Abstract
Human CYP1A2 is an important drug-metabolizing enzyme, similar in sequence to CYP1A1 but with distinct substrate specificity. We have previously shown that residue 382 affected CYP1A1 and CYP1A2 specificities with alkoxyresorufins. To determine whether this residue is also important for the metabolism of other substrates, we have investigated phenacetin oxidation by single (T124S, T223N, V227G, N312L, and L382V) and multiple (L382V/T223N, L382V/N312L, L382V/T223N/N312L, and L382V/T124S/N312L) mutants of CYP1A2. The enzymes were expressed in Escherichia coli and purified. All the CYP1A2 mutants that contained the L382V substitution displayed much higher activities than the wild-type enzyme, with kcat values 3-fold higher, in contrast to other mutants, for which kcat decreased. Likewise, a significant increase in specificity, expressed as the kcat/Km ratio, was observed for the mutants containing the L382V substitution. The efficiency of coupling of reducing equivalents to acetaminophen formation was decreased for all the single mutants except L382V, for which the coupling increased. This effect was also observed with multiple CYP1A2 mutants containing the L382V substitution. Low activities of the four other single mutants were likely caused by dramatically increased uncoupling to water. In contrast, the increase in activity of the L382V-containing mutants resulted from decreased water formation. This finding is consistent with molecular dynamics results, which showed decreased phenacetin mobility leading to increased product formation. The results of these studies confirm the importance of residue 382 in CYP1A2-catalyzed oxidations and show that a single residue substitution can dramatically affect enzymatic activity.
Cytochromes P450 (P450s) are heme-containing monooxygenase enzymes, which are involved in the metabolism of numerous exogenous and endogenous compounds. P450s are ubiquitous in living organisms, with at least 50 families and 82 subfamilies found in different species. Human CYP1A subfamily has two major isoforms: CYP1A1 and CYP1A2. CYP1A2, one of the major P450s in the human liver, was first characterized as a phenacetin O-deethylase (Distlerath et al., 1985). Currently, it is estimated that this enzyme metabolizes approximately 11% of all drugs in humans (Shimada et al., 1994). Despite the fact that CYP1A2 participates in the deactivation and detoxification of xenobiotics, the main interest in this enzyme is because of the metabolic activation of a large number of chemical carcinogens (Guengerich and Shimada, 1991; Lewis et al., 1994).
In humans, CYP1A2 shares 72% amino acid sequence identity with CYP1A1, but the substrate specificities and inhibitor susceptibilities of these enzymes are different. For example, substrates such as phenacetin and 7-methoxyresorufin are primarily metabolized by CYP1A2 with high catalytic efficiency, whereas CYP1A1 displays weak capability to oxidize those substrates. On the other hand, 7-ethoxyresorufin is preferentially oxidized by CYP1A1 (Nerurkar et al., 1993; Burke et al., 1994). The structural basis for such functional differences between highly related enzymes can be investigated using a variety of techniques, including molecular modeling and experimental methods, such as site-directed mutagenesis and NMR. More recently, the crystal structure of CYP1A2 was solved by X-ray crystallography (Sansen et al., 2007); thus, it provides a practical model for structure-function studies.
Our previous studies on structure-function relationships of CYP1A1 indicated that Val382 played an important role in binding of alkoxyresorufin substrates (Liu et al., 2003). The sequence alignment between CYP1A1 and CYP1A2 indicated that five active site residues that are different between these enzymes (Ser122, Asn221, Gly225, Leu312, and Val382 in CYP1A1 and the corresponding residues in CYP1A2: Thr124, Thr223, Val227, Asn312, and Leu382) might be involved in determining substrate specificity (Liu et al., 2004). This result was confirmed by the finding that five reciprocal mutations in CYP1A1 and CYP1A2 altered enzymatic activity with alkoxyresorufins as substrates. Moreover, mutations at position 382 in both CYP1A1 and CYP1A2 shifted substrate specificity from one enzyme to another (Liu et al., 2004). Further computational and experimental studies with multiple CYP1A2 mutants confirmed the importance of this residue for alkoxyresorufin oxidation (Tu et al., 2008). Therefore, it would be of interest to examine the effect of these mutations on oxidation of other types of substrates.
In the current study, we chose phenacetin as a substrate. This compound has been used as the most common marker for CYP1A2 activity in the in vitro studies of 45% of new drugs by investigators in the pharmaceutical industry (Yuan et al., 2002). The objective of the present study was to investigate whether any reciprocal mutations in CYP1A2 may alter phenacetin oxidation and to examine the potential mechanism(s) that might be involved. Five single mutants and four multiple mutants containing the L382V substitution were evaluated using a combination of molecular modeling and experimental methods. These included enzyme kinetics and stoichiometry studies, as well as molecular dynamics (MD) simulations of phenacetin in the active site of CYP1A2 mutants to facilitate the interpretation of experimental results. This study should provide an increased understanding of the biochemical aspects of substrate specificity in the P450 family of enzymes.
Materials and Methods
Materials.
Phenacetin, acetaminophen, 2-hydroxy acetanilide, sodium dithionite, NADPH, ampicillin, isopropyl-β-d-thiogalactopyranoside, δ-aminolevulinic acid, CHAPS, dilauroyl-l-3-phosphatidyl choline (DLPC), and phenylmethanesulfonyl fluoride were from Sigma-Aldrich (St. Louis, MO). Nickel-nitrilotriacetic acid agarose and a gel extraction kit were purchased from QIAGEN (Valencia, CA). Potassium phosphate, EDTA, acetic acid, and high-performance liquid chromatography-grade methanol were purchased from Thermo Fisher Scientific (Waltham, MA). All the other chemicals used were of analytical grade and were obtained from standard commercial sources.
Protein Expression and Purification.
The clones of CYP1A1 wild type (WT), CYP1A2 WT, and CYP1A2 single mutants, T124S, T223N, V227G, N312L, and L382V, all of them containing a His-tag for easy purification, were constructed earlier (Liu et al., 2003, 2004). CYP1A2 His-tag multiple mutants, L382V/T223N, L382V/N312L, L382V/T223N/N312L, and L382V/T124S/N312L, were also constructed previously (Tu et al., 2008). The P450 enzymes were expressed in Escherichia coli DH5α cells and purified essentially as described previously (Liu et al., 2004; Tu et al., 2008). During the purification, the addition of 5 mM caffeine in the purification buffers helped to stabilize CYP1A2 proteins. The substrate caffeine was removed completely from the enzyme preparation during the ultrafiltration stage, as verified by HPLC. Rat P450 reductase was expressed in E. coli and purified according to an established procedure (Liu et al., 2003). The final purity of the enzymes was assessed by SDS-polyacrylamide gel electrophoresis. Western blots were performed using anti-human CYP1A1/1A2 (Oxford Biomedical Research, Oxford, MI), and P450 proteins were visualized as described previously (Kedzie et al., 1991). P450 content was determined by reduced CO/reduced difference spectra (Omura and Sato, 1964), and protein was measured using Folin phenol reagent (Lowry et al., 1951).
P450 Activity Assay.
Phenacetin O-dealkylase activities of CYP1A2 WT and mutants were determined by HPLC measurements as described previously (von Moltke et al., 1996) with some modifications. The reaction mixtures contained 0.5 μM CYP1A2, 1 μM P450 reductase, and 45 μM DLPC in 100 mM potassium phosphate buffer, pH 7.5. The enzymes and DLPC were preincubated for 2 min at 37°C before the dilution. For kinetic assays, phenacetin was added at concentrations ranging from 0 to 1000 μM, and the mixture was incubated for another 3 min at 37°C. The reaction was initiated by adding NADPH to a final concentration of 1 mM in a total volume of 1 ml and conducted for 30 min. The reaction was terminated by the addition of 5 μl of 60% HClO4, and the reaction mixture was put on ice for 10 min. Ten microliters of 2-hydroxy acetanilide (100 μM) was then added as an internal standard for HPLC determination. The reaction mixture was centrifuged at 1000g for 5 min, and 100 μl of the supernatant was removed and used directly for HPLC analysis. The product acetaminophen was eluted from a C18 column (Alltech Associates, Deerfield, IL) with a mobile phase of methanol/0.1% acetic acid (30:70, v/v; flow rate, 1.5 ml/min) and monitored at 254 nm. The product was quantified using acetaminophen standards. The kinetic parameters (Vmax and kcat) were calculated using nonlinear regression with GraphPad Software Inc. (San Diego, CA) Prism software.
Binding Constant Determination.
Spectral binding constants for phenacetin bound in the active site of CYP1A1 WT and CYP1A2 enzymes were obtained using difference visible spectroscopy (Modi et al., 1995). Solutions (800 μl) contained 0.5 μM CYP1A2 WT or the mutants in 100 mM phosphate buffer, containing 20% glycerol and 0.1 mM EDTA, pH 7.4. Two microliters of different concentrations of solutions of phenacetin in menthol was added to the sample cuvette, and the same volume of menthol was added to the reference, and UV spectra were then recorded. The data were analyzed by nonlinear regression analysis using Microsoft (Redmond, WA) Excel software.
NADPH Oxidation.
The rate of NADPH oxidation was determined spectrophotometrically at 340 nm in a cuvette thermostated at 37°C. The reaction mixture was similar to that used for the phenacetin assay and contained 0.5 μM P450 enzyme, 1 μM P450 reductase, 45 μM DLPC, and 1 mM phenacetin in a 100 mM potassium phosphate buffer, pH 7.5, in a volume of 980 μl. The reaction was initiated by the addition of 20 μl of 50 mM NADPH. The NADPH oxidation rates were recorded for approximately 3 min at 340 nm from the beginning of the reaction. The molar extinction coefficient of 6.22 per mM/cm for NADPH at 340 nm was used to obtain oxidation rates in nanomole per minute per nanomole of P450. Three 50-μl aliquots of the reaction mixture were removed after 1, 2, and 3 min and quenched with 50 μl of 10% CF3COOH. The triplicate-quenched reaction mixture was then used to measure hydrogen peroxide (H2O2).
H2O2 Production.
The reaction mixtures from the NADPH oxidation assay were used to measure the production of H2O2 using the xylenol orange iron (III) assay (Jiang et al., 1990; Fang et al., 1997) with slight modifications. The coloring agent was prepared by mixing 100 volumes of 125 μM xylenol orange in 100 mM sorbitol and 1 volume of 25 mM fresh ferrous (Fe2+) ammonium sulfate in 2.5 M H2SO4. The calibration curve was prepared using the quenched reaction mixture, which was supplemented with H2O2 at concentrations ranging from 0 to 10 μM. The H2O2 standard solutions were prepared fresh on the day of the assay by dilution of a 30% H2O2 stock solution. The reaction mixture was incubated at room temperature for 1 h. Absorbance was recorded using a Beckman Coulter, Inc. (Fullerton, CA) spectrophotometer set at 560 nm to obtain the concentration of H2O2 produced in nanomole per minute per nanomole of P450.
Oxygen Consumption.
The reaction was conducted using a Mitocell (Strathkelvin Instruments Ltd., Glasgow, UK), which was connected to a water bath thermostated at 37°C. The reaction mixture was prepared in a similar way to that for the NADPH oxidation assay. Nine hundred eighty microliters of the sample was placed in the chamber of the Mitocell, and once a steady baseline was established, the reaction was initiated by the addition of 20 μl of NADPH. The oxygen consumption was recorded over 5 min as micromolar per hour, which was then converted to nanomole per minute per nanomole of P450.
General Molecular Modeling Methods.
Molecular modeling simulations were conducted using a Silicon Graphics Octane workstation with Insight II software (Accelrys, San Diego, CA). The crystal structure of CYP1A2 (Protein Data Bank code 2hi4) was obtained courtesy of Dr. Eric F. Johnson (The Scripps Research Institute, La Jolla, CA) (Sansen et al., 2007). The heme cofactor was removed and replaced with the oxoheme cofactor. Substrate phenacetin was constructed with Insight II/Builder module and optimized. The models of CYP1A2 single and multiple mutants were constructed from the crystal structure of CYP1A2 WT by the replacement of selected amino acid(s) and further refinement of the structures according to the previously established procedure (Liu et al., 2003, 2004; Tu et al., 2008). MD simulations and energy minimization were carried out using the Insight II/Discover module with the consistent valence force field supplemented with parameters for heme and ferryl oxygen, as described earlier (Paulsen and Ornstein, 1991, 1992). The nonbond cutoff was 16 Å, and all the other parameters were set at their default values. Structural refinement of CYP1A2 WT and mutants involved 1000 steps of minimization using steepest descent gradient followed by 10-ps MD and then another 1000 steps of steepest descent minimization. The optimized structures were used for the subsequent docking studies.
Docking of Phenacetin into the Active Site of CYP1A2 WT and Mutants.
Initially, phenacetin was manually placed into the active site of CYP1A2 WT and the mutants on the distal side of the oxoheme. Docking of phenacetin was performed with Insight II (Accelrys)/Affinity module using default parameters, as described previously (Liu et al., 2004; Ericksen and Szklarz, 2005; Tu et al., 2008). Residues within 10 Å of the initial phenacetin position comprised the flexible region of the receptor (CYP1A2 WT and mutants) during all the docking runs. The Affinity docking method uses both the Monte Carlo search technique and simulated annealing approach, followed by the minimization protocol to generate low-energy substrate binding orientations. A distance-dependent dielectric constant was applied to simulate charge screening by water molecules. The 10 lowest-energy phenacetin binding orientations obtained from Affinity docking were selected for further analysis.
MD Simulations of Enzyme-Substrate Complexes.
MD simulations were performed to investigate phenacetin mobility in the active site and the effect of mutations on substrate orientation. The starting configuration for MD simulations chosen from Affinity docking represented the productive binding orientation of phenacetin leading to its O-dealkylation and had the lowest potential energy rank. The MD simulations of each phenacetin-enzyme complex were performed at 310 K in vacuo essentially as described earlier (Liu et al., 2004; Tu et al., 2008). The substrate, heme, and protein residues within 10 Å from the initial substrate position were flexible without any restraints, whereas the remainder of the protein was fixed. A distance-dependent dielectric constant was used to simulate aqueous environment, and the nonbond cutoff distance was 16 Å. After 5-ps MD equilibration phase, the MD simulations were continued for 100 ps, and 400 frames obtained every 250 fs were extracted to record the snapshots of each enzyme-substrate complex.
In addition, to evaluate the effect of explicit solvent on phenacetin dynamics in the active site, we performed 100-ps MD simulations on solvated enzyme-substrate complexes using a similar protocol. The enzymes chosen were CYP1A2 WT and the L382V mutant. The enzyme-substrate complexes were solvated using crystallographic water molecules from CYP1A2 crystal structure and optimized before MD with 1000 steps of steepest descent minimization using a nonbond cutoff of 16 Å and dielectric constant of 1. Similar to previous MD simulations, only protein residues, heme, substrate, and solvent within a 10-Å radius of the initial substrate position were permitted to move, whereas the remainder of the protein and other water molecules were fixed. This ensures that none of the moving solvent molecules escape from the vicinity of the active site. In contrast to previous MD simulations, the flexible region also included 21 water molecules surrounding phenacetin. For the aqueous simulations, the dielectric constant was set to 1. All the other parameters were the same as for the previous enzyme-substrate simulations without explicit solvent present.
All the MD trajectories were examined using Insight II (Accelrys)/Analysis module. To score the likelihood of hydroxylation, each sampled frame of a given enzyme-substrate complex was evaluated using the following geometric criterion: r ≤ 3.5 Å and θ ≥ 120°, where r represents the distance between the ferryl oxygen and the hydrogen of the substrate to be abstracted; θ represents the angle between ferryl oxygen, the hydrogen atom to be abstracted, and the carbon at the oxidation site, as described previously (Ericksen and Szklarz, 2005; Tu et al., 2008). Trajectory data from 100-ps MD simulations were extracted and graphed using Microsoft Excel. The MD frames where the geometric criterion, r ≤ 3.5 Å and θ ≥ 120°, was satisfied were counted as hits. The number of hits is a useful indicator of whether a CYP1A2-mediated phenacetin O-deethylation occurred.
Results
Kinetics of Phenacetin O-Deethylation by CYP1A2 WT and Mutants.
CYP1A2 enzymes were expressed in E. coli and purified. The overall yield of the procedure was approximately 20 to 40%, similar to that reported previously (Liu et al., 2004; Tu et al., 2008). The purity of CYP1A2 WT and mutants verified by SDS-polyacrylamide gel electrophoresis and Western blots indicated that they were at least 95% pure. The spectrum of the FeII-CO complex exhibited a characteristic peak at 450 nm, with little or no P420 formation. The holoenzyme content of the enzymes was usually in the range of 40 to 60%, as previously observed in our laboratory.
Kinetic parameters, kcat, Km, and substrate specificity (kcat/Km), were determined for purified CYP1A2 WT and mutants using a range of substrate phenacetin concentrations. Phenacetin undergoes O-deethylation to form acetaminophen as the main product of the reaction. Kinetic parameters for CYP1A2 WT and mutants are shown in Table 1. Compared with CYP1A2 WT, the mutations affected both kcat and Km. The CYP1A2 L382V mutant and multiple mutants containing the L382V mutation displayed 2- or 3-fold higher kcat than the WT enzyme. Four other single mutants, namely, T124S, T223N, V227G, and N312L, exhibited much lower kcat than the WT.
TABLE 1.
Kinetic parameters for phenacetin O-deethylation by purified CYP1A2 WT and mutants
| CYP1A2 | Phenacetin O-Deethylation |
|||
|---|---|---|---|---|
| kcata | Kma | kcat/Kmb | [(kcat/Km)mutant/(kcat/Km)WT] | |
| min | μM | μM/min | ||
| WT | 1.28 ± 0.08 | 59.70 ± 1.82 | 0.02 ± 0.01 | 1.00 |
| T124S | 0.29 ± 0.03 | 28.90 ± 1.10 | 0.01 ± 0.01 | 0.47 |
| T223N | 0.30 ± 0.01 | 26.99 ± 0.01 | 0.01 ± 0.01 | 0.52 |
| V227G | 0.38 ± 0.02 | 32.36 ± 2.93 | 0.01 ± 0.01 | 0.55 |
| N312L | 0.31 ± 0.01 | 25.26 ± 0.01 | 0.01 ± 0.01 | 0.57 |
| L382V | 3.03 ± 0.06 | 5.75 ± 0.57 | 0.53 ± 0.04 | 24.77 |
| L382V/T223N | 3.91 ± 0.10 | 7.77 ± 0.80 | 0.51 ± 0.06 | 23.65 |
| L382V/N312L | 3.05 ± 0.15 | 16.14 ± 2.05 | 0.19 ± 0.02 | 8.87 |
| L382V/T223N/N312L | 3.67 ± 0.10 | 32.42 ± 11.31 | 0.12 ± 0.04 | 5.61 |
| L382V/T124S/N312L | 2.81 ± 0.03 | 10.13 ± 2.40 | 0.28 ± 0.07 | 13.34 |
Data are means of triplicate determinations.
kcat/Km represents substrate specificity.
As shown in Table 1, the Km values for CYP1A2 WT and mutants varied greatly. In general, the Km values for all the mutants were more than 50% lower than for the WT (∼60 μM). The lowest values of Km were observed for several mutants containing the L382V substitution, including L382V (∼6 μM), L382V/T223N (∼8 μM), and L382V/T124S/N312L (∼10 μM), which suggests that the L382V mutation significantly increased the binding affinity of the enzyme for phenacetin. The substrate specificities of the L382V mutant and multiple mutants containing the L382V mutation, expressed as kcat/Km, were at least 5-fold higher than that of the WT (Table 1). In particular, the relative substrate specificities of L382V and L382V/T223N mutants were more than 20-fold higher, which is extremely high. Incidentally, phenacetin is less efficiently metabolized by CYP1A1 WT (kcat 0.5/min; KM, 66 μM), with kcat close to one third of the value observed with CYP1A2 WT (see Table 1) and the kcat/Km ratio lower than 0.01.
Phenacetin binding constants were also determined for CYP1A2 WT and some mutants. CYP1A2 WT and the N312L mutant showed similar phenacetin binding, with binding constants of 17.1 and 10.2 μM, respectively. In contrast, the L382V and the L282V/N312L mutants displayed much lower values for binding constants, namely, 0.7 and 3.5 μM, indicating tighter substrate binding. CYP1A1 WT exhibited much weaker binding, with a binding constant of 57 μM.
Stoichiometry of Phenacetin O-Deethylation.
To assess whether the L382V mutation affected CYP1A2 coupling efficiency of reducing equivalents to acetaminophen formation, stoichiometry experiments were conducted. The rates of NADPH oxidation, hydrogen consumption, product formation, H2O2, and excess water production for phenacetin oxidation by CYP1A2 WT and mutants are shown in Table 2. The excess water formation was calculated from the difference between the rates of NADPH oxidation and rates of H2O2 and product formation (excess H2O = NADPH − H2O2 − product). The amount of water was also obtained from the difference between the rate of oxygen consumption and rates of H2O2 plus product formation [H2O = 2(O2 − H2O2 − product)], as reported by others (Fang et al., 1997), giving values that were very similar (within 5%) to those derived from the previous equation (data not shown). The coincubation of phenacetin with CYP1A2 L382V and three multiple mutants, L382V/T223N, L382V/N312L, and L382V/T223N/N312L, resulted in consumption of both NADPH and oxygen by the mutants at rates ∼2-fold greater than those with the WT enzyme. Likewise, the formation of product acetaminophen, as well as byproducts such as H2O2 and water, was 2 to 3 times higher in the case of these mutants. The exception was the L382V/T124S/N312L mutant, which seemed to use NADPH at a rate similar to the WT enzyme but displayed increased consumption of oxygen, along with increased product and H2O2 formation. On the other hand, four single mutants, T124S, T223N, V227G, and N312L, are similar or less efficient than the WT with respect to NADPH oxidation, oxygen consumption, H2O2, and water production but exhibit a substantial decrease in product formation.
TABLE 2.
Rates [nmol/min/(nmol of P450)] determined for phenacetin metabolism by CYP1A2 WT and mutantsa
| CYP1A2 | NADPH Oxidized | O2 Consumed | Product Formed | H2O2 Produced | Excess H2Ob |
|---|---|---|---|---|---|
| WT | 43 ± 4 | 22.7 ± 0.9 | 1.28 ± 0.08 | 14 ± 2 | 15 ± 2 |
| T124S | 27 ± 2 | 14.8 ± 0.4 | 0.29 ± 0.03 | 10 ± 0 | 9 ± 1 |
| T223N | 41 ± 3 | 17.3 ± 0.7 | 0.30 ± 0.00 | 10 ± 1 | 14 ± 2 |
| V227G | 40 ± 3 | 16.9 ± 0.5 | 0.38 ± 0.02 | 9 ± 1 | 15 ± 2 |
| N312L | 32 ± 2 | 18.3 ± 0.3 | 0.31 ± 0.00 | 13 ± 2 | 10 ± 1 |
| L382V | 84 ± 6 | 47.0 ± 1.3 | 3.03 ± 0.06 | 29 ± 3 | 30 ± 4 |
| L382V/T223N | 98 ± 7 | 57.9 ± 2.5 | 3.91 ± 0.10 | 38 ± 3 | 32 ± 5 |
| L382V/N312L | 70 ± 4 | 41.0 ± 2.0 | 3.05 ± 0.15 | 29 ± 3 | 18 ± 3 |
| L382V/T223N/N312L | 72 ± 5 | 41.7 ± 1.1 | 2.18 ± 0.03 | 28 ± 3 | 23 ± 3 |
| L382V/T124S/N312L | 42 ± 3 | 34.2 ± 0.9 | 3.67 ± 0.10 | 26 ± 2 | 9 ± 2 |
Data are means of triplicate determinations.
Excess water (H2O) was calculated from the equation: H2O = NADPH − H2O2 − product.
Table 3 presents the effects of mutations on the coupling efficiency of CYP1A2, both overall and at specific P450 uncoupling branching points. The ratios of product formation to NADPH oxidation, accounting for the overall efficiency of CYP1A2 coupling of reducing equivalents to product, were higher for L382V and multiple mutants containing the L382V substitution than for the WT. In contrast, other single mutants, T124S, T223N, V227G, and N312L, exhibited significantly decreased coupling efficiencies compared with the WT enzyme (20–30% of WT). All of the enzymes displayed similar ratios of H2O2 production to O2 consumption, which suggests that the mutations had no effect on uncoupling at the first and second branching points of the P450 cycle. On the other hand, significant differences between the enzymes were observed with respect to the H2O/product ratios used to measure uncoupling at the third branching point. Thus, for the L382V mutant and multiple mutants containing the L382V substitution, these ratios were generally lower than that for the WT, whereas for the other single mutants, these ratios were 3-fold higher. Therefore, low activities of the four single mutants were likely the result of dramatically increased uncoupling to water, whereas the increase in activity in the L382V-containing mutants resulted from decreased water formation.
TABLE 3.
Effect of mutations on coupling efficiency of CYP1A2 at different branching points of P450 cyclea
| CYP1A2 | Product/NADPHb | H2O2/O2c | H2O/Productd |
|---|---|---|---|
| WT | 0.030 | 0.62 | 11.72 |
| T124S | 0.011 | 0.68 | 31.03 |
| T223N | 0.007 | 0.58 | 46.67 |
| V227G | 0.010 | 0.53 | 39.47 |
| N312L | 0.010 | 0.71 | 32.26 |
| L382V | 0.036 | 0.62 | 9.9 |
| L382V/T223N | 0.040 | 0.66 | 8.18 |
| L382V/N312L | 0.044 | 0.71 | 5.9 |
| L382V/T223N/N312L | 0.030 | 0.67 | 10.55 |
| L382V/T124S/N312L | 0.087 | 0.76 | 2.45 |
Ratios were calculated from parameters in Table 2.
Efficiency of coupling reducing equivalents to product.
Effect on uncoupling at first or second branch point.
Effect on uncoupling at third branch point.
Molecular Modeling Analyses.
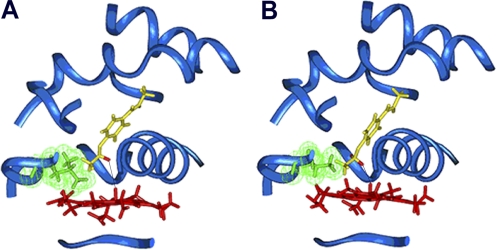
Using the crystal structure of CYP1A2, molecular modeling studies have been conducted to understand the effects of single and multiple mutations on enzyme-substrate interactions and substrate mobility, as well as to explain the alterations of catalytic efficiency. Figure 1 depicts binding orientations of phenacetin within the active sites of CYP1A2 WT and the L382V mutant. In general, the binding orientation of phenacetin in both enzymes was very similar. However, the replacement of Leu382 by a smaller Val increased the volume of the active site and allowed the hydrogens at the oxidation site of phenacetin to approach closer to the ferryl oxygen of the heme, which facilities hydrogen abstraction. The average distance between the hydrogens at the oxidation site of phenacetin and the ferryl oxygen of the L382V mutant was 3.1 Å, compared with 3.7 Å for the WT enzyme. Similar results were also obtained for the L382V/T223N and L382V/T223N/N312L mutants. In contrast, both hydrogens at the oxidation site of phenacetin were much farther away from the ferryl oxygen in the active sites of T124S, T223N, V227G, and N312L mutants (data not shown). These findings correlate well with the results of kinetic and stoichiometric studies described previously and suggest that the L382V substitution not only increases catalytic efficiency (kcat) of CYP1A2 but also decreases Km and enzyme uncoupling.
Fig. 1.
Binding orientation of phenacetin within the active site of CYP1A2 WT (A) and the L382V mutant (B). The protein backbone is depicted as a blue ribbon; the side chain of residue 382 is green, with van der Waals surface displayed; heme is red; and phenacetin is yellow, with hydrogens at the oxidation site shown in red. The distance between the hydrogen to be abstracted and the ferryl oxygen (marked with a black line) is 3.6 Å in the WT and 2.9 Å in the L382V mutant.
To examine the tendency of phenacetin to remain in the productive binding orientation in the active site of the enzyme, 100 ps of MD simulations were performed as described under Materials and Methods. After the 5-ps MD equilibration phase, the energy of the system remained constant throughout the simulations. The mobility of the substrate varied with the mutant: we observed fairly low mobility for the WT enzyme, L382V, T124S, and L382V/T223N/N312L mutants, with root mean square deviation (RMSD) from the initial substrate orientation of 1 to 2 Å, moderate mobility for V227L and N312L mutants with RMSD of 2 to 4 Å, and a high phenacetin mobility in the case of T223N and L382V/T223N mutants (RMSD, >4 Å).
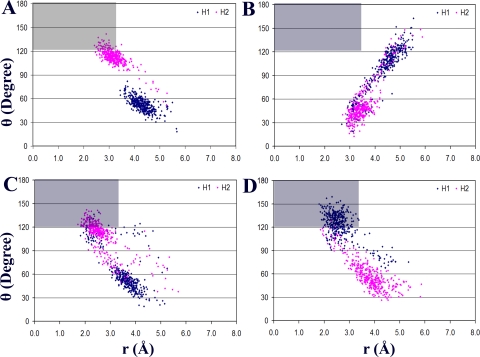
The productive binding orientations of phenacetin were determined using geometric parameters, distance r and angle θ, as described under Materials and Methods. These parameters were then plotted based on 400 recorded snapshots for each enzyme-substrate complex. The representative plots, those for CYP1A2 WT, N312L, L382V, and L382V/T223N mutants, showing the ensembles of substrate orientations, are presented in Fig. 2. The region where the geometric criterion (r ≤ 3.5 Å and θ ≥ 120°) is satisfied is gray, and the points (or frames) located within represent all the snapshots of each enzyme-substrate complex where phenacetin was bound in the productive binding orientation. The distribution of the MD frames for each enzyme-substrate complex displayed varied. Overall, phenacetin showed higher occupancy within a productive binding region (counted as hits) in the L382V and L382V/T223N mutants than in the WT. Similar results were also seen in the case of other multiple mutants containing L382V (data not shown), whereas few hits, if any, were observed for the remaining four single mutants, as shown for the N312L mutant (Fig. 2B). A quantitative analysis of the hits for each enzyme-substrate complex revealed that the number of hits for L382V, L382V/T223N, and L382V/T223N/N312L mutants were 2-, 4-, and 5-fold higher, respectively, than that for the WT (Table 4). In contrast, zero or few hits were obtained for T124S, T223N, V227G, and N312L mutants during 100-ps dynamics. It is possible that some hits might be recorded during a longer simulation time. Overall, the results of MD simulations are, like those from docking experiments, consistent with the kinetic and stoichiometric analyses of CYP1A2 WT and mutants.
Fig. 2.
Ensembles of substrate orientations obtained from 100-ps MD simulations of phenacetin-enzyme complexes described by geometric parameters, distance r and angle θ. H1 in -OCH2- group of phenacetin is blue, and H2 is red. Gray regions, where the criterion (r ≤ 3.5 Å and θ ≥ 120°) is satisfied, represent productive binding orientations of phenacetin within the active site. CYP1A2 enzymes were WT (A), N312L mutant (B), L382V mutant (C), and L382V/T223N mutant (D).
TABLE 4.
Geometric analysis of MD results for CYP1A2 WT and mutants
Hits represent the MD frames where phenacetin was in the productive binding orientation, as evaluated by the geometric criterion: r ≤ 3.5 Å and θ ≥ 120°.
| CYP1A2 | Number of Hits |
|---|---|
| WT | 64 |
| T124S | 0 |
| T223N | 0 |
| V227G | 4 |
| N312L | 0 |
| L382V | 134 |
| L382V/T223N | 252 |
| L382V/T223N/N312L | 308 |
A possible drawback of the simulations described previously might have been the use of the distance-dependent dielectric constant instead of explicit solvent. Therefore, we have also conducted MD simulations of phenacetin docked in the active site of CYP1A2 WT and the L382V mutant with the solvent molecules present and the dielectric constant of 1. In the presence of water, 70 hits were recorded for the WT enzyme and 144 hits for the L382V mutant. Thus, in both cases, the percentage of hits increased less than 10% and to a very similar extent (9.4% for the WT and 7.5% for the mutant) compared with the results from MD simulations without water (see Table 4). This result indicates that, although some changes may be observed with solvent present, simulations with distance-dependent dielectric provide a reasonable approach to explain the observed experimental findings.
Discussion
Our previous studies indicated that residue 382 plays an important role in controlling the specificity of alkoxyresorufin O-dealkylation by CYP1A1 and CYP1A2 (Liu et al., 2003, 2004; Tu et al., 2008). Because phenacetin is another important probe substrate for CYP1A2, the studies to address the effects of reciprocal mutations, particularly L382V, on phenacetin oxidation may broaden our understanding of the role of this residue in substrate specificity. In the present study, nine single and multiple mutants were investigated using a battery of complementary approaches, such as kinetic assays, stoichiometry measurements, and molecular modeling methods. The results showed that the L382V substitution resulted in a significant increase in catalytic activity and substrate specificity of CYP1A2. All of the multiple mutants that contained this substitution displayed very similar kinetics, stoichiometry, and dynamic mobility as the single L382V mutant. Thus, the presence of this single residue was critical for dramatically improving the efficiency of phenacetin oxidation by CYP1A2.
To date, many residues of CYP1A2 have been identified that may play a role in enzyme-ligand interactions by site-directed mutagenesis and/or molecular modeling studies (Yun et al., 2000; Liu et al., 2004; Zhou et al., 2009). In the case of single mutants, most of the mutations, including T124S, T223N, V227G, and N312L in the present study, resulted in decreased catalytic activity and substrate specificity compared with the WT enzyme (Parikh et al., 1999; Liu et al., 2004). A number of CYP1A2 mutants obtained from random mutagenesis showed increased catalytic activities and substrate specificities toward phenacetin (Parikh et al., 1999), but none of them was as highly active as the L382V mutant reported in this study (Table 1). It is worth mentioning that the kinetic parameters for phenacetin O-dealkylation by CYP1A2 WT determined in the present investigation were very similar to those reported by Parikh et al. (1999). Although the L382V mutation dramatically increased oxidation of phenacetin, it led to a significant decrease in 7-methoxyresorufin O-dealkylation (Liu et al., 2004). Thus, the functional effect of residue substitution appears to be dependent on the substrate (Zhou et al., 2009).
Moreover, the binding constants determined for CYP1A2 WT and mutants showed that the L382V substitution leads to tighter phenacetin binding in the active site of the L382V-containing mutants, consistent with enzyme kinetics results. This effect may increase phenacetin residence time, resulting in higher activity.
To better explain the effects of mutations on catalytic activity and substrate specificity of CYP1A2, stoichiometry studies were performed. The coupling efficiencies of different mutants, expressed in terms of product/NADPH, H2O2/O2, and H2O/product ratios, were compared with those of WT. A similar approach has been used by Fang et al. (1997) and Kobayashi et al. (1998) to evaluate coupling efficiencies of CYP2B1 mutants. Frequently, the mutation decreases the coupling efficiency of the P450, as observed with CYP2B1 (Fang et al., 1997; Kobayashi et al., 1998) and P450cam (French et al., 2002), but the effect depends on the substrate. Previously studied CYP1A2 mutants showed only a small increase in coupling efficiency with phenacetin as a substrate (Yun et al., 2000), in contrast to our results with the L382V-containing mutants. In the present studies, the L382V mutant and multiple mutants containing the L382V substitution were similar or more efficient at coupling reducing equivalents to acetaminophen formation than the WT (Table 3). In general, the increased ratios of product/NADPH for the L382V mutant and multiple mutants and the decreased ratios for T124S, T223N, V227G, and N312L mutants were in agreement with the kinetic data regarding phenacetin turnover rates. Although no significant changes of uncoupling to H2O2 at the first and the second branching points were observed in all the mutants, those that contained the L382V substitution showed less uncoupling to water (decreased H2O/product ratio; Table 3). Thus, it seems reasonable to suggest that the L382V substitution in CYP1A2 yields the mutants that use NADPH and O2 more efficiently to oxidize phenacetin to products and display less uncoupling to water, so that the overall coupling efficiency of the enzyme increases, which is consistent with enzyme kinetics results (Table 1).
Molecular modeling studies provide another possible explanation of the effects of mutations on phenacetin specificity, as indicated by changes in kinetic parameters. For these simulations, we used the X-ray structure of CYP1A2, and the structures of the mutants were derived from the crystal. As reported by Sansen et al. (2007), this CYP1A2 structure has a closed compact active site, without clear solvent or substrate access channels, with a relatively small volume of the cavity, estimated at 375 Å3. The active site of CYP1A2 is approximately 44% larger than that of CYP2A6 (260 Å3) and significantly smaller than that of CYP3A4 (1385 Å3) (Sansen et al., 2007). Consequently, only planar compounds, such as α-naphthoflavone, and typical CYP1A2 substrates such as phenacetin, 7-ethoxyresorfin, caffeine, tacrine, or theophylline can be fitted well with the narrow and flat active site cavity of enzyme. Leu382 of CYP1A2 is a critical residue located close to the heme iron, and its replacement with a smaller Val increases the volume of the active site near heme. This activity allows phenacetin to move closer to heme, so that one of the hydrogens at the oxidation site is within a hydrogen-bonding distance from the ferryl oxygen (Fig. 1), which promotes hydrogen abstraction. This is consistent with the MD results (Table 4; Fig. 2). Consequently, the movement of phenacetin closer to the ferryl oxygen in the L382V mutants helps to explain not only the substantial increase in kcat and a decrease in Km for the production of acetaminophen (Table 1) but also more efficient coupling of the P450 reaction cycle with less water formation (Table 3).
MD simulations similar to those described in this work have been successfully used in our previous studies for fairly rapid predictions or interpretation of experimental results (Ericksen and Szklarz, 2005; Tu et al., 2008). The present studies suggest that the use of the distance-dependent dielectric constant can be a reasonable substitute for the presence of explicit water molecules. We have observed only a small increase in the percentage of hits (less than 10%) from MD simulations with water. These conclusions may be further verified by more extensive MD simulations of the complete protein-substrate complexes on nanosecond timescales. More recently, longer 2-ns MD simulations have been successfully used to predict the effect of mutations on the catalytic efficiency of CYP2B6 (Nguyen et al., 2008).
In summary, our results show that the L382V substitution in CYP1A2 can alter the binding orientation of phenacetin within the active site of the enzyme, so that the site of metabolism moves closer to the heme iron. This provides a good mechanistic explanation for the increased catalytic efficiency and coupling efficiency of phenacetin O-dealkylation in CYP1A2 mutants containing the L382V substitution. The current studies also show that a combination of several experimental approaches with molecular modeling methods can improve our understanding of P450 catalysis. These different methodologies complement each other well to offer a mechanistic interpretation of P450 function on a molecular level.
Acknowledgments.
We thank Dr. Rahul Deshmukh for help and advice concerning expression and purification of P450 enzymes. Molecular modeling studies were performed at the Computational Chemistry and Molecular Modeling Laboratory, Department of Basic Pharmaceutical Sciences, School of Pharmacy, West Virginia University, Morgantown, West Virginia.
This work was supported in part by the National Institutes of Health National Center for Research Resources [Grant RR16440]; and the National Institutes of Health National Institute of General Medical Sciences [Grant GM079724].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.109.030767.
- P450
- cytochrome P450
- MD
- molecular dynamics
- DLPC
- dilauroyl-l-3-phosphatidyl-choline
- HPLC
- high-performance liquid chromatography
- WT
- wild type
- RMSD
- root mean square deviation.
References
- Burke MD, Thompson S, Weaver RJ, Wolf CR, Mayer RT. (1994) Cytochrome P450 specificities of alkoxyresorufin O-dealkylation in human and rat liver. Biochem Pharmacol 48:923–936 [DOI] [PubMed] [Google Scholar]
- Distlerath LM, Reilly PE, Martin MV, Davis GG, Wilkinson GR, Guengerich FP. (1985) Purification and characterization of the human liver cytochromes P-450 involved in debrisoquine 4-hydroxylation and phenacetin O-deethylation, two prototypes for genetic polymorphism in oxidative drug metabolism. J Biol Chem 260:9057–9067 [PubMed] [Google Scholar]
- Ericksen SS, Szklarz GD. (2005) Regiospecificity of human cytochrome P450 1A1-mediated oxidations: the role of steric effects. J Biomol Struct Dyn 23:243–256 [DOI] [PubMed] [Google Scholar]
- Fang X, Kobayashi Y, Halpert JR. (1997) Stoichiometry of 7-ethoxycoumarin metabolism by cytochrome P450 2B1 wild-type and five active-site mutants. FEBS Lett 416:77–80 [DOI] [PubMed] [Google Scholar]
- French KJ, Rock DA, Rock DA, Manchester JI, Goldstein BM, Jones JP. (2002) Active site mutations of cytochrome p450cam alter the binding, coupling, and oxidation of the foreign substrates (R)- and (s)-2-ethylhexanol. Arch Biochem Biophys 398:188–197 [DOI] [PubMed] [Google Scholar]
- Guengerich FP, Shimada T. (1991) Oxidation of toxic and carcinogenic chemicals by human cytochrome P-450 enzymes. Chem Res Toxicol 4:391–407 [DOI] [PubMed] [Google Scholar]
- Jiang ZY, Woollard AC, Wolff SP. (1990) Hydrogen peroxide production during experimental protein glycation. FEBS Lett 268:69–71 [DOI] [PubMed] [Google Scholar]
- Kedzie KM, Balfour CA, Escobar GY, Grimm SW, He YA, Pepperl DJ, Regan JW, Stevens JC, Halpert JR. (1991) Molecular basis for a functionally unique cytochrome P450IIB1 variant. J Biol Chem 266:22515–22521 [PubMed] [Google Scholar]
- Kobayashi Y, Fang X, Szklarz GD, Halpert JR. (1998) Probing the active site of cytochrome P450 2B1: metabolism of 7-alkoxycoumarins by the wild type and five site-directed mutants. Biochemistry 37:6679–6688 [DOI] [PubMed] [Google Scholar]
- Lewis DF, Moereels H, Lake BG, Ioannides C, Parke DV. (1994) Molecular modeling of enzymes and receptors involved in carcinogenesis: QSARs and compact-3D. Drug Metab Rev 26:261–285 [DOI] [PubMed] [Google Scholar]
- Liu J, Ericksen SS, Besspiata D, Fisher CW, Szklarz GD. (2003) Characterization of substrate binding to cytochrome P450 1A1 using molecular modeling and kinetic analyses: case of residue 382. Drug Metab Dispos 31:412–420 [DOI] [PubMed] [Google Scholar]
- Liu J, Ericksen SS, Sivaneri M, Besspiata D, Fisher CW, Szklarz GD. (2004) The effect of reciprocal active site mutations in human cytochromes P450 1A1 and 1A2 on alkoxyresorufin metabolism. Arch Biochem Biophys 424:33–43 [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275 [PubMed] [Google Scholar]
- Modi S, Primrose WU, Boyle JM, Gibson CF, Lian LY, Roberts GC. (1995) NMR studies of substrate binding to cytochrome P450 BM3: comparisons to cytochrome P450 cam. Biochemistry 34:8982–8988 [DOI] [PubMed] [Google Scholar]
- Nerurkar PV, Park SS, Thomas PE, Nims RW, Lubet RA. (1993) Methoxyresorufin and benzyloxyresorufin: substrates preferentially metabolized by cytochromes P4501A2 and 2B, respectively, in the rat and mouse. Biochem Pharmacol 46:933–943 [DOI] [PubMed] [Google Scholar]
- Nguyen TA, Tychopoulos M, Bichat F, Zimmermann C, Flinois JP, Diry M, Ahlberg E, Delaforge M, Corcos L, Beaune P, et al. (2008) Improvement of cyclophosphamide activation by CYP2B6 mutants: from in silico to ex vivo. Mol Pharmacol 73:1122–1133 [DOI] [PubMed] [Google Scholar]
- Omura T, Sato R. (1964) The carbon monoxide-binding pigment of liver microsomes. II. Solubilization, purification, and properties. J Biol Chem 239:2379–2385 [PubMed] [Google Scholar]
- Parikh A, Josephy PD, Guengerich FP. (1999) Selection and characterization of human cytochrome P450 1A2 mutants with altered catalytic properties. Biochemistry 38:5283–5289 [DOI] [PubMed] [Google Scholar]
- Paulsen MD, Ornstein RL. (1991) A 175-psec molecular dynamics simulation of camphor-bound cytochrome P-450cam. Proteins 11:184–204 [DOI] [PubMed] [Google Scholar]
- Paulsen MD, Ornstein RL. (1992) Predicting the product specificity and coupling of cytochrome P450cam. J Comput Aided Mol Des 6:449–460 [DOI] [PubMed] [Google Scholar]
- Sansen S, Yano JK, Reynald RL, Schoch GA, Griffin KJ, Stout CD, Johnson EF. (2007) Adaptations for the oxidation of polycyclic aromatic hydrocarbons exhibited by the structure of human P450 1A2. J Biol Chem 282:14348–14355 [DOI] [PubMed] [Google Scholar]
- Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. (1994) Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther 270:414–423 [PubMed] [Google Scholar]
- Tu Y, Deshmukh R, Sivaneri M, Szklarz GD. (2008) Application of molecular modeling for prediction of substrate specificity in cytochrome P450 1A2 mutants. Drug Metab Dispos 36:2371–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke LL, Greenblatt DJ, Duan SX, Schmider J, Kudchadker L, Fogelman SM, Harmatz JS, Shader RI. (1996) Phenacetin O-deethylation by human liver microsomes in vitro: inhibition by chemical probes, SSRI antidepressants, nefazodone and venlafaxine. Psychopharmacology (Berl) 128:398–407 [DOI] [PubMed] [Google Scholar]
- Yuan R, Madani S, Wei XX, Reynolds K, Huang SM. (2002) Evaluation of cytochrome P450 probe substrates commonly used by the pharmaceutical industry to study in vitro drug interactions. Drug Metab Dispos 30:1311–1319 [DOI] [PubMed] [Google Scholar]
- Yun CH, Miller GP, Guengerich FP. (2000) Rate-determining steps in phenacetin oxidations by human cytochrome P450 1A2 and selected mutants. Biochemistry 39:11319–11329 [DOI] [PubMed] [Google Scholar]
- Zhou SF, Yang L-P, Wei MQ, Duan W, Chan E. (2009) Insights into the structure, function, and regulation of human cytochrome P450 1A2. Curr Drug Metab 10:713–729 [DOI] [PubMed] [Google Scholar]