Abstract
STAT3 (signal transducer and activator of transcription 3) is a transcription factor activated by cytokines, growth factors and oncogenes, whose activity is required for cell survival/proliferation of a wide variety of primary tumours and tumour cell lines. Prominent among its multiple effects on tumour cells is the stimulation of cell migration and metastasis, whose functional mechanisms are however not completely characterized. RhoU/Wrch1 (Wnt-responsive Cdc42 homologue) is an atypical Rho GTPase thought to be constitutively bound to GTP. RhoU was first identified as a Wnt-1-inducible mRNA and subsequently shown to act on the actin cytoskeleton by stimulating filopodia formation and stress fibre dissolution. It was in addition recently shown to localize to focal adhesions and to Src-induced podosomes and enhance cell migration. RhoU overexpression in mammary epithelial cells stimulates quiescent cells to re-enter the cell cycle and morphologically phenocopies Wnt-1-dependent transformation. In the present study we show that Wnt-1-mediated RhoU induction occurs at the transcriptional level. Moreover, we demonstrate that RhoU can also be induced by gp130 cytokines via STAT3, and we identify two functional STAT3-binding sites on the mouse RhoU promoter. RhoU induction by Wnt-1 is independent of β-catenin, but does not involve STAT3. Rather, it is mediated by the Wnt/planar cell polarity pathway through the activation of JNK (c-Jun N-terminal kinase). Both the so-called non-canonical Wnt pathway and STAT3 are therefore able to induce RhoU, which in turn may be involved in mediating their effects on cell migration.
Keywords: chromatin immunoprecipitation (ChIP), mouse embryonic fibroblast, promoter analysis, RhoU, signal transducer and activator of transcription 3 (STAT3), Wnt-1
INTRODUCTION
STATs (signal transducers and activators of transcription) are transcription factors that play a major role in the signalling from cytokine receptors. STAT transcription factors become activated via tyrosine phosphorylation by receptor-associated JAK (Janus kinase) kinases [1], dimerize, concentrate into the nucleus and bind to STAT-responsive elements on the promoters of cytokine-induced genes. STAT3 was first isolated as the mediator of acute-phase gene induction in the liver in response to IL (interleukin)-6 and subsequently was identified as the main STAT transcription factor activated by all gp130 cytokines [2,3]. STAT3 is also activated by a wide variety of other cytokines and growth factors [e.g. leptin, IL-12, IL-17, IL-10, interferons, G-CSF (granulocyte colony-stimulating factor), EGF (epidermal growth factor) and PDGF (platelet-derived growth factor)] and by a number of oncogenic proteins such as c-Src, c-Abl, v-Sis, v-Fps, v-Ros, HER-2/neu and Met [4]. A Stat3-null mutation in the mouse leads to early embryonic lethality, and conditional inactivation has revealed complex roles linked to inflammation, regeneration, proliferation, migration and energy homoeostasis [5].
Although STAT activity downstream of cytokines and growth factors is normally tightly controlled in both intensity and duration, STAT3 is found to be constitutively active in the vast majority of human and mouse tumours of both haematologic and solid origin, where its inhibition often leads to growth arrest and apoptotic cell death [6]. STAT3 exerts its pro-oncogenic role both directly, inducing the expression of anti-apoptotic and cell-cycle genes in the tumoral cells, and indirectly, through down-modulation of tumour immune surveillance [7]. STAT3 can also regulate cell movement, contributing to cytoskeleton reorganization and controlling cell adhesion properties, and is thought to play a role in tumour invasion and metastasis by inducing the expression of matrix metalloproteinases and promoting the EMT (epithelial–mesenchymal transition) (recently reviewed in [8]). In the zebrafish embryo, STAT3 activity is required for cell movements during gastrulation, a defect partially rescued by activation of the Wnt/PCP (planar cell polarity) signalling [9].
The evolutionarily conserved Wnt signal transduction pathway plays important roles in development and is strongly implicated in tumorigenesis. As for STAT3, aberrant Wnt signalling occurs in tumours of diverse origin and is likely to represent an important generic step in tumour development, contributing to functions as diverse as resistance to apoptosis, tissue invasion and metastasis, growth-factor-independent growth, evasion from immune response and sustained angiogenesis [10]. Wnt proteins, acting through the frizzled family of seven-pass transmembrane receptors and through recently identified alternative receptors such as Ryk and Ror2 [11], can activate three different pathways: (i) the so-called canonical Wnt signalling pathway, leading to the stabilization and nuclear concentration of β-catenin and to the activation of LEF (lymphoid enhancer factor)/TCF (T-cell factor)-mediated transcription (this is the best characterized pathway mediating the proliferative effects of Wnt ligands); (ii) the Wnt/PCP pathway, involved in controlling cell polarity and movement, particularly during embryogenesis, and mediated by the activation of the JNK (c-Jun N-terminal kinase) pathway; and (iii) the Wnt/Ca2+ pathway, resulting in increased intracellular calcium and activation of PKC (protein kinase C).
Recently, a new Rho family member homologous with Cdc42 named Wrch1 (Wnt-regulated Cdc42 homologue)/RhoU has been isolated as a Wnt-1-responsive mRNA by means of subtraction cloning [12]. RhoU is an atypical Rho GTPase with no detectable GTPase activity and high intrinsic guanine nucleotide exchange, and is thus likely to be constitutively GTP-bound [13,14]. Its regulation occurs both at the level of mRNA expression [12] and through release of the inhibitory effects of the N-terminal domain [14]. RhoU acts on the actin cytoskeleton by stimulating the formation of filopodia, stimulates quiescent cells to re-enter the cell cycle and phenocopies Wnt-1-dependent morphological transformation of mouse mammary epithelial cells [12]. Recently, RhoU was shown to localize to focal adhesions in HeLa cells and fibroblasts and to podosomes in osteoclasts and Src-expressing cells; its depletion causes an increase in focal adhesion numbers and inhibits cell migration [15,16].
In the present study we report that RhoU can also be induced by gp130 cytokines and that both its basal and cytokine-induced transcription in MEF (mouse embryonic fibroblast) cells requires STAT3, which binds to two STAT3-responsive elements on the mouse RhoU promoter. We also show that Wnt-1-mediated RhoU induction is regulated at the transcriptional level but is independent of STAT3, does not involve the canonical β-catenin-dependent pathway and requires JNK activity instead. Finally, we provide evidence for STAT3-mediated RhoU regulation also in human tumour or non-transformed epithelial cell lines.
EXPERIMENTAL
Cell lines and treatments
Stat3+/+, Stat3−/−, Stat3Rev immortalized MEFs [17], HEK (human embronic kidney)-293, HEK-293-Wnt1, HeLa, PC3, DU145 and SKBR3 cells were grown in DMEM (Dulbecco's modified Eagle's medium; Gibco-BRL). T47D, SKOV3, MDAMB-231 and MDA-MB-468 were grown in RPMI 1360 (Gibco-BRL). Both media were supplemented with 10 % (v/v) heat-inactivated FCS (foetal calf serum; Gibco-BRL), 2 mM L-glutamine, 100 units/ml penicillin and 100 μg/ml streptomycin. MCF10A cells were grown in DMEM F12 (Gibco-BRL) supplemented with 5 % (v/v) horse serum (Gibco-BRL), 20 ng/ml EGF, 0.5 μg/ml hydrocortisone and 10 μg/ml insulin. All cell lines were maintained at 37°C in a 5 % CO2 atmosphere. The HEK-293-Wnt1 stable transfectants were generated by stable transfection with a Wnt-1 expression vector. Cells were treated with 20 ng/ml OSM (oncostatin M; R&D Systems) or with recombinant IL-6 (500 ng/ml) plus soluble receptor (250 ng/ml) [18,19] for different lengths of time. MEFs were treated with the following inhibitors: 50 μM SP600125 (Sigma–Aldrich), 10 μM Y-27632 (Sigma–Aldrich) and 100 μM myristoylated PKC peptide inhibitor (Promega) for the times indicated. DU145 and SKBR3 were treated with 50 mM S3I-201 (a STAT3 inhibitor) for the times indicated.
RNA extraction, retrotranscription and qRT-PCR (quantitative real-time PCR)
Total RNA was prepared with the PureLink Micro-to-Midi Total RNA Purification System (Invitrogen) according to the manufacturer's protocol. Samples were treated with DNase I Amp Grade (Invitrogen) before retrotranscription to eliminate genomic DNA contamination. To produce template cDNA, 125 ng of total RNA was reverse-transcribed using the RT High Capacity kit (Applied Biosystems) according to the manufacturer's protocol. qRT-PCR reactions were performed with the ABI Prism 7300 real-time PCR System (Applied Biosystems) using Platinum Quantitative PCR SuperMix-UDG with ROX (Invitrogen). The reactions were carried out in a total volume of 25 μlusing a TaqMan assay (Mm00505976_m1; Applied Biosystems) specific for murine RhoU. Human RhoU was detected by using the Universal Probe Library system (Roche) with the following primers and probe: forward primer, 5′-GACTCCAACTCTGTGACACTGC-3′; reverse primer, 5′-ATGAGGGGCTCACGACACT-3′; and probe #4. Results were analysed with the 2−ΔΔCt method using the 18S rRNA pre-developed TaqMan assay (Applied Biosystem) as an internal control.
EMSA (electrophoretic mobility-shift assay) competition assays
EMSA probes/competitors consisted of double-stranded DNA oligonucleotides formed by a 9 bp long STAT3-binding site flanked by 3 bp on both sides and by a GATC protruding sequence on the 5′ end. Labelling and EMSAs were performed as described previously [2]. Briefly, the HA-SIE (high-affinity sisinducible element) probe (1 pmol) was labelled using [32P]dCTP and Klenow and purified from free nucleotides on a Sephadex spin column. Nuclear extracts (4 μg) were pre-incubated on ice for 10 min in 80 mM KCl, 2 mM MgCl2, 2mM EDTA, 40 mM Hepes, 20 % (v/v) glycerol, 3 μg/reaction poly dI/dC, 5 mM DTT (dithiothreitol) and 1 μg/μl BSA. Competitor (1 μl), corresponding to concentration ratios 10-, 50- or 100-fold higher with respect to that of the labelled probe, was added and left to interact for 10 min on ice followed by incubation with the labelled probe (10000 c.p.m.) for 10 min at room temperature (25 °C). Samples were fractionated on a 5 % (v/v) polyacrylamide gel in 0.25 × TBE buffer (4.65 μg/μl Tris, 30 μg/μl boric acid and 60.5 μg/μl EDTA, pH 8.3) at room temperature. Gels were dried and autoradiographed. The oligonucleotide sequences used are indicated below: site1, upper 5′-GATCCATTTCCAAGACAGC-3′ and lower 5′-GATCGCTGTCTTGGAAATG-3′; site 2, upper 5′-GATCATATTAAAGGAATTC-3′ and lower 5′-GATCGAATTCCTTTAATAT-3′; site 3, upper 5′-GATCGGTTGCTGGGAATTG-3′ and lower 5′-GATCCAATTCCCAGCAACC-3′; site 4, upper 5′-GATCCTTTTCAGATAATAC-3′ and lower 5′-GATCGTATTATCTGAAAAG-3′; site 5, upper 5′-GATCAGGAGCCGGGAAAGG -3′ and lower 5′-GATCCCTTTCCCGGCTCCT-3′; site 6, upper 5′-GATCCGTTTCGAGGCGCCG-3′ and lower 5′-GATCCGGCGCCTCGAAACG-3′; HA-SIE, upper 5′-GGGCATTTCCCGTAAATCGT-3′ and lower 5′-CCCACGATTTACGGGAAATG-3′; and unrelated, upper 5′-GATCGGTTTCCGGACCAGC-3′ and lower 5′-GATCGCTGGTCCGGAAACC-3′.
Western blot analysis
Total protein extracts were obtained in 50 mM Tris/HCl (pH 7.4), 10 % (v/v) glycerol, 1 % (v/v) Nonidet P40, 150 mM NaCl, 2 mM EDTA, 2 mM DTT, 0.4 mM Na3VO4, 10 mM NaF, 0.5 mM PMSF and 40 μg/ml protease inhibitor cocktail (Sigma–Aldrich). Proteins were cleared by centrifugation (12000 g for 10 min at 4°C) and the concentration was measured using the Bradford assay (Bio-Rad). Samples were fractionated on SDS/PAGE and transferred on to PVDF membrane (Millipore) for immunoblotting. The following antibodies were used: anti-phospho-STAT3 (Tyr705; Cell Signaling Technology), total STAT3 (Santa Cruz Biotechnology), actin (Santa Cruz Biotechnology) and GAPDH (glyceraldehyde-3-phosphate dehydrogenase; Millipore).
Generation of luciferase reporter plasmids
RhoU promoter fragments were generated by PCR using a genomic BAC vector as a template. The following primers were used: −1765 forward (5′-ACTAGGTACCGAATTCAGGTGGCCCAAGGGC-3′); −1300 forward (5′-GGCCGGTACCGCATCTCTGTACTTCAGGTTGTCC-3′); −770 forward (5′-GGCCGGTACCCGCTAACAAAGGGTGGTATCCTGC-3′); −200 forward (5′-GGCCGGTACCGGTCTCGGATAGAAGAAGGAGCC-3′); and reverse (5′-TATCTCGAGGCCGCGAGACCAGCTGCC-3′). Amplified fragments were digested with KpnI/XhoI, cloned into pBlueScript (Stratagene), sequenced and subcloned into the promoter-less luciferase reporter vector pGL3basic (Promega). Mutant constructs were made by site-directed mutagenesis using the QuikChange® Mutagenesis Kit (Stratagene) according to the manufacturer's protocol. The following primers were used for mutagenesis (mutated bases are underlined): −1300 M2 sense (5′-GGGTATAAGTAATGCTAATATTAAAGCCCTTCGTTTTGACAGAATGGTTA-3′);−1300 M2 antisense (5′-TAACCATTCTGTCAAAACGAAGGGCTTTAATATTAGCATTACTTATACCC-3′); −1300 M3 sense (5′-CGTGAGTCACCATGTGGTTGCTGGCCCTTGAACTCAGGACC-3′); −1300 M3 antisense (5′-GGTCCTGAGTTCAAGGGCCAGCAACCACATGGTGACTCACG-3′); −1300 M5/−770 M5 sense (5′-GGAAGCTTGGGAAGGAGCCGGCCCAGGGGGGTGG-3′); −1300 M5/−770 M5 antisense (5′-CCACCCCCCTGGGCCGGCTCCTTCCCAAGCTTCC-3′); −1300 MA sense (5′-CAACAACATCTGGGACATAAACCAGGCTCAGTATTTAACTAGCTCTTGTC-3′); −1300 MA antisense (5′GACAAGAGCTAGTTAAATACTGAGCCTGGTTTATGTCCCAGATGTTGTTG-3′); −770 MB sense (5′-CCAAAGAGAGGGAGCGCTAAACCAGGGTGGTATCCTGCATC-3′); −770 MB antisense (5′-GATGCAGGATACCACCCTGGTTTAGCGCTCCCTCTCTTTGG-3′); −770 MC sense (5′-CAAAGGGTGGTATCCTGCATCCAAATGAGAATACCTGATTTTTAACAG-3′); −770 MC antisense (5′-CTGTTAAAAATCAGGTATTCTCATTTGGATGCAGGATACCACCCTTTG-3′). All mutant constructs were controlled by sequencing.
Transient transfections, co-culture and luciferase assays
MEFs and HeLa cells were seeded in 24-well plates and transfected 24 h later with Lipofectamine™ reagent (Invitrogen). Each promoter construct was mixed at a 10:1 ratio with the pSEAP vector [20] as an internal control for transfection efficiency for a total amount of 0.4 μg of DNA and incubated with 5 μlof Lipofectamine™ according to the manufacturer's protocol. In cotransfection experiments, promoter constructs were mixed at a ratio of 5:5:1 (reporter/expression vector/pSEAP). At 24 h after transfection, the cells were treated with OSM or detached and re-seeded with an equal amount of Wnt1-expressing HEK-293 or wild-type control cells for co-culture experiments. At 48 h after transfection, the cells were washed twice with PBS and lysed in 50 μl of passive lysis buffer (Promega). Luciferase assays were performed using a luciferase assay system (Promega) according to the manufacturer's protocol. SEAP assays were performed using the phospha-light system (Applied Biosystems) according to the manufacturer's protocol. Luciferase and SEAP chemiluminescent activity were quantified using a Mithras LB940 multilabel reader (Berthold Technologies). SEAP values were used to normalize for differences caused by unequal transfection efficiency.
ChIP (chromatin immunoprecipitation) assay
Stat3+/+ and Stat3−/− MEFs were treated or not with OSM as described above. ChIP assays were performed with the fast ChIP method [21] with some modifications. Briefly, cross-linking was performed with 1 % (v/v) formaldehyde for 10 min at room temperature. The reaction was quenched by the addition of glycine to a final concentration of 125 mM and incubation for 10 min at room temperature. Cells were lysed and sonicated at 4°Cfor six 10 s rounds followed by a 60 s pause, at an amplitude of 40 % (Ultrasonic Processor VCX-750, Sonics and Materials).
Immunoprecipitations were performed by incubating overnight at 4°C 1 ml of sheared chromatin with anti-STAT3 serum (R&D Systems, 5 μl), anti-acetyl-histone H3 (Upstate Cell Signaling Solutions, 2 μg) or negative control IgG (ChIP-IT Control Kit-Mouse; Active Motif, 2 μg). Primers for PCR amplification were designed using the program Primer3 [22] to obtain an amplicon size of approx. 150 bp. PCR products were fractionated on a 2 % (w/v) agarose gel and stained with ethidium bromide. PCR reactions were performed using 5 μl of immunoprecipitated DNA using the following primers: RhoU site 3, forward 5′-GGCACCAGATCCCATTACAG-3′ and reverse 5′-TTTTGTGCTGCCCTTTCTCT-3′; RhoU site 5, forward 5′-ATCGCAAGGTCAGCTTTCAC-3′ and reverse 5′-CTTGCTTGGCCATTTACCAT-3′; Socs3 −65, forward 5′-GGAGAGACAGCGGTCGTAAG-3′ and reverse 5′-CACAGCCTTTCAGTGCAGAG-3′; and β-globin, forward 5′-CTCCCCCTCACTCTGTTCTG-3′ and reverse 5′-AGGAGGAGGGGAAGCTGATA-3′.
RESULTS
RhoU is a STAT3 transcriptional target induced by gp130 cytokines
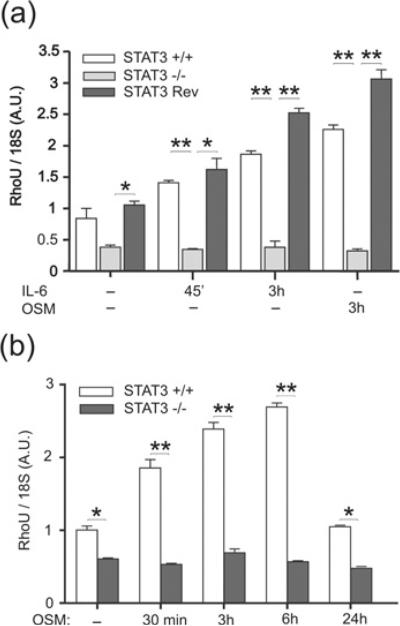
Gene expression profiling of Stat3+/+ or Stat3−/− MEFs either untreated or stimulated with different cytokines has led us to identify, among others, the atypical Rho GTPase RhoU/Wrch1 as a STAT3-dependent target of gp130 cytokine stimulation [23]. Moreover, RhoU displayed a striking degree of conserved co-expression with STAT3 across hundreds of human and mouse microarray experiments [24], further suggesting a close functional correlation. Quantitative PCR analysis of RhoU mRNA levels showed indeed profoundly defective expression in Stat3−/− MEFs already under basal conditions (Figure 1a). In addition, whereas both IL-6 and OSM treatment induced RhoU mRNA in Stat3+/+ MEFs, they were completely uneffective in the absence of Stat3. As a result, RhoU expression levels were 4–5-fold lower in the Stat3−/− MEFs following cytokine stimulation. These defects are indeed STAT3-dependent since reintroducing Stat3 by stable transfection in the Stat3−/− MEFs (revertant MEFs) completely rescued both RhoU basal expression and cytokine inducibility (Figure 1a). RhoU mRNA induction in response to OSM stimulation was already appreciable 30 min after OSM treatment, peaked at 6 h and decreased to basal levels 24 h later (Figure 1b).
Figure 1. RhoU mRNA expression is defective in Stat3−/− MEFs.
Total RNA was extracted from Stat3+/+, Stat3−/− and revertant (rev) MEFs either untreated or stimulated with IL-6 or OSM for the indicated times. RhoU mRNA was measured by qRT-PCR. Expression was normalized to 18S, and results are shown as means ± S.E.M. of two independent experiments performed in duplicate. Asterisks indicate statistically significant differences between selected groups (*P < 0.05 and **P < 0.01). A.U., arbitrary units.
The RhoU promoter contains two functional STAT3-binding sites fully responsible for OSM inducibility
Since RhoU expression is induced by gp130 cytokines/STAT3, we searched the mouse RhoU promoter region for potential STAT3 regulatory elements. Bioinformatics analysis [23] identified six potential STAT3-binding sites (sites 1–6) that were assessed for in vitro binding capacity by EMSA competition assays (Figure 2a). Liver nuclear extracts from LPS-treated mice were used as a source for activated STAT3, with a labelled HASIE [25] double-stranded oligonucleotide as a probe. STAT3 binding to the HA-SIE probe was then competed with non-labelled double-stranded oligonucleotides based on the sequences of the six predicted STAT3-binding sites. Site 3 showed the greatest in vitro binding activity, with full competition at a ratio of 100:1 between non-labelled competitor and labelled probe, and partial competition at the ratios of 50:1 and 10:1. Sites 5 and 2 were also able to compete, albeit with lower efficiency/affinity, whereas sites 1, 4 and 6 did not show any significant competing activity. Self-competition with a unlabelled HA-SIE oligonucleotide or competition with an unrelated sequence were used as positive and negative controls respectively.
Figure 2. Functional STAT3 binding sites in the RhoU promoter.
(a) EMSA competition assays. A radioactively labelled HA-SIE probe was incubated with liver nuclear extracts containing activated STAT3 and the binding was competed with increasing amounts of unlabelled oligonucleotides based on the sequence of the six predicted STAT3-binding sites on the mouse RhoU promoter, whose sequence and position from the TSS (transcription start site) are indicated. Unlabelled unrelated or HA-SIE oligonucleotides were used as negative or positive competing controls respectively. (b and c) Transient transfection assays. The indicated constructs were co-transfected with a pSEAP expression vector as an internal control. At 24 h later, cells were stimulated or not with OSM and cultured for a further 24 h. Luciferase activity was normalized to SEAP activity as a control for transfection efficiency. Results shown are means ± S.E.M. of at least two independent experiments performed in duplicate. (b) Transfections were performed with Stat3+/+ and Stat3−/− MEFs. Numbers over the bars indicate the induced fold-change of stimulated over unstimulated cells. The empty pGL3 vector and the previously characterized Socs3 promoter-luciferase construct [26] were used as controls. (c) Stat3+/+ MEFs were transfected with the indicated wild-type or mutant (M) RhoU promoter constructs. (d) ChIP assays were performed with Stat3+/+ and Stat3−/− MEFs, either unstimulated (NT) or treated with OSM for 30 min. Immunoprecipitations were performed with antibodies against acetylated histone H3 (α-Ac. H3), STAT3 (α-STAT3) antisera or control IgG. Non-immunoprecipitated chromatin was used as the total input control (T.I.). Specific primers were used to amplify regions of approx. 150 bp containing the indicated STAT3-binding sites (see the Materials and methods section). Primers amplifying the second intron of the silent β-globin gene (shown) or the non-coding transcript Xist (inactive X specific transcript) (not shown) were used as negative controls. A.U., arbitrary units.
In order to analyse RhoU promoter activity, mouse RhoU promoter deletion fragments were fused to a luciferase reporter and transiently transfected in Stat3+/+ and Stat3−/− MEFs (Figure 2b). Transfected cells were either stimulated with OSM or left untreated for 24 h before measuring luciferase activity. In keeping with the mRNA expression results, transcriptional activity of all promoter fragments was drastically reduced in Stat3-deficient MEFs as compared with their wild-type controls already under basal conditions. In addition, the constructs −1765, −1300 and −770 were induced by OSM treatment in the Stat3+/+ MEFs, confirming that cytokine-dependent RhoU mRNA induction occurs transcriptionally. In contrast, OSM-dependent induction of all constructs was completely defective in the Stat3-deficient cells. Both basal and OSM-stimulated activity of RhoU promoter constructs were comparable with those of the mouse Socs3 promoter [26], a well-recognized STAT3 target. RhoU −200, the shortest promoter fragment tested, displayed reduced activity in both Stat3−/− and Stat3+/+ MEFs and was unresponsive to OSM treatment, suggesting that the region(s) required for OSM induction must be located upstream of the −200 position. Interestingly, and only in the presence of Stat3, both basal transcription levels and OSM inducibility increased with the progressive shortening of the promoter up to position −770, before dropping at −200. This might suggest the presence of repressor element(s) in the distal portions of the promoter. To assess the functional contribution of the tested STAT3-binding sites (see Figure 2a), sites 2, 3 and 5 were mutagenized in the context of the RhoU −1300 construct and the transcriptional activity of the resultant RhoU −1300 M2, M3 or M5 constructs tested in wild-type MEFs (Figure 2c). The site 2 mutation did not have any effect on either basal or OSM-induced expression, suggesting that this site is not involved in STAT3-mediated regulation. Indeed, site 2 was the one with the lowest in vitro binding activity (Figure 2a). Disruption of sites 3 or 5 significantly decreased transcriptional activation in response to OSM stimulation, and the compound double mutation (−1300 M3 + M5) completely abolished promoter inducibility and decreased its basal activity (Figure 2c). Thus both STAT3-binding sites 3 and 5 co-operate to confer full promoter activation in response to OSM stimulation. On the other hand, site 5 also plays a prominent role to confer OSM inducibility to the RhoU −770 promoter construct, since its mutation in this context (−770 M5) almost completely abolished OSM inducibility (Figure 2c). Site 6 did not appear to contribute to this residual induction since neither a site 6 single mutation nor a compound site 5/site 6 double mutation were able to abolish it (−770 M6 and M5+M5 mutants; Figure 2c).
To further analyse the in vivo functionality of sites 3 and 5 we assessed STAT3 in vivo occupancy by ChIP. ChIP assays were performed with chromatin samples prepared from Stat3+/+ and Stat3−/− MEFs, either untreated or stimulated with OSM for 30 min. With respect to the negative IgG controls, both sites showed weak STAT3 binding already in untreated cells (Figure 2d). Similar to what was observed for the well-characterized SOCS3 −65 STAT3-responsive element [26], STAT3 binding to both site 3 and site 5 was strongly induced by OSM, confirming their in vivo functional role in mediating RhoU promoter activity. Importantly, no STAT3 binding was ever detected with chromatin from Stat3−/− MEFs, thus confirming the specificity of the ChIP assay used in the present study.
RhoU induction by Wnt-1 occurs at the transcriptional level, but does not require STAT3
Since RhoU was initially isolated as a Wnt-1-regulated gene, but its transcriptional regulation was never characterized, we decided to investigate the mechanisms involved in RhoU up-regulation downstream of Wnt-1 and the potential co-operation with the STAT3 pathway. Thus RhoU mRNA expression levels were measured in Stat3+/+ and Stat3−/− MEFs stimulated with OSM and Wnt-1 either alone or in combination (Figure 3a). Wnt-1 stimulation was obtained by co-culturing MEFs with HEK-293 cells stably expressing Wnt-1, whereas control cells were co-cultured with wild-type HEK-293 cells. In keeping with previous observations, RhoU mRNA expression was dependent on STAT3 already under basal conditions and failed to be induced by OSM stimulation in Stat3−/− cells. In contrast, Wnt-1 stimulation could similarly induce RhoU mRNA in both Stat3+/+ and Stat3−/− MEFs. Likewise, Wnt-1 could induce RhoU promoter transcription at similar levels in the presence or absence of Stat3 (Figure 3b). These results suggest that Wnt-1-mediated RhoU induction occurs at the transcriptional level and is independent of STAT3. This was further supported by the observation that the RhoU −770 M5 construct, which has lost OSM inducibility, is still fully responsive to Wnt-1 stimulation (Figure 4a). Thus both the Wnt-1 and the gp130/STAT3 pathways can activate transcription of the RhoU promoter, but they act through independent mechanisms. Of note, RhoU induction could not be obtained using the supernatants of the Wnt-1-expressing HEK-293 cells (results not shown), suggesting that specific stimulation by membrane-bound Wnt-1 and not secretion of soluble mediators by the Wnt-1-expressing cells is involved.
Figure 3. RhoU induction downstream of the Wnt-1 and STAT3 pathways.
(a) qRT-PCR analysis. Stat3+/+ and Stat3−/− MEFs were co-cultured with wild-type or Wnt-1-expressing HEK-293 cells for 24 h with or without OSM stimulation. RhoU mRNA levels were measured by qRT-PCR and normalized to 18S RNA. Results are shown as means ± S.E.M. of two independent experiments performed in duplicate. (b) Transient transfection assays. Stat3+/+ and Stat3−/− MEFs were transfected with the RhoU −770 construct and the pSEAP vector. At 24 h after co-transfection, cells were treated as in (a) for a further 24 h. Results shown are means ± S.E.M. of normalized luciferase activity from at least two independent experiments performed in duplicate. (c) Stat3+/+ MEFs were transfected with the following vectors: the indicated RhoU promoter deletion fragments and TOPFlash and FOPFlash reporters. Transfected MEFs were Wnt-1-stimulated as in (a). Results shown are means ± S.E.M. of normalized luciferase activity from at least two independent experiments performed in duplicate. A.U., arbitrary units.
Figure 4. RhoU induction downstream of Wnt-1 is independent from STAT3 and β-catenin.
(a) Stat3+/+ MEFs were transfected with the RhoU −1300 and −770 vectors, either wild-type or mutated (M) in the indicated LEF/TCF or STAT3-binding sequences. Wnt-1 stimulation and data analysis were performed as described for Figure 3. (b) The RhoU −770 construct and TOPFlash and FOPFlash reporters were co-transfected with a stable mutant β-catenin (S33A) or with an empty control vector in Stat3 wild-type MEFs. Luciferase activity was normalized and reported as above. A.U., arbitrary units.
Similar to what was observed for OSM, Wnt-1-mediated induction progressively increased with promoter shortening, suggesting that the inhibitory sequence(s) already postulated in the case of OSM induction may be also active in the context of Wnt-1 stimulation (Figure 3c). As in the case of OSM responsiveness, the RhoU −200 construct was not anymore able to respond to Wnt-1 stimulation, indicating that the Wnt-1-responsive element(s) must lie in the region between positions −770 and −200. The reporter vector Super8X-TOPFlash and its negative control Super8X-FOPFlash [27] were used in all co-culture experiments to monitor Wnt pathway activation (results not shown).
RhoU promoter induction by Wnt-1 does not involve the canonical β-catenin pathway
The molecular mechanisms leading to RhoU gene activation downstream of Wnt-1 are currently unknown. Among the many signalling events activated by Wnt ligands, the canonical Wnt/β-catenin pathway which leads to the up-regulation of several target genes mediated by the LEF/TCF transcription factor is the best characterized [28,29]. Mouse RhoU promoter analysis using the TRANSFAC open-source program [30] identified three consensus sequences for LEF/TCF (sites A, B, C; Figure 2b). However, none of the three sequences appears to play a role in RhoU promoter activity, as shown by transient transfection assays of the relative promoter mutants −1300 MA, 770 MB or −770 MC (Figure 4a). Moreover, downstream activation of the Wnt-1/β-catenin pathway by expression of a stabilized mutant form of β-catenin did not have any effect on RhoU promoter transcription, whereas it strongly activated the TOPFlash control construct (Figure 4b). Thus the canonical Wnt/β-catenin pathway does not appear to be involved in mediating RhoU transcriptional induction.
A JNK-mediated non-canonical Wnt pathway is responsible for RhoU activation
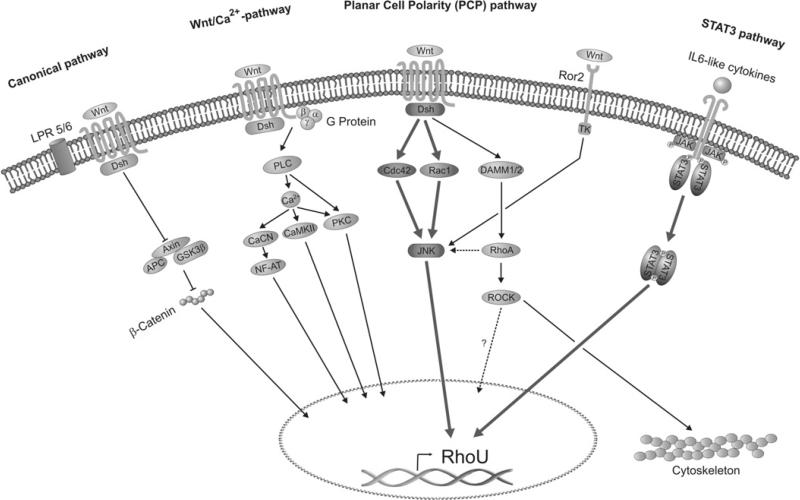
Besides the canonical β-catenin pathway, Wnt ligands are known to activate the Wnt/Ca2+ and the PCP non-canonical pathways (as illustrated in Figure 7). In order to assess their potential involvement in RhoU induction by Wnt-1, we have used available specific chemical inhibitors to interfere with the activities of the main mediators involved.
Figure 7. Schematic representation of the best known Wnt signal transduction pathways.
The Wnt signal branches into three main pathways: the canonical β-catenin-dependent, the non-canonical planar cell polarity (PCP) and Wnt-Ca2+. All of these pathways are thought to occur through the Frizzled receptor and Dishevelled. Other receptors have recently been shown to be able to activate additional non-canonical pathways. In particular the Ror2 receptor, depicted here, converges on to JNK in a similar manner to the Wnt/PCP pathway. In the present study we have demonstrated that RhoU is transcriptionally induced both by Wnt-1, through the PCP pathway involving the Rho GTPases Rac1/Cdc42 and the MAPK JNK, and by gp130 cytokines through the JAK/STAT3 pathway (thick lines). APC, adaptor protein complex; CaCN, calcineurin; CaMKII, Ca2+/calmodulin-dependent protein kinase II; Dsh, Dishevelled; GSK3β, glycogen synthase kinase 3β; LPR 5/6, lipoprotein receptor 5/6; NF-AT, nuclear factor of activated T-cells; PLC, phospholipase C; TK, tyrosine kinase.
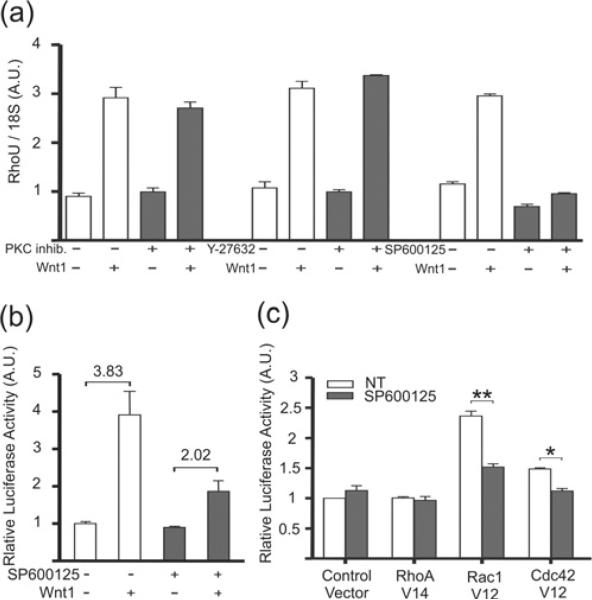
First, RhoU mRNA levels were measured by qRT-PCR in wild-type MEFs treated or not with specific inhibitors for PKC (Wnt/Ca2+ pathway), ROCK (Rho-associated kinase) and JNK (PCP pathway) in the presence or absence of Wnt-1 stimulation (Figure 5a). Wnt-1-mediated RhoU mRNA induction was unaffected by PKC and ROCK inhibitors, whereas it was completely abolished by inhibiting JNK activity (Figure 5a). A similar effect was also observed at the level of RhoU promoter activation, since Wnt-1-mediated induction of the −770 promoter construct was strongly reduced in the presence of the JNK inhibitor (Figure 5b).
Figure 5. Wnt-1-mediated RhoU activation follows a non-canonical pathway involving JNK.
(a) qRT-PCR analysis. Wild-type MEFs were stimulated or not with Wnt-1 as described for Figure 3 and treated with specific inhibitors for PKC (myristoylated peptide inhibitor, 100 μM), ROCK (Y-27632, 10 μM) and JNK (SP600125, 50 μM) for 24 h. RhoU expression levels were measured by qRT-PCR and normalized to 18S RNA. Values are means ± S.E.M. of two independent experiments performed in duplicate. (b and c) Transient transfection assays. Wild-type MEFs were transfected with the promoter fragment −770 alone (b) or in combination with plasmids encoding the constitutively active GTPases RhoA (RhoA V14), Rac1 (Rac1 V12), Cdc42 (Cdc42 V12) or a control empty vector (c). At 24 h later, cells were stimulated or not with Wnt-1 as described for Figure 3 and treated with 50 μM SP600125 (b) or simply treated or not with 50 μM SP600125 (c) and cultured for a further 24 h. Luciferase activity was quantified and normalized as described for Figure 2. A.U., arbitrary units.
Thus RhoU transcriptional activation by Wnt-1 appears to be mainly mediated by the non-canonical PCP pathway, with a central role played by the MAPK (mitogen-activated protein kinase) JNK. Since the activation of JNK downstream of Wnt is known to be mediated by the small GTPases Cdc42 and Rac (see Figure 7), we performed co-transfection experiments of the RhoU promoter with constitutively active Rac1 and Cdc42 mutants (Figure 5c). Both Rac1-Val12 and Cdc42-Val12, but not the control RhoA-Val14, were able to significantly activate the RhoU promoter and this induction was abolished by JNK inhibition, strengthening the idea that Wnt-1 can activate the PCP pathway leading to the induction of the RhoU promoter through Rac1 and Cdc42-mediated JNK activation.
STAT3/gp130-dependent regulation of RhoU in human epithelial cell lines
In order to extend our results to more biologically relevant experimental systems, we analysed RHOU mRNA expression and its correlations with the gp130/STAT3 pathway in human transformed and non-transformed epithelial cell lines. OSM treatment led to RHOU mRNA up-regulation also in HeLa cells, albeit with different kinetics as compared with MEFs (Supplementary Figure S1a at http://www.BiochemJ.org/bj/421/bj4210283add.htm). In the same cells, additionally, both OSM and Wnt-1 stimulation were able to induce RHOU promoter activity (fragment −770) to a similar extent as observed in MEFs (Supplementary Figure S1b and S1c).
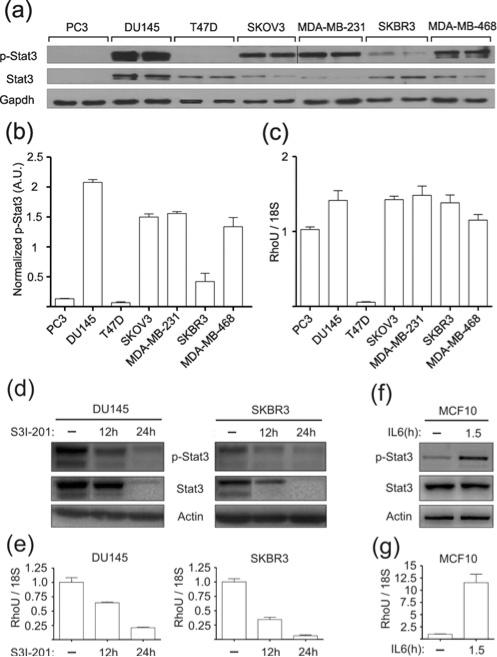
Since RhoU is thought to mediate Wnt-1 oncogenic properties and might therefore play a similar role downstream of STAT3, we decided to correlate STAT3 activation and RHOU expression levels in a panel of human tumour cell lines well-characterized for STAT3 constitutive activation. RHOU expression levels (Figure 6c) were always elevated in cell lines with high (DU145, SKOV3, MDA-MB-231 and MDA-MB-468) or moderate (SKBR3) levels of constitutively active STAT3 (Figures 6a and 6b), all of which, with the exception of SKOV3 cells, have been reported to be addicted to STAT3 activity for survival/proliferation [31–33]. Conversely, cell lines with no detectable phosphorylated STAT3 showed variable RHOU mRNA levels (Figures 6a–6c). In particular, RHOU mRNA was almost undetectable in the breast cancer cells T47D, whereas its levels were comparable with those of the other cell lines in the prostate cancer PC3 cells. Of note, these are known to display constitutive Wnt-1 expression, which may be responsible for RhoU up-regulation in the absence of STAT3 activity [34].
Figure 6. RHOU mRNA regulation by STAT3 in human tumour cell lines.
(a) Total and phospho-STAT3 were measured by Western blot analysis along with GAPDH as a loading control in the indicated tumour cell lines. (b) Quantification of the phospho-STAT3 signal in (a) normalized to GAPDH and reported as means ± S.E.M. of two replicates. (c) RHOU mRNA from the same samples analysed in (a) was measured by qRT-PCR and normalized to 18S. Results shown are means ± S.E.M. of two independent experiments performed in duplicate. (d–g) Total and phospho-STAT3 protein levels (d and f) and RHOU mRNA (e and g) were measured by Western blot analysis and qRT-PCR respectively, in DU145 and SKBR3 cells treated or not with the STAT3 inhibitor S3I-201 (d and e) or in MCF10A cells treated with IL-6 plus soluble IL-6 receptor. A.U., arbitrary units.
In order to assess the functional correlation between STAT3 activity and RHOU expression, DU145 and SKBR3 cells were then treated with the specific STAT3 inhibitor S3I-201 [33]. Both phospho- and total STAT3 levels were drastically reduced in both cell lines after S3I-201 treatment (Figure 6d), correlating with a dramatic decrease of RHOU mRNA levels as determined by qRT-PCR (Figure 6e; 78% and 93% reduction at 24 h in DU145 and SKBR3 cells respectively).
Finally, IL-6 stimulation triggered STAT3 phosphorylation correlating with a strong up-regulation of RHOU mRNA expression in MCF10A cells, a non-transformed human mammary cell line displaying almost undetectable levels of STAT3 phosphorylation under basal conditions (Figures 6f and 6g).
DISCUSSION
The atypical Rho GTPase RhoU was first identified as a Wnt-1-responsive gene by subtractive cloning [12]. RhoU overexpression can trigger cellular transformation and phenocopies the effects of Wnt-1 in mammary epithelial cells, suggesting a potential role in Wnt-mediated tumorigenesis [12]. Rather than being regulated by GDP/GTP exchange like most Rho GTPases, RhoU activity can be modulated by acting on its N-terminal domain, which exerts a partial inhibitory effect on the RhoU ability to activate PAK (p21-activated kinase) and to cause cellular transformation [14]. This inhibition can be relieved by interaction of the PxxP motif in the N-terminal domain with SH3-containing protein adaptors such as Grb2 and Nckβ, suggesting functional interactions with receptor tyrosine kinase-activated pathways. Moreover, unlike most Rho GTPases that are constitutively expressed, RhoU expression is tightly controlled [35,36]. In addition to being induced by Wnt-1, RhoU mRNA has been recently shown to accumulate in osteoclasts in response to RANKL (receptor activator for nuclear factor κB ligand)-induced differentiation [37]. However, the molecular mechanisms for either induction have not been characterized. In the present study, we demonstrate for the first time that RhoU induction by Wnt-1 occurs at the transcriptional level. Moreover, we show that RhoU is also a prominent target for STAT3-mediated transcription downstream of the gp130 cytokines IL-6 and OSM (Figure 7). Intriguingly, although both in vivo functional STAT3-binding sites 3 and 5 are required to confer full OSM inducibility to the −1300 RhoU promoter construct, site 5 alone is sufficient in the context of the shorter −770 promoter fragment, which lacks site 3. Both sites might indeed be required both in vivo and in the longer promoter construct to overcome the observed inhibitory effect of sequence(s) located between the −1300 and −770 positions. Interestingly, STAT3 was required not only for RhoU cytokine-mediated induction, but also for its basal expression, and both functionally tested STAT3-binding sites, site 3 and site 5, displayed a low level of constitutive binding following ChIP analysis. Constitutive STAT3 binding to a subset of sites was recently shown in NIH 3T3 cells [38], although a correlation with basal gene expression was not tested. Low amounts of Tyr705-phosphorylated STAT3 are present in growing MEFs and could be responsible for the constitutive binding observed. Alternatively, non-canonical mechanisms recruiting unphosphorylated STAT3 may be involved. At any rate, the ability of STAT3 to transcriptionally regulate a subset of target genes under basal conditions may have important implications for its physiology.
Both Wnt ligands and STAT3 play surprisingly similar multiple roles in tumorigenesis. In particular, both are involved in stimulating cell movements in tumoral cells as well as during development [8,39]. RhoU is known to act on the actin cytoskeleton, triggering filopodia formation and stress fibre dissolution. In addition, RhoU was recently shown to localize to focal adhesions and podosomes and to regulate their genesis promoting cell migration [15,16]. Intriguingly, STAT3 was also shown to localize to focal adhesions in HeLa and ovary tumour cells, where it interacted with activated focal adhesion kinase and paxillin and promoted cell migration [40], and we could demonstrate that activation of the gp130/STAT3 pathway induces both RhoU mRNA and promoter activity also in HeLa cells. Thus RhoU transcriptional induction might be one of the mechanisms through which STAT3 acts on focal adhesions and ultimately triggers cell migration. Indeed, the same Stat3−/− MEFs used in the present study, exhibiting low basal RhoU levels, also display impaired migration in a wound healing assay [41]. In addition, the convergence of both Wnt-1 and STAT3 signalling on RhoU may represent a point of cross-talk between the two pathways, which has been proposed in several systems. STAT3 expression was induced by the Wnt/β-catenin pathway in oesophageal cancer cells and in embryonic stem cells [42,43]. STAT3 was also proposed to act upstream of the Wnt/PCP pathway during the gastrulation of zebrafish embryos [9] and to promote β-catenin nuclear accumulation in colorectal cancer cells [44]. Moreover, the negative regulator of Wnt signalling Duplin was recently shown to also suppress STAT3 activity [45]. Indeed, both STAT3 and Wnt-1 aberrant activation are often observed in the same tumours, suggesting that the two pathways can co-operate in different ways depending on the cellular context.
However, the convergence of the Wnt-1 and gp130/STAT3 pathways on RhoU induction must involve distinct mechanisms, since STAT3 is not required for Wnt-1-mediated induction of the RhoU promoter. In addition, as previously suggested by Tao et al. [12], RhoU induction by Wnt-1 appears also to occur independently of β-catenin, suggesting the involvement of one of the non-canonical pathways. We could identify this pathway with the Wnt/PCP pathway, since Wnt-1-mediated RhoU induction is abolished by an inhibitor of JNK activity. Interestingly, RhoU itself was reported to be able to activate JNK [12], suggesting the existence of a positive-feedback loop whereby activated RhoU may enhance its own transcription by means of JNK activation. Supporting and extending this observation, we could demonstrate that both constitutively active Rac1 and Cdc42, which lie upstream of JNK in the Wnt/PCP pathway, can activate RhoU transcription in a JNK-dependent way. Thus, at least in MEFs, Wnt-1 can activate both the canonical β-catenin pathway and the non-canonical PCP pathway at the same time, as shown by the simultaneous activation of RhoU and TOPFlash in our system. It is intriguing that the Wnt/PCP pathway is best characterized as regulating cell movements during embryonic gastrulation and that the main functions of RhoU appear to be linked to focal adhesions and cell migration.
At present, the transcription factors mediating the effects of the PCP pathway are unknown, and our system may provide a relatively straightforward way to identify at least some of them. However, bioinformatic analysis of the RhoU promoter region involved in Wnt-1 responsiveness did not disclose any obvious candidate, and co-transfection experiments with transcription factors potentially activated by JNK such as c-fos, c-jun and CREB (cAMP-response-element-binding protein) did not modify promoter activity (D. Schiavone and V. Poli, unpublished work). More detailed promoter analysis will therefore be required in order to identify the PCP-activated transcription factors. The identification of RhoU as a common transcriptional target of both STAT3 and Wnt-1, together with the known roles of both pathways in stimulating cell migration in normal and tumoral cells, suggests the intriguing idea that RhoU may represent a common mediator of STAT3 and Wnt effects on cell motility under both physiological and pathological conditions, being potentially a common therapeutic target. Interestingly, we have observed a positive correlation between STAT3 phosphorylation and RhoU expression levels in all tested tumour-derived cell lines. Indeed, phosphorylated STAT3 was undetectable in T47D cells, the only ones displaying very low RHOU mRNA levels. The metastatic prostate cancer cell line PC3 in contrast, also lacking detectable STAT3 activation, displayed high RHOU mRNA expression. In these cells Wnt-1 expression was reported to be extremely elevated, and could thus play a dominant role in RhoU induction bypassing the need for STAT3 activity [34]. A direct correlation between STAT3 activity and RhoU transcription in tumour cell lines is also supported by our observation that specific STAT3 inhibition triggered a dramatic reduction of RHOU mRNA in DU145 and SKBR3 cells. Accordingly, acute STAT3 activation by IL-6 treatment could efficiently up-regulate RHOU mRNA in the non-transformed mammary epithelial cell line MCF10A. Taken together, these results strengthen the hypothesis that RhoU may represent a common transcriptional and functional target of both the Wnt-1 and STAT3 pathways in physiological, as well as pathological, contexts. Further studies addressing the role played by RhoU in mediating specific features of STAT3 and Wnt-mediated cell transformation will be required to address this issue.
Supplementary Material
SUPPLEMENTARY ONLINE DATA
The RhoU/Wrch1 Rho GTPase gene is a common transcriptional target of both the gp130/STAT3 and Wnt-1 pathways
Davide SCHIAVONE*, Sarah DEWILDE*1, Francesco VALLANIA*2, James TURKSON†, Ferdinando DI CUNTO* and Valeria POLI*3
*Molecular Biotechnology Center and Department of Genetics, Biology and Biochemistry, University of Turin, Via Nizza 52, 10126 Turin, Italy, and
†BioMolecular Science Center and Department of Molecular Biology and Microbiology, University of Central Florida, Orlando, FL 32826, U.S.A.
Figure S1 RhoU induction by OSM and Wnt-1 in HeLa cells
(a) RhoU mRNA was measured by qRT-PCR in HeLa cells treated with OSM for different lengths of time. (b and c) Transient transfection assays. The RhoU −770 and Socs3 (b)or TOPFlash/FOPFlash promoter constructs (c) were co-transfected with−a pSEAP vector in HeLa cells. At 24 h later, cells were either stimulated with OSM (b) or co-cultured with wild-type (NT) or Wnt-1-expressing (Wnt1) HEK-293 cells (c) for 24 h. Results shown are means ± S.E.M. of normalized luciferase activity from at least two independent experiments performed in duplicate.
Received 13 January 2009/1 April 2009; accepted 28 April 2009 Published as BJ Immediate Publication 28 April 2009, doi:10.1042/BJ20090061
1 Present address: BioIndustry Park del Canavese, 10010 Colleretto Giacosa, Italy.
2 Present address: Center for Genome Sciences, Department of Genetics, Washington University in St Louis School of Medicine, 4444 Forest Parkway, St Louis, MO 63108, U.S.A.
3 To whom correspondence should be addressed (valeria.poli@unito.it).
© The Authors Journal compilation © 2009 Biochemical Society
ACKNOWLEDGEMENTS
We wish to thank Dr I. Barbieri for help with the MCF10 cells, Dr C. J. Auernhammer for kindly providing the SOCS3 mouse promoter construct, Professor M. Orlandini for the mutant β-catenin (S33A) and the Wnt-1 constructs, Professor S. Rose-John and Professor C. Toniatti for the gift of recombinant plasmids and Professor G. Merlo, Professor E. Calautti and Professor D. Taverna for critically reading the manuscript prior to submission.
FUNDING
This work was supported by the Italian Ministry of University and Research (MIUR COFIN and FIRB) and by the Italian Cancer Research Association (AIRC).
Abbreviations used
- ChIP
chromatin immunoprecipitation
- DMEM
Dulbecco's modified Eagle's medium
- DTT
dithiothreitol
- EGF
epidermal growth factor
- EMSA
electrophoretic mobility-shift assay
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HA-SIE
high-affinity sis-inducible element
- HEK
human embryonic kidney
- IL
interleukin
- JAK
Janus kinase
- JNK
c-Jun N-terminal kinase
- LEF
lymphoid enhancer factor
- MAPK
mitogen-activated protein kinase
- MEF
mouse embryonic fibroblast
- OSM
oncostatin M
- PCP
planar cell polarity
- PKC
protein kinase C
- qRT-PCR
quantitative real-time PCR
- ROCK
Rho-associated kinase
- STAT
signal transducer and activator of transcription
- TCF
T-cell factor
- Wrch1
Wnt-responsive Cdc42 homologue
REFERENCES
- 1.Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. J. Biol. Chem. 2007;282:20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- 2.Alonzi T, Maritano D, Gorgoni B, Rizzuto G, Libert C, Poli V. Essential role of STAT3 in the control of the acute-phase response as revealed by inducible gene inactivation in the liver. Mol. Cell. Biol. 2001;21:1621–1632. doi: 10.1128/MCB.21.5.1621-1632.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wegenka UM, Buschmann J, Lutticken C, Heinrich PC, Horn F. Acute-phase response factor, a nuclear factor binding to acute-phase response elements, is rapidly activated by interleukin-6 at the posttranslational level. Mol. Cell. Biol. 1993;13:276–288. doi: 10.1128/mcb.13.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turkson J, Jove R. STAT proteins: novel molecular targets for cancer drug discovery. Oncogene. 2000;19:6613–6626. doi: 10.1038/sj.onc.1204086. [DOI] [PubMed] [Google Scholar]
- 5.Poli V, Alonzi T. STAT3 function in vivo. In: Sehgal PB, Levy D, Hirano T, editors. Signal Transducers and Activators of Transcription (STATs): Activation and Biology. Kluwer Academic Publishers; Boston: 2003. pp. 493–512. [Google Scholar]
- 6.Kortylewski M, Jove R, Yu H. Targeting STAT3 affects melanoma on multiple fronts. Cancer Metastasis Rev. 2005;24:315–327. doi: 10.1007/s10555-005-1580-1. [DOI] [PubMed] [Google Scholar]
- 7.Kortylewski M, Yu H. Role of Stat3 in suppressing anti-tumor immunity. Curr. Opin. Immunol. 2008;20:228–233. doi: 10.1016/j.coi.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pensa S, Regis G, Boselli D, Novelli F, Poli V. STAT1 and STAT3 in tumorigenesis: two sides of the same coin? In: Stephanou A, editor. JAK-STAT Pathway in Disease. Landes Bioscience Books; Texas: 2009. pp. 100–121. [Google Scholar]
- 9.Miyagi C, Yamashita S, Ohba Y, Yoshizaki H, Matsuda M, Hirano T. STAT3 non cell-autonomously controls planar cell polarity during zebrafish convergence and extension. J. Cell Biol. 2004;166:975–981. doi: 10.1083/jcb.200403110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ilyas M. Wnt signalling and the mechanistic basis of tumour development. J. Pathol. 2005;205:130–144. doi: 10.1002/path.1692. [DOI] [PubMed] [Google Scholar]
- 11.van Amerongen R, Mikels A, Nusse R. Alternative wnt signaling is initiated by distinct receptors. Sci. Signal. 2008;1:re9. doi: 10.1126/scisignal.135re9. [DOI] [PubMed] [Google Scholar]
- 12.Tao W, Pennica D, Xu L, Kalejta RF, Levine AJ. Wrch-1, a novel member of the Rho gene family that is regulated by Wnt-1. Genes Dev. 2001;15:1796–1807. doi: 10.1101/gad.894301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saras J, Wollberg P, Aspenstrom P. Wrch1 is a GTPase-deficient Cdc42-like protein with unusual binding characteristics and cellular effects. Exp. Cell Res. 2004;299:356–369. doi: 10.1016/j.yexcr.2004.05.029. [DOI] [PubMed] [Google Scholar]
- 14.Shutes A, Berzat AC, Cox AD, Der CJ. Atypical mechanism of regulation of the Wrch-1 Rho family small GTPase. Curr. Biol. 2004;14:2052–2056. doi: 10.1016/j.cub.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 15.Chuang YY, Valster A, Coniglio SJ, Backer JM, Symons M. The atypical Rho family GTPase Wrch-1 regulates focal adhesion formation and cell migration. J. Cell Sci. 2007;120:1927–1934. doi: 10.1242/jcs.03456. [DOI] [PubMed] [Google Scholar]
- 16.Ory S, Brazier H, Blangy A. Identification of a bipartite focal adhesion localization signal in RhoU/Wrch-1, a Rho family GTPase that regulates cell adhesion and migration. Biol. Cell. 2007;99:701–716. doi: 10.1042/BC20070058. [DOI] [PubMed] [Google Scholar]
- 17.Costa-Pereira AP, Tininini S, Strobl B, Alonzi T, Schlaak JF, Is'harc H, Gesualdo I, Newman SJ, Kerr IM, Poli V. Mutational switch of an IL-6 response to an interferon-gamma-like response. Proc. Natl. Acad. Sci. U.S.A. 2002;99:8043–8047. doi: 10.1073/pnas.122236099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackiewicz A, Wiznerowicz M, Roeb E, Karczewska A, Nowak J, Heinrich PC, Rose-John S. Soluble interleukin 6 receptor is biologically active in vivo. Cytokine. 1995;7:142–149. doi: 10.1006/cyto.1995.1019. [DOI] [PubMed] [Google Scholar]
- 19.Fiorillo MT, Toniatti C, Van Snick J, Ciliberto G. Expression of the murine interleukin 6 receptor in hepatoma cells: the intracytoplasmic domain is not required for interleukin 6 signal transduction. Eur. J. Immunol. 1992;22:799–804. doi: 10.1002/eji.1830220325. [DOI] [PubMed] [Google Scholar]
- 20.Rizzuto G, Cappelletti M, Mennuni C, Wiznerowicz M, DeMartis A, Maione D, Ciliberto G, La Monica N, Fattori E. Gene electrotransfer results in a high-level transduction of rat skeletal muscle and corrects anemia of renal failure. Hum. Gene Ther. 2000;11:1891–1900. doi: 10.1089/10430340050129503. [DOI] [PubMed] [Google Scholar]
- 21.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 22.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 23.Vallania F, Schiavone D, Dewilde S, Pupo E, Garbay S, Calogero R, Pontoglio M, Provero P, Poli V. Genome-wide discovery of functional transcription factor binding sites by comparative genomics: The case of Stat3. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5117–5122. doi: 10.1073/pnas.0900473106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pellegrino M, Provero P, Silengo L, Di Cunto F. CLOE: identification of putative functional relationships among genes by comparison of expression profiles between two species. BMC Bioinformatics. 2004;5:179. doi: 10.1186/1471-2105-5-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sadowski HB, Shuai K, Darnell JE, Jr, Gilman MZ. A common nuclear signal transduction pathway activated by growth factor and cytokine receptors. Science. 1993;261:1739–1744. doi: 10.1126/science.8397445. [DOI] [PubMed] [Google Scholar]
- 26.Auernhammer CJ, Bousquet C, Melmed S. Autoregulation of pituitary corticotroph SOCS-3 expression: characterization of the murine SOCS-3 promoter. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6964–6969. doi: 10.1073/pnas.96.12.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr. Biol. 2003;13:680–685. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 28.Moon RT, Bowerman B, Boutros M, Perrimon N. The promise and perils of Wnt signaling through β-catenin. Science. 2002;296:1644–1646. doi: 10.1126/science.1071549. [DOI] [PubMed] [Google Scholar]
- 29.Widelitz R. Wnt signaling through canonical and non-canonical pathways: recent progress. Growth Factors. 2005;23:111–116. doi: 10.1080/08977190500125746. [DOI] [PubMed] [Google Scholar]
- 30.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia R, Bowman TL, Niu G, Yu H, Minton S, Muro-Cacho CA, Cox CE, Falcone R, Fairclough R, Parsons S, et al. Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene. 2001;20:2499–2513. doi: 10.1038/sj.onc.1204349. [DOI] [PubMed] [Google Scholar]
- 32.Mora LB, Buettner R, Seigne J, Diaz J, Ahmad N, Garcia R, Bowman T, Falcone R, Fairclough R, Cantor A, et al. Constitutive activation of Stat3 in human prostate tumors and cell lines: direct inhibition of Stat3 signaling induces apoptosis of prostate cancer cells. Cancer Res. 2002;62:6659–6666. [PubMed] [Google Scholar]
- 33.Siddiquee K, Zhang S, Guida WC, Blaskovich MA, Greedy B, Lawrence HR, Yip ML, Jove R, McLaughlin MM, Lawrence NJ, et al. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7391–7396. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen G, Shukeir N, Potti A, Sircar K, Aprikian A, Goltzman D, Rabbani SA. Up-regulation of Wnt-1 and β-catenin production in patients with advanced metastatic prostate carcinoma: potential pathogenetic and prognostic implications. Cancer. 2004;101:1345–1356. doi: 10.1002/cncr.20518. [DOI] [PubMed] [Google Scholar]
- 35.Aspenstrom P, Ruusala A, Pacholsky D. Taking Rho GTPases to the next level: the cellular functions of atypical Rho GTPases. Exp. Cell Res. 2007;313:3673–3679. doi: 10.1016/j.yexcr.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 36.Wennerberg K, Der CJ. Rho-family GTPases: it's not only Rac and Rho (and I like it). J. Cell Sci. 2004;117:1301–1312. doi: 10.1242/jcs.01118. [DOI] [PubMed] [Google Scholar]
- 37.Brazier H, Stephens S, Ory S, Fort P, Morrison N, Blangy A. Expression profile of RhoGTPases and RhoGEFs during RANKL-stimulated osteoclastogenesis: identification of essential genes in osteoclasts. J. Bone Miner. Res. 2006;21:1387–1398. doi: 10.1359/jbmr.060613. [DOI] [PubMed] [Google Scholar]
- 38.Snyder M, Huang XY, Zhang JJ. Identification of novel direct Stat3 target genes for control of growth and differentiation. J. Biol. Chem. 2008;283:3791–3798. doi: 10.1074/jbc.M706976200. [DOI] [PubMed] [Google Scholar]
- 39.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell. Dev. Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 40.Silver DL, Naora H, Liu J, Cheng W, Montell DJ. Activated signal transducer and activator of transcription (STAT) 3: localization in focal adhesions and function in ovarian cancer cell motility. Cancer Res. 2004;64:3550–3558. doi: 10.1158/0008-5472.CAN-03-3959. [DOI] [PubMed] [Google Scholar]
- 41.Ng DC, Lin BH, Lim CP, Huang G, Zhang T, Poli V, Cao X. Stat3 regulates microtubules by antagonizing the depolymerization activity of stathmin. J. Cell. Biol. 2006;172:245–257. doi: 10.1083/jcb.200503021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hao J, Li TG, Qi X, Zhao DF, Zhao GQ. WNT/β-catenin pathway up-regulates Stat3 and converges on LIF to prevent differentiation of mouse embryonic stem cells. Dev. Biol. 2006;290:81–91. doi: 10.1016/j.ydbio.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 43.Yan S, Zhou C, Zhang W, Zhang G, Zhao X, Yang S, Wang Y, Lu N, Zhu H, Xu N. β-Catenin/TCF pathway upregulates STAT3 expression in human esophageal squamous cell carcinoma. Cancer Lett. 2008;271:85–97. doi: 10.1016/j.canlet.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 44.Kawada M, Seno H, Uenoyama Y, Sawabu T, Kanda N, Fukui H, Shimahara Y, Chiba T. Signal transducers and activators of transcription 3 activation is involved in nuclear accumulation of β-catenin in colorectal cancer. Cancer Res. 2006;66:2913–2917. doi: 10.1158/0008-5472.CAN-05-3460. [DOI] [PubMed] [Google Scholar]
- 45.Yamashina K, Yamamoto H, Chayama K, Nakajima K, Kikuchi A. Suppression of STAT3 activity by Duplin, which is a negative regulator of the Wnt signal. J. Biochem. 2006;139:305–314. doi: 10.1093/jb/mvj033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY ONLINE DATA
The RhoU/Wrch1 Rho GTPase gene is a common transcriptional target of both the gp130/STAT3 and Wnt-1 pathways
Davide SCHIAVONE*, Sarah DEWILDE*1, Francesco VALLANIA*2, James TURKSON†, Ferdinando DI CUNTO* and Valeria POLI*3
*Molecular Biotechnology Center and Department of Genetics, Biology and Biochemistry, University of Turin, Via Nizza 52, 10126 Turin, Italy, and
†BioMolecular Science Center and Department of Molecular Biology and Microbiology, University of Central Florida, Orlando, FL 32826, U.S.A.
Figure S1 RhoU induction by OSM and Wnt-1 in HeLa cells
(a) RhoU mRNA was measured by qRT-PCR in HeLa cells treated with OSM for different lengths of time. (b and c) Transient transfection assays. The RhoU −770 and Socs3 (b)or TOPFlash/FOPFlash promoter constructs (c) were co-transfected with−a pSEAP vector in HeLa cells. At 24 h later, cells were either stimulated with OSM (b) or co-cultured with wild-type (NT) or Wnt-1-expressing (Wnt1) HEK-293 cells (c) for 24 h. Results shown are means ± S.E.M. of normalized luciferase activity from at least two independent experiments performed in duplicate.
Received 13 January 2009/1 April 2009; accepted 28 April 2009 Published as BJ Immediate Publication 28 April 2009, doi:10.1042/BJ20090061
1 Present address: BioIndustry Park del Canavese, 10010 Colleretto Giacosa, Italy.
2 Present address: Center for Genome Sciences, Department of Genetics, Washington University in St Louis School of Medicine, 4444 Forest Parkway, St Louis, MO 63108, U.S.A.
3 To whom correspondence should be addressed (valeria.poli@unito.it).
© The Authors Journal compilation © 2009 Biochemical Society