Abstract
Hereditary hearing impairment (HI) displays extensive genetic heterogeneity. Autosomal recessive (AR) forms of prelingual HI account for ~75% of cases with a genetic etiology. A novel AR non-syndromic HI locus (DFNB47) was mapped to chromosome 2p25.1-p24.3, in two distantly related Pakistani kindreds. Genome scan and fine mapping were carried out using microsatellite markers. Multipoint linkage analysis resulted in a maximum LOD score of 4.7 at markers D2S1400 and D2S262. The three-unit support interval was bounded by D2S330 and D2S131. The region of homozygosity was found within the three-unit support interval and flanked by markers D2S2952 and D2S131, which corresponds to 13.2 cM according to the Rutgers combined linkage-physical map. This region contains 5.3 Mb according to the sequence-based physical map. Three candidate genes, KCNF1, ID2 and ATP6V1C2 were sequenced, and were found to be negative for functional sequence variants.
Introduction
Genetic hearing impairment (HI) can be classified as either syndromic or non-syndromic. Autosomal recessive (AR) inheritance predominates in hereditary non-syndromic HI (NSHI), and accounts for ~75% of the cases, whereas autosomal dominant forms are observed in 15% of cases. Mitochondrial and X-linked NSHI, on the other hand, are less frequent (Morton 1991). HI in AR forms is usually due to a sensorineural defect (Petit 1996), and is generally prelingual, not progressive and all frequencies are affected with severe to profound HI. NSHI is the most heterogeneous trait known. To date 46 loci for AR NSHI have been mapped and 21 genes have been isolated (Van Camp and Smith 2005). A large number of genes with different functions are involved in the etiology of HI because of the complexity of the inner ear, and the various mechanisms that can lead to the HI phenotype (Steel and Bussoli 1999). This article describes the mapping of a novel ARNSHI locus, DFNB47, to chromosome 2p25.1-p24.3 in two distantly related consanguineous Pakistani kindreds.
Materials and methods
Family history
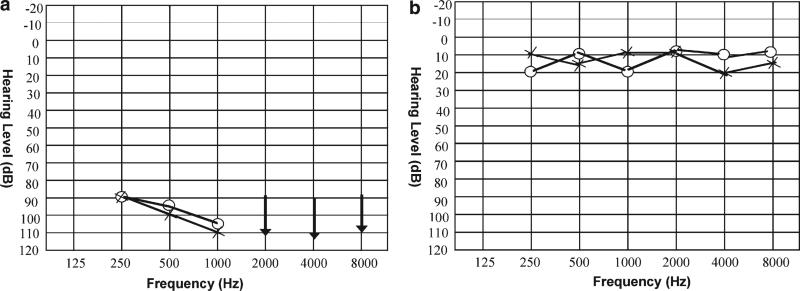
Approval was obtained from the Institutional Review Boards of the Quaid-I-Azam University and Baylor College of Medicine and Affiliated Hospitals prior to the study. Informed consent was obtained from all family members who participated in the study. A number of pedigrees were ascertained including pedigrees 4052a and 4052b, which are distantly related consanguineous families from Sadiq Abad in Pakistan. Information obtained during interviews with multiple family members was used to construct the pedigrees and to clarify consanguineous relationships. Although various pedigree members reported that the two family branches were related, the exact relationship could not be specified. In the two pedigrees, 4052a and 4052b (Fig. 1), the HI displayed an AR mode of inheritance. Affected individuals manifest prelingual profound HI and use sign language for communication. Physical examination of hearing-impaired individuals did not reveal any syndromic features, such as maxillofacial or limb deformities, visual loss and mental deficiency. In addition, no gross vestibular involvement was noted in the clinical history and physical examination. Due to the remote location of the family, further vestibular testing, electroretinography, and temporal bone imaging were not feasible. An audiogram from hearing-impaired individual 14 of pedigree 4052a shows bilateral, profound HI that affects all frequencies (Fig. 2).
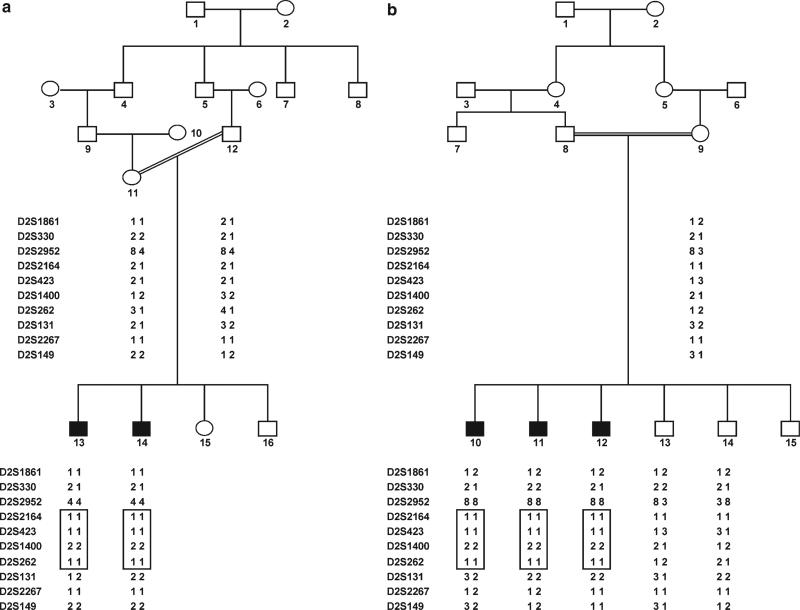
Fig. 1.
Drawing of pedigrees 4052a and 4052b which segregate DFNB47. Black symbols represent individuals with hearing impairment (HI) due to DFNB47. Clear symbols represent unaffected individuals. Haplotypes for the most closely linked STRs are shown below each symbol. The haplotype for DFNB47 is displayed in a box
Fig. 2.
Audiograms from a hearing-impaired individual 14 of pedigree 4052a, and b unaffected individual 16 of pedigree 4052a. Circles and crosses represent air conduction for right and left ear, respectively. Arrows represent residual hearing. The audiogram from the hearing-impaired family member shows that DFNB47 is associated with bilateral, profound HI that affects all frequencies
Extraction of genomic DNA and genotyping
Venous blood samples were collected from a total of ten individuals, including five who are hearing-impaired. Genomic DNA was extracted from whole blood following a standard protocol (Grimberg et al. 1999), quantified by spectrophotometric readings at optical density 260, and diluted to 40 ηg/μl for polymerase chain reaction (PCR) amplification. A genome scan was carried out on all the DNA samples at the Center for Inherited Disease Research (CIDR). A total of 396 fluorescently labeled short tandem repeat (STR) markers were genotyped. These markers were spaced ~10 cM apart and are located on the 22 autosomes and the X and Y chromosomes.
For fine mapping, PCR for microsatellite markers were performed according to standard procedure in a total volume of 25 μl with 40 ηg of genomic DNA, 0.3 μl of each primer, 200 μM dNTP and 1× PCR buffer (Fermentas Life Sciences, Burlington, ON, Canada). PCR was carried out for 35 cycles: 95°C for 1 min, 57°C for 1 min and 72°C for 1 min in a thermal cycler (PerkinElmer Life and Analytical Sciences, Boston, MA, USA). PCR products were resolved on 8% non-denaturing polyacrylamide gel and genotypes were assigned by visual inspection.
Linkage analysis
The order of genome scan markers and fine mapping markers were determined based on the National Center for Biotechnology Information (NCBI) Build 34 sequence-based physical map (International Human Genome Sequence Consortium 2001). The Rutgers combined linkage-physical map of the human genome (Kong et al. 2004) was utilized for genetic map distances in multipoint linkage analysis for the fine mapping and genome scan markers. The fine-mapping marker D2S1861 was not found on the NCBI sequence-based physical map and was placed on the sequence-based physical map using e-PCR (Schuler 1997), then the genetic map position was deduced by interpolation on the Rutgers combined linkage-physical map. PEDCHECK (O'Connell and Weeks 1998) was used to identify Mendelian inconsistencies while the MERLIN (Abecasis et al. 2002) program was utilized to detect potential genotyping errors that did not produce a Mendelian inconsistency. Haplotypes were constructed using SIM-WALK2 (Weeks et al. 1995; Sobel and Lange 1996). Two-point linkage analysis was carried out on all autosomal markers from the genome scan using the MLINK program of the FASTLINK computer package (Cottingham et al. 1993). Multipoint linkage analysis was performed using ALLEGRO (Gudbjartsson et al. 2002). An AR mode of inheritance with complete penetrance and a disease allele frequency of 0.001 were used for the analysis. For the genome scan markers, allele frequencies were estimated from the founders and reconstructed genotypes of founders from the two pedigrees (4052a and 4052b) and 43 additional pedigrees that underwent a genome scan at CIDR at the same time. Equal allele frequencies were used for the fine mapping markers, because it was not possible to estimate allele frequencies from the founders, since these markers were only genotyped in the two families. A sensitivity analysis was carried out in order to evaluate whether a false positive result had occurred due to using incorrect allele frequencies (Freimer et al. 1993). Multipoint linkage analysis was performed by varying the allele frequency for the allele segregating with the disease allele from 0.2 to 0.8 for the fine mapping markers.
Sequencing of candidate genes
Primers were designed for the exons and 1,000 bp upstream of the first exon (promoter region) of the KCNF1 (MIM 603787; NM_002236), ID2 (MIM 600386; NM_002166) and ATP6V1C2 (NM_144583) genes using Primer3 software (Rozen and Skaletsky 2000). DNA from hearing-impaired individuals 14 of pedigree 4052a and 12 of pedigree 4052b, plus DNA from unaffected individual 13 of family 4052b, were diluted to 5 μg/ηl, amplified by PCR under standard conditions, then purified with ExoSAP-IT (USB Corporation, Cleveland, OH, USA). Sequencing was performed with the BigDye Terminator v3.1 Cycle Sequencing Kit together with an Applied Biosystems 3700 DNA Analyzer (Applera Corporation, Foster City, CA, USA). Sequence variants were identified via Sequencher™ Version 4.1.4 software (Gene Codes Corporation, Ann Arbor, MI, USA).
Results
Two-point linkage analysis of the genome scan markers gave a maximum LOD score of 1.3 (θ=0) at marker D2S1360 for pedigree 4052a, and a LOD score of 1.5 (θ=0) at marker D2S2952 for pedigree 4052b. The maximum multipoint LOD score for the genome scan markers was 3.0 when the scores for the two branches of the family were summed, and it was obtained at marker D2S2952. In order to fine map the region on chromosome 2, twelve additional markers were selected from the Marshfield genetic map (Broman et al. 1998). Two markers (D2S1861, D2S330) were proximal to genome scan marker D2S2952, another two (D2S2164, D2S423) were between D2S2952 and genome scan marker D2S1400, while eight markers (D2S262, D2S131, D2S2267, D2S149, D2S2346, D2S272, D2S320 and D2S2375) lie distal to D2S1400. Analysis of the marker genotypes within this region with PEDCHECK and MERLIN did not elucidate any genotyping errors. Genotypes for the fine mapping markers were analyzed using two-point and multipoint linkage analysis. The maximum two-point LOD score was at marker D2S2952 with a value of 3.0 at θ=0 (Table 1). Multipoint linkage analysis gave a maximum LOD score of 4.7 at markers D2S1400 and D2S262. When the marker allele frequencies were varied for the fine mapping markers from 0.2 to 0.8, the maximum multipoint LOD score varied from 4.7 to 4.5, respectively, and remained at markers D2S1400 and D2S262. The three-unit support interval is flanked by markers D2S330 and D2S131. This region is 17.9 cM according to the Rutgers combined linkage-physical map of the human genome, and corresponds to 6.6 Mb on the sequence-based physical map (Table 1).
Table 1.
Two-point LOD score results between DFNB47 and chromosome 2 fine mapping markers
| Markera | Genetic map positionb | Physical map positionc | LOD score at θ |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0.0 | 0.01 | 0.02 | 0.04 | 0.05 | 0.1 | 0.2 | 0.3 | |||
| D2S1861 | 10.17 | 4,955,741 | –1.14 | –0.49 | –0.27 | –0.06 | 0 | 0.14 | 0.14 | 0.08 |
| D2S330 | 13.94 | 6,802,851 | – ∞ | – 3.40 | – 2.65 | – 1.87 | – 1.62 | – 0.91 | – 0.34 | – 0.13 |
| D2S2952 | 18.57 | 8,099,709 | 2.99 | 2.91 | 2.84 | 2.68 | 2.60 | 2.20 | 1.45 | 0.79 |
| D2S2164 | 19.11 | 8,248,730 | 1.12 | 1.09 | 1.05 | 0.98 | 0.94 | 0.77 | 0.45 | 0.21 |
| D2S423 | 23.72 | 9,962,317 | 2.56 | 2.49 | 2.42 | 2.28 | 2.21 | 1.87 | 1.22 | 0.65 |
| D2S1400 | 28.51 | 11,628,666 | 2.52 | 2.45 | 2.38 | 2.24 | 2.18 | 1.84 | 1.21 | 0.66 |
| D2S262 | 28.51 | 11,920,777 | 2.82 | 2.74 | 2.67 | 2.52 | 2.45 | 2.08 | 1.38 | 0.77 |
| D2S131 | 31.80 | 13,389,278 | – ∞ | – 2.71 | – 1.85 | – 1.06 | – 0.82 | – 0.18 | 0.19 | 0.21 |
| D2S2267 | 33.08 | 13,757,406 | – ∞ | –2.18 | –1.66 | –1.14 | –0.98 | –0.53 | –0.20 | –0.08 |
| D2S149 | 33.25 | 14,414,735 | – ∞ | –3.33 | –2.47 | –1.65 | –1.39 | –0.69 | –0.18 | –0.02 |
LOD scores are sums of two-point LOD scores for pedigrees 4052a and 4052b
Genome scan markers are shown in italics. Markers in bold type flank the three-unit support interval
Sex-averaged Kosambi map distance (cM) from the Rutgers combined linkage-physical map (Kong et al. 2004)
Sequence-based physical map distance in bases according to Build 34 of the human reference sequence (International Human Genome Sequence Consortium 2001)
Haplotypes were constructed to determine the critical linkage interval. A historic recombination event between markers D2S262 and D2S131 defined the centromeric boundary of this interval, and it was observed in hearing-impaired individual 13 of pedigree 4052a and individuals 11 and 12 of pedigree 4052b. All hearing-impaired individuals were homozygous at genome scan marker D2S2952, but for different alleles for each pedigree (Fig. 1). Because these two pedigrees are historically related and are thus assumed to inherit the disease haplotype from common founders, the telomeric boundary is then assigned between markers D2S2952 and D2S2164. The region of homozygosity was therefore contained within the three-unit support interval and flanked by markers D2S2952 and D2S131. This narrowed down the DFNB47 interval to a physical map distance of 5.3 Mb, and to 13.2 cM according to the Rutgers combined linkage-physical map of the human genome.
Three candidate genes within the DFNB47 interval, namely KCNF1, ID2 and ATP6V1C2, were screened in three family members, and were found to be negative for functional sequence variants.
Discussion
The linkage data presented here map DFNB47 to a 13.2 cM-interval on chromosomal region 2p25.1-p24.3, according to the Rutgers combined linkage-physical map of the human genome. Two loci for AR NSHI have previously been localized to chromosome 2, DFNB9 (2p22-p23) and DFNB27 (2q23-q31), while two autosomal dominant NSHI loci, DFNA16 (2q24) and DFNA43 (2p12), have been mapped. Only the gene for DFNB9, OTOF (MIM 603681), has been identified (Van Camp and Smith 2005). OTOF (NM_194248; 26, 654, 606-26, 756, 101) is positioned ~20 Mb distal to the DFNB47 interval according to Build 34 of the human genome sequence (International Human Genome Sequence Consortium 2001). By interpolation onto the Rutgers map, this places OTOF approximately 17 cM centromeric to DFNB47.
To date 23 known genes lie in the 5.3 Mb-region that contains DFNB47. One of the genes in this region, KCNF1, is a strong candidate for DFNB47. This gene codes for potassium voltage-gated channel subfamily F member 1. Potassium ion channels are a diverse family of plasma membrane proteins that play an essential role in various cellular processes, including maintenance of membrane potential and cell signaling (Su et al. 1997). KCNQ4 (MIM 603537), a voltage-gated K+ channel gene expressed in the cochlea, has been previously mapped to the DFNA2 locus (1p34.2) for a form of autosomal dominant NSHI (Kubisch et al. 1999). Voltage-gated K+ channel genes have been shown to be responsible for various hereditary diseases. For instance, mutations in the KVLQT1 gene (MIM 607542) (a voltage-gated K+ channel gene) result in Jervell and Lange-Nielsen syndrome (JLNS) and Long QT syndrome, which are inherited AR diseases, with congenital HI being one of their characteristics (Ilhan et al. 1999). JLNS can also result from mutations in another voltage-gated K+ channel gene, KCNE1 (MIM 176261). Although KCNF1 is the strongest candidate gene within the DFNB47 interval, this gene was screened in family 4052 and was found to be negative for sequence variants.
Another good candidate gene is inhibitor of DNA binding 2 (ID2), which is a member of the ID family of genes that promotes cell proliferation. In embryonic mouse, ID2 expression was detected in the vestibular and acoustic ganglia, and also in the epithelium of the otic vesicle and surrounding mesenchyme (Jen et al. 1997). However, no functional sequence variants were found in ID2 for family 4052. Other genes that are expressed in the inner ear (The Hearing Research Group at Brigham & Women's Hospital 2002; Holme et al. 2002) include: (1) cleavage and polyadenylation specific factor 3 (CPSF3; MIM 606029), an important regulator of viral and cellular gene expression (Calzado et al. 2004); (2) tyrosine 3/tryptophan 5-monooxygenase (YWHAQ; MIM 609009), which is also expressed in the spinal cord of patients with amyotrophic lateral sclerosis (Malaspina et al. 2000); and (3) ornithine decarboxylase 1 (ODC1; MIM 165640), the rate-limiting enzyme in polyamine synthesis.
ATP6V1C2 (ATPase, H+ transporting, lysosomal 42 kDa, V1) is an isoform of an H+-ATPase subunit (Smith et al. 2002), which has been implicated in AR distal renal tubular acidosis (dRTA). This gene was screened in eight dRTA families, two of whom had sensorineural HI, but no functional sequence variants were reported to have been found (Smith et al. 2002). Likewise, in family 4052, ATP6V1C2 had negative results after sequencing.
No functional sequence variants were identified in the three candidate genes KCNF1, ID2 and ATP6V1C2. Because only the exonic and promoter regions were sequenced, the possibility of a functional variant in the intronic regions cannot be ruled out. On the other hand, 20 other known genes and 7 expressed sequence tags that encode hypothetical proteins are contained within the DFNB47 interval. Further fine-mapping and sequencing work are required in order to identify the DFNB47 gene which causes ARNSHI.
Acknowledgements
We wish to thank the family members for their invaluable participation and cooperation. This work was supported by the Higher Education Commission, Pakistan, the American Hearing Research Foundation, and the National Institutes of Health—National Institute of Deafness and other Communication Disorders grant R01-DC03594. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, Contract Number N01-HG-65403.
Contributor Information
Muhammad Jawad Hassan, Department of Biological Sciences, Quaid-I-Azam University, Islamabad, Pakistan.
Regie Lyn P. Santos, Department of Molecular and Human Genetics, Baylor College of Medicine, One Baylor Plaza, Alkek Building N1619.01, Houston, TX 77030, USA
Muhammad Arshad Rafiq, Department of Biosciences, COMSATS Institute of Information Technology, Islamabad, Pakistan.
Maria H. Chahrour, Department of Molecular and Human Genetics, Baylor College of Medicine, One Baylor Plaza, Alkek Building N1619.01, Houston, TX 77030, USA
Thanh L. Pham, Department of Molecular and Human Genetics, Baylor College of Medicine, One Baylor Plaza, Alkek Building N1619.01, Houston, TX 77030, USA
Muhammad Wajid, Department of Biological Sciences, Quaid-I-Azam University, Islamabad, Pakistan.
Nadine Hijab, Department of Molecular and Human Genetics, Baylor College of Medicine, One Baylor Plaza, Alkek Building N1619.01, Houston, TX 77030, USA.
Michael Wambangco, Department of Molecular and Human Genetics, Baylor College of Medicine, One Baylor Plaza, Alkek Building N1619.01, Houston, TX 77030, USA.
Kwanghyuk Lee, Department of Molecular and Human Genetics, Baylor College of Medicine, One Baylor Plaza, Alkek Building N1619.01, Houston, TX 77030, USA.
Muhammad Ansar, Department of Biosciences, COMSATS Institute of Information Technology, Islamabad, Pakistan.
Kai Yan, Department of Molecular and Human Genetics, Baylor College of Medicine, One Baylor Plaza, Alkek Building N1619.01, Houston, TX 77030, USA.
Wasim Ahmad, Department of Biological Sciences, Quaid-I-Azam University, Islamabad, Pakistan.
Suzanne M. Leal, Department of Molecular and Human Genetics, Baylor College of Medicine, One Baylor Plaza, Alkek Building N1619.01, Houston, TX 77030, USA
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Broman K, Murray JC, Scheffield VC, White RL, Weber JL. Comprehensive human genetic maps: individual and sex specific variation in recombination. Am J Hum Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzado MA, Sancho R, Munoz E. Human immunodeficiency virus type 1 Tat increases the expression of cleavage and polyadenylation specificity factor 73-kilodalton subunit modulating cellular and viral expression. J Virol. 2004;78:6846–6854. doi: 10.1128/JVI.78.13.6846-6854.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham R, Indury RM, Schaffer AA. Faster sequential genetic linkage computations. Am J Hum Genet. 1993;53:252–263. [PMC free article] [PubMed] [Google Scholar]
- Freimer NB, Sandkuijl LA, Blower SM. Incorrect specification of marker allele frequencies: effects on linkage analysis. Am J Hum Genet. 1993;52:1102–1110. [PMC free article] [PubMed] [Google Scholar]
- Grimberg J, Nawoschik S, Bellusico L, McKee R, Turck A, Eisenberg A. A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res. 1999;17:83–90. doi: 10.1093/nar/17.20.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Jonasson K, Frigge ML, Kong A. Allegro, a new computer program for multipoint linkage analysis. Nat Genet. 2002;25:12–13. doi: 10.1038/75514. [DOI] [PubMed] [Google Scholar]
- Holme RH, Bussoli TJ, Steel KP. Table of gene expression in the developing ear. 2002 URL: http://www.ihr.mrc.ac.uk/hereditary/genetable/search.shtml.
- Ilhan A, Tuncer C, Komsuoglu SS, Kali S. Jervell and Lange-Nielsen syndrome: neurologic and cardiologic evaluation. Pediatr Neurol. 1999;21:809–813. doi: 10.1016/s0887-8994(99)00100-9. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequence Consortium Initial sequence and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Jen Y, Manova K, Benezra R. Each member of the Id gene family exhibits a unique expression pattern in mouse gastrulation and neurogenesis. Dev Dyn. 1997;208:92–106. doi: 10.1002/(SICI)1097-0177(199701)208:1<92::AID-AJA9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Kong X, Murphy K, Raj T, He C, White PS, Matise TC. A combined linkage-physical map of the human genome. Am J Hum Genet. 2004;75:1143–1148. doi: 10.1086/426405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubisch C, Schroeder BC, Friedrich T, Lutjohann B, El-Amraoui A, Marlin S, Petit C, Jentsch TJ. KCNQ4, a novel potassium channel expressed in sensory outer hair cells, is mutated in dominant deafness. Cell. 1999;96:437–446. doi: 10.1016/s0092-8674(00)80556-5. [DOI] [PubMed] [Google Scholar]
- Malaspina A, Kaushik N, de Belleroche J. A 14–3-3 mRNA is up-regulated in amyotrophic lateral sclerosis spinal cord. J Neurochem. 2000;75:2511–2520. doi: 10.1046/j.1471-4159.2000.0752511.x. [DOI] [PubMed] [Google Scholar]
- Morton NE. Genetic epidemiology of hearing impairment. Ann N Y Acad Sci. 1991;630:16–31. doi: 10.1111/j.1749-6632.1991.tb19572.x. [DOI] [PubMed] [Google Scholar]
- Petit C. Genes responsible for human hereditary deafness: symphony of a thousand. Nat Genet. 1996;14:385–391. doi: 10.1038/ng1296-385. [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics methods and protocols: methods in molecular biology. Humana, NJ: 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Schuler GD. Sequence mapping by electronic PCR. Genome Res. 1997;7:541–550. doi: 10.1101/gr.7.5.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AN, Borthwick KJ, Karet FE. Molecular cloning and characterization of novel tissue-specific isoforms of the human vacuolar H(+)-ATPase C, G and d subunits, and their evaluation in autosomal recessive distal renal tubular acidosis. Gene. 2002;297:169–177. doi: 10.1016/s0378-1119(02)00884-3. [DOI] [PubMed] [Google Scholar]
- Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- Steel KP, Bussoli TJ. Deafness genes: expressions of surprise. Trends Genet. 1999;15:207–211. doi: 10.1016/s0168-9525(99)01753-9. [DOI] [PubMed] [Google Scholar]
- Su K, Kyaw H, Fan P, Zeng Z, Shell BK, Carter KC, Li Y. Isolation, characterization, and mapping of two human potassium channels. Biochem Biophys Res Commun. 1997;241:675–681. doi: 10.1006/bbrc.1997.7830. [DOI] [PubMed] [Google Scholar]
- The Hearing Research Group at Brigham & Women's Hospital Human Cochlear cDNA library and EST database. 2002 URL: http://www.hearing.bwh.harvard.edu/estinfo.htm.
- Van Camp G, Smith RJH. Hereditary Hearing Loss Homepage. 2005 URL: http://www.webhost.ua.ac.be/hhh/
- Weeks DE, Sobel E, O'Connell JR, Lange K. Computer programs for multilocus haplotyping of general pedigrees. Am J Hum Genet. 1995;56:1506–1507. [PMC free article] [PubMed] [Google Scholar]