Abstract
Autosomal recessive non-syndromic hearing impairment (ARNSHI) is the most common form of prelingual inherited hearing impairment (HI). Here is described the mapping of a novel ARNSHI locus in a consanguineous Pakistani family with profound congenital HI. Two-point and multipoint linkage analyses were performed for the genome scan and fine mapping markers. Haplotypes were constructed to determine the region of homozygosity. At θ = 0, the maximum two-point LOD score of 4.0 was obtained at marker AAC040. A maximum multipoint LOD score of 5.3 was derived at marker D12S320, with the three-unit support interval demarcated by D12S89 and D12S1042. The region of homozygosity is flanked by markers D12S358 and D12S1042, which corresponds to 22.4 cM according to the Rutgers combined linkage-physical map of the human genome and spans 15.0 Mb on the sequence-based physical map. A novel ARNSHI locus DFNB62 was mapped to chromosome 12p13.2-p11.23. DFNB62 represents the second ARNSHI locus to map to chromosome 12.
Keywords: 12p13.2-p11.23, autosomal recessive non-syndromic hearing impairment, DFNB62, Pakistan
Hearing impairment (HI) is the most common hereditary sensorineural disease in humans. Among cases of hereditary non-syndromic hearing impairment (NSHI), the autosomal recessive form accounts for approximately 77% (1) and usually exhibits a more severe hearing phenotype which is prelingual in onset (2). It has been estimated that at least 1% of human proteincoding genes are involved in the hearing process (3). Thus far >60 autosomal recessive nonsyndromic hearing impairment (ARNSHI), loci have been mapped and 21 ARNSHI genes have been identified (4). In most cases, ARNSHI loci have been mapped either in families with consanguineous marriages or in endogamous populations (3). In this report, a consanguineous Pakistani family segregating a novel ARNSHI locus, DFNB62, is described. This locus was mapped to a 15 Mb region on chromosome 12p13.2-p11.23.
Materials and methods
Family history
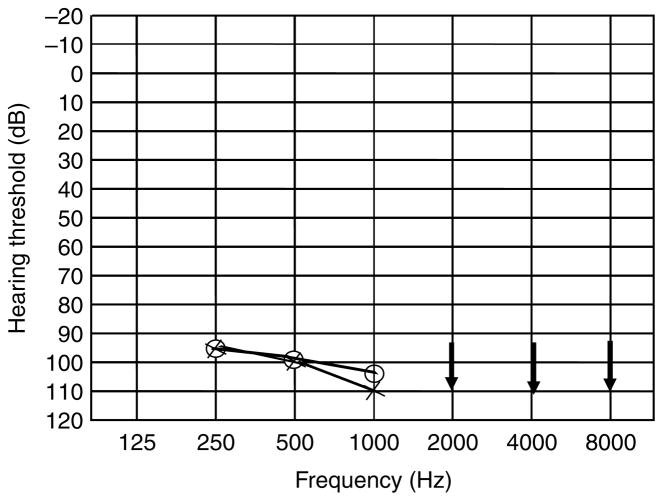
The study was approved by the Institutional Review Boards of Quaid-I-Azam University and Baylor College of Medicine. Signed informed consent was obtained from all family members who participated in the study. The family described here resides in the Punjab province of Pakistan. Owing to strict social customs, the family members rarely marry outside their community. Pedigree 4134 was constructed based on information obtained from family members. All affected individuals have a history of prelingual profound HI involving all frequencies and use sign language for communication. The transmission of HI within the family is consistent with autosomal recessive inheritance (Fig. 1). Regardless of age, affected family members display the same level of profound HI, implying that the hearing loss is not progressive. Medical history and physical examination of the affected individuals were performed by trained otolaryngologists affiliated with government hospitals. The hearing-impaired individuals underwent careful examination for balance problems, mental retardation, defects in ear morphology, dysmorphic facial features, eye disorders including night blindness and tunnel vision and were not found to have any syndromic features or vestibular disorders. Owing to logistic constraints, test battery for vestibular disease and temporal bone imaging are not available. The audiogram of affected individual VI-6 is shown in Fig. 2, thus demonstrating that DFNB62 causes profound HI that affects all frequencies.
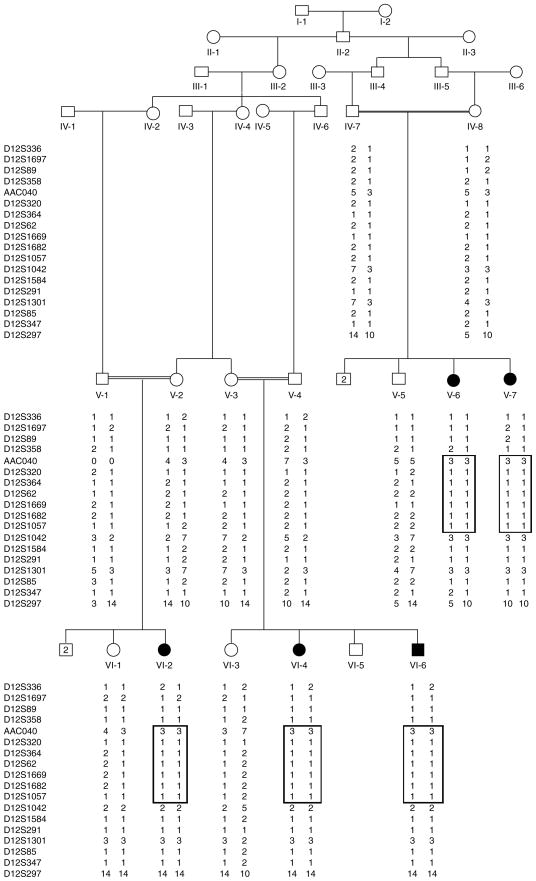
Fig. 1.
Pedigree of family 4134 with autosomal recessive non-syndromic hearing impairment due to DFNB62. Filled symbols represent hearing-impaired individuals. Clear symbols represent unaffected family members. For genotyped individuals, haplotypes are shown beneath each symbol, with the disease-associated haplotype in a box. The genotype for individual V-1 at marker AAC040 is unknown and has been designated ‘0 0’.
Fig. 2.
Audiogram of affected individual VI-6 demonstrating profound hearing impairment that involves all frequencies in both ears. Circles and crosses represent air conduction for right and left ear, respectively. Arrows indicate residual hearing at 2–8 kHz.
DNA isolation and genotyping
Genomic DNA was extracted from whole blood by a standard protocol (5), quantified by spectrophotometry at optical density 260, and diluted to 40 ηg/μt for polymerase chain reaction (PCR) amplification. A genome scan was carried out on all DNA samples at the National Heart, Lung and Blood Institute (NHLBI) Mammalian Genotyping Service (Center for Medical Genetics, Marshfield, WI). A total of 410 fluorescently labeled short tandem repeat (STR) markers were genotyped. These markers are spaced ~10 cm apart and are located on 22 autosomes and on the X and Y chromosomes. For fine mapping, PCR amplification of polymorphic STR markers was performed by using 40 ηg of genomic DNA in 25 μl reaction mixture and resolved on 8% non-denaturing polyacrylamide gel. Genotypes were assigned by visual inspection.
Linkage analysis
The pedigree and genotype data were checked with PEDCHECK (6) and MERLIN (7) software. Two-point linkage analysis was carried out using the MLINK program of the FASTLINK computer package (8). Multipoint linkage analysis was performed using ALLEGRO (9). The order of the genome scan and fine mapping markers were determined following the National Center for Biotechnology Information (NCBI) Build 34 sequence-based physical map (10), then genetic map distances were derived from the Rutgers combined linkage-physical map of the human genome (11). Haplotypes were constructed using SIMWALK2 (12,13). An autosomal recessive mode of inheritance with complete penetrance and a disease allele frequency of 0.001 were used for the analysis. For the genome scan markers, allele frequencies were estimated from the founders and reconstructed genotypes of founders from pedigree 4134 and 35 other pedigrees that underwent a genome scan at the same time. Equal allele frequencies were used for the fine mapping markers, because it was not possible to estimate allele frequencies from the founders, since these markers were only genotyped in this family. To evaluate whether false positive results were obtained because of incorrect allele frequencies (14), a sensitivity analysis was carried out. This was done by carrying out the multipoint linkage analysis using varying allele frequencies between 0.2 and 0.7 for the fine mapping markers for the alleles that was segregating with the disease allele.
Results
Two-point linkage analysis of the genome scan markers produced a maximum LOD score of 4.0 (θ = 0) at marker AAC040 while the maximum multipoint LOD score was 4.8 at the same marker. To fine-map the DFNB62 locus, 31 additional markers were selected from the Marshfield (15) and deCode genetic maps (16). Nineteen of these markers were informative for linkage (Table 1). After genotyping these markers, the data was reanalyzed using two-point and multipoint linkage analysis. The maximum two-point LOD score of 4.0 (θ = 0) remained at marker AAC040 (Table 1). A maximum multipoint LOD score of 5.3 was achieved at D12S320. The three-unit support interval extended from marker D12S89 to marker D12S1042, spanning a 24.3-cM region according to the Rutgers combined linkage-physical map of the human genome (11). This interval includes 15.7 Mb on the sequence-based physical map (10).
Table 1.
Two point LOD score results between the DFNB62 locus and chromosome 12 markers
| Markera | Genetic map positionb | Physical map positionc | LOD score at θ = |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0.0 | 0.01 | 0.02 | 0.04 | 0.05 | 0.1 | 0.2 | 0.3 | |||
| D12S336 | 24.51 | 9,385,296 | −4.60 | −2.16 | −1.62 | −1.09 | −0.92 | −0.47 | −0.14 | −0.05 |
| D12S1697 | 26.72 | 11,685,470 | −3.47 | −1.19 | −0.67 | −0.20 | −0.07 | 0.25 | 0.30 | 0.18 |
| D12S89 | 27.00 | 11,793,257 | −1.43 | −0.37 | −0.12 | 0.10 | 0.16 | 0.29 | 0.27 | 0.17 |
| D12S358 | 28.89 | 12,530,590 | 1.75 | 1.70 | 1.65 | 1.55 | 1.50 | 1.26 | 0.82 | 0.44 |
| AAC040 | 32.19 | 13,065,278 | 4.02 | 3.94 | 3.85 | 3.67 | 3.58 | 3.13 | 2.21 | 1.30 |
| D12S320 | 32.19 | 13,513,310 | 1.15 | 1.12 | 1.08 | 1.01 | 0.98 | 0.82 | 0.53 | 0.29 |
| D12S364 | 32.19 | 13,724,570 | 1.65 | 1.60 | 1.56 | 1.47 | 1.42 | 1.21 | 0.80 | 0.45 |
| D12S62 | 34.32 | 15,219,370 | 2.31 | 2.24 | 2.18 | 2.06 | 2.00 | 1.71 | 1.14 | 0.63 |
| D12S1669 | 37.83 | 19,429,661 | 1.62 | 1.58 | 1.53 | 1.44 | 1.40 | 1.18 | 0.77 | 0.42 |
| D12S1682 | 40.34 | 20,570,897 | 2.31 | 2.24 | 2.18 | 2.05 | 1.98 | 1.67 | 1.06 | 0.53 |
| D12S1057 | 46.46 | 24,568,387 | 2.31 | 2.24 | 2.18 | 2.06 | 2.00 | 1.71 | 1.14 | 0.63 |
| D12S1042 | 51.27 | 27,538,599 | −∞ | 0.29 | 0.69 | 1.07 | 1.17 | 1.36 | 1.18 | 0.78 |
| D12S1584 | 55.41 | 31,610,488 | 2.31 | 2.24 | 2.18 | 2.06 | 2.00 | 1.71 | 1.14 | 0.63 |
| D12S291 | 59.08 | 41,688,390 | 1.55 | 1.51 | 1.48 | 1.40 | 1.36 | 1.17 | 0.80 | 0.48 |
| D12S1301 | 59.08 | 42,348,809 | −∞ | 2.69 | 2.91 | 3.03 | 3.03 | 2.87 | 2.21 | 1.41 |
| D12S85 | 61.52 | 45,622,954 | 2.98 | 2.91 | 2.84 | 2.69 | 2.61 | 2.24 | 1.52 | 0.86 |
| D12S347 | 65.70 | 50,298,255 | 1.58 | 1.53 | 1.49 | 1.41 | 1.37 | 1.17 | 0.80 | 0.46 |
| D12S297 | 66.82 | 50,899,108 | −∞ | −1.86 | −1.15 | −0.46 | −0.25 | 0.28 | 0.49 | 0.38 |
Markers in bold type flank the haplotype. Genome scan markers are shown in italics.
Cumulative sex-averaged Kosambi genetic map distances (cM) from the Rutgers combined linkage-physical map of the human genome (11).
Sequence-based physical map distances in bases according to Build 34 of the human reference sequence (10).
Using SIMWALK2, haplotypes were constructed to determine the critical recombination events (Fig. 1). The disease haplotype (region of homozygosity) is flanked by markers D12S358 and D12S1042 and is smaller than the three-unit support interval. It is 22.4 cM long and contains 15.0 Mb. The critical recombination defining the cosegregating interval occurred in the affected individuals. The telomeric boundary of this interval was defined by a recombination between markers D12S358 and AAC040 observed in individual V-6. The affected individuals in both branches of the family were homozygous at genome scan marker D12S1042 but for different alleles in each family branch. Therefore the centromeric boundary of the region of homozygosity was assigned between markers D12S1057 and D12S1042.
Discussion
Large consanguineous families such as those found in the Pakistani population have been instrumental in mapping autosomal recessive HI loci. In this study, evidence for linkage was found in a consanguineous Pakistani family to the novel HI locus, DFNB62. The DFNB62 interval was mapped to a 22.4-cM region on chromosome 12p13.2-p11.23. Four loci for NSHI, DFNB50 (12q23) (4), DFNA25 (12q22-q24.11) (17), DFNA41 (12q24.32-qter) (18), and DFNA48 (12q13.13-q15) (19,20), have been previously mapped on chromosome 12. For these four loci, only one gene, MYO1A, has been identified for DFNA48 (20). MYO1A (MIM 601478) is 28.2 Mb distal to the DFNB62 locus and was excluded as the cause of ARNSHI in this family by both linkage analysis and direct sequencing.
Currently, the DFNB62 linkage interval contains 75 known genes, 15 of which encode hypothetical proteins. Among the known genes is a gene for Keutel syndrome, which is caused by mutations in matrix Gla protein (MGP; MIM 154870). Patients with Keutel syndrome are found mostly in consanguineous families and have the following as cardinal features: midface hypoplasia, flat nasal bridge, cartilage calcification, and brachytelephalangism or ‘drumstick’ fingers (21). Additional clinical signs include heart defects (e.g., pulmonary artery stenosis), developmental delay, and respiratory abnormalities. About 70% of cases report HI with or without otitis media. Family 4134 was screened for the MGP gene through direct sequencing and no functional variants were discovered.
Besides MGP, there are at least nine other genes in the DFNB62 interval that are expressed in the inner ear (22). Notable among these is EMP1 (epithelial membrane protein 1; MIM 602333), a member of the peripheral myelin protein 22 (PMP22) family. Mutations in PMP22 (MIM 601097) have been shown to cause autosomal dominant Charcot-Marie-Tooth (CMT) disease with deafness (23), presumably due to dysfunctional Schwann cells thus resulting in hypomyelination of the auditory nerve. In one CMT family, otoacoustic emission testing suggested cochlear abnormalities in addition to the auditory nerve dysfunction seen with auditory brainstem response (24). EMP1 has not been associated with inner ear disorders, but it is highly expressed in the developing murine nervous system including the peripheral (cranial) nerves (25). The EMP1 gene was negative for functional variants in one unaffected and two affected individuals from family 4134.
Identification of the DFNB62 gene will enhance our knowledge of the molecular mechanisms behind the HI phenotype.
Acknowledgments
We thank the family members for their invaluable participation and cooperation. The work was funded by the Higher Education Commission, Islamabad, Pakistan, the American Hearing Research Foundation, the NIH National Institute of Deafness and other Communication Disorders grant R01-DC03594. Genotyping services were provided by the NHLBI Mammalian Genotyping Service.
References
- 1.Morton NE. Genetic epidemiology of hearing impairment. Ann N Y Acad Sci. 1991;630:16–31. doi: 10.1111/j.1749-6632.1991.tb19572.x. [DOI] [PubMed] [Google Scholar]
- 2.Cohen MM, Gorlin RJ. Epidemiology, etiology and genetic patterns. In: Gorlin RJ, Toriello HV, Cohen MM, editors. Hereditary hearing loss and its syndromes. Oxford: Oxford University Press; 1995. pp. 43–61. [Google Scholar]
- 3.Friedman TB, Griffith AJ. Human nonsyndromic sensorineural deafness. Annu Rev Genomics Hum Genet. 2003;4:341–402. doi: 10.1146/annurev.genom.4.070802.110347. [DOI] [PubMed] [Google Scholar]
- 4.Van Camp G, Smith RJH. Hereditary hearing loss homepage. Retrieved 13 January 2006, from http://webhost.ua.ac.be/hhh/
- 5.Grimberg J, Nawoschik S, Bellusico L, McKee R, Turck A, Eisenberg A. A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res. 1989;17:83–90. doi: 10.1093/nar/17.20.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin – rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 8.Cottingham R, Indury RM, Schaffer AA. Faster sequential genetic linkage computations. Am J Hum Genet. 1993;53:252–263. [PMC free article] [PubMed] [Google Scholar]
- 9.Gudbjartsson DF, Jonasson K, Frigge ML, Kong A. Allegro, a new computer program for multipoint linkage analysis. Nat Genet. 2001;25:12–13. doi: 10.1038/75514. [DOI] [PubMed] [Google Scholar]
- 10.International Human Genome Sequence Consortium. Initial sequence and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. July 2003 reference sequence as viewed in: Genome Bioinformatics Group of UC Santa Cruz. UCSC Genome Browser. Retrieved 13 January 2006, from http://genome.ucsc.edu/cgi-bin/hgGateway. [DOI] [PubMed]
- 11.Kong X, Murphy K, Raj T, He C, White PS, Matise TC. A combined linkage-physical map of the human genome. Am J Hum Genet. 2004;75:1143–1148. doi: 10.1086/426405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weeks DE, Sobel E, O’Connell JR, Lange K. Computer programs for multilocus haplotyping of general pedigrees. Am J Hum Genet. 1995;56:1506–1507. [PMC free article] [PubMed] [Google Scholar]
- 13.Sobel E, Lange K. Descent graphs in pedigree analysis. Applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- 14.Freimer NB, Sandkuijl LA, Blower SM. Incorrect specification of marker allele frequencies: effects on linkage analysis. Am J Hum Genet. 1993;52:1102–1110. [PMC free article] [PubMed] [Google Scholar]
- 15.Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong A, Gudbjartsson DF, Sainz J, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 17.Greene CC, McMillan PM, Barker SE, et al. DFNA25, a novel locus for dominant nonsyndromic hereditary hearing impairment, maps to 12q21–24. Am J Hum Genet. 2001;68:254–260. doi: 10.1086/316925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanton SH, Liang CY, Cai MW, et al. A novel locus for autosomal dominant non-syndromic deafness (DFNA41) maps to chromosome 12q24-qter. J Med Genet. 2002;39:567–570. doi: 10.1136/jmg.39.8.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D’Adamo P, Pinna M, Capobianco S, et al. A novel autosomal dominant non-syndromic deafness locus (DFNA48) maps to 12q13-q14 in a large Italian family. Hum Genet. 2003;112:319–320. doi: 10.1007/s00439-002-0880-6. [DOI] [PubMed] [Google Scholar]
- 20.Donaudy F, Ferrara A, Esposito L, et al. Multiple mutations of MYO1A, a cochlear-expressed gene, in sensori-neural hearing loss. Am J Hum Genet. 2003;72:1571–1577. doi: 10.1086/375654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hur DJ, Raymond GV, Kahler SG, Riegert-Johnson DL, Cohen BA, Boyadjiev SA. A novel MGP mutation in a consanguineous family: review of the clinical and molecular characteristics of Keutel syndrome. Am J Med Genet A. 2005;135:36–40. doi: 10.1002/ajmg.a.30680. [DOI] [PubMed] [Google Scholar]
- 22.The Hearing Research Group at Brigham & Women’s Hospital. Human cochlear cDNA library and EST database. 2002 http://hearing.bwh.harvard.edu/estinfo.HTM.
- 23.Sambuughin N, de Bantel A, McWilliams S, Sivakumar K. Deafness and CMT disease associated with a novel four amino acid deletion in the PMP22 gene. Neurology. 2003;60:506–508. doi: 10.1212/01.wnl.0000044048.27971.fc. [DOI] [PubMed] [Google Scholar]
- 24.Kovach MJ, Campbell KC, Herman K, et al. Anticipation in a unique family with Charcot–Marie–Tooth syndrome and deafness: delineation of the clinical features and review of the literature. Am J Med Genet. 2002;108:295–303. doi: 10.1002/ajmg.10223. [DOI] [PubMed] [Google Scholar]
- 25.Wulf P, Suter U. Embryonic expression of epithelial membrane protein 1 in early neurons. Brain Res Dev Brain Res. 1999;116:169–180. doi: 10.1016/s0165-3806(99)00092-9. [DOI] [PubMed] [Google Scholar]