Oriented sample (OS) solid-state NMR is capable of determining the three-dimensional structures of proteins in their native functional environments when they are immobilized and aligned in supramolecular assemblies1, such as virus particles or membranes. Examples include gramicidin2, AchR M2 domain3, M2 domain of the influenza A virus4, fd5 and Pf16 phage coat proteins, phospholamban7, Vpu (from HIV-1)8, and MerF9.
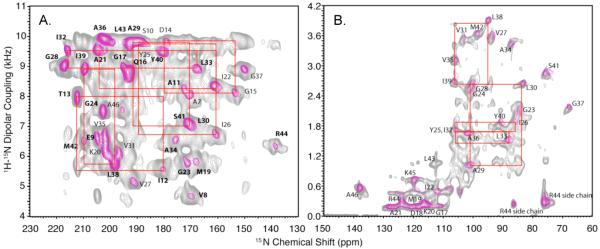
With improved sample preparation methods and the implementation of experiments that yield high-resolution separated local field (SLF) spectra, the principal roadblock to atomic-resolution structure determination is obtaining sequence-specific assignments of resolved resonances. While the shot-gun approach10 simultaneously assigns spectra and measures structural constraints, it relies on the preparation of multiple selectively isotopically labeled samples, and is restricted to residues in regular secondary structures whether α-helix or β-sheet. The goal is to utilize uniformly labeled samples for all steps of the structure determination process. The feasibility of using dilute-spin-exchange to identify signals from proximate sites has been demonstrated for both virus particles1 and membrane proteins11. However, this is limited by the requirement for lengthy intervals (several seconds) for the spin-exchange to occur among the weakly coupled nuclei. As an alternative to the recent cross-relaxation driven method12, a proton-mediated dilute-spin exchange experiment has been implemented13, which has the potential to accelerate data acquisition. The latter method is based on the transfer of magnetization between the low-gamma spins via the proton bath under mismatched Hartmann-Hahn (MMHH) conditions. 15N-15N correlations for distances of up to 6.7 Å can be identified13. In principle, the method is applicable to any dilute spin system bridged by a strong proton dipolar network (e.g., amide backbone 15N spins) thus providing a strategy for sequential assignment of resonances in OS solid-state NMR of proteins. The cross peaks are established within a few milliseconds dramatically shortening the overall experiment time compared to conventional dilute spin exchange experiments11, 14 based on spin diffusion15. The MMHH method has been extended to measure the heteronuclear dipolar couplings16 by including the SAMPI4 pulse sequence in the indirect dimension17, thus yielding a spin-exchanged high-resolution SLF spectrum. Solid-state NMR spectra of the structural form of Pf1 coat protein in magnetically aligned bacteriophage particles and of the membrane-bound form reconstituted in magnetically aligned bicelles have been previously measured and assigned6, 18, 19 and, therefore can be used as a test system for the applicability of the MMHH method to biological samples. Figure 1a shows the PISEMA20 spectrum of uniformly 15N-labeled Pf1 bacteriophage. The PISEMA spectrum is overlaid with its spin-exchanged version acquired using the pulse sequence shown in Fig. 2a. For the Pf1 bacteriophage spectra PISEMA has been used instead of SAMPI4 in order to measure larger (> 5 kHz) dipolar couplings due to relatively low radiofrequency B1 fields (< 50 kHz) available on the spectrometer. For the spin-exchanged spectrum of Fig. 1a, the proton mismatch amplitude was set at 22% larger than the Hartmann-Hahn condition during the exchange period, the mixing time was set to 4 ms, and a 1 sec z-filter was incorporated in order to eliminate residual proton magnetization and let the probe cool down before a long period of continuous irradiation is re-applied. The mismatch amplitude of about 20% was found to be near optimal for the magnetization transfer among the backbone 15N spins. In contrast, a 10% mismatch amplitude was previously found16 to be optimal for the more distant 15N spins in a NAL crystal (separated by as much as 6.7 Å as opposed to about 2.8 Å for the 15N spins in adjacent residues in an α-helix. By overlaying the spectra with and without spin-exchange in Fig. 1a it is possible to distinguish the cross peaks from the main peaks, and establish sequential connectivity among the latter. The red boxes depicted in fig. 1 connect two main peaks and two crosspeaks for each pair of interacting residues. For clarity, the results for residues I12-T13, D14-G15, A21-I22, A29-L30, Y25-I26, I32-L33, L38-I39, are highlighted; many others are established by the displayed data. Notably, these assignments are in agreement with those obtained previously by using a combination of the selective isotopic labeling and the “shotgun approach”18, and the crosspeaks generally follow the i, i+1 connectivity pattern. Additional weaker crosspeaks may arise from more distant 15N-15N interactions (e.g. i, i+3) due to their proximity in helical structures, which merits additional investigation.
Figure 1.
A. PISEMA spectrum of Pf1 phage (magenta contours) overlaid with the spin-exchanged PISEMA spectrum (gray contours) acquired using pulse sequence of Fig. 2a. Data were acquired on a Bruker Avance II spectrometer operating at 500 MHz, temperature T = −3 °C, 64 scans, 50 kHz B1 field, 80 t1 points (magenta lines); and 1k scans, 64 t1 points, 4 ms contact time 22% MMHH (gray lines). Only the helical part of the whole Pf1 spectrum is shown. Red boxes (depicted here for a number of representative residues) establish connectivity between the adjacent amide sides. B. SAMPI4 spectrum of Pf1 coat protein reconstituted in magnetically aligned bicelles (magenta contours) overlaid with its spin-exchanged version depicted in Fig. 2b (gray lines). Data were acquired on a Bruker Avance III spectrometer operating at 700 MHz, 32 scans, 50 kHz B1 field, 82 linear t1 points (magenta lines); and 1k scans, 55 kHz 1H B1 field during the MMHH period and 50 kHz B1 field everywhere else, 0.1 sec z-filter, 82 linear t1 points (gray lines). Representative connectivities for some of the residues as in Fig 1a are shown as an illustrative example.
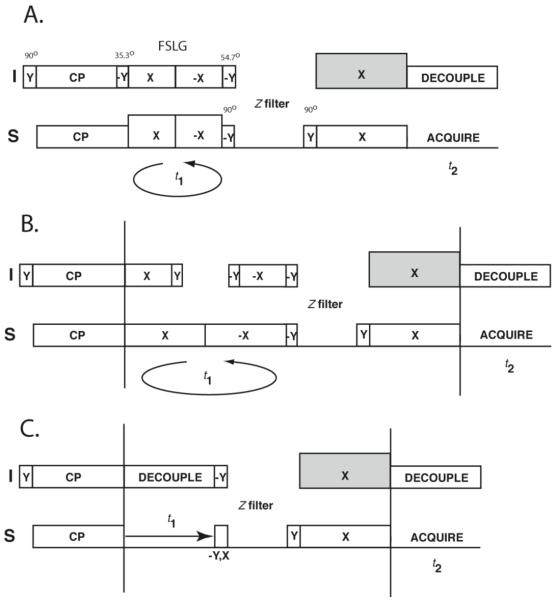
Figure 2.
A-B. Spin-exchanged SLF pulse sequences for spectroscopic assignment in solid-state NMR of oriented samples. The t1 dimension is evolved using either the frequency-switched Lee-Goldburg scheme (A) or SAMPI4 (B), followed by the z-filter and the proton-mediated spin exchange, during which the proton rf amplitude is set at 10%-25% above the Hartmann-Hahn matching condition. C. Homonuclear 15N-15N exchange employing mismatched Hartmann-Hahn magnetization transfer. The intermediate pulse before the z-filter selects either real or imaginary component of the t1 evolution.
To demonstrate the feasibility of applying the MMHH method to membrane proteins, the membrane-bound form of the uniformly 15N-labeled Pf1 coat protein was reconstituted into magnetically aligned bicelles as previously described19, 21. Figure 1b shows the SAMPI4 spectrum of Pf1 coat protein in bicelles processed by the Maximum Entropy Method (MEM)22, magenta contour lines. The SAMPI4 spectrum is overlaid with its exchanged version processed by MEM (gray lines) and acquired using the pulse sequence of Fig. 2b. Due to the perpendicular orientation of the normal relative to the magnetic field and the slightly reduced order parameter of the bicelle, the dipolar couplings are scaled by approximately a factor of 0.4 compared to the magnetically aligned Pf1 phage. Therefore, the MMHH amplitude was reduced to about +10% to retain the same average Hamiltonians13 that result in the maximum magnetization transfer as in the case of magnetically aligned phage. Representative connectivities for some of the residues as in Fig. 1a are shown, which are also consistent with earlier assignments19. This demonstrates that the MMHH spin exchange can be applied to assign spectra of membrane proteins in lipid bilayer environments.
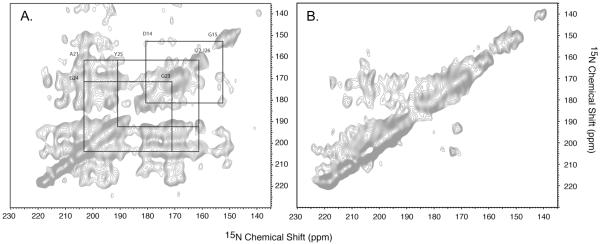
In practice, homonuclear 15N-15N spin exchange spectrum acquired using the pulse sequence of Fig. 2c can provide independent validation of the assignment and aid in resolving ambiguities when the main peaks in the exchanged PISEMA spectrum have similar dipolar couplings. Figure 3a shows the 15N-15N exchange spectrum of magnetically aligned Pf1 phage that provides sequential assignment for residues A21 through I26 via cross-validation of the peaks in the corresponding exchanged PISEMA spectrum (Fig. 1a). Other connectivities can also be established.
Figure 3.
A. 15N-15N exchange spectrum used to assist in the assignment process of magnetically aligned Pf1 phage (500 MHz 1H field, 256 scans, 128 complex t1 points, 1 sec z-filter, 5 ms MMHH). Using the exchanged PISEMA spectra alone for spectral assignment may give rise to ambiguities when the main peaks have similar 1H-15N dipolar couplings, thus the crosspeaks are produced near the location of the main peaks. As an illustrative example, sequential assignments of fig. 1a for residues A21 through I26 are validated using the 15N-15N crosspeaks. B. 15N-15N exchange spectra acquired without using the MMHH block with 1 sec exchange time (z-filter). Many of the cross peaks of fig. 3a are missing from the spectrum.
To further illustrate the efficiency of the MMHH transfer, a control experiment for Pf1 phage has been done without the MMHH block, whose pulse sequence essentially corresponds to the dilute spin-exchange experiment11 employing proton-driven spin diffusion. As can be seen from Fig. 3b, only very few cross-peaks are established after 1 sec mixing time (corresponding to the 1 sec z-filter in the MMHH experiment), and for such a short mixing time the cross peaks lack symmetry with respect to the main diagonal. More cross peaks can be established when the mixing time is made longer than 3 sec (results not shown); however, this considerably lengthens the overall time of the experiment.
The combination of MMHH spin-exchange and high-resolution SLF spectroscopy accelerates the assignment and analysis of OS solid-state NMR spectra of uniformly labeled proteins. Moreover, the inclusion of the additional dipolar coupling dimension helps eliminate ambiguities when the main peaks have overlapping chemical shifts. It has been shown that the method can be applied for various alignment media including magnetically oriented bacteriophage and membrane proteins reconstituted in bicelles. Moreover, this purely spectroscopic technique is applicable to proteins of arbitrary topology. While selective labeling and “shotgun approach” are still invaluable for the initial steps of assignment and eliminating potential ambiguities, this further development of the method improves the practicality of determining the three-dimensional structures of membrane proteins in their native phospholipid bilayer environment.
ACKNOWLEDGMENT
We thank Lena Jairam for assistance with sample preparation. This research was supported by grants MCB 0843520 from NSF and North Carolina Biotechnology Center to A.A.N. and by grants from the National Institutes of Health to S.J.O. and utilized the Biomedical Technology Resource for NMR Molecular Imaging of Proteins at the University of California, San Diego, which is supported by grant P41EB002031.
REFERENCES
- (1).Cross TA, Opella SJ. J. Am. Chem. Soc. 1983;105:306–308. [Google Scholar]
- (2).Ketchem RR, Hu W, Cross TA. Science. 1993;261:1457–1460. doi: 10.1126/science.7690158. [DOI] [PubMed] [Google Scholar]
- (3).Opella SJ, Marassi FM, Gesell JJ, Valente AP, Kim Y, Oblatt-Montal M, Montal M. Nat. Struct. Biol. 1999;6(4):374–379. doi: 10.1038/7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Wang J, Kim S, Kovacs F, Cross TA. Prot. Sci. 2001;10:2241–2250. doi: 10.1110/ps.17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Zeri AC, Mesleh MF, Nevzorov AA, Opella SJ. Proc. Natl. Acad. Sci. USA. 2003;100(11):6458–6463. doi: 10.1073/pnas.1132059100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Thiriot DS, Nevzorov AA, Zagyanskiy L, Wu CH, Opella SJ. J Mol Biol. 2004;341:869–879. doi: 10.1016/j.jmb.2004.06.038. [DOI] [PubMed] [Google Scholar]
- (7).Traaseth NJ, Buffy JJ, Zamoon J, Veglia G. Biochemistry. 2006;45(46):13827–13834. doi: 10.1021/bi0607610. [DOI] [PubMed] [Google Scholar]
- (8).Park SH, De Angelis AA, Nevzorov AA, Wu CH, Opella SJ. Biophys. J. 2006;91:3032–3042. doi: 10.1529/biophysj.106.087106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).De Angelis AA, Howell SC, Nevzorov AA, Opella SJ. J. Am. Chem. Soc. 2006;128(37):12256–12267. doi: 10.1021/ja063640w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Marassi FM, Opella SJ. J Biomol NMR. 2002;23(3):239–42. doi: 10.1023/a:1019887612018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Marassi FM, Gesell JJ, Valente AP, Kim Y, Oblatt-Montal M, Montal M, Opella SJ. J. Biomol. NMR. 1999;14:141–148. doi: 10.1023/a:1008391823293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Xu J, Struppe JS, Ramamoorthy A. J. Chem. Phys. 2008;128(5):052308. doi: 10.1063/1.2826323. [DOI] [PubMed] [Google Scholar]
- (13).Nevzorov AA. J. Am. Chem. Soc. 2008;130:11282–11283. doi: 10.1021/ja804326b. [DOI] [PubMed] [Google Scholar]
- (14).Cross TA, Opella SJ. J. Am. Chem. Soc. 1983;105:7471–7473. [Google Scholar]
- (15).Suter D, Ernst RR. Phys. Rev. B. 1985;32(9):5608–5627. doi: 10.1103/physrevb.32.5608. [DOI] [PubMed] [Google Scholar]
- (16).Nevzorov AA. J. Magn. Reson. 2009;201(1):111–114. doi: 10.1016/j.jmr.2009.08.006. [DOI] [PubMed] [Google Scholar]
- (17).Nevzorov AA, Opella SJ. J. Magn. Reson. 2007;185:59–70. doi: 10.1016/j.jmr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- (18).Thiriot DS, Nevzorov AA, Opella SJ. Protein Sci. 2005;14:1064–1070. doi: 10.1110/ps.041220305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Opella SJ, Zeri AC, Park SH. Annu. Rev. Phys. Chem. 2008;59:635–57. doi: 10.1146/annurev.physchem.58.032806.104640. [DOI] [PubMed] [Google Scholar]
- (20).Wu CH, Ramamoorthy A, Opella SJ. J. Magn. Reson. A. 1994;109:270–272. [Google Scholar]
- (21).Park SH, Loudet C, Marassi FM, Dufourc EJ, Opella SJ. J. Magn. Reson. 2008;193(1):133–138. doi: 10.1016/j.jmr.2008.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Jones DH, Opella SJ. J. Magn. Reson. 2006;179(1):105–113. doi: 10.1016/j.jmr.2005.11.014. [DOI] [PubMed] [Google Scholar]