Abstract
Mutations in SEPN1 result in a spectrum of early onset muscle disorders referred to as SEPN1-related myopathy. The SEPN1 gene encodes selenoprotein N (SelN), which contains the amino acid selenocysteine (Sec). Incorporation of Sec occurs due to redefinition of a UGA codon during translation. Efficient insertion requires a Selenocysteine Insertion Sequence in the 3’ UTR and, for at least a subset of selenoprotein genes, a Selenocysteine Redefinition Element (SRE) located adjacent to the UGA codon. We report the effect of three novel and one previously reported point mutation in the SelN SRE element on Sec insertion efficiency. Notably, the previously reported mutation c.1397G>A (p.R466Q), which weakens the secondary structure of the SRE element, reduces Sec insertion efficiency and SelN RNA levels. Muscle from patients with this mutation have negligible levels of SelN protein. This data highlights the importance of the SRE element during SelN expression and illustrates a novel molecular mechanism by which point mutations may lead to SEPN1-related myopathy.
Keywords: Selenocysteine, SEPN1, myopathy, Selenium, Recoding, selenoprotein N, SelN
Introduction
SEPN1-related myopathy consist of 4 autosomal recessive disorders originally considered to be separate entities: rigid spine muscular dystrophy (RSMD1) (Flanigan, et al., 2000; Moghadaszadeh, et al., 2001), the classical form of multiminicore disease (Ferreiro, et al., 2002), desmin related myopathy with Mallory-body like inclusions (Ferreiro, et al., 2004), and congenital fiber-type disproportion (Clarke, et al., 2006). All are clinically characterized by poor axial muscle strength, scoliosis and neck weakness, and a variable degree of spinal rigidity. Early ventilatory insufficiency can lead to death by respiratory failure.
Selenoprotein N (SelN; MIM# 606210) expression occurs early in embryogenesis and is required for normal muscle development in Zebrafish (Petit, et al., 2003; Thisse, et al., 2003)[Jurynec et al., In Press]. A unique feature common to all selenoproteins is the presence of the amino acid Sec, which has a lower pKa than cysteine, producing a highly reactive group at physiological pH. Recent studies demonstrate that SelN can affect the redox state and is physically associated with the Ryanodine Receptor intracellular calcium release channel (RyR) [Jurynec et al., In Press]. The simplest interpretation is that SelN modifies the regulation of RyR mediated calcium mobilization required for normal muscle development and differentiation.
Sec insertion during decoding of eukaryotic selenoprotein mRNA requires a cis-acting selenocysteine insertion sequence (SECIS) located in the 3’ UTR (Berry, et al., 1991; Krol, 2002). During decoding of the UGA-Sec codon, a SECIS RNA binding protein (SBP2) recruits the Sec elongation factor and Sec tRNA[Ser]Sec to the ribosome (Driscoll and Copeland, 2003; Hatfield and Gladyshev, 2002). Illustrating the importance of this process for SelN expression and normal muscle function is the finding of a single homozygous disease causing point mutation in the 3’ UTR SECIS of a patient with RSMD1 (Allamand, et al., 2006). This mutation is sufficient to prevent Sec incorporation and significantly reduces both SelN mRNA and protein levels.
A second cis-acting selenocysteine codon redefinition element (SRE) has been described in a subset of selenoprotein genes including SEPN1 (Howard, et al., 2005; Howard, et al., 2007). The SEPN1 SRE consists of a highly conserved stem-loop structure that starts 6 nucleotides downstream of the UGA codon. Experimental evidence illustrates that both the stem-loop structure and the length and sequence of the spacer separating it from the UGA-Sec codon are important for Sec incorporation efficiency.
To date, four missense mutations have been identified in the SRE element of SEPN1. Three are previously unpublished point mutations: c.1388G>T occurs in the SEPN1 SRE between the UGA codon and the stem-loop structure, c.1405C>T and c.1406G>A occur within, but maintain the base pairing potential of the SRE stem-loop. One previously reported point mutation c.1397G>A alters the base pairing resulting in a C:A mismatch near the base of the SRE stem-loop (Moghadaszadeh, et al., 2001). To determine if these mutations may be disease causing due to disruption of the Sec insertion pathway, the effect of each mutation on Sec insertion efficiency was determined.
Methods
Cloning of SEPN1 sequences into the p2luc reporter vector
The generation of reporter constructs UGA2 with and without the SECIS element has been previously described (Howard, et al., 2005). Directed mutagenesis of the SRE element was performed via PCR mutagenesis using GeneTailor site-directed mutagenesis system (Invitrogen). All numbering is based on cDNA sequence with the first nucleotide being the A of the ATG initiation codon. GenBank reference sequence for SEPN1 is NM_020451.2 The predicted ΔG of the changes within the SRE stem- loop is given in kcals/mole (Markham and Zuker, 2005).
In Vitro Translations and Cell Culture Transfections
RRL in vitro transcription and translation reactions were done according to manufacturer’s specifications using the TNT Quick Coupled Transcription/Translation system obtained from Promega in a final volume of 10uls supplemented with varying amounts of SBP2-CT (amino acids 399–846) and readthrough efficiency determined as described (Howard, et al., 2007).
293FT cell line was maintained in DMEM + 5% FBS (Hyclone). Cells were transfected using Lipofectamine 2000 (Invitrogen), and the 1-day protocol. Briefly, suspension cells were added directly to the DNA complexes in 96-well plates. Transfected cells were incubated overnight at 37° C in 5% CO2, then an equal volume of DMEM + 10% FBS were added to each well, and the plates were incubated for an additional 48 hours prior to luciferase analysis as described (Howard, et al., 2005).
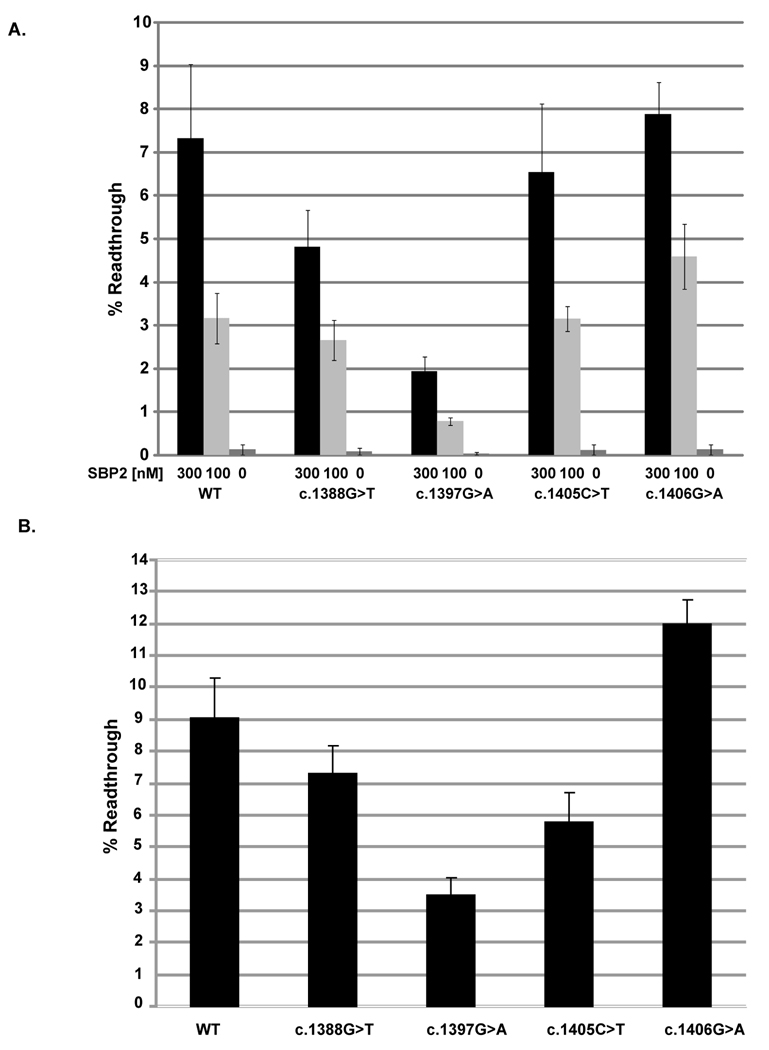
Statistical significance of the differences in mean readthrough efficiency observed between the wildtype reporter construct and each mutation was assessed by a Mann-Whitney Rank Sum Test (SPSS 16.0). P values calculated for the in vitro translation system supplemented with 300nM SBP2-CT (Figure 2A) were as follows: c.1388G>T P=0.065, c.1397G>A P=0.002, c.1405C>T P=0.24, c.1406G>A P=0.48. P-values calculated in the same way for the same constructs tested in transfected cells (Figure 2B) were all less than 0.001.
Fig. 2.
The effect of SEPN1 SRE mutations on UGA redefinition. A) Percent readthrough of the UGA codon in the wildtype (WT) or mutant SRE contexts (c.1388G>T, c.1397G>A, c.1405C>T, and c.1406G>A) were measured using the 2luc reporter system in vitro using a RRL transcription and translation system supplemented with increasing amounts of SBP2-CT (SBP2) at the indicated concentrations. B) Percent readthrough of the UGA codon in the wildtype (WT) or mutant SRE context was measured following expression of the 2luc reporter constructs in transfected 293FT cells.
Fibroblast culturing
Primary skin fibroblasts were grown in DMEM + 10% FCS (Life Technologies), 20 U/ml streptomycin and penicillin at 37°C in a humidified atmosphere with 5% CO2.
Western blot analysis
Protein was extracted from fibroblasts or muscle cryosections for western blot analysis by homogenizing fibroblasts or muscle tissue in a buffer containing 80 mM Tris–HCl pH 6.8, 10% SDS, 0.12 M sucrose, 10 mM EDTA, 1 mM PMSF, 1 mM benzamidine and incubating for 10 min at 55°C. The protein concentration was determined using the BCA protein Assay (Pierce Chemical Company).
Extracts were separated on 4–15% tris-glycine SDS polyacrylamide gels, transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon-P, Millipore), and blocked with PBS + 0.2% tween-20, 1.25% SDS and 5% nonfat dry milk. SelN protein was detected by chemiluminescence (Immobilon Western, Millipore) of membranes probed with a polyclonal anti-selN antibody (Ab 137; (Petit, et al., 2003); 1:200 incubated overnight at 4°C) and a horseradish peroxidase (HRP)-conjugated secondary antibody (Invitrogen). Anti-tubulin antibodies (Sigma) were used to ensure equal loading. Blots were scanned with an imaging densitometer and protein bands were quantified using Image J 1.37v software.
RNA extraction and quantitative RT-PCR
Total RNAs were extracted from 2 independent fibroblast cultures with Trizol as described (Allamand et al., 2006). Briefly, cDNA was produced from 1µg of total RNA using the Superscript II RT kit as recommended (InVitrogen). Quantitative real time PCR (7–8 independent experiments total) were conducted by amplification of a 290 bp SEPN1 product and a 248 bp HPRT product using SyBR Green and the LightCycler 480 apparatus (Roche Diagnostics, Germany). Results were normalized to control fibroblasts. Statistical significance was assessed by a Mann-Whitney Rank Sum Test using SigmaStat (Systat, Germany).
Results
Four missense mutations in the SEPN1 SRE
We identified missense mutations located in the SRE of 10 patients: c.1388G>T (p.G463V; 1 heterozygous), c.1405C>T (p.R469W; 1 homozygous), c.1406G>A (p.R469Q; 2 homozygous), and c.1397G>A (p.R466Q; 6 heterozygous). All numbering is based on cDNA sequence with the first nucleotide being the A of the ATG initiation codon. GenBank reference sequence for SEPN1 is NM_020451.2. All patients presented with the characteristic SEPN1-related myopathy phenotype, marked by congenital muscle weakness involving the axial muscles, spinal rigidity, scoliosis, and respiratory insufficiency generally appearing at the beginning of the second decade. The patient heterozygous for the c.1388G>T mutation and an out-of-frame 94-bp deletion (c.1- 25_69del) showed a particularly severe phenotype. She walked at 14 months but developed scoliosis from the first year of life and died at 5.5 years of restrictive respiratory failure despite ventilatory assistance. Her eldest sister died prior to this study at age 7, with a similar phenotype. The remaining patients are currently aged 13 to 47 years. Most of the adults underwent spinal fusion; all require nasal ventilation but remain ambulant.
Quantitative analysis of selenocysteine insertion
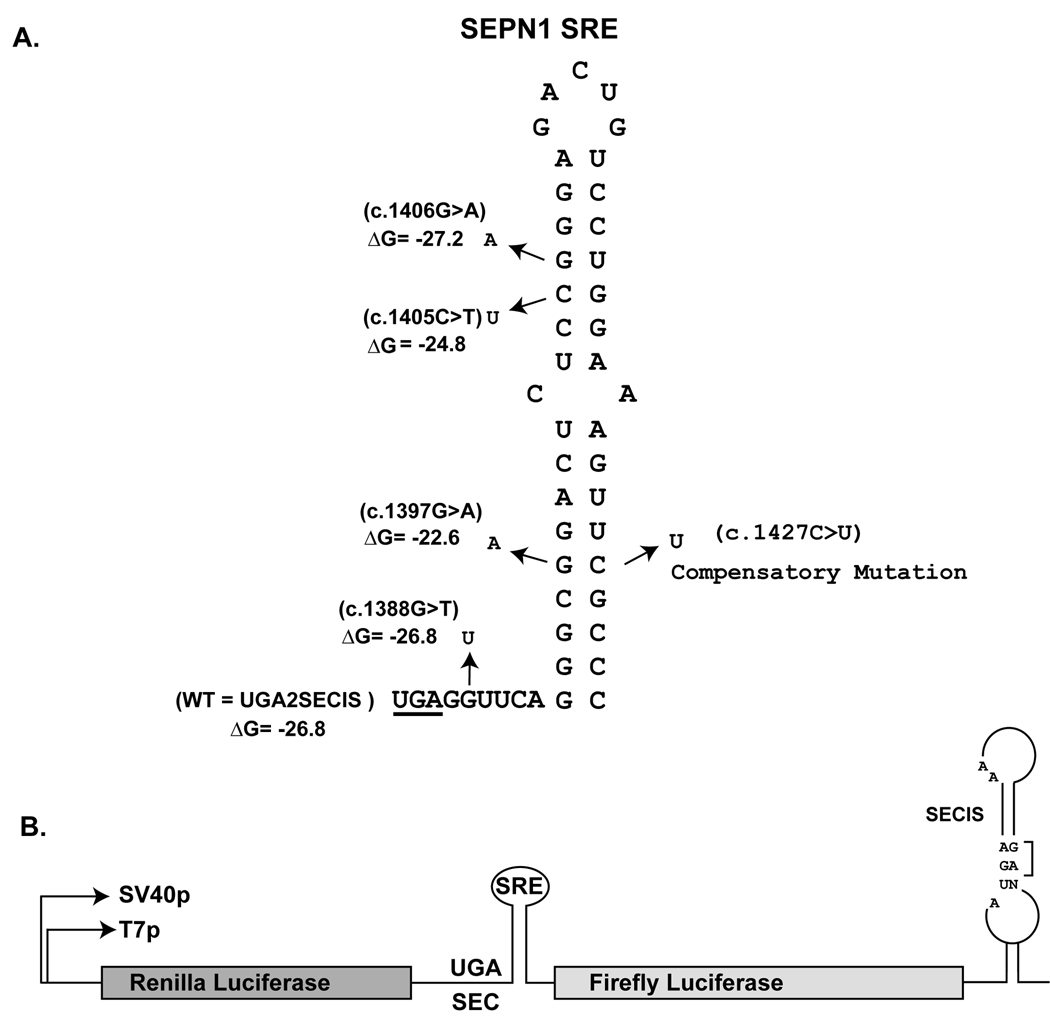
We modified a dual luciferase (2luc) reporter system (Grentzmann, et al., 1998) to quantitatively measure UGA-Sec decoding in vitro and in cultured mammalian cells as described (Howard, et al., 2005; Howard, et al., 2007). The UGA-Sec codon and surrounding SRE sequences were cloned between the two reporter genes and the SECIS element was included downstream of the firefly luciferase coding sequences to produce UGA2SECIS (Fig. 1A and 1B) (Howard, et al., 2005). The point mutations corresponding to c.1388G>T, c.1397G>A, c.1405C>T, and c.1406G>A were introduced into this dual luciferase reporter vector, both with and without the 3’ UTR SECIS. Fig. 1A shows the SEPN1 SRE element, the location of each mutation and the effect on the predicted free energy of the stem-loop structure (Markham and Zuker, 2005).
Fig. 1.
Stem-loop structure of the SEPN1 SRE. A) The UGA codon is underlined. The location and nucleotide identity of the four RSMD patient mutations are indicated by arrows. The predicted ∆G of the stem-loop is given in kcals/mole. The identity of the compensatory mutation introduced to restore base pairing between c.1397 and c.1427 is indicated. All numbering is based on cDNA sequence with the first nucleotide being the A of the ATG initiation codon. GenBank reference sequence for SEPN1 is NM_020451.2 B) Illustration of the Dual Luciferase (2luc) reporter vector. Renilla and Firefly coding sequences flank the SEPN1 SRE sequence and the SEPN1 SECIS is downstream of the Firefly open reading frame in the 3’UTR. UGA2SECIS referred to as WT in Figure 2 is the reporter vector with wildtype SEPN1 SRE and SECIS sequences. Essential features of the SECIS element such as the GA quartet and unpaired As in the loop are indicated.
The 2Luc plasmids containing either UGA2SECIS (WT), or UGA2SECIS with each patient mutation were transcribed and translated in Rabbit Reticulocyte Lysate (RRL) supplemented with increasing amounts of a fully functional C-terminal portion of SBP2 (SBP2-CT), SBP2 is the limiting factor for Sec insertion in RRL (Mehta, et al., 2004) and readthrough efficiency increases upon SBP2-CT addition up to a maximum of 300nM (Fig. 2A). Firefly luciferase activity was normalized to renilla luciferase expression, and compared to an in-frame control construct in which the UGA codon has been altered to UGC to determine readthrough efficiency. WT UGA decoding efficiency in the presence of 300nM SBP2-CT, a concentration allowing maximal Sec insertion efficiency, was approximately 7%. Mutation of the spacer region (c.1388G>T) or a mutation which alters the sequence of the SRE and weakens the stem-loop (c.1397G>A) reduced readthrough levels to 5% (P=0.065) and 2% (P=0.002) respectively. To determine if the c.1397G>A mutation reduces readthrough efficiency by weakening the SRE stem-loop or alteration of the sequence, a compensatory mutation was made to restore base pairing potential (Fig.1A). An approximate two fold increase in readthrough efficiency was observed when the double mutant c.1397G>A/c.1427C>T was tested relative to the c.1397G>A single mutation. The second mutation restores base pairing potential to form an A:U base pair in the SRE stem-loop (the wildtype base pair is G:C). Although readthrough efficiency was only partially restored (approximately 2 fold more readthrough), this result implies that both the potential to base pair and the identity of the base pair at this position is important for SRE function in UGA decoding. The mutations c.1405C>T and c.1406G>A, which maintain base pairing within the SRE stem loop, did not have a significant effect on Sec insertion efficiency. Readthrough levels were <1% when the 2luc constructs lacked the 3’ UTR SECIS or the UGA-Sec codon was changed to UAG (data not shown). The very low level of readthrough observed in the absence of the SECIS or exogenously added SBP2-CT (Fig. 2A) supports that readthrough is due to Sec insertion rather than decoding of the UGA codon by a near cognate aminoacylated tRNA.
To confirm these results in cultured mammalian cells, the same constructs were transfected into 293FT cells. After allowing for expression, cells were lysed and the firefly and renilla luciferase activities determined. Readthrough efficiency was 9% for the WT UGA2SECIS (Fig. 2B). As observed in in vitro RRL translation assays, the mutation (c.1397 G>A) showed close to threefold reduction in decoding efficiency (P<0.001). The mutation c.1406G>A, which is predicted to increase the stability of the stem-loop, resulted in a 3% increase in the readthrough levels (P<0.001). To determine if the change in amino acid composition at these positions might affect the protein stability, the UGA codon in each was changed to a UGC codon. No significant difference in either luciferase activity was detected following expression in 293FT cells when compared to the UGC control constructs (data not shown). In addition, Renilla luciferase activity was compared between the wildtype UGA containing constructs and corresponding constructs containing a point mutation in the SRE. No statistically significant reduction in Renilla activity was observed providing evidence that the SRE point mutations did not affect RNA levels. Consequently, changes in protein stability or RNA levels could not account for the observed differences in selenocysteine insertion efficiency in this reporter system.
The effect of the c.1397G>A point mutation on SelN expression in patients
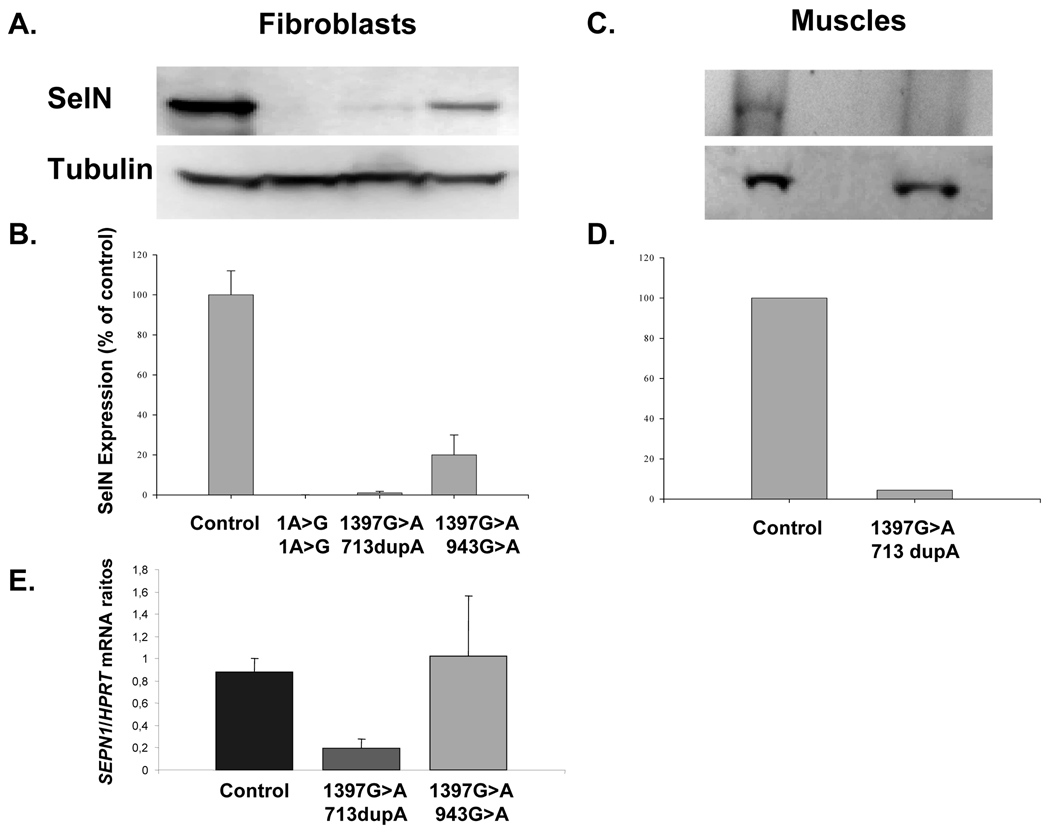
Human primary fibroblasts from two SEPN1-related myopathy patients carrying the mutations: c.1397G>A/c.713dupA and c.1397G>A/c.943G>A were tested for SelN protein levels by western blot analysis of protein extracts, probed with the anti-SelN antibody Ab 137 (Fig. 3A). The c.713dupA frameshift mutation induces a premature termination codon at position 299, whereas c. 943G>A is a missense mutation located far upstream of the UGA-Sec codon. Fibroblasts from a patient carrying a homozygous mutation of the start codon (c.1A>G) were utilized as a null control. All values were normalized to tubulin and standardized against SelN levels in normal control fibroblasts (100%) (Fig. 3B). SelN levels in the fibroblasts with the 1397G>A/713dupA and 1397G>A/943G>A mutations were <5% and approximately 20% of the normal control respectively. Immunoblot analysis of archived frozen muscle tissue from a biopsy of a third patient with the heterozygous c.1397G>A/c.713dupA mutation demonstrated that, as with fibroblasts, only very low levels of SelN were detectable (Fig. 3C and 3D). SelN mRNA levels were determined by quantitative RT-PCR of RNA isolated from primary fibroblasts cultures containing the c.1397G>A/c.713dupA, and c.1397G>A/c.943G>A (Fig. 3E). In c.1397G>A/c.713dupA fibroblasts, SelN RNA levels were reduced to approximately 20% of normal levels (P<0.001). However, in c.1397G>A/c.943G>A fibroblasts, SelN mRNA levels were near normal levels (P=0.694). It should be noted that c.713dupA homozygote fibroblasts have only 5% residual SelN mRNA levels (unpublished data), therefore the c.1397G>A allele may be less sensitive to degradation than c.713dupA. Given that missense alleles are unlikely to affect mRNA stability, the detection limit of the system may be insufficient to distinguish changes in mRNA levels in the heterozygous cells harboring one missense mutation (c.943G>A) and the c.1397G>A mutation. Nevertheless, it is clear from the c.1397G>A/c.713dupA results that the c.1397G>A mutation does effect the stability of SelN mRNA leading to a reduction in mRNA levels.
Fig. 3.
Analysis of SelN protein levels in patient fibroblasts and muscle. A) Western blot analysis of SelN and tubulin expression in fibroblasts or muscle from individuals with the SelN alleles: wildtype (Control), c.1A>G homozygote, c.1397G>A/c.713dupA and c.1397G>A/c.943G>A heterozygotes. B) Quantification of SelN expression normalized to tubulin and expressed as a percentage of the control. C) Western blot analysis of protein extracted from wild type control and c.1397G>A/c.713dupA muscle. D) Quantification of SelN protein from C.(c.1397G>A/c.713dupA Protein). E) Quantitative real-time PCR analysis of SelN RNA from Wildtype (Control), c.1397G>A/c.713dupA, and c.1397G>A/c.943G>A fibroblasts.
These results are consistent with 1397G>A/713dupA fibroblasts having two inactive SelN alleles, and the 1397G>A/943G>A having one inactive allele and one allele that expresses reduced levels of SelN. We infer that alleles carrying the missense mutation c.1397G>A express only negligible levels of SelN protein due to both reduced Sec insertion and SelN mRNA levels.
Discussion
Recent studies have implicated mutations affecting both cis- and trans- acting factors of the Sec insertion pathway in several human disorders (Dumitrescu, et al., 2005; Hu, et al., 2001; Kumaraswamy, et al., 2000). Of direct relevance to this study is the recent finding that a single homozygous point mutation in the SEPN1 3’ UTR SECIS is sufficient to cause RSMD1 (Allamand, et al., 2006).
The nine amino acids on either side of the SelN Sec residue are highly conserved in all chordates (Howard, et al., 2005). This is potentially significant since Sec is often at the catalytic center of selenoproteins. In three of the RSMD1 patients described here, the conserved Arg residues downstream of Sec have been mutated to uncharged amino acids (p.466Arg>Gln/c.1397G>A; p.469Arg>Trp/c.1405C>T; and p.469Arg>Gln/c.1406G>A). These changes may compromise the activity or stability of the SelN protein expressed, and thus further contributes to the observed phenotype. These amino acid changes however had no observable effect on the stability of the expressed reporter in 293FT cells.
Here we demonstrate that one of the missense mutation (c.1397G>A) in the SEPN1 SRE significantly reduces Sec insertion efficiency, and weakens the mRNA structure. It is possible that one role of the SRE is to stabilize SEPN1 RNA and that mutations in this element may be responsible for RNA degradation. Several mRNA “quality control” pathways exist to degrade mRNAs which are not properly translated (Isken and Maquat, 2007). One mechanism of mRNA degradation is via the nonsense mediated decay (NMD) pathway [review (Chang, et al., 2007)]. Inefficient redefinition of the UGA codon may lead to the recognition of this codon as a stop, thus activating NMD. An alternate explanation is activation of the No-go decay pathway whereby mRNAs with paused ribosomes due to inefficient UGA decoding may be cleaved and subsequently degraded by RNA nucleases (Doma and Parker, 2006). Regardless of the mechanism, the combined effect of reduced Sec insertion and SelN mRNA levels creates an effectively null allele.
Previous experiments have shown that alteration of only three base pairs of the SRE significantly impairs Sec insertion (Howard, et al., 2005). As shown here, altering one nucleotide which weakens the SRE stemloop (c.1397G>A) is sufficient to truncate the SRE stimulatory effect on UGA decoding. A compensatory mutation which restored base pairing potential at this position to an A:U resulted in a partial restoration of readthrough efficiency (less than 2 fold; data not shown). The partial restoration of readthrough efficiency is consistent with the results reported in our original description of the SRE (Howard, et al., 2005) which indicated that at some positions not only the base pairing but also the sequence of the base pair can be an important determinant of readthrough efficiency. Interestingly, the mutation c.1406G>A, predicted to increase the stability of the stemloop structure, resulted in an increase of Sec insertion efficiency in transfected cells. In addition, a mutation (c.1388G>T) within the six nucleotide spacer region separating the UGA-Sec codon from the stem-loop structure was shown to reduce Sec insertion efficiency.
The results presented here suggest that interference with SRE function represents a novel mechanism by which a missense mutation in the coding region of the SEPN1 gene may prevent SelN expression, and further illustrate the importance of the SRE element in the expression of SelN protein in vivo.
Acknowledgments
The authors acknowledge the patients, their families, and the clinicians who provided DNA and cell lines for this study: Drs C. Bönneman, C. Castiglioni, B. Estournet, A. Fidzianska, F. Muntoni, S. Quijano-Roy, C. Sewry, A. Urtizberea. This work was supported by NIH R01GM077462 (MTH), NIH R01NS043264 (KMF), the Institut National de la Santé et de la Recherche Médicale (INSERM), the Association Française contre les Myopathies (AFM), and the GIS-Institut des Maladies Rares (France).
References
- Allamand V, Richard P, Lescure A, Ledeuil C, Desjardin D, Petit N, Gartioux C, Ferreiro A, Krol A, Pellegrini N, et al. A single homozygous point mutation in a 3'untranslated region motif of selenoprotein N mRNA causes SEPN1-related myopathy. EMBO Rep. 2006;7(4):450–454. doi: 10.1038/sj.embor.7400648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry MJ, Banu L, Chen YY, Mandel SJ, Kieffer JD, Harney JW, Larsen PR. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3' untranslated region. Nature. 1991;353(6341):273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Clarke NF, Kidson W, Quijano-Roy S, Estournet B, Ferreiro A, Guicheney P, Manson JI, Kornberg AJ, Shield LK, North KN. SEPN1: associated with congenital fiber-type disproportion and insulin resistance. Ann Neurol. 2006;59(3):546–552. doi: 10.1002/ana.20761. [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440(7083):561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll DM, Copeland PR. Mechanism and regulation of selenoprotein synthesis. Annu Rev Nutr. 2003;23:17–40. doi: 10.1146/annurev.nutr.23.011702.073318. [DOI] [PubMed] [Google Scholar]
- Dumitrescu AM, Liao XH, Abdullah MS, Lado-Abeal J, Majed FA, Moeller LC, Boran G, Schomburg L, Weiss RE, Refetoff S. Mutations in SECISBP2 result in abnormal thyroid hormone metabolism. Nat Genet. 2005;37(11):1247–1252. doi: 10.1038/ng1654. [DOI] [PubMed] [Google Scholar]
- Ferreiro A, Ceuterick-de Groote C, Marks JJ, Goemans N, Schreiber G, Hanefeld F, Fardeau M, Martin JJ, Goebel HH, Richard P, et al. Desmin-related myopathy with Mallory body-like inclusions is caused by mutations of the selenoprotein N gene. Ann Neurol. 2004;55(5):676–686. doi: 10.1002/ana.20077. [DOI] [PubMed] [Google Scholar]
- Ferreiro A, Quijano-Roy S, Pichereau C, Moghadaszadeh B, Goemans N, Bonnemann C, Jungbluth H, Straub V, Villanova M, Leroy JP, et al. Mutations of the selenoprotein N gene, which is implicated in rigid spine muscular dystrophy, cause the classical phenotype of multiminicore disease: reassessing the nosology of early-onset myopathies. Am J Hum Genet. 2002;71(4):739–749. doi: 10.1086/342719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanigan KM, Kerr L, Bromberg MB, Leonard C, Tsuruda J, Zhang P, Gonzalez-Gomez I, Cohn R, Campbell KP, Leppert M. Congenital muscular dystrophy with rigid spine syndrome: a clinical, pathological, radiological, and genetic study. Ann Neurol. 2000;47(2):152–161. [PubMed] [Google Scholar]
- Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. Rna. 1998;4(4):479–486. [PMC free article] [PubMed] [Google Scholar]
- Hatfield DL, Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol Cell Biol. 2002;22(11):3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard MT, Aggarwal G, Anderson CB, Khatri S, Flanigan KM, Atkins JF. Recoding elements located adjacent to a subset of eukaryal selenocysteine-specifying UGA codons. Embo J. 2005;24(8):1596–1607. doi: 10.1038/sj.emboj.7600642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard MT, Moyle MW, Aggarwal G, Carlson BA, Anderson CB. A recoding element that stimulates decoding of UGA codons by Sec tRNA[Ser]Sec. Rna. 2007;13(6):912–920. doi: 10.1261/rna.473907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YJ, Korotkov KV, Mehta R, Hatfield DL, Rotimi CN, Luke A, Prewitt TE, Cooper RS, Stock W, Vokes EE, et al. Distribution and functional consequences of nucleotide polymorphisms in the 3'-untranslated region of the human Sep15 gene. Cancer Res. 2001;61(5):2307–2310. [PubMed] [Google Scholar]
- Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21(15):1833–3856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- Krol A. Evolutionarily different RNA motifs and RNA-protein complexes to achieve selenoprotein synthesis. Biochimie. 2002;84(8):765–774. doi: 10.1016/s0300-9084(02)01405-0. [DOI] [PubMed] [Google Scholar]
- Kumaraswamy E, Malykh A, Korotkov KV, Kozyavkin S, Hu Y, Kwon SY, Moustafa ME, Carlson BA, Berry MJ, Lee BJ, et al. Structure-expression relationships of the 15-kDa selenoprotein gene. Possible role of the protein in cancer etiology. J Biol Chem. 2000;275(45):35540–35547. doi: 10.1074/jbc.M004014200. [DOI] [PubMed] [Google Scholar]
- Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33(Web Server issue):W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Rebsch CM, Kinzy SA, Fletcher JE, Copeland PR. Efficiency of mammalian selenocysteine incorporation. J Biol Chem. 2004;279(36):37852–37859. doi: 10.1074/jbc.M404639200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghadaszadeh B, Petit N, Jaillard C, Brockington M, Roy SQ, Merlini L, Romero N, Estournet B, Desguerre I, Chaigne D, et al. Mutations in SEPN1 cause congenital muscular dystrophy with spinal rigidity and restrictive respiratory syndrome. Nat Genet. 2001;29(1):17–18. doi: 10.1038/ng713. [DOI] [PubMed] [Google Scholar]
- Petit N, Lescure A, Rederstorff M, Krol A, Moghadaszadeh B, Wewer UM, Guicheney P. Selenoprotein N: an endoplasmic reticulum glycoprotein with an early developmental expression pattern. Hum Mol Genet. 2003;12(9):1045–1053. doi: 10.1093/hmg/ddg115. [DOI] [PubMed] [Google Scholar]
- Thisse C, Degrave A, Kryukov GV, Gladyshev VN, Obrecht-Pflumio S, Krol A, Thisse B, Lescure A. Spatial and temporal expression patterns of selenoprotein genes during embryogenesis in zebrafish. Gene Expr Patterns. 2003;3(4):525–532. doi: 10.1016/s1567-133x(03)00054-1. [DOI] [PubMed] [Google Scholar]