Abstract
Successful cancer therapy requires the elimination or incapacitation of all tumor cells capable of regenerating a tumor. Therapeutic advances therefore necessitate the characterization of the cells that are able to propagate a tumor in vivo. We show an important link between tumor genotype and isolation of tumor-propagating cells (TPCs). Three mouse models of the most common form of human lung cancer each had TPCs with a unique cell surface phenotype. The cell surface marker Sca1 did not enrich for TPCs in tumors initiated with oncogenic Kras, and only Sca1-negative cells propagated EGFR mutant tumors. In contrast, Sca1-positive cells were enriched for tumor-propagating activity in Kras tumors with p53 deficiency. Primary tumors that differ in genotype at just one locus can therefore have tumor-propagating cell populations with distinct markers. Our studies show that the genotype of tumor samples must be considered in studies to identify, characterize, and target tumor-propagating cells.
Summary
Depending on the tumor initiating genetic event in three tractable mouse models of lung adenocarcinoma, the surface maker Sca1 had variable success in identifying cells with tumor-propagating activity. These findings uncover the impact that tumor genotype can have on the phenotype of tumor-propagating cells, which may have important therapeutic significance.
Highlights
-identification of the first lung tumor-propagating cell population
-lung cancers of different genotype have tumor-propagating cells with distinct markers
-tumor samples should be separated by genotype to study tumor-propagating cells
Introduction
Identification of functionally important tumor cell populations with the ability to propagate the tumor phenotype in vivo (tumor-propagating cells, or TPCs) promises to elucidate mechanisms of tumorigenesis and potential new therapeutic targets. Molecularly-defined tumor cell populations with such capacity (often referred to as cancer stem cells or tumor-initiating cells) have been identified in a variety of malignancies, yet in some cancers, nearly every cell has tumor-propagating ability (reviewed in Shackleton et al., 2009; Visvader and Lindeman, 2008). While tumor genotype is known to influence response to targeted therapies, the identity of tumor-propagating cells in cancers with different genotypes has not been compared.
We used genetically defined murine models to identify lung tumor-propagating cells. In two murine models of the most common form of lung cancer, adenocarcinoma, tumors are initiated using conditional activation of oncogenic K-rasG12D from the Lox-Stop-Lox-KrasG12D allele alone (hereafter, Kras) (Jackson et al., 2001) or with conditional p53 deficiency in p53flox/flox mice (hereafter, Kras;p53-flox) (Jackson et al., 2005). Both models yield non-small-cell lung cancers, specifically adenocarcinomas, and recapitulate key aspects of human lung cancers (Jackson et al., 2005; Jackson et al., 2001). Mice bearing a mutant human epidermal growth factor receptor transgene, EGFRT790M-L858R (hereafter, EGFR), develop adenocarcinomas representative of lung cancer found in nonsmokers (Li et al., 2007). Importantly, EGFR mutant tumors are histologically similar to the tumors in the Kras and Kras;p53-flox mice.
We hypothesized that isolating cells positive for the cell surface marker of stem/progenitor cells in the corresponding normal tissue would prove useful in identification of lung TPCs. While CD133-positive human lung cancer cells have propagating activity in sphere cultures and subcutaneous injections (Chen et al., 2008; Eramo et al., 2008; Jiang et al., 2009), lung TPCs with demonstrable self-renewal and differentiation in the lung microenvironment have not been reported. We and others have used Sca1 (Ly6a) cell surface expression to identify stem cells in the normal murine lung (Kim et al., 2005; McQualter et al., 2009; McQualter et al., 2010; Teisanu et al., 2009), making it an interesting marker to test in murine lung cancer.
Orthotopic transplantations of lung adenocarcinoma cells yielded secondary tumors that recapitulated the features of the primary lung tumors. Sca1-positive cells from Kras;p53-flox tumors were highly enriched for tumor-propagating activity. Sca1 status did not enrich for TPCs in Kras tumors and only Sca1-negative cells propagated EGFR tumors. Thus, primary tumor genotype is an important determinant of the cell surface phenotype of lung tumor-propagating cell populations.
Results
Orthotopic lung tumor transplantation assay
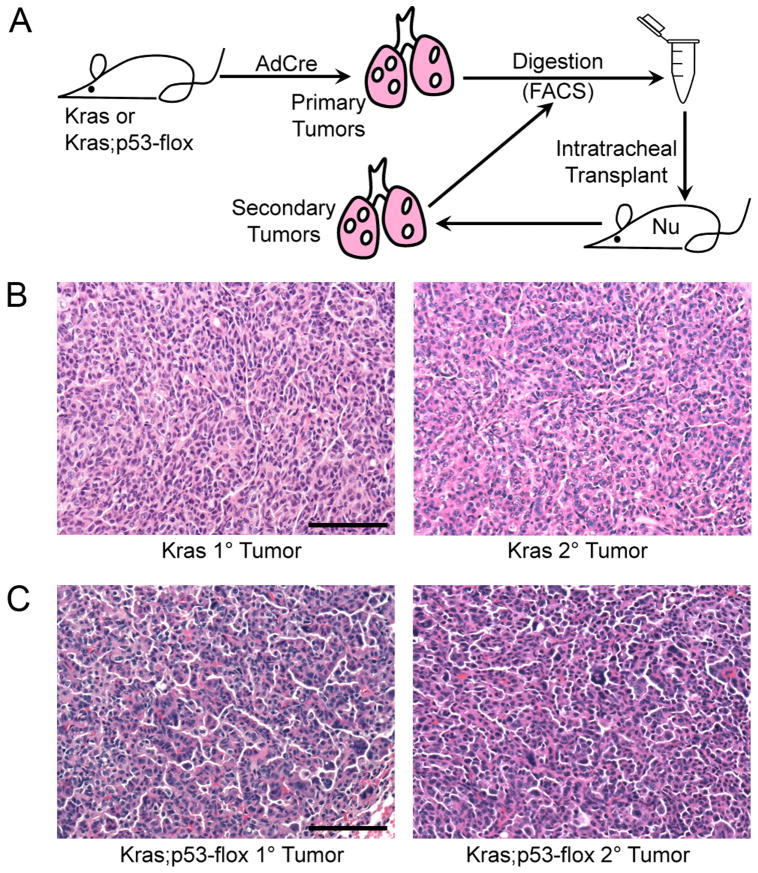
We first established an orthotopic transplantation assay for propagating lung tumor cells in vivo. Primary adenocarcinomas were dissected from the lungs of Kras mice, dissociated into single cell suspensions and transplanted intratracheally (IT) into recipient mice (Figure 1A). Secondary tumors formed in recipient mice four to eight months (average 5.5 months) after IT transplant of 103 to 106 cells. The secondary adenocarcinomas recapitulated the histopathology of the primary tumors (Figure 1B), validating this assay for lung cancer cell function. Secondary Kras;p53-flox tumors also recapitulated the adenocarcinoma histopathology of the corresponding primary tumors (Figure 1C). Limiting dilution analysis performed by injecting various numbers of tumor cells indicated that the frequency of TPCs was not significantly different between the two models (1 out of 10,000 Kras;p53-flox tumor cells gave rise to a tumor versus 1 out of 12,000 Kras tumor cells, p = 0.78, data not shown). Thus, the ability to isolate and functionally test TPCs in these two models was similar using our transplantation assay.
Figure 1.
Orthotopic transplantation of Kras and Kras;p53-flox tumor cells recapitulates the primary tumor phenotype.
(A) Cartoon of the transplantation scheme used to assay lung tumor-propagating cells (TPCs) through serial transplantation. (B) H&E staining of primary (left) and secondary (right) Kras lung adenocarcinomas showing similar pathological grade, nuclear features, and general tumor architecture. (C) H&E staining of primary (left) and secondary (right) Kras;p53-flox lung tumors showing similar histopathological characteristics of advanced adenocarcinoma, including pleomorphic nuclei and rare giant cells. All images, 200× magnification. Scale bar = 100μM.
Prospective isolation of tumor-propagating cells
We next sought to determine if lung tumor-propagating cells could be identified prospectively from Kras;p53-flox tumors using expression of Sca1, a marker of normal lung stem/progenitor cells. The fraction of CD45-negative CD31-negative Sca1-positive cells (hereafter, Sca1+ cells) within Kras;p53-flox tumors ranged from 0.56% to 1.74% (average 1.11%) of tumor cells (Figure 2A and data not shown). The majority of the remaining non-hematopoietic and non-endothelial cells consisted of Sca1-negative, highly auto-fluorescent cells (hereafter, Sca1- cells).
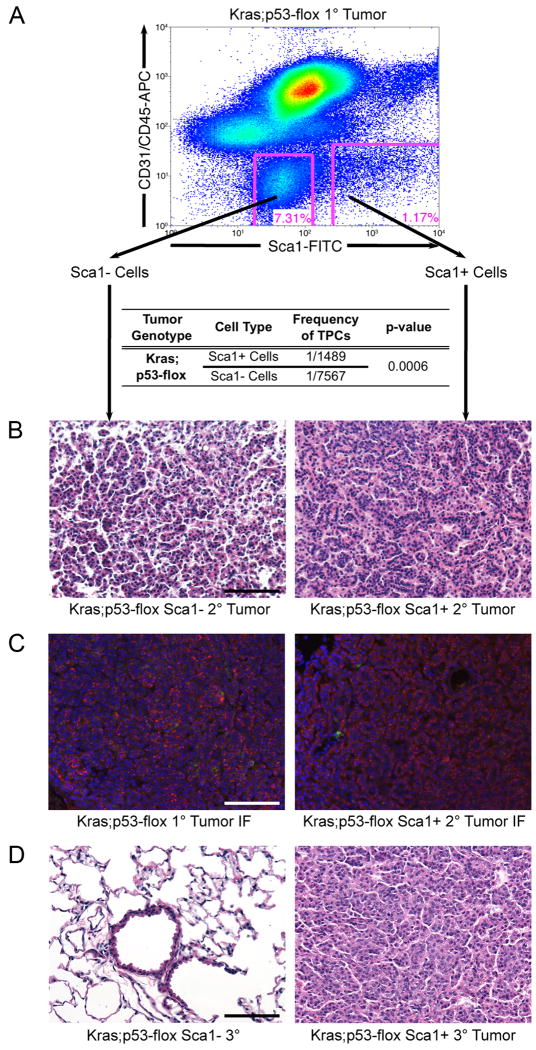
Figure 2.
Sca1 + cells from Kras;p53-flox tumors are lung tumor-propagating cells.
(A) Representative FACS analysis of Kras;p53-flox tumor cells used for transplantation (top). Limiting dilution transplantation of the sorted cells indicated that the Sca1 + population was significantly enriched for TPCs (table). (B) Sca1- cell transplants yielded smaller, more diffuse lesions (left), whereas Sca1 + cell transplants yielded secondary tumors that recapitulated the histopathology of primary Kras;p53-flox tumors (right, compare to Figure 1C). (C) Immunofluorescence (IF) staining with antisera raised against SP-C (red), CCSP (green), and counterstain DAPI (blue) showed that primary Kras;p53-flox lung adenocarcinomas (left) are mainly composed of SP-C+ cells, a pattern recapitulated in secondary tumors from Sca1 + cell transplants (right). (D) Serial transplantation of secondary tumor cell populations revealed a lack of tumor formation from Sca1 - cells (left), in contrast to tertiary tumor development from Sca1 + cells (right). All images, 200× magnification. Scale bar = 100μM. See also Figure S1 and Table S1 for additional data.
To compare the tumor-propagating capacity of each population, we performed limiting dilution transplantations with Kras;p53-flox Sca1+ or Sca1- cells (Figure 2A, Tables 1, S1). We observed a highly significant 5-fold enrichment for tumor-propagating potential in the Sca1+ population compared to the Sca1- population (p = 0.0006); 1 in 1,489 Sca1+ cells had tumor-propagating activity, compared to 1 in 7,567 Sca1- cells. Secondary tumors from Sca1+ cells recapitulated the primary lesions (compare Figures 2B, 1C), whereas the few secondary tumors that formed from Sca1- cell transplants exhibited a smaller, more diffuse phenotype (Figures 2B, S1A, p=0.046). Most cells in murine lung adenocarcinomas express the alveolar type II (AT2) cell differentiation marker surfactant protein C (SP-C) and do not stain for the bronchiolar Clara cell differentiation marker Clara cell secretory protein (CCSP) with the exception of rare SP-C+ CCSP+ cells (Jackson et al., 2001; Kim et al., 2005). Secondary tumors arising from Sca1+ cells shared the same staining pattern for these lung cell markers (Figure 2C), indicating a similar differentiation pattern of the primary tumors and the Sca1+ cell derived secondary tumors. Further demonstrating the differentiation capacity of Sca1+ tumor cells, the overall abundance of Sca1+ and Sca1- cells was re-established in the secondary tumors derived from injection of Sca1+ cells (Figure S1B). In contrast, only Sca1- cells were present in a secondary tumor derived from Sca1- cells (Figure S1B). Finally, expression analysis for several other markers suggested that the primary Sca1-cells were more differentiated than Sca1+ cells (Figure S1C).
Table 1. Transplantation results of Tumor Cell Populations.
| Number of cells transplanted | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| <100 | 100-1K | 1K-5K | 5K-10K | 10K-100K | >100K | ||||
| Tumor Genotype | Cell Type | Number of mice with tumors / Number transplanted (%) |
Frequency of TPCs* | p-value | |||||
| Kras; p53-flox | Sca1+Cells | 3/5 (60%) |
2/3 (67%) |
10/14 (71%) |
2/2 (100%) |
ND | ND | 1/1489 | 0.0006† |
| Sca1-Cells | ND | 2/9 (22%) |
6/17 (35%) |
2/4 (50%) |
ND | ND | 1/7567 | ||
| Kras | Sca1+Cells | ND | 1/4 (25%) |
3/10 (30%) |
5/11 (45%) |
5/8 (63%) |
ND | 1/13990 | 0.7311‡ |
| Sca1-Cells | ND | 1/3 (33%) |
4/8 (50%) |
14/18 (78%) |
4/6 (67%) |
7/8 (88%) |
1/16037 | ||
Varied numbers of tumor cells as indicated were transplanted in sterile PBS into Foxn1nu (nude) recipients.
The number of recipient mice with at least one secondary tumor was determined by histological analysis.
Results shown are the combination of 14 and 9 different transplantation experiments with Kras and Kras;p53-flox tumor cells, respectively.
See also Tables S1 and S2 for more details.
TPCs: Tumor-propagating cells. The frequency of cells with tumor-propagating capacity in the population was determined by limiting dilution analysis with L-calc software using the transplanted cell numbers shown in Tables S1 and S2.
Kras;p53-flox Sca1 + cells vs. Kras;p53-flox Sca1- cells.
Kras Sca1 + cells vs. Kras Sca1- cells.
ND: not determined.
We next used serial transplantation to determine if the Sca1+ Kras;p53-flox tumor cells had the stem cell property of self-renewal. Secondary tumor cells derived from Sca1+ Kras;p53-flox tumor cell transplants gave rise to tertiary tumors that were similar to the corresponding primary and secondary tumors (Figure 2D), and the Sca1+ fraction of secondary tumors more frequently yielded tumors after transplantation than the Sca1-fraction (Figure S1D). Importantly, secondary tumor cells derived from Sca1- Kras;p53-flox tumor cell transplants were unable to yield tertiary tumors (Figures 2D, S1D). Therefore, the Sca1+ cells from Kras;p53-flox tumors were capable of differentiation and self-renewal in serial transplantations, whereas Sca1- cells appeared more differentiated and did not self-renew.
We confirmed that our ability to detect an enrichment of TPCs in the Sca1+ population from Kras;p53-flox tumors did not result from a lack of tumor cells in the Sca1- fraction using several different assays. First, we performed one-lox recombination specific PCR, in which a PCR product is obtained only from the recombined allele after effective Cre activity, to assess the status of the floxed Kras and p53 loci in the sorted cell populations and confirm the presence of tumor cells. While we did observe tumor-to-tumor variability, the amount of recombination of the Kras and p53 alleles was similar in the Sca1+ and Sca1- cells within each tumor sample (Figure S1E). Second, we compared the transplantation efficiency of the Kras;p53-flox tumor cell populations using a GFP allele to mark and identify transplanted tumor cells. The percentage of GFP+ cells detected after transplantation of Sca1+ cells was not significantly different than after transplantation of Sca1- cells (Figure S1F, 0.84% and 0.62%, respectively, p=0.501). Notably, these transplantation efficiencies were much greater than that of normal lung cells (0.015% - 0.033%, D. Raiser, C. Kim, unpublished), suggesting that both populations were capable of initial expansion in vivo as expected for tumor cells. Finally, the cell cycle status of Sca1+ cells did not significantly differ from that of Sca1-cells (Figure S1G).
Comparison of lung tumor-propagating cell phenotype between mouse models
Having identified Sca1+ cells as the TPCs in the Kras;p53-flox model, we next wished to determine if the same marker could identify TPCs in Kras lung tumors. The frequency of Sca1+ cells in Kras tumors was similar to that in Kras;p53-flox tumors (range, 0.4% to 2.9%, average 0.95%, p = 0.69) (Figures 3A, 2A, data not shown). Sca1+ cells from Kras and Kras;p53-flox tumors expressed CCSP and SP-C, whereas Sca1- cells from both tumor genotypes expressed very high levels of SP-C and virtually no CCSP (Figure S2A), confirming that Sca1 staining was valid for sorting similar cells from Kras and Kras;p53-flox tumors.
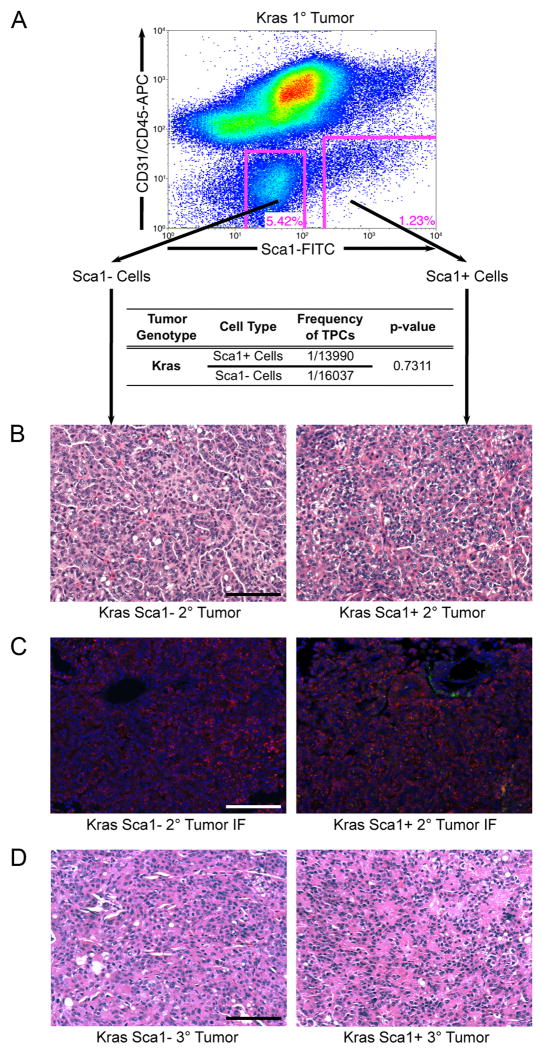
Figure 3.
The Sca1 + and Sca1- populations from Kras lung tumors are equally capable of propagating tumors.
(A) Representative FACS analysis of Kras tumors showing a pattern of Sca1 staining similar to Kras;p53-flox tumors (top, compare to Figure 2A). Limiting dilution transplantation indicated that Sca1+ and Sca1- Kras tumor cells are identical in tumor-propagating potential (table). (B) Secondary tumors from Sca1- (left) and Sca1+ (right) Kras tumor cell transplants displayed similar pathological features. (C) IF analysis of secondary tumors (as in Figure 2) revealed similar SP-C and CCSP marker status in secondary tumors from Sca1- cells (left) and Sca1+ cells (right). (D) Both Sca1- Kras tumor cells (left) and Sca1+ tumor cells (right) were capable of serial transplantation to form tertiary tumors. All images, 200× magnification. Scale bar = 100μM. See also Figure S2 and Table S2 for additional data.
Surprisingly and in contrast to the Kras;p53-flox tumors, the Sca1+ and Sca1- cells from Kras tumors were equally capable of forming secondary tumors (Figure 3A, Tables 1, S2). The TPC frequencies were 1 out of 13,990 Sca1+ cells and 1 out of 16,097 Sca1-cells (p= 0.7311). The secondary tumors derived from Sca1+ cells and Sca1- cells were similar to the primary tumors (compare Figures 3B, 1B) and were indistinguishable by histology, differentiation status (determined by FACS analysis for Sca1 and the pattern of staining for CCSP and SP-C), size, and proliferative index (Figures 3C, S2B, data not shown). Finally, Sca1+ and Sca1- cells isolated from Kras secondary tumors were capable of yielding tertiary and quaternary tumors in serial transplant assays (Figures 3D, S2C, data not shown), suggesting that both populations were capable of self-renewal. In summary, Sca1 provided no enrichment for tumor-propagating capacity in Kras lung tumors, whereas Sca1+ cells were a uniquely self-renewing tumor-propagating cell population in Kras mutant tumor cells lacking p53.
Importantly, we confirmed the difference in tumor cell genotype in fractionated cells from Kras and Kras;p53-flox tumors. First, recombination analysis confirmed that Kras tumors retained p53 (Figure S2D). Second, we verified that p53 expression was retained in Sca1+ and Sca1- Kras tumor cells and was not detectable in most of the fractionated Kras;p53-flox tumor cells (Figure S2E).
Our results with Kras and Kras;p53-flox tumors suggested that lung cancers of different genotypes contain TPCs with distinct phenotypes, so we next tested this hypothesis in a third model of lung adenocarcinoma: EGFR mutant tumors. EGFR tumors had a Sca1, CD31 and CD45 staining pattern nearly identical to the Kras and Kras;p53-flox tumors; approximately 1% of the cells were Sca1+ (Figure S2F). Strikingly, only the Sca1- cells were capable of propagating EGFR tumors (Figure S2G) when either 5,000 cells (0/2 for Sca1+ and 3/3 for Sca1-) or 25,000 cells (0/2 for Sca1+ and 4/4 for Sca1-) were transplanted. Importantly, both populations expressed the EGFR transgene at equal levels (Figure S2H). Thus, EGFR tumors contained TPCs distinct from those in Kras-driven lung tumors.
Discussion
Our work indicates that lung adenocarcinomas of differing genotypes have tumor-propagating cells with distinct markers. In Kras;p53-flox tumors, positive selection for the surface marker Sca1 enriches for cancer cells capable of self-renewal and differentiation. In contrast, Sca1 does not select for TPCs in Kras or EGFR mutant lung tumors. The study of functionally important cells in lung cancers therefore requires a different strategy for isolation of tumor-propagating cells from each model. On a broader scale, the genotype of tumor samples must be taken into account in studies to identify, characterize and target tumor-propagating cells in lung cancers and other malignancies.
It is clear that histologically distinct tumors contain tumor-propagating populations with different markers. For example, cells with a granulocyte-macrophage progenitor phenotype are the candidate tumor-propagating cells in chronic myelogenous leukemia (Jamieson et al., 2004), whereas in acute myelogenous leukemia, the tumor-propagating cells more closely resemble hematopoietic stem cells or multipotent progenitors (Bonnet and Dick, 1997). Murine mammary tumorigenesis driven by Wnt leads to the development of heterogeneous adenocarcinomas with various features of differentiation, while the Her2/neu oncogene promotes the development of more homogenous luminal epithelial tumors in mice, and these two distinct subtypes of breast cancer appear to have different tumor-propagating cell populations (Cho et al., 2008; Liu et al., 2007; Vaillant et al., 2008). Finally, human and murine brain cancers also appear to have distinct TPC markers (Read et al., 2009; Ward et al., 2009). Importantly, prior to the present study no direct comparisons had been made between tumor-propagating cell populations within tumors of the same tissue origin and same histopathological subtype with human or murine samples.
Our results indicate that the same marker or set of markers may not identify tumor-propagating cells in all patient samples of a specific tumor type. CD44-positive, CD24-negative/low tumor cells propagated tumors from invasive lobular carcinomas and adenocarcinomas from breast cancer patients (Al-Hajj et al., 2003). In another study, however, a marker signature including positive CD24 status identified breast cancer-propagating cells in patient samples (Pece et al., 2010). The utility of seemingly different marker sets in these studies may be due to differences in the genotype of breast tumor specimens as we observed for lung tumors. It has been possible to identify tumor-propagating cells with the same marker across multiple patient samples without regard to tumor genotype in some tissues. For example, CD133+ cells were the tumor-propagating population in medulloblastomas and glioblastomas (Singh et al., 2004). Therefore, it is possible that our results indicate a role for genotype in TPC phenotype that is specific to lung cancer or that our results are attributable to important distinctions between genetically defined mouse models and tumors found in patients. On the other hand, it is clear that currently known TPC populations are not homogenous. We reason that more precise purification of TPCs will need to take into account the genotype of tumor samples. Separating specimens according to genotype may reveal important patterns in the tumor-propagating activity of isolated cell populations that cannot be observed by combining results from all tumors. A retrospective analysis of the predominant genetic changes found in the patient samples from which TPCs have been identified may stratify the utility of marker combinations and be a first step to addressing this possibility.
It will be particularly important to consider genotype in the definition of tumor-propagating cells in human lung cancer, given the diversity of genetic mutations and tumor subtypes found in patient lung cancers. Our study has focused on three murine models of adenocarcinoma, representing the three most commonly mutated genes in human lung cancer: amongst smokers, TP53 and KRAS mutations are the most common (71% and 21% of patients, respectively), whereas EGFR mutations are more commonly found in never-smokers (58% of patients) (reviewed in Sun et al., 2007). At least twenty-six different genes are mutated at high frequency in patient lung adenocarcinomas, and additional mutations likely exist (Ding et al., 2008). These separate genotypes of adenocarcinoma are usually indistinguishable at the histological level and until recently were largely treated with the same chemotherapeutics, albeit unsuccessfully. Identification of tumor-propagating cell populations to target in these patient subsets, as in the mouse models shown here, may offer new therapeutic approaches. While each human lung tumor sample is expected to be genetically distinct, there are predominant mutations in human lung adenocarcinomas that are known to predict response to targeted therapies and which may be the most informative for studies of TPCs. We suggest that those seeking to identify human lung TPCs genotype their tumor samples for these loci (KRAS, BRAF, EGFR, HER2, ALK fusion, and PIK3CA) (Herbst et al., 2008) and determine if their tumor propagation results correlate with mutation status. Finally, several other subtypes of lung cancer, such as squamous cell carcinoma, small cell lung cancer and large cell carcinoma, have known, common genetic mutations distinct from those found in adenocarcinomas and these subtypes remain to be examined for tumor-propagating cells in murine or human samples.
Our findings have important implications for the development of novel directed therapeutics. Successful cancer therapy necessitates the eradication or permanent incapacitation of all tumor cells with the ability to generate a tumor. It is well established that primary tumor genotype can influence therapeutic response in lung cancer. Patients with lung adenocarcinomas bearing EGFR mutations exhibit a beneficial response to treatment with EGFR inhibitors but unfortunately develop drug resistance, whereas few patients lacking EGFR mutations have response to such treatment (reviewed in Sun et al., 2007). Furthermore, patients with KRAS-mutant lung cancers are particularly resistant to virtually all therapeutic options (Riely et al., 2009). Our data now extend the idea that lung tumor genotype influences therapeutic response. Knowledge of each patient's tumor genotype and TPC phenotype may be needed in combination to optimize therapeutic selection for any tumor type. Future therapeutics designed to target tumor-propagating cells found in some patients' tumors might not be equally effective against other patients' tumors if their genotype is distinct.
Our data establish concepts that will be invaluable for further work to define the mechanisms that regulate tumor propagation in lung cancer and diverse malignancies. This is the first report of lung cancer cells with self-renewal and differentiation capacity in an orthotopic transplantation assay. It will be important to test additional markers for TPCs in the different lung tumor genotypes (especially including markers that can be used in human lung cancer samples), to identify markers for further enrichment of Kras;p53-flox lung TPCs, to compare TPC function in lung tumors over time with tumor progression, and to determine if and how discrete lung cancer cell populations differentially respond to therapeutics. It will also be interesting to determine the degree to which the cell of origin of genetically distinct lung tumors overlap and how these cells relate in phenotype to TPCs. Further understanding of the influence of genotype on tumor propagation could lend insight toward developing better therapeutic approaches for cancer patients. Whether the tumor-propagating cells within a given tumor type are a rare subpopulation with unique molecular features, stochastic in nature yet rare tumor cells, or virtually all cells within a tumor, it will be crucial to further understand the genetic and epigenetic contributions that drive these important tumor cells.
Experimental Procedures
Mice and Tissues
Lox-Stop-Lox-KrasG12D mice and Lox-Stop-Lox-KrasG12D;p53fl/fl mice (Jackson et al., 2005; Jackson et al., 2001) were maintained in viral-free conditions on 129 SvJae background. 6- to 12-week-old mice were used for intranasal adenoviral-Cre infections (Jackson et al., 2001). 8- to 16-week-old Foxn1nu (nude) mice (Harlan, Jax, or Taconic) were used as recipients. All mouse experiments were approved by the CHB Animal Care and Use Committee, accredited by AAALAC, and were performed in accordance with relevant institutional and national guidelines and regulations. Tissue preparation was as described (Jackson et al., 2001). Five H&E sections spaced 50 μm apart were analyzed for tumor formation by at least two investigators including a pathologist with expertise in murine lung cancer. Immunostaining of tissues was as described (Kim et al., 2005) using antisera raised against mouse CC10 (Santa Cruz, sc-9773) and proSP-C (Chemicon, AB3786) with donkey Alexa Fluor secondary antibodies (Molecular Probes). Triple-color microscopy and imaging was performed with a Nikon 90i and a Roper Scientific CCD camera and NIS-Elements software and processed with NIS-Elements and Adobe Photoshop.
FACS Analysis of tumor cells
Mice were euthanized with avertin overdose and lungs were dissected and examined grossly for tumor formation. Tumors were dissected from the lungs of Kras mice 6 to 10 months (average 7.5 months) or Kras;p53-flox mice 4 to 7 months (average 5.5 months) after adenoviral-Cre infection with as little contribution from surrounding normal tissue as possible. Tumor tissue was prepared as described for normal lung (Kim et al., 2005) using Sca1-FITC, CD45.2-Biotin, Pecam-Biotin, and Streptavidin-APC (Pharmingen) with 7AAD (Molecular Probes) staining to eliminate dead cells. Cell sorting was performed with a Cytomation MoFlo or a BD FACS Aria, and data were analyzed with FloJo software (Tree Star, Inc.).
Tumor Transplants
Intratracheal transplants were as described (McLemore et al., 1987) with modifications. Mice were anesthetized with avertin and supported on an intubation stand. A halogen light was focused on the sternum, the trachea was visualized by oral transillumination, and a 22G, 1-inch, IV catheter (Fisher) was inserted into the trachea. A 40-50 μL cell suspension in sterile PBS was pipetted into the catheter and the mouse inhaled the cell suspension. Mice were monitored for signs of lung tumor onset and euthanized for gross and histological analysis upon signs of distress or at 6 months to 1 year post transplant.
Statistical Analysis
TPC frequency was calculated using limiting dilution analysis with L-Calc Software (Stemcell Technologies, Inc.). Fold enrichment for tumor-propagating activity was determined by comparing the TPC frequency in populations of interest, L-Calc provided p-values. Except where indicated, Student's t-test was used to compare measurements between 2 conditions. GraphPad Prism (GraphPad Software, Inc.) and Mstat (McArdle Laboratory for Cancer Research, University of Wisconsin) were used for graphing and statistical analyses. p-values less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank S. Lawrence, K. Mercer, E. Jackson, A. Dooley, S. Perera, the DFCI, CHB HemOnc, and MIT FACS facilities, and members of the Kim Lab for technical assistance and discussions, E. Passegue for cell cycle protocol, R. Bronson for histopathology, T. Jacks for mouse strains and helpful discussions, G. Evan, S. Ryeom, L. Zon, C. Cepko, S. Elledge, S. Armstrong, H. Hock and K. Cichowski for critical reading. This work was supported by a DoD, Air Force Office of Scientific Research, National Defense Science and Engineering Graduate Fellowship, 32 CFR 168a (to S.J.C.), PF-09-121-01-DDC from the American Cancer Society (to K.W.S.), Dana Farber Harvard Cancer Center Lung Cancer SPORE grant P50 CA090578, R01 AG2400401, R01 CA122794, R01 CA140594 (to K-K.W.), the V Foundation, American Cancer Society Research Scholar Grant #RSG-08-082-01-MGO, the Harvard Stem Cell Institute, NIH/NCI 2P50CA090578 and The Lung Cancer Alliance (to C.F.K.). We apologize for not citing primary references due to space constraints.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nature medicine. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Chen YC, Hsu HS, Chen YW, Tsai TH, How CK, Wang CY, Hung SC, Chang YL, Tsai ML, Lee YY, et al. Oct-4 expression maintained cancer stem-like properties in lung cancer-derived CD133-positive cells. PLoS ONE. 2008;3:e2637. doi: 10.1371/journal.pone.0002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RW, Wang X, Diehn M, Shedden K, Chen GY, Sherlock G, Gurney A, Lewicki J, Clarke MF. Isolation and molecular characterization of cancer stem cells in MMTV-Wnt-1 murine breast tumors. Stem Cells. 2008;26:364–371. doi: 10.1634/stemcells.2007-0440. [DOI] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, Conticello C, Ruco L, Peschle C, De Maria R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell death and differentiation. 2008;15:504–514. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, Jacks T. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes & development. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, Gotlib J, Li K, Manz MG, Keating A, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- Jiang F, Qiu Q, Khanna A, Todd NW, Deepak J, Xing L, Wang H, Liu Z, Su Y, Stass SA, et al. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res. 2009;7:330–338. doi: 10.1158/1541-7786.MCR-08-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT, Jacks T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Li D, Shimamura T, Ji H, Chen L, Haringsma HJ, McNamara K, Liang MC, Perera SA, Zaghlul S, Borgman CL, et al. Bronchial and peripheral murine lung carcinomas induced by T790M-L858R mutant EGFR respond to HKI-272 and rapamycin combination therapy. Cancer Cell. 2007;12:81–93. doi: 10.1016/j.ccr.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Liu JC, Deng T, Lehal RS, Kim J, Zacksenhaus E. Identification of tumorsphere- and tumor-initiating cells in HER2/Neu-induced mammary tumors. Cancer Res. 2007;67:8671–8681. doi: 10.1158/0008-5472.CAN-07-1486. [DOI] [PubMed] [Google Scholar]
- McLemore TL, Liu MC, Blacker PC, Gregg M, Alley MC, Abbott BJ, Shoemaker RH, Bohlman ME, Litterst CC, Hubbard WC, et al. Novel intrapulmonary model for orthotopic propagation of human lung cancers in athymic nude mice. Cancer Res. 1987;47:5132–5140. [PubMed] [Google Scholar]
- McQualter JL, Brouard N, Williams B, Baird BN, Sims-Lucas S, Yuen K, Nilsson SK, Simmons PJ, Bertoncello I. Endogenous fibroblastic progenitor cells in the adult mouse lung are highly enriched in the Sca-1 positive cell fraction. Stem Cells. 2009;27:623–633. doi: 10.1634/stemcells.2008-0866. [DOI] [PubMed] [Google Scholar]
- McQualter JL, Yuen K, Williams B, Bertoncello I. Evidence of an epithelial stem/progenitor cell hierarchy in the adult mouse lung. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1414–1419. doi: 10.1073/pnas.0909207107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Read TA, Fogarty MP, Markant SL, McLendon RE, Wei Z, Ellison DW, Febbo PG, Wechsler-Reya RJ. Identification of CD15 as a marker for tumor-propagating cells in a mouse model of medulloblastoma. Cancer Cell. 2009;15:135–147. doi: 10.1016/j.ccr.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riely GJ, Marks J, Pao W. KRAS mutations in non-small cell lung cancer. Proc Am Thorac Soc. 2009;6:201–205. doi: 10.1513/pats.200809-107LC. [DOI] [PubMed] [Google Scholar]
- Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 2009;138:822–829. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers--a different disease. Nat Rev Cancer. 2007;7:778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- Teisanu RM, Lagasse E, Whitesides JF, Stripp BR. Prospective isolation of bronchiolar stem cells based upon immunophenotypic and autofluorescence characteristics. Stem Cells. 2009;27:612–622. doi: 10.1634/stemcells.2008-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant F, Asselin-Labat ML, Shackleton M, Forrest NC, Lindeman GJ, Visvader JE. The mammary progenitor marker CD61/beta3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res. 2008;68:7711–7717. doi: 10.1158/0008-5472.CAN-08-1949. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- Ward RJ, Lee L, Graham K, Satkunendran T, Yoshikawa K, Ling E, Harper L, Austin R, Nieuwenhuis E, Clarke ID, et al. Multipotent CD15+ cancer stem cells in patched-1-deficient mouse medulloblastoma. Cancer Res. 2009;69:4682–4690. doi: 10.1158/0008-5472.CAN-09-0342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.