Abstract
Efavirenz primary and secondary metabolism was investigated in vitro and in vivo. In human liver microsome (HLM) samples, 7- and 8-hydroxyefavirenz accounted for 22.5 and 77.5% of the overall efavirenz metabolism, respectively. Kinetic, inhibition, and correlation analyses in HLM samples and experiments in expressed cytochrome P450 show that CYP2A6 is the principal catalyst of efavirenz 7-hydroxylation. Although CYP2B6 was the main enzyme catalyzing efavirenz 8-hydroxylation, CYP2A6 also seems to contribute. Both 7- and 8-hydroxyefavirenz were further oxidized to novel dihydroxylated metabolite(s) primarily by CYP2B6. These dihydroxylated metabolite(s) were not the same as 8,14-dihydroxyefavirenz, a metabolite that has been suggested to be directly formed via 14-hydroxylation of 8-hydroxyefavirenz, because 8,14-dihydroxyefavirenz was not detected in vitro when efavirenz, 7-, or 8-hydroxyefavirenz were used as substrates. Efavirenz and its primary and secondary metabolites that were identified in vitro were quantified in plasma samples obtained from subjects taking a single 600-mg oral dose of efavirenz. 8,14-Dihydroxyefavirenz was detected and quantified in these plasma samples, suggesting that the glucuronide or the sulfate of 8-hydroxyefavirenz might undergo 14-hydroxylation in vivo. In conclusion, efavirenz metabolism is complex, involving unique and novel secondary metabolism. Although efavirenz 8-hydroxylation by CYP2B6 remains the major clearance mechanism of efavirenz, CYP2A6-mediated 7-hydroxylation (and to some extent 8-hydroxylation) may also contribute. Efavirenz may be a valuable dual phenotyping tool to study CYP2B6 and CYP2A6, and this should be further tested in vivo.
Efavirenz-based antiretroviral therapy continues to be the preferred initial therapy in the treatment of naive HIV-1/AIDS patients, but its use is associated with variable treatment response and adverse effects in most part because of the large differences in pharmacokinetics (Marzolini et al., 2001; Csajka et al., 2003). Efavirenz is predominantly cleared by hepatic metabolism (Mutlib et al., 1999b). The metabolites identified in human plasma and urine (almost exclusively as glucuronide or sulfate conjugates) were 7- and 8-hydroxyefavirenz (primary metabolites) and 8,14-dihydroxyefavirenz (secondary metabolite). Thus, factors that alter efavirenz clearance could influence efficacy or toxicity of the drug.
Cytochrome P450 (P450) 2B6 is the main enzyme catalyzing the major clearance mechanism of efavirenz, 8-hydroxylation to 8-hydroxyefavirenz, in vitro (Ward et al., 2003; Desta et al., 2007). Clinical studies in HIV patients have repeatedly shown that CYP2B6 genetic variants with functional consequences are associated with higher efavirenz exposure and in some studies with increased risk for adverse central nervous system effects compared with those without variants (Zanger et al., 2007). However, not all efavirenz pharmacokinetic variability could be explained by the 8-hydroxylation pathway or CYP2B6 alone because a large intersubject variability in efavirenz exposure remains even after accounting for known CYP2B6 genetic variations (Rotger et al., 2007; Arab-Alameddine et al., 2009). Other P450s that include expressed CYP3A4/5 and CYP1A2 show activity toward efavirenz 8-hydroxylation in vitro (Ward et al., 2003), but the in vivo contribution of these enzymes, if any, appears to be marginal (Mouly et al., 2002; Tsuchiya et al., 2004; Bristol-Myers Squibb Company, 2009). Metabolic pathways other than efavirenz 8-hydroxylation may also contribute to efavirenz clearance. Efavirenz 7-hydroxylation to 7-hydroxyefavirenz has been shown in in vitro (Ward et al., 2003; Desta et al., 2007) and in vivo animal and human studies (Mutlib et al., 1999b), although the contribution of this route to the overall clearance of efavirenz and the enzymes involved remains poorly defined. Correlation analysis between the activity of P450 enzymes and formation rates of 7-hydroxyefavirenz in a small panel of human liver microsomes (HLMs) (Ward et al., 2003) implicated CYP2A6 compared with other P450s (r = 0.45; p = 0.19). A significant correlation between formation rate of 7-hydroxyefavirenz and CYP2A6 protein (Spearman r = 0.395; p < 0.0001) and activity (r = 0.583; p < 0.0001) was observed in a subsequent study involving a large panel of human liver samples (Desta et al., 2007). However, it was difficult to ascertain the contribution of this enzyme in efavirenz 7-hydroxylation because a significant correlation between CYP2A6 protein and activity with CYP2B6 protein and activity, as well as formation rate of 8-hydroxyefavirenz, was also observed (Desta et al., 2007). Recent association studies in HIV patients support the role CYP2A6 may play in efavirenz clearance (Arab-Alameddine et al., 2009; di Iulio et al., 2009; Kwara et al., 2009a,b). However, direct evidence linking CYP2A6 or any other enzyme in efavirenz metabolism is lacking.
Based largely on in vitro studies, CYP2B6, which shows highly variable expression and activity among individuals in part as a result of genetic variation and exposure to inhibitors or inducers, metabolizes a growing list of clinically important drugs, environmental chemicals, and endogenous compounds (for reviews, see Ekins and Wrighton, 1999; Hodgson and Rose, 2007; Zanger et al., 2007; Wang and Tompkins, 2008; Mo et al., 2009). However, information on the clinical relevance of this enzyme has been generally limited because of the lack of suitable in vivo substrate probe. Bupropion 4-hydroxylation has been increasingly used as an in vitro and in vivo probe of activity, but its in vivo utility has important limitations (Kharasch et al., 2008). Efavirenz has been proposed to be an alternative probe of CYP2B6 activity to evaluate the clinical relevance of this enzyme (Ward et al., 2003; US Food and Drug Administration, 2006). To identify and select appropriate markers (e.g., metabolic ratios) that can best describe CYP2B6 activity in vivo, a comprehensive characterization of the primary and secondary metabolism of efavirenz and the specific enzymes involved is important.
The main aims of this study were to use human liver cellular fractions, expressed P450s, and clinical samples collected from efavirenz-treated subjects to 1) assess the contribution of efavirenz 7-hydroxylation in efavirenz elimination and to identify the specific P450 enzyme(s) catalyzing this reaction; 2) characterize the metabolism of efavirenz via the 8-hydroxylation pathway to further confirm previous findings and use this as a positive control; and 3) characterize more closely efavirenz secondary metabolism and identify the P450s involved.
Materials and Methods
Chemicals.
Efavirenz, 8-hydroxyefavirenz, 7-hydroxyefavirenz, 8,14-dihydroxyefavirenz, and nevirapine were obtained from Toronto Research Chemicals Inc. (North York, ON, Canada). Diethyldithiocarbamate, ketoconazole, furafylline, omeprazole, pilocarpine, quercetin, quinidine, thioTEPA, ticlopidine, troleandomycin, glucose 6-phosphate, glucose-6-phosphate dehydrogenase, and NADP+ were purchased from Sigma-Aldrich (St. Louis, MO). Letrozole was purchased from U.S. Pharmacopeia (Rockville, MD). Sulfaphenazole was obtained from SAFC Corp. (St. Louis, MO). 8-Hydroxyefavirenz glucuronide was a gift from Dr. David Christ (DuPont Pharmaceuticals, Wilmington, DE). All the other chemicals were of high-performance liquid chromatography (HPLC) grade.
Microsomal Preparations.
HLM samples were prepared from human liver tissues that were medically unsuitable for transplantation as described elsewhere (Jeong et al., 2009a), or characterized HLM samples were purchased from CellzDirect (Dallas, TX) and BD Biosciences (San Jose, CA). Baculovirus-insect cell-expressed human P450s (CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4, and CYP3A5) (with oxidoreductase) were purchased from BD Biosciences. HLM samples and P450s were stored at −80°C until used.
Liquid Chromatography/Tandem Mass Spectrometry Assay Method.
A new selective and sensitive liquid chromatography/tandem mass spectrometry (LC/MS/MS) method was developed to detect and quantify efavirenz and its known metabolites in microsomal incubates because a previously used HPLC method (Ward et al., 2003) lacked sensitivity to quantify metabolites produced at low rate. The MS/MS system was an API 2000 MS/MS triple quadruple system (Applied Biosystems, Foster City, CA) equipped with a turbo ion spray and was coupled with a Shimadzu (Columbia, MD) HPLC system consisting of an LC-20AB pump and SIL-20A HT autosampler, all controlled by Analyst 1.4.2 software (Applied Biosystems/MDS Sciex, Foster City, CA) in conjunction with Windows 2000 (Microsoft, Redmond, WA). Separation of compounds was achieved using a reverse-phase chromatography using a Phenomenex (Torrance, CA) Luna C18 column (100 × 2 mm; 3 μm) and a mobile phase that consisted of acetonitrile and 0.1% formic acid (50:50; v/v) (isocratic flow rate 0.15 ml/min).
Stock solutions of efavirenz, 7-hydroxyfefavirenz, 8-hydroxyefavirenz, 8,14-dihydroxyefavirenz, and nevirapine (internal standard) were prepared separately in polypropylene tubes by adding methanol to a concentration of 1 mg/ml. These stock solutions were stored at −20°C. Serial dilutions were performed from the stock solutions in methanol and mobile phase for MS/MS optimization experiments. Parent and fragment ions were detected in multiple reaction monitoring (MRM) mode. All the analytes (nevirapine, efavirenz, 7- and 8-hydroxyefavirenz, and 8,14-dihydroxyefavirenz) were observed under unit resolution for quadrupoles 1 and 3. Mass spectrometry (MS) optimization was performed by adjustments of the source- and compound-dependent parameters as listed in Supplemental Table 1. Optimal gas pressures for all the analytes were nitrogen nebulizer gas, 10 psi; curtain gas, 10 psi; collision gas, 3.0 psi; ion source gas (1), 24 psi; and source gas (2), 41 psi. Other parameter settings were ion spray voltage, −4500 V in negative mode (4200 V in positive mode); and source temperature of 470°C. Efavirenz and its metabolites were measured with the quantifier MRM and confirmed with the qualifier MRM transition (Supplemental Table 1). The MRM scan was performed in positive ion mode for the internal standard (nevirapine) and in negative ion mode for efavirenz and its metabolites. The quantifier and qualifier transitions were selected based on the signal intensity of the metabolite peaks (Supplemental Table 1). The parent and daughter ions were the same for 7- and 8-hydroxyefavirenz. Therefore, the two metabolites were quantified after chromatographic separation.
Identification/Confirmation of Efavirenz Metabolites.
Pilot incubation experiments were performed as described previously with slight modification (Ward et al., 2003; Desta et al., 2007) to identify potentially new metabolites and/or confirm the presence of known efavirenz primary and secondary metabolites. Efavirenz was used as a substrate to determine primary metabolites, whereas 7- and 8-hydroxyefavirenz and 8-hydroxyefavirenz glucuronide were used as substrates to assess secondary metabolism. Substrates (in methanol) were added to a tube, and solvent was evaporated. Residue was reconstituted with phosphate reaction buffer (0.2 M Na2HPO4 titrated with 0.2 M NaH2PO4 to a pH of 7.4), and a mixture of efavirenz (20 μM; or 7- or 8-hydroxyefavirenz, 5 μM; 8-hydroxyefavirenz glucuronide, 50 μM) and HLMs (1 mg/ml) was allowed to equilibrate for 5 min at 37°C (final volume, 250 μl). The reaction was initiated by adding an NADPH-generating system (cofactors) (13 mM NADP, 33 mM glucose 6-phosphate, 33 mM MgCl2, and 0.4 U/ml glucose-6-phosphate dehydrogenase) and allowed to proceed for 30 min at 37°C. After terminating the reaction by immediately adding 500 μl of acetonitrile and placing tubes on ice, nevirapine was added as an internal standard for the incubation sample of efavirenz and 7- and 8-hydroxyefavirenz. Ritonavir (25 μl of 0.01 mg/ml) was used as an internal standard for incubation consisting of 8-hydroxyefavirenz glucuronide. All the samples were vortex-mixed for 30 s and centrifuged at high speed for 5 min in an Eppendorf model 5415D centrifuge (Brinkmann Instruments, Westbury, NY).
The supernatant was removed, transferred to another tube, and extracted with ethyl acetate under alkaline pH as described previously (Ward et al., 2003). After centrifugation for 15 min at 2500 rpm in a Beckman Coulter (Brea, CA) Allegra 6R Benchtop Centrifuge, the organic layer was removed, transferred to another tube, and evaporated to dryness. The residue was reconstituted with 50 μl of mobile phase from which 10 μl was injected onto the LC/MS/MS system described below (efavirenz, and 7- and 8-hydroxyefavirenz incubations). For 8-hydroxyefavirenz glucuronide incubation, the residue was reconstituted in 150 μl of mobile phase from which 100 μl was injected onto HPLC/UV/visible system (Ward et al., 2003) that consisted of a Zorbax SB-C18 column (150 × 4.6 mm; 3.5-μm particle size; Phenomenex), a Luna C18 Guard column (30 × 4.6 mm; 5 μm; Phenomenex), and a mobile phase composed of 65% 10 mM KH2PO4 (adjusted to pH 2.4 with 1% phosphoric acid) and 35% (v/v) acetonitrile (flow rate, 0.8 ml/min). Negative control incubations consisting of no substrate, no cofactors, or no microsomes (bovine serum albumin was used instead) were run in parallel and processed as noted previously.
Retention times, precursor ion (Q1) scan data obtained under negative electrospray ionization conditions ([M + H]− m/z 330 for 7- and 8-hydroxyefavirenz, m/z 314 for efavirenz, and m/z 346 for 8,14-dihydroxyefavirenz), and product ion scan (Q3) spectral data of the known synthetic metabolite standards were compared with those metabolite peaks obtained from microsomal incubates to confirm previously reported efavirenz primary and secondary metabolites (7- and 8-hydroxyefavirenz and 8,14-dihydroxyefavirenz). Precursor ion (Q1) scans were also conducted in both negative and positive ion modes under different MS settings to search for potentially new, structurally predicted, oxidative metabolites of efavirenz. If any potential metabolite peak was detected by Q1 scan, then sample and negative control incubations were reinjected into the LC/MS/MS and subjected to a product ion scan (Q3) range of m/z 100 to 400. For the 8-hydroxyefavirenz glucuronide incubations, the analysis was made using HPLC with UV detection because LC/MS/MS analysis did not yield useful information.
Linear conditions for metabolite formation with respect to protein and time were assessed before the subsequent studies. A final protein concentration of 0.25 mg/ml and 10-min incubation represented linear conditions and were used. In addition, these incubation conditions were selected to minimize sequential metabolism while ensuring assay sensitivity. The same conditions were used for determination of efavirenz sequential metabolism.
Kinetic Analyses.
Rates of efavirenz metabolism to 7- and 8-hydroxyefavirenz were determined by incubating a range of efavirenz concentrations (1–150 μM) in duplicate for 10 min at 37°C with characterized HLM samples (n = 7; 0.25 mg of protein/ml) and cofactors. The reaction was terminated and processed as described previously.
Correlation Analyses.
Correlation between formation rates of efavirenz metabolism to 7- and 8-hydroxyefavirenz and the activity of specific isoforms was tested by incubating efavirenz (10 μM) with a panel of 15 characterized HLM samples (0.25 mg/ml) and cofactors in duplicate for 10 min at 37°C. The activity of each P450 isoform in each HLM sample, determined by isoform-specific reaction markers, was as provided by the supplier (BD Biosciences, http://www.bdbiosciences.com/ptDatabaseList.jsp).
Inhibition Analyses.
Efavirenz (10 μM) was incubated with HLMs (0.25 mg/ml) and the NADPH-generating system at 37°C for 10 min in the absence (control) and presence of the following known isoform-specific inhibitors: pilocarpine (50 μM) and letrozole (50 μM) for CYP2A6, quercetin (50 μM) for CYP2C8, sulfaphenazole (25 μM) for CYP2C9, ticlopidine (5 μM) for CYP2C19 and CYP2B6, quinidine (1 μM) for CYP2D6, and ketoconazole (1 μM) for CYP3A. Inhibition by furafylline (20 μM) for CYP1A2, thioTEPA (50 μM) for CYP2B6, diethyldithiocarbamate (50 μM) for CYP2E1, and troleandomycin (50 μM) for CYP3A has been shown to be time-dependent. Therefore, a preincubation protocol was implemented in which the inhibitor was first incubated with HLM sample (0.25 mg of protein/ml) and cofactors for 15 min at 37°C before the reaction was initiated by addition of the respective selective substrate probe. The specific conditions for use of these inhibitors have been described in detail in previous publications (Newton et al., 1995; Bourrié et al., 1996; Rae et al., 2002; Ward et al., 2003; Jeong et al., 2009a,b). Sample processing was as described previously. The percentage formation rate remaining after inhibition relative to the uninhibited controls (without inhibitors or vehicle controls) was calculated.
Additional experiments were performed to evaluate the role CYP2A6 and CYP2B6 play in catalyzing efavirenz 7- and 8-hydroxylation and to assess the relative selectivity of pilocarpine and thioTEPA toward these enzymes. Efavirenz (10 μM) was incubated for 10 min at 37°C with HLM samples (0.25 mg/ml) and cofactors in the absence (control) and presence of an inhibitor. A single concentration of thioTEPA (50 μM) or pilocarpine (50 μM) or multiple concentrations of thioTEPA (0–100 μM) or pilocarpine (0–100 μM) were used in these inhibition experiments.
Efavirenz Primary and Secondary Metabolism by Expressed P450 Isoforms.
Efavirenz (10 and 100 μM) was incubated with expressed CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4, or CYP3A5 (13–26 pmol) and cofactors (same composition as previously) at 37°C for 10 min. The lower concentration of efavirenz is close to the Km values, whereas the higher concentration represents concentration at saturation (Vmax in Michaelis-Menten curve). For those isoforms that showed substantial activity toward efavirenz metabolism, namely, expressed CYP2A6 and CYP2B6, full kinetic analyses were performed by incubating efavirenz (1–150 μM) with cofactors and expressed CYP2A6 (26 pmol) or CYP2B6 (13 pmol) for 10 min. In addition, inhibition of expressed CYP2A6 and CYP2B6 by thioTEPA and pilocarpine was tested by incubating efavirenz (10 μM) with expressed CYP2A6 (26 pmol) or CYP2B6 (13 pmol) in the absence (control) and presence of thioTEPA (0–100 μM) or pilocarpine (0–100 μM). Further processing of incubate was as described previously.
Efavirenz sequential metabolism by expressed P450s was determined by incubating 5 μM 7-hydroxyefavirenz, 5 μM 8-hydroxyefavirenz, and 50 μM 8-hydroxyefavirenz glucuronide for 10 min with 13 to 26 pmol of P450 and cofactors (30-min incubation was used for the glucuronide). The kinetics for the metabolism of 7- or 8-hydroxyefavirenz (1–150 μM) was determined in expressed CYP2B6 (13 pmol). HPLC with UV detection (Ward et al., 2003) and/or LC/MS/MS as described previously were used to monitor metabolites formed.
In Vivo Studies.
To determine efavirenz metabolism and pharmacokinetics in vivo, efavirenz and its metabolites were measured in plasma samples obtained from healthy volunteers who participated in a clinical trial and received a single 600-mg oral dose of efavirenz. The clinical trial was approved by the Institutional Review Board of the Indiana University School of Medicine. Eligible subjects participated in the trial after signing a written informed consent form. Plasma samples were collected at baseline and 0.5 to 72 h after efavirenz dosing.
A pilot study has shown that nearly all the efavirenz metabolites exist predominantly as conjugates, consistent with in vivo evidence (Mutlib et al., 1999b). Therefore, plasma samples were analyzed after enzymatic hydrolysis (deconjugation by incubating overnight with β-glucuronidase). Plasma samples (200 μl) were incubated with 2 ml of sodium acetate buffer (0.2 M), pH 5.0, and 100 μl of 10,000-unit β-glucuronidase at 37°C for 17 h. After adding the internal standard, the samples were extracted with 5 ml of ethyl acetate under alkaline pH (0.1 M sodium carbonate buffer, pH 9.4). The organic layer was evaporated to dryness in SpeedVac (Thermo Fisher Scientific, Waltham, MA). Residue was reconstituted in 100 μl of mobile phase and analyzed by the LC/MS/MS system described below.
The LC/MS/MS assay described in vitro was modified to measure plasma concentrations of efavirenz and its metabolites in clinical samples. Chromatographic analysis was performed using an Agilent 1100 series HPLC (Agilent Technologies, Santa Clara, CA) and a LEAP Technologies model HTS PAL autosampler (Carrboro, NC) coupled with an Applied Biosystems API 3200 triple-quadrupole mass spectrometer equipped with a turbo ion spray in positive ionization mode and controlled by Analyst software version 1.4.1 (Applied Biosystems/MDS Sciex) in conjunction with Windows 2000 (Microsoft). The separation column used was the same as that described for in vitro studies. A gradient elution profile was used: initial mobile phase, 99% of 0.01% formic acid in water and 1% of methanol (v/v); and the secondary mobile phase consisted of 99% methanol and 1% of 0.01% formic acid in water. The secondary mobile phase was increased from 50% to 90% linearly between 0 and 16 min; the initial mobile phase conditions were resumed after 16 min and remained constant for an additional 4 min, allowing the column to equilibrate. The eluate was introduced, without splitting, at 0.800 ml/min to the turbo ion source. MS optimization was achieved via adjustments of both the compound-dependent and compound-independent parameters for efavirenz, 8-hydroxyefavirenz, 7-hydroxyefavirenz, and 8,14-dihydroxyefavirenz. The analytes were optimized at a source temperature of 400°C, under unit resolution for quadrupoles 1 and 3, and were given a dwell time of 100 ms and a settling time of 75 ms. Optimal gas pressures for all five analytes, including the internal standard, were collision gas, 6 psi; curtain gas, 10 psi; ion source gas (1), 30 psi; and ion source gas (2), 25 psi. The compound-independent mass spectrometer parameters, as well as the parent and fragment ions that were used in MRM mode for detection, are listed in Supplemental Table 2. The limit of quantification of efavirenz and its metabolites in the in vitro and in vivo LC/MS/MS method was 3 ng/ml. The standard curve was linear over the range of 3 to 10,000 ng/ml. The day-to-day and within-day coefficient of variation was less than 21% at the lowest (5 ng/ml) and highest (5000 ng/ml) quality control concentrations used.
Data Analysis.
Apparent kinetic constants (Vmax and Km) were estimated by fitting formation rates of metabolites versus substrate concentrations to appropriate kinetic equations by nonlinear regression analysis using GraphPad Software Inc. (San Diego, CA; www.graphpad.com) Prism version 5.00 for Windows. In vitro intrinsic clearances (CLint) represent Vmax/Km. IC50 values were determined by analysis of the plot of the logarithm of the inhibitor concentration versus the percentage of activity remaining after inhibition using GraphPad software. Correlation analysis was performed by a nonparametric test (Spearman's rank correlation test) using GraphPad software. A p value less than 0.05 was considered statistically significant. Data are presented as mean ± S.D. or as averages of duplicate experiments.
Results
Primary Metabolism of Efavirenz In Vitro.
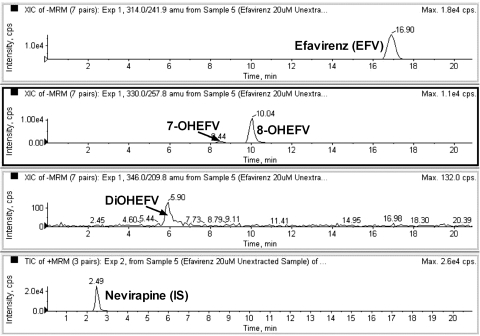
Two metabolites (Fig. 1) that were consistent with monohydroxyefavirenz were positively identified as 7- and 8-hydroxyefavirenz based on comparison of their retention times, precursor ion scan data (molecular mass of 331), and MS/MS fragmentation patterns with synthetic metabolite standards.
Fig. 1.
MRM trace chromatograms of efavirenz (EFV) in human liver microsomal incubates. Efavirenz (20 μM) was incubated with HLM samples and cofactors. Subsequent sample processing and LC/MS/MS conditions were as described under Materials and Methods. Metabolites that were consistent with monohydroxylated (7- and 8-hydroxyEFV) and secondary metabolite (dihydroxylated EFV) were identified in the microsomal incubates. Nevirapine was used as an internal standard.
Kinetic analyses.
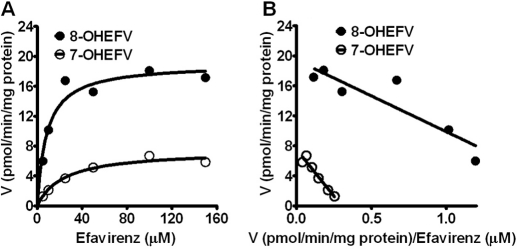
The kinetics for the formation of 7- and 8-hydroxyefavirenz from efavirenz was determined in several HLM samples. Representative Michaelis-Menten kinetic and Eadie-Hofstee plots in an HLM sample are depicted in Fig. 2, and the kinetic parameters of seven HLM samples are summarized in Table 1. The average Km values for the formation of 7- and 8-hydroxyefavirenz were comparable (average 14.7 versus 12.7 μM, respectively). The in vitro intrinsic clearances, CLint (Vmax/Km), show high variability among HLMs (∼35- and ∼13-fold for 7- and 8-hydroxyefavirenz formation, respectively) (Table 1). Because there was no marked difference in Km between the HLM samples tested, the interindividual difference in CLint is largely driven from differences in Vmax. The CLint for formation of 8-hydroxyefavirenz was on average 3.2-fold higher than that for 7-hydroxyefavirenz. Assuming that sequential metabolism is minimal (present study) and that the contributions of other elimination pathways such as nonhepatic clearance (Mutlib et al., 1999b) and efavirenz N-glucuronidation (Bélanger et al., 2009) to overall elimination of efavirenz are minimal, 7- and 8-hydroxylation appear to account for 22.5% (range, 9.8–38.5%) and 77.5% (range, 61.5–90.2%) of the overall efavirenz metabolism in vitro, respectively. In the limited number of HLMs, CLint values of 8-hydroxyefavirenz correlated significantly with CLint values of 7-hydroxyefavirenz (Spearman r = 0.86; p = 0.024), and this correlation appears to be mainly derived from Vmax values (r = 0.71; p = 0.088) rather than Km values (r = 0.21; p = 0.66).
Fig. 2.
Representative kinetics for the metabolism of efavirenz (EFV) to 7-hydroxyEFV and 8-hydroxyEFV in human liver samples (IU42). Increasing concentrations of EFV (5–150 μM) were incubated with HLM samples (0.25 mg/ml) and cofactors for 10 min at 37°C. Formation rates of metabolites (pmol/min/mg protein) versus EFV concentration (μM) were best fit to a one-site hyperbolic Michaelis-Menten equation (A) (see Data Analysis). The corresponding Eadie-Hofstee plots are shown (B). Each point represents the average of duplicate incubations. Kinetic parameters derived from these and another six HLM samples are listed in Table 1.
TABLE 1.
Kinetic parameters for the formation of 8-hydroxyefavirenz and 7-hydroxyefavirenz from efavirenz in seven different HLM samples and expressed CYP2A6 and CYP2B6
Increasing concentrations of efavirenz (1–150 μM) were incubated with HLM samples (0.25 mg/ml) and cofactors at 37°C (see Materials and Methods). Kinetic parameters for the formation of 8-hydroxyefavirenz and 7-hydroxyefavirenz were estimated by fitting the velocity versus substrate concentrations to the Michaelis-Menten equation as described in the Data Analysis section. Kinetic parameters in the individual HLM samples (S.E. of parameter estimates) are presented. In vitro CLint was calculated. Percentage contribution to the overall clearance was estimated as follows: CLint (metab) · 100/Clint (total). CLint (total) is the sum of CLint of 7- and 8-hydroxyefavirenz. Assumptions: 1) these two pathways are the main clearance mechanisms; 2) efavirenz N-glucuronidation is minor; and 3) under the incubation conditions, sequential metabolism is minimal.
| HLMs | 8-Hydroxyefavirenz |
7-Hydroxyefavirenz |
||||||
|---|---|---|---|---|---|---|---|---|
| Vmax | Km | CLint | % of Total CLint | Vmax | Km | CLint | % of Total CLint | |
| Vmax/Km | Vmax/Km | |||||||
| IU6 | 84.4 | 19.4 | 4.37 | 62.7 | 48.7 | 18.8 | 2.60 | 37.3 |
| IU31 | 51.8 | 7.6 | 6.86 | 83.4 | 10.7 | 7.8 | 1.37 | 16.6 |
| IU42 | 17.9 | 7.1 | 2.52 | 89.0 | 7.5 | 23.9 | 0.31 | 11.0 |
| IU59 | 30.9 | 7.9 | 3.90 | 79.0 | 8.6 | 8.3 | 1.04 | 21.0 |
| IU58 | 28.7 | 25.2 | 1.14 | 90.2 | 2.2 | 17.5 | 0.12 | 9.8 |
| IU65 | 24.8 | 12.4 | 2.00 | 61.5 | 16.8 | 13.4 | 1.25 | 38.5 |
| HLT | 139.6 | 9.5 | 14.75 | 77.1 | 57.0 | 13.0 | 4.38 | 22.9 |
| Mean | 54.0 | 12.7 | 5.10 | 77.5 | 21.6 | 14.7 | 1.58 | 22.5 |
| SD | 43.9 | 7.0 | 4.66 | 11.6 | 21.9 | 5.8 | 1.47 | 11.6 |
| Expressed P450s | ||||||||
| CYP2B6 | 1.4 | 9.0 | 0.16 | |||||
| CYP2A6 | 0.6 | 7.7 | 0.08 | 0.4 | 7.6 | 0.05 | ||
Vmax, pmol/min/mg protein or pmol/min/pmol P450; Km, μM; CLint, μl/min/mg protein or μl/min/pmol P450.
Correlation analyses.
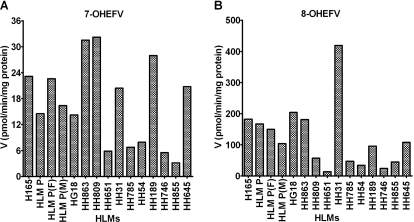
The metabolism of efavirenz to 7- and 8-hydroxyefavirenz was tested in a panel of characterized HLM samples (n = 15). Rates of formation of 7- and 8-hydroxyefavirenz varied widely among the HLMs [formation rate of 7-hydroxyefavirenz, 16.9 ± 9.7 (range, 3.2–32.2) pmol/min/mg protein, 10-fold difference; formation rate of 8-hydroxyefavirenz, 122.1 ± 103.6 (range, 13.7–419.2) pmol/min/mg protein, 31-fold difference] (Fig. 3). The average formation rates (pmol/min/mg protein) of efavirenz metabolism to 8-hydroxyefavirenz at 10 μM efavirenz was 7.7-fold higher (range, 1.8–20.5-fold difference) than the formation rates of 7-hydroxyefavirenz. Comparing the rates of metabolite formation from efavirenz (10 μM), efavirenz 7-hydroxylation accounts for 15.6 ± 8.7% (range, 4.7–35.9%), whereas efavirenz 8-hydroxylation accounts for the remaining (84.5 ± 8.7%; range, 64.1–93.5%). These results are consistent with the CLint data in Table 1.
Fig. 3.
Efavirenz (EFV) metabolism in a panel of 15 characterized HLMs. EFV (10 μM) was incubated with microsomes from different human livers (0.25 mg/ml) and cofactors for 15 min at 37°C. Formation rates (pmol product/min/mg protein) of 7-hydroxyEFV (7-OHEFV) (A) and of 8-hydroxyEFV (8-OHEFV) (B) are shown. Rates represent average of duplicate incubation measurements. Correlations between formation rates of 7-OHEFV or 8-OHEFV and the activity of P450 enzymes are illustrated in Table 2.
Correlations between rates of efavirenz metabolism and the activities of P450 isoforms in the HLMs are summarized in Table 2. A highly significant correlation between formation rates of 8-hydroxyefavirenz and CYP2B6 activity and between formation rates of 7-hydroxyefavirenz and CYP2A6 was observed. The activities of certain other isoforms showed modest but significant correlation (p < 0.05) with formation rates of 7-hydroxyefavirenz (CYP2B6, CYP2C8, and CYP3A) and formation rates of 8-hydroxyefavirenz (CYP1A2, CYP2A6, and CYP2C8) (Table 2). Formation rates of 7-hydroxyefavirenz correlated weakly but significantly with formation rates of 8-hydroxyefavirenz (Spearman r = 0.54; p = 0.038) (Table 2), consistent with the CLint data described previously, which may reflect the significant correlation between CYP2A6 and CYP2B6 activity in the panel of HLMs studied (r = 0.76; p = 0.0011).
TABLE 2.
Correlation of formation rates of 8-hydroxyefavirenz and 7-hydroxyefavirenz from efavirenz (10 μM) with the activities of different P450 enzymes in HLM samples (n = 15)
Efavirenz (10 μM) was incubated with microsomes from different human livers (0.25 mg/ml) and an NADPH-generating system for 15 min at 37°C. Data were analyzed using the nonparametric correlation test (Spearman r). The activity of each isoform was determined using the respective specific substrate probe reaction (see Materials and Methods). CYP3A was assayed using testosterone (T).
| P450 Isoforms | Efavirenz Metabolites |
|||
|---|---|---|---|---|
| 7-OHEFV |
8-OHEFV |
|||
| Spearman r | p Value† | Spearman r | p Value† | |
| CYP1A2 | 0.31 | 0.26 | 0.53 | 0.044 |
| CYP2A6 | 0.80 | 0.0003 | 0.62 | 0.014 |
| CYP2B6 | 0.57 | 0.03 | 0.81 | 0.0003 |
| CYP2C8 | 0.58 | 0.02 | 0.58 | 0.03 |
| CYP2C9 | 0.30 | 0.27 | 0.37 | 0.17 |
| CYP2C19 | 0.28 | 0.32 | 0.03 | 0.93 |
| CYP2D6 | −0.20 | 0.48 | 0.08 | 0.79 |
| CYP2E1 | 0.01 | 0.98 | 0.22 | 0.43 |
| CYP3A (T) | 0.61 | 0.015 | 0.37 | 0.18 |
| CYP4A11 | 0.25 | 0.36 | 0.004 | 0.99 |
| FMO | 0.21 | 0.45 | 0.06 | 0.84 |
p < 0.05 is considered statistically significant.
Efavirenz metabolism by a panel of expressed enzymes.
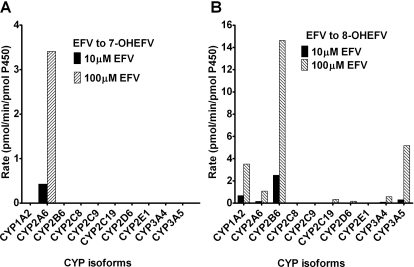
To obtain qualitative information as to which isoforms might catalyze efavirenz primary metabolism, the ability of 10 expressed P450 enzymes to catalyze 7- and 8-hydroxylation of efavirenz (10 and 100 μM) was tested. The lower concentration of efavirenz is close to the Km values, whereas the higher concentration represents concentration at saturation (Vmax in Michaelis-Menten curve). Efavirenz 7-hydroxylation was solely catalyzed by expressed CYP2A6 at both 10 and 100 μM efavirenz (Fig. 4A). None of the other isoforms tested showed any detectable 7-hydroxylase activity. As expected, CYP2B6 catalyzed efavirenz 8-hydroxylation at the highest rate at both concentrations (Fig. 4B). Other isoforms (CYP1A2, CYP3A5, CYP2A6, CYP3A4, CYP2C19, and CYP2D6) also showed activity toward this reaction. Kinetic studies with expressed CP2B6 and CYP2A6 revealed that CYP2B6 did not produce 7-hydroxyefavirenz at any of the concentrations tested (Table 1), confirming that this enzyme is not involved in efavirenz 7-hydroxylation, whereas CYP2A6 was involved in catalyzing 7- and 8-hydroxylation of efavirenz, with similar Km but slightly higher Vmax for 7-hydroxylation (Table 1). CLint for CYP2A6-catalyzed 8-hydroxylation was lower than that of CYP2A6-catalyzed efavirenz 7-hydroxylation (by 1.5-fold) and CYP2B6-catalyzed efavirenz 8-hydroxylation (by 3.2-fold).
Fig. 4.
Efavirenz (EFV) metabolism by a panel of P450 enzymes. EFV (10 or 100 μM) was incubated with expressed P450s (13 or 26 pmol of P450) and cofactors for 10 min at 37°C. Rates (pmol product/min/pmol P450) of 7-hydroxyEFV (7-OHEFV) (A) and 8-hydroxyEFV (8-OHEFV) (B) are presented. Each point is an average of duplicate incubation measurements.
Inhibition of efavirenz metabolism by P450 isoform-specific inhibitors in HLM samples.
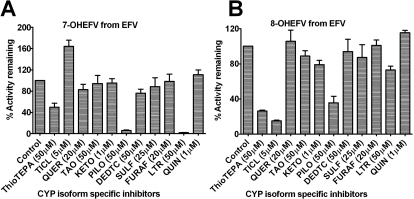
As shown in Fig. 5A, pilocarpine was the most potent inhibitor of efavirenz 7-hydroxylation (by 94%) followed by thioTEPA (by 50.3%), whereas ticlopidine increased formation of 7-hydroxyefavirenz (by 64%). In addition, formation of 7-hydroxyefavirenz was potently inhibited by letrozole (by 98.1%) (Fig. 5A), consistent with a recent finding that letrozole is a relatively selective inhibitor of CYP2A6 (Jeong et al., 2009b). As expected, thioTEPA and ticlopidine inhibited formation of 8-hydroxyefavirenz by more than 70% (Fig. 5B). Pilocarpine also inhibited formation of 8-hydroxyefavirenz by more than 60%; ketoconazole inhibited formation by 21%, as did letrozole by 27% (Fig. 5B).
Fig. 5.
Inhibition of efavirenz (EFV) metabolism in HLM samples. EFV (10 μM) was incubated with HLMs (0.25 mg/ml) and cofactors for 10 min at 37°C in the absence (control) and presence of P450 isoform-specific inhibitor (see Materials and Methods for details). Data (percentage activity remaining after inhibition relative to the vehicle control) represent mean ± S.D. of three duplicate measurements or three different HLMs. TICL, ticlopidine; QUER, quercetin; TAO, troleandomycin; KETO, ketoconazole; PILO, pilocarpine; DEDTC, diethyldithiocarbamate; SULF, sulfaphenazole; FURAF, furafylline; LTR, letrozole; and QUIN, quinidine. The final concentrations of these inhibitors used are indicated in brackets. A preincubation protocol was used for FURAF, TAO, thioTEPA, and DEDTC (see Materials and Methods).
P450-selective chemical inhibitors are often used to dissect the contribution of the corresponding P450 to drug metabolism. Thus, thioTEPA and pilocarpine have been considered selective inhibitors of CYP2B6 (Rae et al., 2002) and CYP2A6 (Bourrié et al., 1996), respectively. Although not specific, ticlopidine is also a potent inhibitor of CYP2B6 (Richter et al., 2004). Additional inhibition experiments in HLM samples and expressed enzymes (CYP2A6 and CYP2B6) were performed to test whether inhibition of CYP2A6 by thioTEPA and of CYP2B6 by pilocarpine represents lack of selectivity of these inhibitors or involvement of both enzymes in the pathways studied, as well as to test whether ticlopidine's effect may involve activation of CYP2A6. The inhibition data are summarized in Table 3. ThioTEPA inhibited formation of 8-hydroxyefavirenz by 68 and 55% in HLMs (IC50, 9.8 μM) and expressed CYP2B6, respectively (IC50, 6.6 μM); of 7-hydroxylefavirenz by 45% in HLMs (IC50, 31.7 μM) and by 83% in expressed CYP2A6; and of 8-hydroxyefavirenz by 83% in expressed CYP2A6 (IC50, 6.9 μM) (Table 3). Pilocarpine potently inhibited 7-hydroxylation of efavirenz (by >94%) in HLM samples (IC50, 2.3 μM) and expressed CYP2A6; and of 8-hydroxylation in HLMs, expressed CYP2A6, and CYP2B6 by 67.3% (IC50, 29.5 μM), 93.8, and 24.4% (IC50, 49.2 μM), respectively (Table 3). The potency of inhibition by pilocarpine of 8-hydroxylation in HLMs was approximately 2.8-fold higher than that observed with expressed CYP2B6 (Table 3). As shown in Table 3, ticlopidine increased formation of 7-hydroxyefavirenz (to 151%) in samples. However, when expressed CYP2A6-catalyzed 7- and 8-hydroxylation were assessed, no increase in formation of 7-hydroxyefavirenz was observed; instead, this reaction was slightly inhibited by ticlopidine (by <23%) (Table 3). No metabolite of ticlopidine that coelutes with 7-hydroxefavirenz was noted when ticlopidine was incubated alone with HLM samples and cofactors. As expected, ticlopidine potently inhibited efavirenz 8-hydroxylation (81.9 and 73%, respectively) in HLM samples and CYP2B6 (Table 3). Because ticlopidine minimally inhibited CYP2A6 in expressed enzyme, it is possible that there is a metabolic shift toward 7-hydroxylation in HLM samples.
TABLE 3.
Inhibition of efavirenz 7- and 8-hydroxylation by thioTEPA, pilocarpine, and ticlopidine in HLM samples and expressed CYP2A6 and CYP2B6
Efavirenz (10 μM) was incubated for 10 min at 37°C in duplicate with HLM samples (0.25 mg/ml) (or 26 pmol of CYP2A6 or 13 pmol of CYP2B6) and cofactors in the absence (control) and presence of an inhibitor. For determination of percentage activity remaining, a single concentration of thioTEPA (50 μM), pilocarpine (50 μM), or ticlopidine (5 μM) was used. To determine the IC50 values, multiple concentrations of thioTEPA (0–100 μM) or pilocarpine (0–100 μM) were used.
| Inhibitor | Microsomes | 7-Hydroxylation |
8-Hydroxylation |
||
|---|---|---|---|---|---|
| %Activity Remaining | IC50 | %Activity Remaining | IC50 | ||
| μM | μM | ||||
| ThioTEPA | |||||
| HLMs | 55.0 ± 1.4 | 31.7 | 32.3 ± 4.3 | 9.8 | |
| CYP2A6 | 17.4 ± 0.6 | 6.6 | 17.5 ± 1.4 | 6.9 | |
| CYP2B6 | 44.9 ± 0.2 | ||||
| Pilocarpine | |||||
| HLMs | 4.3 ± 1.9 | 2.3 | 32.7 ± 0.8 | 29.5 | |
| CYP2A6 | 5.8 ± 0.5 | 6.2 ± 0.4 | |||
| CYP2B6 | 75.6 ± 4.2 | 135.6 | |||
| Ticlopidine | |||||
| HLMs | 150.8 ± 11.1 | 18.1 ± 0.6 | |||
| CYP2A6 | 77.5 ± 6.4 | 77.8 ± 6.2 | |||
| CYP2B6 | 27.0 ± 0.5 | ||||
Secondary (Sequential) Metabolism of Efavirenz.
The data presented in this section should be viewed as preliminary findings because the precise structural identities of the metabolites were not unequivocally established.
Metabolite identification in HLM samples.
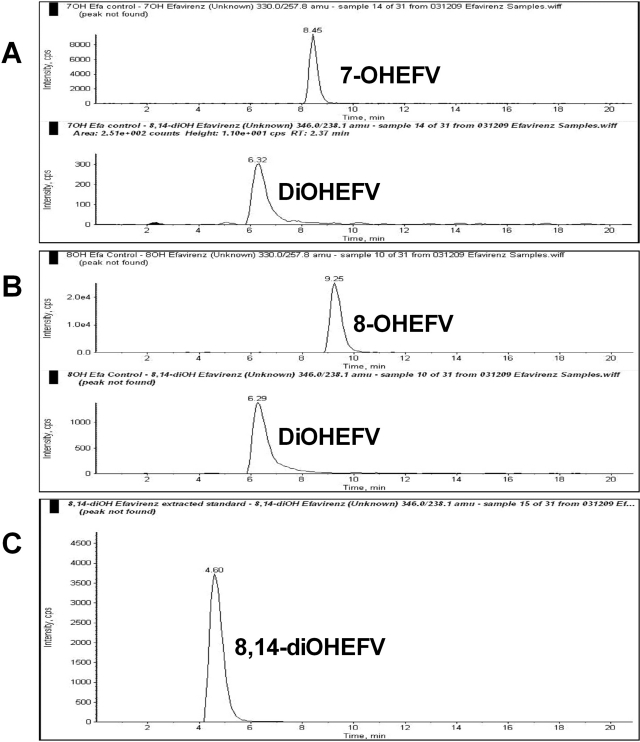
In addition to the two monohydroxylated metabolites of efavirenz (7- and 8-hydroxyefavirenz), a third metabolite peak that was consistent with a dihydroxyefavirenz (molecular mass of 347; with [M-H]− at m/z 346) was identified when efavirenz was incubated with HLMs and cofactors (but not in negative controls) (Fig. 1). Because previous studies have suggested that 8-hydroxyefavirenz is 14-hydroxylated to form 8,14-dihydroxyefavirenz (Mutlib et al., 1999b; Ward et al., 2003), the possibility that the dihydroxylated metabolite of efavirenz might be 8,14-dihydroxyefavirenz was explored. The major fragment ions of synthetic 8,14-dihydroxyefavirenz standard, determined by infusing a standard solution onto the MS/MS system set in the quantitative optimization mode, were 238 and 210 (Supplemental Table 1). The major MS/MS fragment ions of the metabolite peak observed in efavirenz HLM sample incubation were determined by injecting an aliquot of an efavirenz (20 μM) incubation mixture onto the LC/MS/MS system using the product ion mode; the signal generated by the parent ion mass m/z 346 produced fragmentation product ions of 238 and 210, consistent with those of synthetic 8,14-dihydroxyefavirenz (Supplemental Fig. 1). When the retention times of the metabolite peak produced in efavirenz incubates were compared with that of synthetic standard of 8,14-dihydroxyefavirenz, a marked difference was observed in the LC/MS/MS (Supplemental Fig. 1) [and HPLC with UV detection (data not shown)]. Therefore, because the secondary metabolite observed in efavirenz microsomal incubates appears to be different from 8,14-dihydroxyefavirenz, the unknown metabolite is designated as only dihydroxylated efavirenz.
To test whether this dihydroxylated metabolite was also formed from the primary metabolites of efavirenz (7- or 8-hydroxyefavirenz), each metabolite was incubated with HLM samples and cofactors. Incubation of 7- (Fig. 6A) and 8-hydroxyefavirenz (Fig. 6B) resulted in the appearance of a peak that coeluted chromatographically with the dihydroxylated efavirenz. This peak was not observed in the negative control experiments. Again, the precursor and product ion scans of the metabolite formed from these substrates were the same as those from synthetic 8,14-dihydroxyefavirenz standard, but the retention time (6.8 min) of the dihydroxylated efavirenz was markedly different from that of 8,14-dihydroxyefavirenz (4.66 min) (Fig. 6C).
Fig. 6.
MRM trace chromatograms of 7-hydroxyefavirenz (7-OHEFV) and 8-hydroxyefavirenz (8-OHEFV) in human liver microsomal incubates. Each substrate (5 μM) was incubated in HLM samples and cofactors for 10 min at 37°C. A metabolite peak that was consistent with dihydroxylated efavirenz was formed in microsomal incubations of 7-OHEFV (A) and 8-OHEFV (B). Synthetic 8,14-dihydroxyefavirenz (8,14-diOHEFV) standard was directly injected (C).
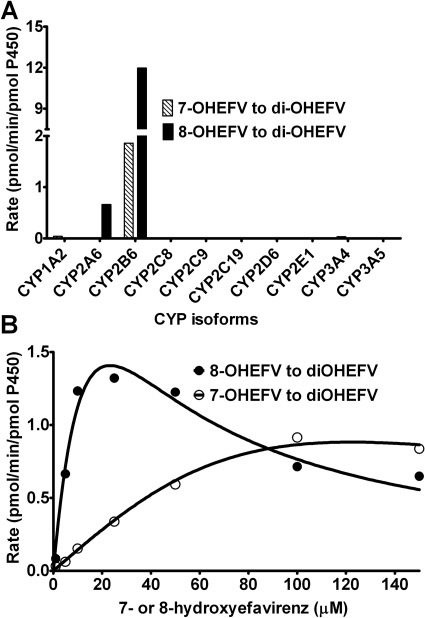
Secondary metabolism in expressed P450s.
To identify the P450 isoforms involved in efavirenz sequential metabolism, 7- and 8-hydroxyefavirenz were incubated with a panel of expressed P450 enzymes. Because the MS/MS characteristics of the dihydroxylated metabolite were the same as those of 8,14-dihydroxyefavirenz, quantification of the dihydroxyefavirenz was made using standard curves of 8,14-dihydroxyefavirenz. CYP2B6 catalyzed formation of dihydroxyefavirenz from 7- and 8-hydroxyefavirenz at the highest rate compared with other P450s (Fig. 7A). Kinetics for the metabolism of 8-hydroxyefavirenz in expressed CYP2B6 (Fig. 7B) was characterized by substrate inhibition kinetic model (Vmax, 4.21 pmol/min/pmol P450; Km, 23.2 μM; Ks, 23.4 μM), whereas that of 7-hydroxyefavirenz was best described by the Michaelis-Menten equation (Vmax, 1.3 pmol/min/pmol P450; Km, 62.4 μM). At lower substrate concentrations, 8-hydroxyefavirenz was more efficiently oxidized by CYP2B6 than 7-hydroxyefavirenz.
Fig. 7.
Secondary metabolism of efavirenz (EFV) by a panel of expressed P450s. Sequential metabolism was tested using 7-hydroxyEFV (7-OHEFV) and 8-hydroxyEFV (8-OHEFV) as substrates. 7-OHEFV (5 μM) and 8-OHEFV (5 μM) were incubated with expressed P450s (13 or 26 pmol of P450) and cofactors for 10 min at 37°C, and the formation of a dihydroxylated efavirenz was monitored (A). Kinetics for the metabolism of 7-OHEFV and 8-OHEFV to dihydroxylated efavirenz was determined by incubating each substrate (1–150 μM) with 13 pmol of CYP2B6 and cofactors for 10 min at 37°C (B). Kinetic analysis was performed by fitting to a one-site hyperbolic Michaelis-Menten equation (7-OHEFV metabolism) or using substrate inhibition equation (8-OHEFV) (see Data Analysis). Each point represents average picomole of product per minute per picomole of CYP2B6 of duplicate incubation measurements.
As described previously, 8,14-dihydroxyefavirenz was not detected in vitro when efavirenz and 8-hydroxyefavirenz were used as substrates. However, this metabolite was detected and quantified in plasma samples obtained from subjects taking a single 600-mg oral dose of efavirenz (see below). In vivo, 8-hydroxyefavirenz is rapidly conjugated (glucuronidated or sulfated) (Mutlib et al., 1999b). The possibility that 8-hydroxyefavirenz glucuronide might undergo 14-hydroxylation was tested by incubating 8-hydroxyefavirenz glucuronide with HLM samples and cofactors. A unique peak was noted (this was not observed in negative control experiments) at a retention time of 8.6 min in this incubation (data not shown). This peak was consistent with involvement of P450-mediated reaction. In a panel of expressed P450s, 8-hydroxyefavirenz glucuronide was converted to a metabolite peak mainly by expressed CYP2A6 (2.29 pmol/min/pmol P450) and to some extent by CYP1A2 (1.19 pmol/min/pmol P450) more than CYP2B6 (0.21 pmol/min/pmol P450) (Supplemental Fig. 2). This metabolite is likely to be 14-hydroxylated of 8-hydroxyefavirenz glucuronide. However, attempts to confirm this suggestion through treatment of the microsomal incubates with β-glucuronidase and assay the metabolite hydrolyzed did not provide useful information, probably because its formation in vitro was low.
Identification and Quantification of Efavirenz Metabolites In Vivo.
Efavirenz and its metabolites were analyzed in plasma samples obtained from healthy volunteers administered a single 600-mg oral dose of efavirenz. In Fig. 8, MRM trace chromatograms of extracted blank plasma samples (Fig. 8A) and of extracted plasma samples 3 h after efavirenz dosing (Fig. 8B) are shown. All the primary (7- and 8-hydroxyefavirenz) and secondary (dihydroxylated efavirenz) metabolites that were characterized in vitro were detected in the clinical samples (Fig. 8B). Efavirenz and metabolites were not detected in (blank) plasma samples obtained from the same subjects before efavirenz administration (data not shown). A metabolite that had the same retention time and MS/MS characteristics as those of synthetic standard 8,14-dihydroxyefavirenz was identified in vivo (Fig. 8). This metabolite was not formed (detected) in vitro from any of the substrates tested (efavirenz, 7-, or 8-hydroxyefavirenz).
Fig. 8.
Representative MRM trace chromatograms of efavirenz (EFV) and its monohydroxylated (7- and 8-hydroxyefavirenz) and secondary metabolites (8,14-dihydroxyefavirenz and dihydroxyefavirenz) in plasma samples obtained from subjects administered a single 600-mg oral dose of efavirenz. Extracted blank samples spiked with authentic standards of efavirenz and its metabolites (A), and extracted plasma sample obtained 3 h after administration of a single 600-mg oral dose of efavirenz to healthy volunteers (B). Plasma samples were treated with β-glucuronidase, and the peaks represent total (free + conjugated).
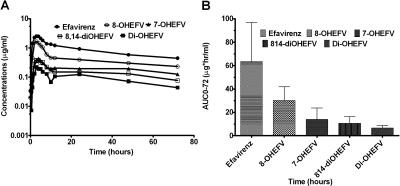
The concentration versus time curves of efavirenz and it metabolites in healthy volunteers (n = 5) administered a single 600-mg oral dose of efavirenz are shown in Fig. 9A. The pharmacokinetic data provided are total concentrations (free + conjugated) of efavirenz and its metabolites quantified after incubation of the samples with β-glucuronidase (“deconjugation”). The β-glucuronidase that was used also contained sulfatase; thus, it could not reliably distinguish between glucuronide and sulfate conjugates. Although it is likely that these conjugates are mostly glucuronides (Mutlib et al., 1999b), conjugates are used in this manuscript to reflect nonselectivity of the enzymatic hydrolysis used. The plasma area under the plasma concentration-time curves (AUC0–72) of efavirenz and metabolites are provided in Fig. 9B. Based on the AUC0–72 in plasma, efavirenz was the most abundant, followed by 8-hydroxyefavirenz > 7-hydroxyefavirenz > 8,14-dihydroxyefavirenz > dihydroxylated efavirenz.
Fig. 9.
Pharmacokinetics of efavirenz (EFV) and its primary and secondary metabolites in healthy volunteer subjects (n = 5) administered a single 600-mg oral dose of EFV. Plasma concentrations versus time curves (A) and area under the concentration-time curve (AUC0–72) (B) of efavirenz and metabolites are shown. 8-OHEFV, 8-hydroxyEFV; 7-OHEFV, 7-hydroxyEFV; 8,14-diOHEFV, 8,14-dihydroxyefavirenz; diOHEFV, dihydroxylated EFV.
Discussion
The major new findings of the present study were that CYP2A6-mediated efavirenz 7-hydroxylation accounts for ∼23% of efavirenz metabolism; CYP2A6 is a partial contributor toward efavirenz 8-hydroxylation; efavirenz is metabolized sequentially to novel dihydroxylated metabolite(s), via CYP2B6-mediated 7- and 8-hydroxyefavirenz hydroxylation as intermediary; and 8,14-dihydroxyefavirenz is formed in vivo but not in vitro, suggesting novel metabolic reactions and challenging previous notion that it is formed through direct 14-hydroxylation of 8-hydroxyefavirenz (Mutlib et al., 1999b; Ward et al., 2003). The identification and quantification of all the efavirenz primary (7- and 8-hydroxyefavirenz) and secondary (8,14-dihydroxyefavirenz and a dihydroxylated) metabolites and the first demonstration of their full pharmacokinetics in plasma of healthy subject taking a single 600-mg oral dose of efavirenz confirm clinical relevance of the in vitro findings. Finally, the role CYP2B6 plays in efavirenz 8-hydroxylation, a metabolic route accounting for ∼77% of efavirenz clearance, was further confirmed (Ward et al., 2003). These data should help to predict determinants of efavirenz metabolism and response. They should also form the scientific basis with which to select the most appropriate pharmacokinetic parameters when efavirenz is used to assess the activity of CYP2B6 (and possibly CYP2A6) in vivo.
The in vitro data reported here show that 7- and 8-hydroxyefavirenz are the primary metabolites of efavirenz and concur with previous reports (Mutlib et al., 1999b; Ward et al., 2003; Desta et al., 2007). Based on the in vitro CLint in HLM samples, 7- and 8-hydroxylation on the average account for ∼22.5 and ∼77.5% of efavirenz overall metabolism (Table 1). Provided the contribution of other elimination routes [e.g., efavirenz N-glucuronidation (Bélanger et al., 2009) and nonhepatic clearance (Mutlib et al., 1999b)] is minimal, 7- and 8-hydroxylation represent minor and major clearance mechanisms of efavirenz.
The identification of new and novel dihydroxylated efavirenz in microsomal incubates of efavirenz, 7-, and 8-hydroxyefavirenz suggests that efavirenz is sequentially oxidized, with monohydroxylated efavirenz as intermediary. Efavirenz secondary metabolism via 8-hydroxyefavirenz to 8,14-dihydroxyefavirenz has been previously proposed in vitro (Ward et al., 2003) and in vivo (Mutlib et al., 1999b). However, 14-hydroxylation of 8-hydroxyefavirenz was not observed in the present in vitro study because the dihydroxylated metabolite(s) formed was not consistent with 8,14-dihydroxyefavirenz. In addition, efavirenz secondary metabolism can undergo via 7-hydroxyefavirenz to dihydroxylated efavirenz. The precise structural identity of the dihydroxylated metabolite(s) remains unknown. It is possible that the secondary hydroxylation site may occur at the C15 or C16 portion of the cyclopropyl part of the molecule, resulting in MS/MS properties indistinguishable from those of 8,14-dihydroxyefavirenz. It is also unclear whether the metabolite formed from 7-hydroxyefavirenz is the same as that formed from 8-hydroxyefavirenz. Because attempts to separate the metabolites formed from 7- and 8-hydroxyefavirenz chromatographically did not yield useful information, the metabolites formed may have similar physicochemical characteristics. In theory, 7- and 8-hydroxyefavirenz can undergo 8-hydroxylation and 7-hydroxyefavirenz, respectively, resulting in the formation of 7,8-dihydroxyefavirenz. This possibility cannot be ruled out but may be questionable because the most abundant MS/MS fragments of the dihydroxylated metabolite were similar to those from 8,14-dihydroxyefavirenz.
The in vivo relevance of the in vitro findings is shown by the fact that all the primary and secondary metabolites of efavirenz that were identified in vitro were confirmed in plasma samples of healthy volunteers after deconjugation. These data represent the first comprehensive description of efavirenz metabolite pharmacokinetics and mirror the relative differences in the in vitro CLint of 7- and 8-hydroxyefavirenz, providing support to the in vivo relevance of the in vitro findings. 8,14-Dihydroxyefavirenz, a metabolite that was not detected in vitro, was positively identified and quantified in the clinical samples treated with β-glucuronidase. The reason for the lack of in vitro and in vivo correlation is not clear. In vivo, 8-hydroxyefavirenz is rapidly conjugated (glucuronidated and/or sulfated) by phase II enzymes (Mutlib et al., 1999b). It is possible that the conjugated 8-hydroxyefavirenz undergoes 14-hydroxylation in vivo, from which 8,14-dihydroxyefavirenz could be released on treatment of efavirenz plasma samples with β-glucuronidase as observed in our study. Consistent with this suggestion, Mutlib et al. (1999a) reported a glucuronide (C14)-sulfate (C8) diconjugate of dihydroxylated efavirenz in urine and bile of rats administered efavirenz, 8-hydroxyefavirenz, or 8-hydroxyefavirenz sulfate. Other examples show that sulfate or glucuronide metabolites can undergo oxidation by P450s (Mutlib et al., 1999a; Delaforge et al., 2005, and references therein). Preliminary proof of concept in support of oxidation of a conjugated 8-hydroxyefavirenz was obtained in the present study where a metabolite peak that was consistent with involvement of P450s (CYP2A6, CYP1A2, and CYP2B6) was noted when 8-hydroxyefavirenz glucuronide was incubated with HLM samples and cofactors. Together, these data begin to provide a mechanistic understanding by revealing novel metabolic reactions of efavirenz.
A comprehensive set of in vitro experiments was conducted to identify the P450s involved in efavirenz primary and secondary metabolism. Formation rate of 7-hydroxyefavirenz was substantially inhibited by selective CYP2A6 inhibitor pilocarpine (Bourrié et al., 1996) and CYP2A6 inhibitor letrozole (Jeong et al., 2009b) (Fig. 5), correlated significantly with CYP2A6 activity (Table 2), and catalyzed exclusively by expressed CYP2A6 (Fig. 4). The Km values derived from HLM samples and expressed CYP2A6 (Table 1) were very close, suggesting that the enzyme catalyzing 7-hydroxylation of efavirenz in HLM samples is CYP2A6. Efavirenz 7-hydroxylation in HLM samples was inhibited by thioTEPA (Table 3). It is likely that this is mediated by the ability of thioTEPA to inhibit CYP2A6 rather than involvement of CYP2B6 in this reaction because expressed CYP2B6 was not found to be capable of efavirenz 7- hydroxylation (see below). ThioTEPA potently inhibits CYP2B6 (Rae et al., 2002; Ward et al., 2003; Harleton et al., 2004; Richter et al., 2005), but its selectivity has been questioned recently by other authors (Turpeinen et al., 2004; Obach et al., 2007; Walsky and Obach, 2007). The present data provide additional support that thioTEPA may inhibit CYP2A6 in addition to CYP2B6.
The kinetic, inhibition, and correlation analyses in HLM samples and data from expressed P450s confirm previous findings that CYP2B6 is the principal catalyst of efavirenz 8-hydroxylation (Ward et al., 2003; Desta et al., 2007). However, these data also implicate for the first time that CYP2A6 is catalyzing efavirenz 8-hydroxylation. First, pilocarpine inhibited efavirenz 8-hydroxylation in HLMs and expressed CYP2B6. It is clear that pilocarpine is frequently used as a selective inhibitor of CYP2A6 in vitro, but its selectivity has been poorly validated. Its ability to inhibit CYP2B6-catalyzed reaction (present data) and previous report showing its inhibitory effect on CYP2C9 (Bourrié et al., 1996) suggest that pilocarpine may not be highly selective toward CYP2A6. However, all the effects of pilocarpine on efavirenz 8-hydroxylation might not be explained by nonselectivity alone. The involvement of CYP2A6 in efavirenz 8-hydroxylation is supported by the fact that pilocarpine inhibited 8-hydroxylation of efavirenz more in HLM samples than in expressed CYP2B6 (Table 3); letrozole modestly inhibited efavirenz 8-hydroxylation (by 27%) (Fig. 5); expressed CYP2A6 catalyzes efavirenz 8-hydroxylation (Fig. 4); and CYPA6 activity is significantly correlated with formation rate of 8-hydroxyefavirenz (Table 2). Therefore, CYP2A6 appears to contribute to the overall clearance of efavirenz by being the sole catalyst of efavirenz 7-hydroxylation, a pathway that accounts on average for ∼23% of efavirenz metabolism with large interindividual variability, and by participating (∼20–30%) in efavirenz 8-hydroxylation.
Efavirenz has been recently described as a preferred marker of CYP2B6 activity in vitro and in vivo (Ward et al., 2003; US Food and Drug Administration, 2006). The present data also support the notion that it may serve as a CYP2A6 probe. Although the primary and secondary metabolites of efavirenz are pharmacologically inactive, identifying P450s that catalyze them is important to select the most appropriate metabolic ratios that best describe CYP2B6 and CYP2A6 activities in vivo. That CYP2B6 was found to be the primary catalyst of 7- and 8-hydroxyefavirenz to the respective dihydroxylated metabolite (Fig. 7B) provides important information in this respect. Moreover, the data show that monohydroxylated efavirenz metabolites, as with efavirenz, are efficient substrates of CYP2B6, irrespective of the site of hydroxylation. This information may help to model and better understand the active site of CYP2B6.
The findings presented here may have important implications. First, they suggest that CYP2B6 and CYP2A6 activity may influence efavirenz exposure and response. CYP2B6 metabolic status as a key determinant of efavirenz exposure, and probably response, in HIV patients has been established (e.g., Haas et al., 2004; Tsuchiya et al., 2004; Rotger et al., 2007; Zanger et al., 2007). CYP2A6 activity exhibits large interindividual variation (Pelkonen et al., 2000; Di et al., 2009), mostly because of CYP2A6 gene polymorphisms but also as a result of exposure to inducers and inhibitors of the enzyme (Nakajima et al., 2006; Di et al., 2009; Mwenifumbo and Tyndale, 2009). More recently, CYP2A6 genetic variation has been shown to influence efavirenz exposure in HIV patients (Arab-Alameddine et al., 2009; di Iulio et al., 2009; Kwara et al., 2009a,b). Second, efavirenz may serve as an effective probe of CYP2B6 and CYP2A6 activity in vitro and in vivo. The identification of novel metabolic pathways and P450s involved in efavirenz primary and secondary metabolism should facilitate identification of efavirenz pharmacokinetic parameters that best reflect CYP2B6 and CYP2A6 activities in vivo. Clinical studies are ongoing to clarify the precise contribution of CYP2A6 and CYP2B6 in efavirenz metabolism using genetics and drug interactions as markers.
This work was supported by the National Institutes of Health National Institute of General Medical Sciences [Grants GM078501, GM067308].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.109.031393.
 The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
- P450
- cytochrome P450
- HLM
- human liver microsome
- HPLC
- high-performance liquid chromatography
- LC/MS/MS
- liquid chromatography/tandem mass spectrometry
- MRM
- multiple reaction monitoring
- MS
- mass spectrometry
- CLint
- intrinsic clearance
- AUC0–72
- plasma area under the plasma concentration-time curve.
References
- Arab-Alameddine M, Di Iulio J, Buclin T, Rotger M, Lubomirov R, Cavassini M, Fayet A, Décosterd LA, Eap CB, Biollaz J, et al. (2009) Pharmacogenetics-based population pharmacokinetic analysis of efavirenz in HIV-1-infected individuals. Clin Pharmacol Ther 85:485–494 [DOI] [PubMed] [Google Scholar]
- Bélanger AS, Caron P, Harvey M, Zimmerman PA, Mehlotra RK, Guillemette C. (2009) Glucuronidation of the antiretroviral drug efavirenz by UGT2B7 and an in vitro investigation of drug-drug interaction with zidovudine. Drug Metab Dispos 37:1793–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourrié M, Meunier V, Berger Y, Fabre G. (1996) Cytochrome P450 isoform inhibitors as a tool for the investigation of metabolic reactions catalyzed by human liver microsomes. J Pharmacol Exp Ther 277:321–332 [PubMed] [Google Scholar]
- Bristol-Myers Squibb Company, 2009.Bristol-Myers Squibb Company (2009) Package insert of efavirenz (Sustiva) at http://packageinserts.bms.com/pi/pi_sustiva.pdf (updated September 2009), New York [Google Scholar]
- Csajka C, Marzolini C, Fattinger K, Décosterd LA, Fellay J, Telenti A, Biollaz J, Buclin T. (2003) Population pharmacokinetics and effects of efavirenz in patients with human immunodeficiency virus infection. Clin Pharmacol Ther 73:20–30 [DOI] [PubMed] [Google Scholar]
- Delaforge M, Pruvost A, Perrin L, André F. (2005) Cytochrome P450-mediated oxidation of glucuronide derivatives: example of estradiol-17beta-glucuronide oxidation to 2-hydroxy-estradiol-17beta-glucuronide by CYP 2C8. Drug Metab Dispos 33:466–473 [DOI] [PubMed] [Google Scholar]
- Desta Z, Saussele T, Ward B, Blievernicht J, Li L, Klein K, Flockhart DA, Zanger UM. (2007) Impact of CYP2B6 polymorphism on hepatic efavirenz metabolism in vitro. Pharmacogenomics 8:547–558 [DOI] [PubMed] [Google Scholar]
- di Iulio J, Fayet A, Arab-Alameddine M, Rotger M, Lubomirov R, Cavassini M, Furrer H, Günthard HF, Colombo S, Csajka C, et al. (2009) In vivo analysis of efavirenz metabolism in individuals with impaired CYP2A6 function. Pharmacogenet Genomics 19:300–309 [DOI] [PubMed] [Google Scholar]
- Di YM, Chow VD, Yang LP, Zhou SF. (2009) Structure, function, regulation and polymorphism of human cytochrome P450 2A6. Curr Drug Metab 10:754–780 [DOI] [PubMed] [Google Scholar]
- Ekins S, Wrighton SA. (1999) The role of CYP2B6 in human xenobiotic metabolism. Drug Metab Rev 31:719–754 [DOI] [PubMed] [Google Scholar]
- Haas DW, Ribaudo HJ, Kim RB, Tierney C, Wilkinson GR, Gulick RM, Clifford DB, Hulgan T, Marzolini C, Acosta EP. (2004) Pharmacogenetics of efavirenz and central nervous system side effects: an Adult AIDS Clinical Trials Group study. AIDS 18:2391–2400 [PubMed] [Google Scholar]
- Harleton E, Webster M, Bumpus NN, Kent UM, Rae JM, Hollenberg PF. (2004) Metabolism of N,N′,N″-triethylenethiophosphoramide by CYP2B1 and CYP2B6 results in the inactivation of both isoforms by two distinct mechanisms. J Pharmacol Exp Ther 310:1011–1019 [DOI] [PubMed] [Google Scholar]
- Hodgson E, Rose RL. (2007) The importance of cytochrome P450 2B6 in the human metabolism of environmental chemicals. Pharmacol Ther 113:420–428 [DOI] [PubMed] [Google Scholar]
- Jeong S, Nguyen PD, Desta Z. (2009a) Comprehensive in vitro analysis of voriconazole inhibition of eight cytochrome P450 (CYP) enzymes: major effect on CYPs 2B6, 2C9, 2C19, and 3A. Antimicrob Agents Chemother 53:541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Woo MM, Flockhart DA, Desta Z. (2009b) Inhibition of drug metabolizing cytochrome P450s by the aromatase inhibitor drug letrozole and its major oxidative metabolite 4,4′-methanol-bisbenzonitrile in vitro. Cancer Chemother Pharmacol 64:867–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharasch ED, Mitchell D, Coles R. (2008) Stereoselective bupropion hydroxylation as an in vivo phenotypic probe for cytochrome P4502B6 (CYP2B6) activity. J Clin Pharmacol 48:464–474 [DOI] [PubMed] [Google Scholar]
- Kwara A, Lartey M, Sagoe KW, Kenu E, Court MH. (2009a) CYP2B6, CYP2A6 and UGT2B7 genetic polymorphisms are predictors of efavirenz mid-dose concentration in HIV-infected patients. AIDS 23:2101–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwara A, Lartey M, Sagoe KW, Rzek NL, Court MH. (2009b) CYP2B6 (c.516G–>T) and CYP2A6 (*9B and/or *17) polymorphisms are independent predictors of efavirenz plasma concentrations in HIV-infected patients. Br J Clin Pharmacol 67:427–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzolini C, Telenti A, Decosterd LA, Greub G, Biollaz J, Buclin T. (2001) Efavirenz plasma levels can predict treatment failure and central nervous system side effects in HIV-1-infected patients. AIDS 15:71–75 [DOI] [PubMed] [Google Scholar]
- Mo SL, Liu YH, Duan W, Wei MQ, Kanwar JR, Zhou SF. (2009) Substrate specificity, regulation, and polymorphism of human cytochrome P450 2B6. Curr Drug Metab 10:730–753 [DOI] [PubMed] [Google Scholar]
- Mouly S, Lown KS, Kornhauser D, Joseph JL, Fiske WD, Benedek IH, Watkins PB. (2002) Hepatic but not intestinal CYP3A4 displays dose-dependent induction by efavirenz in humans. Clin Pharmacol Ther 72:1–9 [DOI] [PubMed] [Google Scholar]
- Mutlib AE, Chen H, Nemeth G, Gan LS, Christ DD. (1999a) Liquid chromatography/mass spectrometry and high-field nuclear magnetic resonance characterization of novel mixed diconjugates of the non-nucleoside human immunodeficiency virus-1 reverse transcriptase inhibitor, efavirenz. Drug Metab Dispos 27:1045–1056 [PubMed] [Google Scholar]
- Mutlib AE, Chen H, Nemeth GA, Markwalder JA, Seitz SP, Gan LS, Christ DD. (1999b) Identification and characterization of efavirenz metabolites by liquid chromatography/mass spectrometry and high field NMR: species differences in the metabolism of efavirenz. Drug Metab Dispos 27:1319–1333 [PubMed] [Google Scholar]
- Mwenifumbo JC, Tyndale RF. (2009) Molecular genetics of nicotine metabolism. Handb Exp Pharmacol 192:235–259 [DOI] [PubMed] [Google Scholar]
- Nakajima M, Fukami T, Yamanaka H, Higashi E, Sakai H, Yoshida R, Kwon JT, McLeod HL, Yokoi T. (2006) Comprehensive evaluation of variability in nicotine metabolism and CYP2A6 polymorphic alleles in four ethnic populations. Clin Pharmacol Ther 80:282–297 [DOI] [PubMed] [Google Scholar]
- Newton DJ, Wang RW, Lu AY. (1995) Cytochrome P450 inhibitors. Evaluation of specificities in the in vitro metabolism of therapeutic agents by human liver microsomes. Drug Metab Dispos 23:154–158 [PubMed] [Google Scholar]
- Obach RS, Walsky RL, Venkatakrishnan K. (2007) Mechanism-based inactivation of human cytochrome P450 enzymes and the prediction of drug-drug interactions. Drug Metab Dispos 35:246–255 [DOI] [PubMed] [Google Scholar]
- Pelkonen O, Rautio A, Raunio H, Pasanen M. (2000) CYP2A6: a human coumarin 7-hydroxylase. Toxicology 144:139–147 [DOI] [PubMed] [Google Scholar]
- Rae JM, Soukhova NV, Flockhart DA, Desta Z. (2002) Triethylenethiophosphoramide is a specific inhibitor of cytochrome P450 2B6: implications for cyclophosphamide metabolism. Drug Metab Dispos 30:525–530 [DOI] [PubMed] [Google Scholar]
- Richter T, Mürdter TE, Heinkele G, Pleiss J, Tatzel S, Schwab M, Eichelbaum M, Zanger UM. (2004) Potent mechanism-based inhibition of human CYP2B6 by clopidogrel and ticlopidine. J Pharmacol Exp Ther 308:189–197 [DOI] [PubMed] [Google Scholar]
- Richter T, Schwab M, Eichelbaum M, Zanger UM. (2005) Inhibition of human CYP2B6 by N,N′,N″-triethylenethiophosphoramide is irreversible and mechanism-based. Biochem Pharmacol 69:517–524 [DOI] [PubMed] [Google Scholar]
- Rotger M, Tegude H, Colombo S, Cavassini M, Furrer H, Décosterd L, Blievernicht J, Saussele T, Günthard HF, Schwab M, et al. (2007) Predictive value of known and novel alleles of CYP2B6 for efavirenz plasma concentrations in HIV-infected individuals. Clin Pharmacol Ther 81:557–566 [DOI] [PubMed] [Google Scholar]
- Tsuchiya K, Gatanaga H, Tachikawa N, Teruya K, Kikuchi Y, Yoshino M, Kuwahara T, Shirasaka T, Kimura S, Oka S. (2004) Homozygous CYP2B6 *6 (Q172H and K262R) correlates with high plasma efavirenz concentrations in HIV-1 patients treated with standard efavirenz-containing regimens. Biochem Biophys Res Commun 319:1322–1326 [DOI] [PubMed] [Google Scholar]
- Turpeinen M, Nieminen R, Juntunen T, Taavitsainen P, Raunio H, Pelkonen O. (2004) Selective inhibition of CYP2B6-catalyzed bupropion hydroxylation in human liver microsomes in vitro. Drug Metab Dispos 32:626–631 [DOI] [PubMed] [Google Scholar]
- US Food and Drug Administration, 2006.US Food and Drug Administration (2006) Draft Guidance for Industry: Drug Interaction Studies—Study Design, Data Analysis and Implications for Dosing and Labeling, US Food and Drug Administration, Silver Spring, MD [Google Scholar]
- Walsky RL, Obach RS. (2007) A comparison of 2-phenyl-2-(1-piperidinyl)propane (ppp), 1,1′,1″-phosphinothioylidynetrisaziridine (thioTEPA), clopidogrel, and ticlopidine as selective inactivators of human cytochrome P450 2B6. Drug Metab Dispos 35:2053–2059 [DOI] [PubMed] [Google Scholar]
- Wang H, Tompkins LM. (2008) CYP2B6: new insights into a historically overlooked cytochrome P450 isozyme. Curr Drug Metab 9:598–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward BA, Gorski JC, Jones DR, Hall SD, Flockhart DA, Desta Z. (2003) The cytochrome P450 2B6 (CYP2B6) is the main catalyst of efavirenz primary and secondary metabolism: implication for HIV/AIDS therapy and utility of efavirenz as a substrate marker of CYP2B6 catalytic activity. J Pharmacol Exp Ther 306:287–300 [DOI] [PubMed] [Google Scholar]
- Zanger UM, Klein K, Saussele T, Blievernicht J, Hofmann MH, Schwab M. (2007) Polymorphic CYP2B6: molecular mechanisms and emerging clinical significance. Pharmacogenomics 8:743–759 [DOI] [PubMed] [Google Scholar]