Abstract
Electron microscopy has recently revealed striking structural orderliness in kinetochore proteins and protein complexes that associate with microtubules. In addition to their astonishing appearance and intrinsic beauty, the structures are functionally informative. The Dam1 and Ndc80 complexes bind to the microtubule lattice as rings and chevrons, respectively. These structures give insight into how the kinetochore couples to dynamic microtubules, a process critical to the accurate segregation of chromosomes. HURP and kinesin-13 arrange tubulin into sleeves and bracelets surrounding the microtubule lattice. These structures may reflect the ability of these proteins to modulate microtubule dynamics by interacting with specialized tubulin configurations. In this review, we compare and contrast the structure of these proteins and their interactions with microtubules to illustrate how they may attach to and modulate the dynamics of microtubules.
Introduction
The kinetochore is the supramolecular structure that attaches chromosomes to microtubules (MTs). During mitosis it must maintain a floating grip on dynamic microtubule ends in order to achieve accurate chromosome segregation. Similar challenges are faced by cells when they are splicing RNA, polymerizing DNA or holding together replicating chromosomes. In these cases, enzymes that fully or partially enclose the nucleic acid polymer enable processivity, while exhibiting structural flexibility and weak or changing substrate affinity. Does the kinetochore use a similar mechanism to hold on to a microtubule?
The kinetochore maintains attachment to MTs even as thousands of tubulin subunits are added or removed. Higher eukaryotic spindles have multiple microtubules per kinetochore and therefore could let go of one or more microtubules while holding onto others. However, in budding yeast, each kinetochore binds to the tip of one microtubule and remains attached whether the microtubule is polymerizing or depolymerizing (1). These attachments are load bearing as the kinetochore carries the chromosome with it. Two types of models can explain this behavior. One is a biased diffusion model, also called a thermal ratchet or molecular velcro. We use the name 'velcro' to emphasize that this type of model invokes many weak interactions with the MT, analogous to the many hooks and loops of velcro that only together make a robust connection. However, unlike velcro, the kinetochore attachment is dynamic. Any diffusion that positions additional binding sites near the microtubule is favored by the increased affinity generated (2). In the other model, the kinetochore encircles the microtubule and cannot be pulled off because the MT end forms some type of a structural barrier. A ring could satisfy either model but the latter model requires a ring encircling the MT. Moreover, the depolymerizing and polymerizing ends must be wide enough to prevent a ring that fits around a MT from coming off. Depolymerizing MTs are known to have curling protofilaments, which would push a ring along as proposed by the conformational wave model (3). Polymerizing MTs come together as a sheet, which is wider at the end and could also prevent a ring from coming off (4) (See Box 1).
Two kinetochore constituents have been identified that can form rings around microtubules, the Dam1 complex from budding yeast (also named the DASH complex) (Figure 1A) and higher eukaryotic kinesin-13s (Figure 1B) (5–7, 8.). The rings are fundamentally different in that kinesin-13 stabilizes rings of tubulin protofilaments (Box 1), whereas the Dam1 complex comprises the rings itself. Electron microscopy has revealed structures of two other microtubule-binding proteins. HURP, a mitotic spindle protein, promotes the formation of a novel polymer of tubulin that surrounds a microtubule like a sleeve (Figure 1C)(9). Finally, the Ndc80 kinetochore complex does not form rings but instead decorates microtubules as chevrons (Figure 1D) (10). Here, we discuss the evidence that these rings, bracelets, sleeves and chevrons enable kinetochore attachment to a dynamic microtubule, and we point out questions that remain to be resolved.
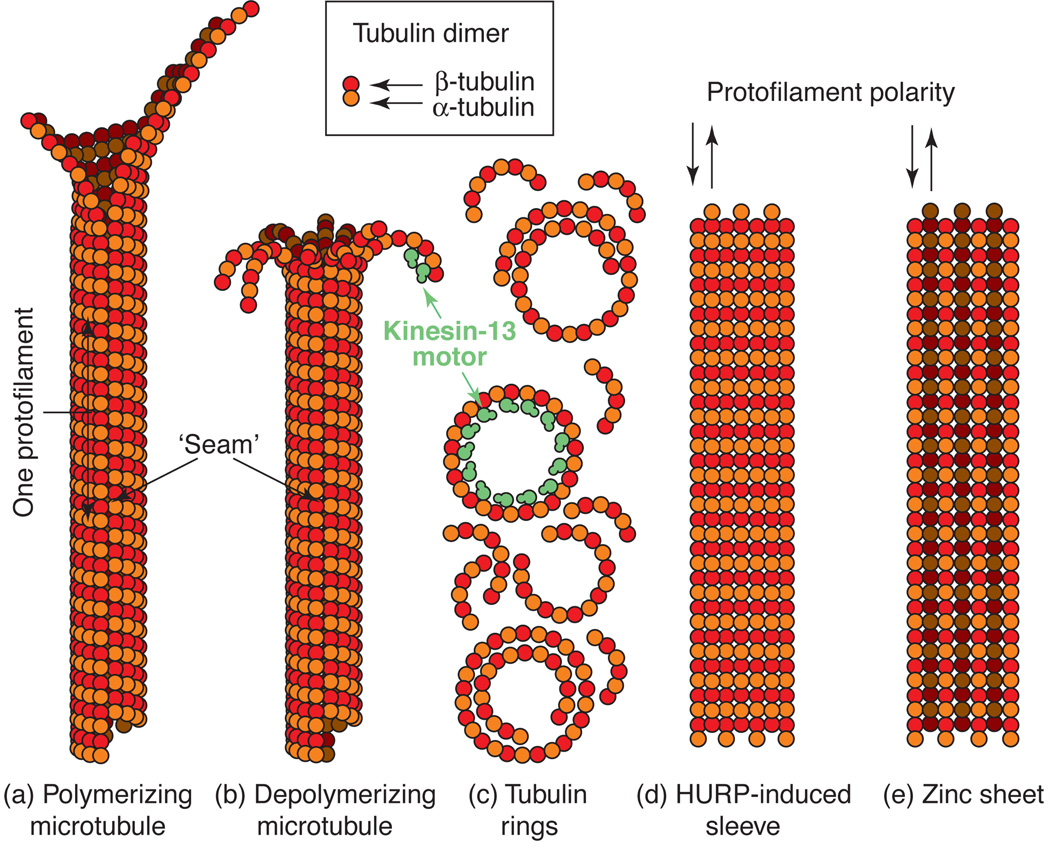
Figure 1. Microtubule Structures.
A. Polymerizing microtubules. Microtubules assemble from a sheet of 13 protofilaments, which roll up to form a tubular structure. The helical pitch intrinsic to the polymerizing tubulin dimers gives rise to a noticeable seam in the microtubule that has been visualized in the electron microscope. The fundamental unit of microtubule assembly is the αβ-tubulin dimer (light green box).
B. Depolymerizing microtubules. Individual protofilaments peel apart during microtubule disassembly. The outwardly curved protofilaments, which form naturally during disassembly due to conformational change in the tubulin dimer triggered by the hydrolysis of bound GTP, can be stabilized by the motor domains of kinesin-13 family members (shown in green). In the presence of ATP the disassembly of microtubules is promoted by kinesin-13s due to their ability to form a high affinity complex with curved protofilaments.
C. Tubulin rings. The products of microtubule disassembly in vitro can be individual tubulin dimers or curved protofilaments of various sizes depending on the buffer constituents. These protofilament rings can be stabilized by motor domains of kinesin-13 family proteins (shown in green) in the presence of non-hydrolyzable ATP. This promotes a strong rigor association of the motor domain preferentially with curved, rather than straight, protofilaments.
D. HURP-induced sheets. Sheets of tubulin that form in the presence of HURP and preformed microtubules consist of antiparallel protofilaments similar to zinc sheets with the exception that the luminal surfaces are all on the same side of the sheet. It is this surface (the back side of the sheet shown here) that interacts with HURP bound to microtubules.
E. Zinc sheets. Microtubules assembled in the presence of zinc ions will form antiparallel, interdigitating straight protofilament sheets (sometimes referred to as macrotubes). Neighboring protofilaments show opposite surfaces; a protofilament showing what would be the luminal surface in a MT (shaded dark) is neighbor to protofilaments showing what would be the outer surface in a MT (not shaded). These structures are useful for structural studies and as unusual substrates for motor activity.
The Dam1 complex: Rings around microtubules
The Dam1 complex is an excellent candidate for forming attachments because it oligomerizes to form rings around microtubules (Figure 1A). A single complex is a heterodecamer with one copy of each of 10 proteins, has a molecular weight of 204 kDa and is approximately 70 Å × 105 Å (7, 8). The heterodecamers fit together longitudinally to form the ring around a MT (Figure 1A). The rings apparently do not contain tubulin although that has not been finally resolved. When seen end on by EM, the Dam1 ring has approximately16 subunits (presumably 16 heterodecamers) surrounding the 13 protofilaments of the microtubule (7). Scanning transmission electron microscopy measures the mass of the ring to be equivalent to 25 heterodecamers (11). Rings are difficult to detect in vitro in the absence of microtubules suggesting that microtubules might guide the assembly of these structures. The lack of 1:1 stoichiometry between the number of Dam1 complexes and the number of protofilaments argues that Dam1 does not bind tightly to one part of the tubulin dimer and thus could have a loose grip that allows free movement along the microtubule (12). Recent results suggest that flexible tethers on two of the proteins (Dam1 and Duo1) bind to the microtubule (11). The Kd of the entire Dam1 complex for microtubules is reported to be 200 nM (7).
Recombinant Dam1 complex attaches to MTs and reproduces many of the behaviors of the kinetochore. It can bind to depolymerizing microtubule ends (13) and stay attached to the end of polymerizing or depolymerizing microtubules even against a force of up to 3 pN (12). The level of force is similar to that generated by ATP-driven motors that move organelles inside cells. Thus, it can form physiologically relevant load-bearing tip attachments even as thousands of tubulin subunits are removed or added to a microtubule. Moreover, like the kinetochore, the Dam1 complex changes the dynamics of the plus-end in ways that promote lengthening (14).
Genetic evidence also supports the role of the Dam1 complex in kinetochore attachment to microtubules. All the components of the Dam1 complex are essential in budding yeast. Temperature-sensitive mutants in Dam1 complex proteins show defects in biorientation and attachment (15, 16). Phosphorylation of Dam1 on a single residue by Mps1 kinase is required to target kinetochores to the plus-end of microtubules in budding yeast (17). If the phosphorylation site is mutated, kinetochores are attached laterally to MTs by the Dam1 complex, providing additional evidence that Dam1 is required in vivo for stable attachment of kinetochores to the plus ends of MTs. Surprisingly, accurate chromosome segregation can still occur.
Although enticing to propose, the ring has not been proven as the physiologically important organization of the Dam1 complex. In yeast, the Dam1 complex has been measured by immunoblot analysis to be present at 650 copies per cell, just enough for one 16-mer ring around each of 32 kinetochore microtubules (8). In fact, fluorescence microscopy measures 16–20 Dam1 complexes in each budding yeast kinetochore at metaphase (18). The quantification is consistent with a single ring around each kinetochore microtubule, but a ring has never been seen near the plus-end of the microtubule even when examined by electron tomography with a resolution of ~5 nm. Instead thin fibrils of unknown composition are seen connected to both the outside and inside of a microtubule (19). The ring may be sensitive to freeze substitution and therefore not visualized by these methods. At anaphase, each kinetochore only contains 10 –11 Dam1 complexes, suggesting the 16-mer ring is not needed during anaphase when microtubules undergo rapid depolymerization (18), a time when a ring would seem to be most important. The nature of the Dam1 structure in cells remains to be determined and could be altered by binding to the Ndc80 complex and other kinetochore components.
Another question is what performs the function of Dam1 in other organisms. In fission yeast, the Dam1 complex is required only if kinesin-8 is absent, suggesting kinesin-8 performs a redundant function (20). Kinesin-8 is a plus-end directed motor that acts as a length-dependent depolymerase (21). Its ability to substitute for the Dam1 complex is especially interesting considering that, evolutionarily, the kinesin-8s are the next closely related clade to the kinesin-13s (see below) and may possess mechanistic similarities (21–23). Homologs of the Dam1 complex have not been found in other eukaryotic cells but lack of sequence homology does not preclude a structural and functional match. Thus, great interest accompanied the discovery that kinesin-13 forms rings around microtubules.
Kinesin-13: Bracelets of tubulin
GDP-tubulin rings have long been observed as products of microtubule disassembly (24). There are a number of enzymes that bind preferentially to protofilament rings rather than straight protofilaments in the microtubule lattice. This promotes the transition from the polymerizing to the depolymerizing state (see Box 1) thus serving as a controlled method of rapidly disassembling microtubules. The most mechanistically well-characterized of these enzymes is the kinesin-13 family of microtubule motors, which includes mammalian MCAK and Drosophila melanogaster Klp10A (reviewed in (25)). Kinesin-13’s core motor domain actively promotes the formation of protofilament rings from assembled, stabilized MTs in the presence of non-hydrolyzable ATP analogs such as AMP-PNP. Relative to the motile kinesins, the MT binding interface of the kinesin-13 prefers to form a high affinity complex with a curved, protofilament rather than a straight protofilament as would be found in the intact MT lattice (26, 27). The opportunity to form this tight complex in AMP-PNP (which locks the motor in the ATP bound state) is only available at the MT ends where the protofilaments can be bent outward. No further catalytic disassembly is possible because the motor cannot hydrolyze the analog. This results in the formation of rings with stoichiometrically bound kinesin-13 motor domains (see Box 1).
Recently two papers have described another novel property of kinesin-13 motors. When added to stabilized MTs at high stoichiometry in AMP-PNP, they promote the assembly of curved longitudinal protofilaments that assemble around the MT forming helical rings or “bracelets” (Figure 1B) that consist of a protofilament spiral plus (most likely) attached kinesin-13 motors. In some cases the structure is of such high order that strong diffractions at 80 Å (the length of one tubulin dimer) and 160 Å (the pitch of the spiral) are obtained (5). These rings have been proposed to be the higher eukaryotic counterpart of the Dam1 rings. However, the rings are seen only in the presence of AMP-PNP (as opposed to other nucleotides), which is not found in cells. Furthermore, the “ring” part of the spiral is actually a curved protofilament of tubulin dimers that remain longitudinally associated possibly by virtue of the artificial stabilization of the microtubule by taxol or GMP-CPP. The symmetry more closely matches that of the underlying microtubule, which may preclude the loose attachment required to maintain a floating grip on a dynamic microtubule and hints at an ordered interaction between the bound kinesin-13 motor domains on the longitudinal protofilament spiral and the wall of the microtubule lattice. Finally, recent tomography of the mammalian kinetochore found no rings (28).
Although kinesin-13 may not provide the higher eukaryotic version of the Dam1 ring, in vivo, the discovery of the ring is still informative. The rings can be induced with core kinesin-13 motor domain alone (6) or core motor plus neck (5). The motor appears to mediate an association between the protofilament spiral and the MT surface implicating two opposed MT binding sites on the core motor domain. Recent studies on D. melanogaster Klp10A (6) suggest that two motor domains, one bound to the protofilament and one bound to the MT lattice, subsequently interact with each other to mediate ring formation. One problem with this hypothesis is that the dimerization domains are not present in the core motor domain, which has been shown to be a monomer in solution (29, 30). However, weak interactions between motors might be templated by the MT lattice. Such unexpected core motor domain dimerizations have been seen before, particularly (and inexplicably) with proteins isolated from cold-blooded animals such as Drosophila and squid (31). Alternatively, as is thought to occur with mammalian kinesin-13 family member, MCAK, one motor may crosslink the protofilament to the MT from either side. Interestingly, there are a number of positively charged residues that are specifically conserved in the kinesin-13 depolymerases yet are located on the side of the motor domain opposite the known MT binding domain. This suggests an alternative hypothesis in which kinesin-13s possess two distinct MT binding domains within the core motor. The canonical kinesin motor MT binding domain is known to bind tightly to curved protofilaments in the presence of non-hydrolyzable ATP analogs. The exposed side of the motor domain may be capable of interacting with the surface of the assembled MT in a relatively ordered manner as well. While it is unclear whether the rings are functionally relevant they do illustrate the ability of the MT lattice and MT-binding proteins to template potentially informative structures.
HURP: Formation of a tubulin sleeve
Another example of the ability of the MT to template novel structures is the sleeve formed by HURP (hematoma up-regulated protein), a component of the Ran-importin β-regulated spindle assembly pathway (32, 33). Cryoelectron micrographs of recombinant HURP bound to MTs show a novel polymer of tubulin forming a sleeve around a normal MT (9) (Figure 1C). The HURP is on the inside of the sleeve between the MT and the HURP-induced tubulin polymer. The tubulin in the sleeve is arranged in antiparallel protofilaments with P2 symmetry (Box 1). This means that every protofilament has the same side out, normally the outer surface. (The outer surface can be distinguished from the luminal surface in vitro because the outer surface binds kinesin heads.) In contrast, in Zn2+ sheets of tubulin, the protofilaments exhibit P21 symmetry (Box 1). The protofilaments are antiparallel, but adjacent filaments expose alternating outer and luminal surfaces. Similarly to the kinesin-13 bracelets, the HURP protein is located between the surface of the MT and the wrapped sheet. HURP possesses significant regions of positive charge that are postulated to interact with MTs and protofilaments in an oriented manner. An intriguing difference is that the luminal side of antiparallel protofilaments is interacting with the MT surface-bound HURP. Both HURP and kinesin-13s appear to favor the formation of these templated structures in contrast to simple decoration of the surface of the MT. Both these proteins may actively participate in the structural modulation of the MT lattice in conjunction with crosslinking to other tubulin structures.
The sleeves are unlikely to attach the kinetochore to microtubules. Although HURP associates with kinetochore MTs, it does not appear to be part of the kinetochore but instead forms a gradient or comet tail extending out from the kinetochore. Because HURP stabilizes kinetochore MTs (32–34) it is attractive to hypothesize that the sleeve forms on kinetochore microtubules and impedes depolymerization. HURP may use its MT stabilizing properties to promote congression to the metaphase plate and antagonize the MT destabilizing effect of kinetochore-associated kinesin-13s.
The Ndc80 complex decorates microtubules as chevrons
Two reports recently showed that components of the kinetochore besides the Dam1 complex bind MTs, the Ndc80 complex and KNL-1 (known as Spc105 in budding yeast). The Ndc80 complex contains four proteins: Ndc80 (also known as Hec1 in humans and Tid3 in budding yeast), Nuf2, Spc24 and Spc25. The complex is a long coiled coil rod connecting two globular domains (35, 36). The C-terminal regions of Spc24 and Spc25 bind tightly to form the domain facing the inner kinetochore (37). The N-terminal regions of Ndc80 and Nuf2 form the domain facing the MT. Until recently, this domain was thought to interface with the Dam1 complex and not bind microtubules (38). Two reports have now shown that the Ndc80 complex binds MTs directly (10, 39). The affinity of the entire complex for MTs is low with an apparent Kd of 3 µM and was enhanced by other kinetochore proteins, KNL-1 and the Mis12 complex (10). Together, the Ndc80 complex, KNL-1 and the Mis12 complex showed half-maximal binding at 750 nM polymerized tubulin. The N-terminal region of Hec1 (human Ndc80) is a calponin-homology domain as is found in the EB1 MT tip-binding protein (39). Curiously, as seen by cryoEM, EB1 binds only to the seam of MTs, (seen in Box 1) (40). In contrast, the Ndc80/Nuf2 N-terminal domain and the entire Ndc80 complex bind all round the MT (10, 39). Binding curves plateau at 1.5 Ndc80/Nuf2 N-terminal domains bound per tubulin dimer instead of the 1 per 13 tubulin dimers expected for a protein that binds to MT seams (39).
Cryo-electron micrographs of the Ndc80 complex shows chevrons decorating the MTs (10) (Figure 1D). Each complex extends off of the MT like a porcupine quill. On a given MT, all the quills point in the same direction suggesting the Ndc80 heads bind in a well-defined orientation with respect to the MT lattice. It is not known whether the quills point to the plus or minus end of the MT.
The low affinity of the Ndc80 complex for MTs and the fact that each complex binds individually are key features expected for the molecular velcro model for the kinetochore. Moreover, genetic analyses show that it is important for attachment. It is essential in fission and budding yeast. Mutations in the Ndc80 complex in the absence of active Aurora B do not detach the kinetochores from the microtubules in metaphase but weaken the attachment such that the kinetochores fall off during anaphase (41) (42). siRNA against the Ndc80 complex dramatically reduces but does not eliminate attachment (43). Although it is in the right place with the right characteristics, no evidence has yet shown that Ndc80 can form dynamic load-bearing attachments like the Dam1 complex.
Conclusion
The Dam1 ring, the kinesin-13 ring, and the HURP sleeve are distinctly different structures but display interesting similarities. The microtubule may, by virtue of its repeating subunit construction, have a tendency to template ordered structures, such as rings or sheets, or to promote weak interactions in proteins that associate with tubulin. This is undoubtedly occurring during the formation of the kinesin-13 bracelets and the HURP sleeve, but the microtubule may also exert a templating effect on the formation of the Dam1 ring. The Ndc80 quills are not templated into an oligomer by the MT, however the stereospecificity of the interaction is clear by their uniform orientation. In conclusion, the Dam1 complex, the Ndc80 complex, and the depolymerizing motors likely work together to bind the kinetochore to a dynamic MT. These proteins assemble unique MT structures that reveal the complex nature of their MT binding domains. The kinetochore may actually be a ring of molecular velcro that handles dynamic microtubules with a light but firm touch.
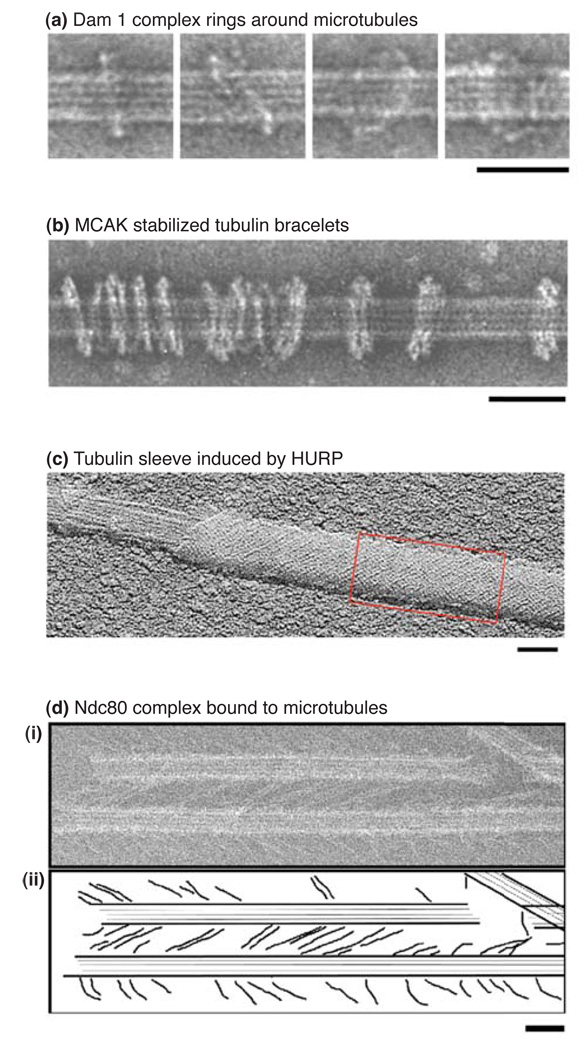
Figure 2. Rings, bracelets, sleeves and chevrons formed by kinetochore proteins.
A. Dam1 complex rings around microtubules. Recombinant Dam1 complex bound to paclitaxel-stabilized microtubule, Images are electron micrographs of negative stained material. Bar represents 50 nm. Reprinted from (8) with permission from Nature.
B. MCAK stabilized tubulin bracelets. Paclitaxel stabilized microtubule in the presence of recombinant MCAK (core motor domain plus 64 residues of the neck domain) and AMPPNP. Image is an electron micrograph of negative stained material. Bar represents 50 nm. Reprinted from (5) with permission from Landes.
C. HURP-induced tubulin sheets. Recombinant HURP was incubated with tubulin in the presence of GTP. Vitrification by quick-freezing was followed by freeze-drying and then unidirectional metal shadowing before cryo elecron microscopy. The red box indicates a region of the microtubule covered with a sleeve. Bar represents 50 nm. Reprinted from (9) with permission from Elsevier.
D. Ndc80 chevrons bound to microtubules. Recombinant Ndc80 complex bound to microtubules stabilized with GMPCPP. Image is an electron micrograph of negative stained material. Bar represents 50 nm. Reprinted from (10) with permission from Elsevier.
References
- 1.Pearson CG, Yeh E, Gardner M, Odde D, Salmon ED, Bloom K. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr Biol. 2004 Nov 9;14(21):1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 2.Hill TL. Theoretical problems related to the attachment of microtubules to kinetochores. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4404–4408. doi: 10.1073/pnas.82.13.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koshland DE, Mitchison TJ, Kirschner MW. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature. 1988 Feb 11;331(6156):499–504. doi: 10.1038/331499a0. [DOI] [PubMed] [Google Scholar]
- 4.Mandelkow EM, Mandelkow E, Milligan RA. Microtubule dynamics and microtubule caps: a time-resolved cryo-electron microscopy study. J Cell Biol. 1991 Sep;114(5):977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moores CA, Cooper J, Wagenbach M, Ovechkina Y, Wordeman L, Milligan RA. The role of the kinesin-13 neck in microtubule depolymerization. Cell Cycle. 2006 Aug;5(16):1812–1815. doi: 10.4161/cc.5.16.3134. [DOI] [PubMed] [Google Scholar]
- 6.Tan D, Asenjo AB, Mennella V, Sharp DJ, Sosa H. Kinesin-13s form rings around microtubules. J Cell Biol. 2006 Oct 9;175(1):25–31. doi: 10.1083/jcb.200605194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westermann S, Avila-Sakar A, Wang HW, Niederstrasser H, Wong J, Drubin DG, et al. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005 Jan 21;17(2):277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 8.Miranda JJ, De Wulf P, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol. 2005 Feb;12(2):138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- 9.Santarella RA, Koffa MD, Tittmann P, Gross H, Hoenger A. HURP wraps microtubule ends with an additional tubulin sheet that has a novel conformation of tubulin. J Mol Biol. 2007 Feb 2;365(5):1587–1595. doi: 10.1016/j.jmb.2006.10.064. [DOI] [PubMed] [Google Scholar]
- 10.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006 Dec 1;127(5):983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 11.Miranda JJ, King DS, Harrison SC. Protein Arms in the Kinetochore-Microtubule Interface of the Yeast DASH Complex. Mol Biol Cell. 2007 Apr 25; doi: 10.1091/mbc.E07-02-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci U S A. 2006 Jun 27;103(26):9873–9878. doi: 10.1073/pnas.0602249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westermann S, Wang HW, Avila-Sakar A, Drubin DG, Nogales E, Barnes G. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006 Mar 23;440(7083):565–569. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- 14.Franck AD, Powers AF, Gestaut DR, Davis TN, Asbury CL. Tension promotes microtubule elongation in vitro: A direct mechanism for length control in mitosis. Nature Cell Biol. 2007 doi: 10.1038/ncb1609. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka K, Mukae N, Dewar H, van Breugel M, James EK, Prescott AR, et al. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005 Apr 21;434(7036):987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- 16.Cheeseman IM, Enquist-Newman M, Muller-Reichert T, Drubin DG, Barnes G. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J Cell Biol. 2001;152(1):197–212. doi: 10.1083/jcb.152.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimogawa MM, Graczyk B, Gardner MK, Francis SE, White EA, Ess M, et al. Mps1 phosphorylation of Dam1 couples kinetochores to microtubule plus-ends at metaphase. Curr Biol. 2006 August 8;16:1489–1501. doi: 10.1016/j.cub.2006.06.063. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006 Jun;8(6):581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McIntosh JR. Rings around kinetochore microtubules in yeast. Nat Struct Mol Biol. 2005 Mar;12(3):210–212. doi: 10.1038/nsmb0305-210. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez-Perez I, Renwick SJ, Crawley K, Karig I, Buck V, Meadows JC, et al. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. Embo J. 2005 Aug 17;24(16):2931–2943. doi: 10.1038/sj.emboj.7600761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006 Sep;8(9):957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- 22.Dagenbach EM, Endow SA. A new kinesin tree. J Cell Sci. 2004 Jan 1;117(Pt 1):3–7. doi: 10.1242/jcs.00875. [DOI] [PubMed] [Google Scholar]
- 23.Ovechkina Y, Wordeman L. Unconventional motoring: an overview of the Kin C and Kin I kinesins. Traffic. 2003 Jun;4(6):367–375. doi: 10.1034/j.1600-0854.2003.00099.x. [DOI] [PubMed] [Google Scholar]
- 24.Nogales E, Wang HW, Niederstrasser H. Tubulin rings: which way do they curve? Curr Opin Struct Biol. 2003 Apr;13(2):256–261. doi: 10.1016/s0959-440x(03)00029-0. [DOI] [PubMed] [Google Scholar]
- 25.Moores CA, Milligan RA. Lucky 13-microtubule depolymerisation by kinesin-13 motors. J Cell Sci. 2006 Oct 1;119(Pt 19):3905–3913. doi: 10.1242/jcs.03224. [DOI] [PubMed] [Google Scholar]
- 26.Moores CA, Yu M, Guo J, Beraud C, Sakowicz R, Milligan RA. A mechanism for microtubule depolymerization by KinI kinesins. Mol Cell. 2002 Apr;9(4):903–909. doi: 10.1016/s1097-2765(02)00503-8. [DOI] [PubMed] [Google Scholar]
- 27.Ogawa T, Nitta R, Okada Y, Hirokawa N. A common mechanism for microtubule destabilizers-M type kinesins stabilize curling of the protofilament using the class-specific neck and loops. Cell. 2004 Feb 20;116(4):591–602. doi: 10.1016/s0092-8674(04)00129-1. [DOI] [PubMed] [Google Scholar]
- 28.Dong Y, Vanden Beldt KJ, Meng X, Khodjakov A, McEwen BF. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat Cell Biol. 2007 May;9(5):516–522. doi: 10.1038/ncb1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maney T, Wagenbach M, Wordeman L. Molecular dissection of the microtubule depolymerizing activity of mitotic centromere-associated kinesin. J Biol Chem. 2001 Sep 14;276(37):34753–34758. doi: 10.1074/jbc.M106626200. [DOI] [PubMed] [Google Scholar]
- 30.Hertzer KM, Ems-McClung SC, Kline-Smith SL, Lipkin TG, Gilbert SP, Walczak CE. Full-length dimeric MCAK is a more efficient microtubule depolymerase than minimal domain monomeric MCAK. Mol Biol Cell. 2006 Feb;17(2):700–710. doi: 10.1091/mbc.E05-08-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoenger A, Thormahlen M, Diaz-Avalos R, Doerhoefer M, Goldie KN, Muller J, et al. A new look at the microtubule binding patterns of dimeric kinesins. J Mol Biol. 2000 Apr 14;297(5):1087–1103. doi: 10.1006/jmbi.2000.3627. [DOI] [PubMed] [Google Scholar]
- 32.Koffa MD, Casanova CM, Santarella R, Kocher T, Wilm M, Mattaj IW. HURP is part of a Ran-dependent complex involved in spindle formation. Curr Biol. 2006 Apr 18;16(8):743–754. doi: 10.1016/j.cub.2006.03.056. [DOI] [PubMed] [Google Scholar]
- 33.Sillje HH, Nagel S, Korner R, Nigg EA. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr Biol. 2006 Apr 18;16(8):731–742. doi: 10.1016/j.cub.2006.02.070. [DOI] [PubMed] [Google Scholar]
- 34.Wong J, Fang G. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J Cell Biol. 2006 Jun 19;173(6):879–891. doi: 10.1083/jcb.200511132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci U S A. 2005 Apr 12;102(15):5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ciferri C, De Luca J, Monzani S, Ferrari KJ, Ristic D, Wyman C, et al. Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J Biol Chem. 2005 Aug 12;280(32):29088–29095. doi: 10.1074/jbc.M504070200. [DOI] [PubMed] [Google Scholar]
- 37.Wei RR, Schnell JR, Larsen NA, Sorger PK, Chou JJ, Harrison SC. Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure. 2006 Jun;14(6):1003–1009. doi: 10.1016/j.str.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 38.Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR, 3rd, et al. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002 Oct 18;111(2):163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- 39.Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007 Jan;14(1):54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- 40.Sandblad L, Busch KE, Tittmann P, Gross H, Brunner D, Hoenger A. The Schizosaccharomyces pombe EB1 homolog Mal3p binds and stabilizes the microtubule lattice seam. Cell. 2006 Dec 29;127(7):1415–1424. doi: 10.1016/j.cell.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 41.Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol. 2006 Jan;8(1):78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- 42.Kotwaliwale C, Biggins S. Microtubule capture: a concerted effort. Cell. 2006 Dec 15;127(6):1105–1108. doi: 10.1016/j.cell.2006.11.032. [DOI] [PubMed] [Google Scholar]
- 43.DeLuca JG, Dong Y, Hergert P, Strauss J, Hickey JM, Salmon ED, et al. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol Biol Cell. 2005 Feb;16(2):519–531. doi: 10.1091/mbc.E04-09-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]