DOI] [PubMed] [Google Scholar]
- 8.Sawabe T, Makino H, Tatsumi M, Nakano K, Tajima K, Iqbal MM, Yumoto I, Ezura Y, Christen R. International Journal of Systematic Bacteriology. 1998;48:769–774. doi: 10.1099/00207713-48-3-769. [DOI] [PubMed] [Google Scholar]
- 9.Short FT, Muehlstein LK, Porter D. Biological Bulletin. 1987;173:557–562. doi: 10.2307/1541701. [DOI] [PubMed] [Google Scholar]
- 10.Kohlmeyer J. Marine Biology. 1971;8:344–350. [Google Scholar]
- 11.Porter D. Mycoses of marine organisms: An overview of pathogenic fungi. New York: Cambridge University Press; 1986. [Google Scholar]
- 12.Engel S, Puglisi MP, Jensen PR, Fenical W. Marine Biology. 2006;149:991–1002. [Google Scholar]
- 13.Puglisi M, Engel S, Jensen P, Fenical W. Marine Biology. 2007;150:531–540. [Google Scholar]
- 14.Kjelleberg S, Steinberg P, Givskov M, Gram L, Manefield M, deNys R. Aquatic Microbial Ecology. 1997;13:85–93. [Google Scholar]
- 15.Nylund GM, Cervin G, Persson F, Hermansson M, Steinberg PD, Pavia H. Marine Ecology-Progress Series. 2008;369:39–50. [Google Scholar]
- 16.Kubanek J, Jensen PR, Keifer PA, Sullards MC, Collins DO, Fenical W. Proceedings of the National Academy of Sciences of the United States of America. Vol. 100. 2003. pp. 6916–6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen PR, Jenkins KM, Porter D, Fenical W. Applied and Environmental Microbiology. 1998;64:1490–1496. doi: 10.1128/aem.64.4.1490-1496.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puglisi MP, Tan LT, Jensen PR, Fenical W. Tetrahedron. 2004;60:7035–7039. [Google Scholar]
- 19.Jiang RW, Lane AL, Mylacraine L, Hardcastle KI, Fairchild CR, Aalbersberg W, Hay ME, Kubanek J. Journal of Natural Products. 2008;71:1616–1619. doi: 10.1021/np800307h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lane AL, Nyadong L, Galhena AS, Shearer TL, Stout EP, Parry RM, Kwasnik M, Wang M, Hay ME, Fernandez FM, Kubanek J. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7314–7319. doi: 10.1073/pnas.0812020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolser RC, Hay ME. Ecology. 1996;77:2269–2286. [Google Scholar]
- 22.Cronin G, Hay ME. Ecology. 1996;77:2287–2301. [Google Scholar]
- 23.Paul VJ, Puglisi MP. Natural Product Reports. 2004;21:189–209. doi: 10.1039/b302334f. [DOI] [PubMed] [Google Scholar]
- 24.McPhail KL, France D, Cornell-Kennon S, Gerwick WH. Journal of Natural Products. 2004;67:1010–1013. doi: 10.1021/np0400252. [DOI] [PubMed] [Google Scholar]
- 25.Talpir R, Rudi A, Kashman Y, Loya Y, Hizi A. Tetrahedron. 1994;50:4179–4184. [Google Scholar]
- 26.Silverstein RM, Webster FX. Spectrometric Identification of Organic Compounds. 6th ed. New York: John Wiley & Sons, Inc.; 1998. [Google Scholar]
- 27.Yong KWL, Jankam A, Hooper JNA, Suksamrarn A, Garson MJ. Tetrahedron. 2008;64:6341–6348. [Google Scholar]
- 28.Littler DS, Littler MM. South Pacific Reef Plants. Washington, D.C: Offshore Graphics, Inc.; 2003. [Google Scholar]
- 29.Saunders GW, Kraft GT. Canadian Journal of Botany. 1994;72:1252–1263. [Google Scholar]
- 30.Hall TA. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- 31.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, F V, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 32.Motulsky HJ, Christopoulos A. Fitting models to biological data using linear and nonlinear regression. A practical guide to curve fitting. San Diego: GraphPad Software, Inc.; 2003. [Google Scholar]
- 33.Lee FYF, Borzilleri R, Fairchild CR, Kim SH, Long BH, Reventos-Suarez C, Vite GD, Rose WC, Kramer RA. Clinical Cancer Research. 2001;7:1429–1437. [PubMed] [Google Scholar]
- 34.Smilkstein M, Sriwilaijaroen N, Kelly JX, Wilairat P, Riscoe M. Antimicrobial Agents and Chemotherapy. 2004;48:1803–1806. doi: 10.1128/AAC.48.5.1803-1806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bennett TN, Paguio M, Gligorijevic B, Seudieu C, Kosar AD, Davidson E, Roepe PD. Antimicrobial Agents and Chemotherapy. 2004;48:1807–1810. doi: 10.1128/AAC.48.5.1807-1810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kubanek J, Prusak AC, Snell TW, Giese RA, Hardcastle KI, Fairchild CR, Aalbersberg W, Raventos-Suarez C, Hay ME. Organic Letters. 2005;7:5261–5264. doi: 10.1021/ol052121f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collins LA, Franzblau SG. Antimicrobial Agents and Chemotherapy. 1997;41:1004–1009. doi: 10.1128/aac.41.5.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Species identification, phylogenetic analysis, and antimicrobial chemical defense potencies for individual algal collections; 1H and 13C NMR spectra for 1–2; COSY, HSQC, HMBC, and NOE NMR spectral data for 1–2.