Abstract
Objectives
Although statins are efficacious for lowering LDL-cholesterol (LDLC), there is wide inter-individual variation in response. We tested the extent to which combined effects of common alleles of LDLR and HMGCR can contribute to this variability.
Methods and Results
Haplotypes in the LDLR 3′-untranslated region (3UTR) were tested for association with lipid-lowering response to simvastatin treatment in the Cholesterol and Pharmacogenetics (CAP) trial (335 African-Americans and 609European-Americans). LDLR haplotype 5 (L5)was associated with smaller simvastatin-induced reductions in LDLC, total cholesterol, non-HDL cholesterol, and apolipoprotein B (P=0.0002–0.03)in African-Americans, but not European-Americans. The combined presence of L5 and previously described HMGCR haplotypes in African-Americans was associated with significantly attenuated apoB reduction(−22.4±1.5% N=89) both compared to noncarriers (−30.6±1.5% N=78, P=0.0001) and to carriers of either individual haplotype (−28.2±1.1% N=158, P=0.001). We observed similar differences when measuring simvastatin-mediated induction of LDLR surface expression using lymphoblast cell lines (P=0.03).
Conclusions
We have identified a common LDLR 3UTR haplotype that is associated with attenuated lipid-lowering response to simvastatin treatment. Response was further reduced in individuals with both LDLR and previously described HMGCR haplotypes. Previously identified racial differences in statin efficacy were partially explained by increased prevalence of these combined haplotypes in African-Americans.
Keywords: LDLR, HMGCR, statin, LDL-cholesterol, pharmacogenomics
Statins are prescribed for the prevention and treatment of cardiovascular disease (CVD) and work primarily to lower LDL-cholesterol (LDLC) 1–4. Despite widespread use of statins, lipid-lowering response is variable and many patients fail to meet lipid-lowering goals 5–6. Variation in response is influenced by multiple factors including age, gender, and racial ancestry 7. For example, African-Americans have on average a lesser response to a given statin dose in comparison to European-Americans 7. While common sequence variants in several candidate genes including HMGCR and APOE have been associated with alterations in lipid-lowering response to statin treatment 8–11, each of these associations accounts for a relatively small portion of the total variance in statin response. Hence the genetic contribution to variability in statin-mediated lipid reductions likely results from multiple polymorphisms with small individual effects.
Statins are competitive inhibitors of HMG-CoA reductase (HMGCR), the enzyme that catalyzes the rate-limiting step in cholesterol biosynthesis, and genetic variation in HMGCR has been associated with attenuated lipid-lowering response to statin treatment 8–9, 12–13. In the Cholesterol and Pharmacogenetics (CAP) study, we found that certain HMGCR haplotypes were associated with smaller simvastatin-mediated lipid reductions in African-Americans but not in European-Americans 9 and that genetically-influenced HMGCR alternative splicing contributed substantially to reduced statin response14.
Although reduced cholesterol synthesis may contribute to statin-mediated reduction of LDL-cholesterol (LDLC), the primary mechanism for this effect is increased plasma LDL clearance due to secondary up-regulation of LDL receptor (LDLR) expression15. Variation at the LDLR locus is known to influence LDLC levels 16–17. Rare loss of function LDLR mutations are causative for familial hypercholesterolemia and the functional severity of these mutations has been associated with statin efficacy 18–20. Common LDLR DNA polymorphisms also have been associated with inter-individual variation in LDLC as well as other lipid and lipoprotein traits 16, 21–24 and these variants are thought to mediate changes in LDLR protein expression or regulation. There is recent evidence that the 3′-untranslated region (3UTR) of LDLR is required for berberine-mediated LDLC reduction through a mechanism that increases LDLR mRNA stability 25. Single nucleotide polymorphisms (SNPs) in the 3UTR have been associated with in vivo LDLC levels as well as in vitro LDLR mRNA stability 26 and variation in this region may also influence statin-mediated lipid reduction 27. To further explore this latter possibility, we assessed the contribution of common sequence variations in the LDLR 3UTR to statin-induced changes in lipids and lipoproteins in participants of the CAP trial. We also determined the combined influence of HMGCR and LDLR DNA variation on simvastatin-mediated lipid reductions.
Methods
Study population
The CAP trial enrolled 944 participants in a 6-week simvastatin trial (40mg/d) that was designed to examine genetic factors affecting simvastatin-mediated changes in lipids and lipoproteins 7. This trial is registered at clinicaltrials.gov (NCT00451828). Participants were healthy adult volunteers who self-reported either African-American (N=335) or European-American ancestry (N=609). They were recruited at either the University of California, Los Angeles School of Medicine (Los Angeles, CA) or at San Francisco General Hospital (San Francisco, CA). Informed consent was obtained and approved by the Institutional Review boards at those institutions and included approval for future genetic studies related to statin efficacy. Baseline health and demographic information were obtained at enrollment. Fasting plasma was collected at two pre-treatment time points (screen visit and enrollment visit) and at two post-treatment time points (4 weeks and 6 weeks of treatment). Lipids and lipoproteins were measured at all four time points. Total cholesterol, triglyceride, apolipoprotein B and HDL-cholesterol were measured using an Express 550 Plus analyzer (Ciba Corning, Oberlin, Ohio) in a laboratory that was monitored for consistency by the Centers for Disease Control-National Heart, Lung, and Blood Institute standardization program as described previously 7. LDL-cholesterol was calculated by the Friedewald equation 28. Because total cholesterol, HDLC, and triglycerides were not significantly different between screen and enrollment, the average of these two measurements was used as the pretreatment value in order to minimize technical variation. For the same reason, the average of four and six week measurements was used as the post-treatment value. ApoB was only measured at enrollment and after six weeks.
Identification of tagSNPs and haplotypes
DNA variants in the LDLR 3UTR (2539bp, chr19:11,241,995–11,244,534 on the GRCh37 reference sequence) were identified by Sanger resequencing of genomic DNA samples derived from 24 African-Americans (AA) and 23 European-Americans (EA) from the Coriell repository. Overlapping segments were PCR amplified and sequenced using 5% BDT v3.1 sequencing chemistry (Applied Biosystems) in MJ Tetrad PTC 225 thermocyclers, and chromatograms were generated from these reaction products on ABI3730 capillary sequencers. Sequences were base-called using Phred and assembled into contigs using Phrap. All sequence contigs were assembled and screened using Polyphred and, for confirmation, all variant sites were visually inspected using Consed. We identified twenty-three variable sites (fifteen with minor allele frequency greater than 5%) and these were mapped using the University of California, Santa Cruz Golden Path human genome assembly as the reference sequence. Six tagSNPs were identified using LD select (r2 > 0.80 and MAF > 0.05)29–30 and these were genotyped in all CAP participants using a combination of MassArray technology (rs1433099, rs7254521, rs2738467, rs17249057; Sequenom, San Diego, CA) and Taqman assays (rs14158 and rs5742911; Applied Biosystems, Foster City, CA). The genotyping data for rs17249057 failed quality control and were not included in subsequent analyses. However, rs17249057 was in linkage disequilibrium with a second genotyped SNP, rs14158 (r2=0.72 in EA and r2=1.00 in AA). All other SNPs were in Hardy-Weinberg equilibrium as measured by χ2-test (P<0.05). SNP frequencies are listed in Supplementary Table I. No LDLR 3UTR tagSNPs were significantly associated with statin-mediated changes in lipid traits following correction for multiple testing.
Haplotypes were inferred separately in African-American and European-American participants of the CAP trial using PHASE2.0 31. Six common haplotypes (Table 1, MAF>0.05) were inferred and these were tested for associations with statin-mediated lipid reduction. Cladogram analysis was performed using TCS v1.21 32. Global association of LDLR 3UTR haplotypes were assessed by analysis of covariance both within the entire CAP population and separately in African-Americans and in European-Americans.
Table 1.
Associations of LDLR 3UTR haplotypes with statin-mediated percent change inlipidsin African-Americans.
| N | LDL-cholesterol | Total cholesterol | apoB† | nonHDLC† | ||
|---|---|---|---|---|---|---|
| LDLR L1 | Noncarriers | 286 | −35.7±4.7 | −24.8±3.4 | −27.2±0.9 | −33.8±4.4 |
| Carriers | 49 | −39.3±5.1 | −27.5±3.7 | −29.3±2.0 | −37.2±4.8 | |
| q-value* | 0.23 | 0.11 | 0.36 | 0.23 | ||
| LDLR L2 | Noncarriers | 223 | −35.7±4.7 | −24.6±3.4 | −26.9±1.0 | −33.6±4.4 |
| Carriers | 112 | −37.2±4.9 | −26.7±3.5 | −29.0±1.4 | −35.7±4.5 | |
| q-value* | 0.53 | 0.11 | 0.22 | 0.28 | ||
| LDLR L3 | Noncarriers | 140 | −35.6±4.8 | −24.7±3.5 | −25.9±1.2 | −33.5±4.5 |
| Carriers | 195 | −36.2±4.7 | −25.2±3.4 | −28.8±1.0 | −34.4±4.4 | |
| q-value* | 0.57 | 0.44 | 0.10 | 0.50 | ||
| LDLR L4 | Noncarriers | 256 | −36.1±4.7 | −25.0±3.4 | −27.7±0.9 | −34.2±4.4 |
| Carriers | 79 | −35.7±4.9 | −25.6±3.6 | −27.4±1.6 | −33.8±4.6 | |
| q-value* | 0.58 | 0.44 | 0.71 | 0.66 | ||
| LDLR L5 | Noncarriers | 207 | −36.9±4.7 | −25.9±3.4 | −29.8±1.0 | −35.2±4.4 |
| Carriers | 128 | −33.5±4.8 | −22.8±3.5 | −23.9±1.3 | −31.1±4.5 | |
| q-value* | 0.12 | 0.03 | 0.001 | 0.03 | ||
| LDLR L6 | Noncarriers | 320 | −36.1±4.7 | −25.2±3.4 | −28.0±0.9 | −34.2±4.4 |
| Carriers | 15 | −33.7±5.9 | −21.2±4.2 | −20.9±3.5 | −31.1±5.5 | |
| q-value* | 0.56 | 0.16 | 0.10 | 0.42 |
Associations were adjusted for age, sex, BMI, smoking status and HMGCR H2/H7 haplotype. Q-values represent P-values following adjustment for multiple testing within each phenotype.
Abbreviations: apoB; apolipoprotein B; nonHDLC, nonHDL-cholesterol.
Quantification of LDLR surface protein expression in lymphoblast cell lines
Lymphoblast cell lines (LCLs) were derived from all CAP participants by Epstein-Barr virus transformation of lymphocytes isolated from blood samples collected at enrollment visit 33–35. LCLs were grown at 37°C (95% O2, 5% CO2) in RPMI 1640 media (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), 500 U/ml penicillin/streptomycin, and 2nmol/L GlutaMAX (Invitrogen). Simvastatin was provided by Merck Inc. (Whitehouse Station, NJ) and was 98% converted to active form (beta-hydroxy simvastatin acid, SVA) by heating to 50°C for 1 hour in 0.1N NaOH prior to use 36. SVA concentrations were quantified by liquid chromatography-tandem mass spectrometry using a Kromasil C18 column (Keystone Scientific, Bellfonte, PA) on a Shimadzu LC-10AD HPLC system (Shimadzu, Columbia, MD) connected to a Waters Micromass Quattro LC triple quadrupole mass spectrometer (Waters, Milford, MA) 37. LCLs (N=193) derived from African-Americans were normalized to a uniform cell density and exposed to 2μM simvastatin or sham buffer for twenty-four hours 14. LDLR protein was quantified following exposures aspreviously described 25. Briefly, 4 × 106 cells were incubated with a monoclonal LDLR antibody diluted 1:50, washed, and subsequently incubated with anti-IgG-FITC labeled antibody diluted 1:400. An isotype-matched normal mouse IgG diluted 1:50 was used to quantify background fluorescence. All antibodies were purchased from Santa Cruz Biosciences(Santa Cruz, CA). Fluorescent intensity in 10,000 gated events was measured on a BD FACS Caliber.
Statistical analysis
Associations of haplotypes with lipids, lipoproteins, and LDLR expression were analyzed by analysis of covariance using the dominant model and including age, sex, BMI, and smoking status as covariates. Associations were repeated after additional adjustment for HMGCR haplotypes H2 and H7, which have been previously associated with attenuated simvastatin response in this population 9. To ensure that analyses were not influenced by nonnormal trait distributions, we also tested for associations using nonparametric analyses. Because these did not change any of the statistical findings, the results are reported using parametric analysis. For SNP analyses and haplotype analyses described in Table 2, correction for multiple testing was performed using QVALUE 38. All statistical analyses were performed using JMP version 7.0 software (SAS, Cary, NC).
Table 2. Association of LDLR 3UTR haplotype 5 (L5) with lipids in African-American and European-American CAP participants.
Values, reported in mg/dL, are mean±SEM for change adjusted for age, sex, BMI, smoking status, and HMGCR haplotype.
| N | Baseline | Treatment | Change | Percent change | ||
|---|---|---|---|---|---|---|
| African-Americans | ||||||
| ApoB | Noncarriers | 207 | 87.1±6.4 | 64.7±1.3 | −28.3±1.0 | −29.8±1.0 |
| Carriers | 128 | 83.9±6.5 | 67.9±1.6 | −21.7±1.3 | −23.9±1.3 | |
| p-value | 0.13 | 0.09 | <0.0001 | 0.0002 | ||
| TC | Noncarriers | 207 | 206.0±11.2 | 152.0±9.3 | −54.9±8.1 | −25.9±3.4 |
| Carriers | 128 | 206.3±11.5 | 158.3±9.6 | −47.7±8.3 | −22.8±3.5 | |
| p-value | 0.96 | 0.07 | 0.02 | 0.008 | ||
| LDLC | Noncarriers | 207 | 132.6±11.1 | 83.4±8.7 | −49.2±7.5 | −36.9±4.7 |
| Carriers | 128 | 130.0±11.4 | 86.1±9.0 | −43.9±7.7 | −33.5±4.8 | |
| p-value | 0.49 | 0.45 | 0.03 | 0.03 | ||
| nonHDLC | Noncarriers | 207 | 151.7±11.9 | 97.8±9.6 | −53.8±8.0 | −35.2±4.4 |
| Carriers | 128 | 148.1±12.2 | 101.7±9.8 | −46.4±8.2 | −31.1±4.5 | |
| p-value | 0.38 | 0.23 | 0.006 | 0.006 | ||
| European-Americans | ||||||
| ApoB | Noncarriers | 567 | 95.7±1.8 | 68.6±1.4 | −27.2±1.2 | −27.5±1.0 |
| Carriers | 40 | 102.3±3.8 | 72.7±2.9 | −30.3±2.7 | −27.6±2.2 | |
| p-value | 0.07 | 0.21 | 0.23 | 0.96 | ||
| TC | Noncarriers | 567 | 211.3±2.7 | 153.4±2.2 | −58.0±1.9 | −26.9±0.7 |
| Carriers | 40 | 218.6±5.9 | 158.0±4.6 | −60.6±4.0 | −26.8±1.5 | |
| p-value | 0.20 | 0.28 | 0.49 | 0.70 | ||
| LDLC | Noncarriers | 567 | 132.0±2.5 | 76.6±1.8 | −55.4±1.7 | −41.6±0.9 |
| Carriers | 40 | 136.9±5.4 | 77.9±3.9 | −59.0±3.7 | −41.8±2.0 | |
| p-value | 0.34 | 0.73 | 0.30 | 0.90 | ||
| nonHDLC | Noncarriers | 567 | 158.9±2.8 | 99.0±2.1 | −59.8±1.9 | −37.2±0.9 |
| Carriers | 40 | 167.1±6.2 | 104.3±4.5 | −62.8±4.0 | −36.2±1.9 | |
| p-value | 0.16 | 0.22 | 0.4 | 0.45 | ||
Results
Association of LDLR 3UTR L5 haplotype with in vivo simvastatin-mediated lipid reductions and in vitro LDLR expression
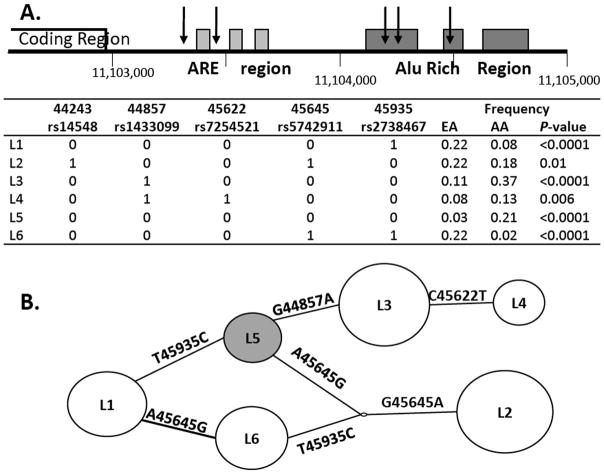
LDLR 3UTR haplotypes were inferred based on tagSNPs selected to cover DNA variation across the entire 3UTR including the AU-rich elements (ARE) and alu-rich regions (Figure 1A). For all six haplotypes, allelic prevalence was significantly different between African-Americans and European-Americans (Figure 1A). LDLR 3UTR haplotype 5 (LDLR L5) was inferred to contain the major allele at all tagSNPs and was more prevalent in African-Americans (Figure 1A and 1B). The alleles represented by this haplotype were also observed in the chimpanzee genome, indicating that they represent the ancestral alleles. Although individual SNPs were not associated with statin-mediated lipid changes in either the CAP population as a whole or within the African-Americans or European-Americans separately, haplotypes within this region were associated with statin-mediated changes in lipids and lipoproteins using an omnibus haplotype test. Within African-Americans, LDLR 3UTR haplotypes were globally associated with statin-mediated changes in apoB (global variance: r2=0.07, P=0.0004), LDLC (r2=0.04, P=0.04), total cholesterol (r2=0.05, P=0.01), and nonHDL-cholesterol (r2=0.05, P=0.0009) by omnibus haplotype test.
Figure 1. LDLR 3UTR SNPs and haplotypes.
A. Inclusion of LDLR 3UTR tagSNPs in inferred haplotypes. Schematic representation of LDLR 3UTR is shown with five tagSNPs indicated by arrows. Chromosomal position labeled is based on genome assembly March 2006. Major alleles are designated by 0 (G, 44243; G, 44857; C, 45622; A, 45645; C, 45935) and minor alleles by 1. B. Cladogram of LDLR 3UTR haplotypes. LDLR 3UTR haplotype 5 (L5) is, shown in grey, was represented by the ancestral allele at all five tagSNPs.
We next assessed the associations of individual LDLR 3UTR haplotypes with statin-mediated lipid changes. There was no evidence for an association of the LDLR 3UTR haplotypes with statin response among European-Americans. Among African-Americans, we observed a significant association of LDLR L5 with attenuated statin-mediated percent changes in TC, apoB, and nonHDLC (Table 1). ApoB reductions were lower in both homozygous LDLR L5 carriers (−19.2±3.8mg/dL apoB N=14, P=0.02) and in heterozygous LDLR L5 carriers (−21.4±1.4mg/dL apoB N=114, P<0.0001) in comparison to noncarriers (−29.2±1.1mg/dL apoB N=207, P-value for trend: 0.0004). Similar associations were observed for TC, LDLC and nonHDLC(data not shown). Associations of LDLR L5 with attenuated statin-mediated changes in all four traits remained significant after adjusting for the previously identified HMGCR haplotypes (H2 and H7) (Table 1) 9. LDLR L5carriers demonstrated a nonsignificant trend towards reduced pre-treatment and elevated post-treatment concentrations for all four traits (Table 2). In European-Americans, no association of LDLR L5 was observed with either pre-treatment lipids or statin-mediated changes in these traits (Table 2).
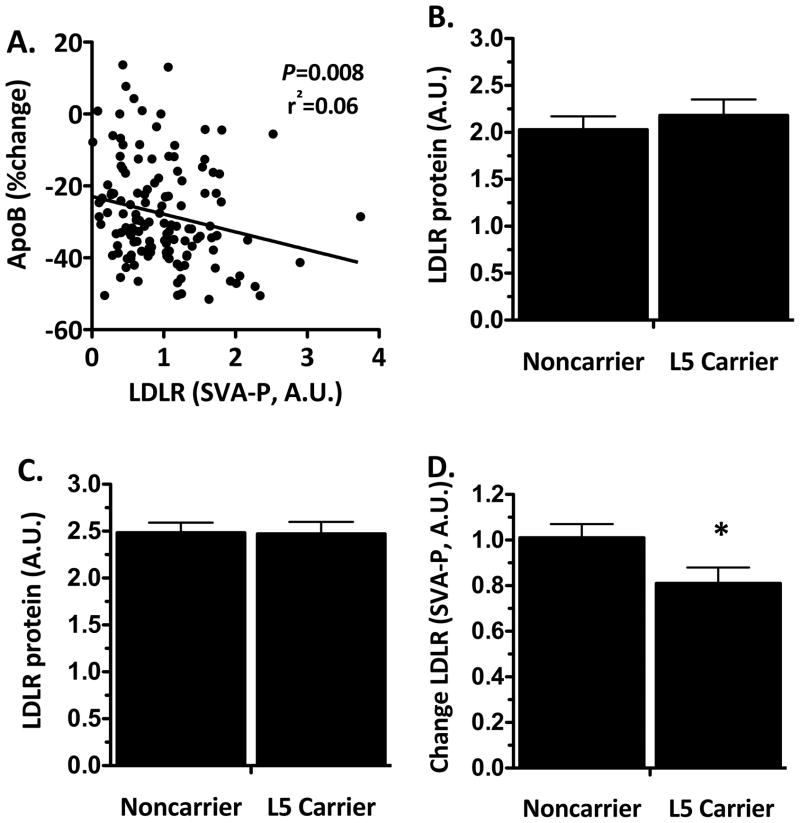
To identify the basis for these findings, we tested for an association of LDLR L5 with change in expression of LDLR surface protein in response to in vitro simvastatin exposure in lymphoblast cell lines derived from African-American CAP participants. LDLR surface protein expression was significantly induced in response to simvastatin exposure (1.45±0.06 fold change, N=193, P=0.002) in a manner that was correlated with in vivo statin-mediated decrease in apoB in the individuals from whom these cell lines were derived (Figure 2A). LDLR surface protein expression was not associated with LDLR L5 following exposure to either sham buffer or simvastatin(Figure 2B and 2C). However, LDLR induction was significantly reduced in cell lines derived from LDLR L5 carriers in comparison to noncarriers (Figure 2D).
Figure 2. Correlation of simvastatin-mediated induction in in vitro LDLR surface expression with in vivo apolipoprotein B reduction and association with LDLR L5 haplotype.
(A) Correlation of in vitro LDLR surface protein induction (simvastatin treated – placebo treated, A.U.) and in vivo apolipoprotein B reductions (simvastatin – baseline, % change) in response to simvastatin exposure. LDLR surface protein expression was measured by FACS following 24 hour exposure to (B) sham buffer or (C) 2μM simvastatin in lymphoblast cell lines derived from 109 LDLR L5 noncarrier and84 LDLR L5 carrier African-American CAP participants.(D) Statin-mediated change in LDLR protein expression is presented as LDLR expression following simvastatin exposure minus LDLR expression following sham exposure(P=0.03 adjusted for age, sex, BMI and smoking status).
Combined influence of LDLR L5 and HMGCR H2/H7 haplotypes on in vivo and in vitro simvastatin response
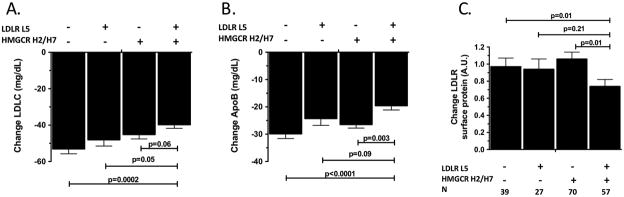
We have previously observed that, similar to the findings for the LDLR L5 haplotype reported here, HMGCR H2 and H7 haplotypes were also associated with reduced LDL response to simvastatin in African Americans. This raises the possibility of additive effects of LDLR L5 and HMGCR H2 or H7(H2/H7)on simvastatin response. Indeed, we observed that carriers of both haplotypes (L5+H2/H7) had significantly attenuated LDLC reductions in comparison to carriers of only the LDLR L5 haplotype (P=0.05) and in comparison to carriers of neither haplotype (P=0.0002, Figure 3A) with a similar trend observed in comparison to HMGCR H2/H7 only carriers (P=0.06). Carriers of both alleles also demonstrated significantly lower simvastatin-mediated apoB reductions in comparison to HMGCR H2/H7 only carriers (P=0.03) and in comparison to carriers of neither haplotype ((P<0.0001, Figure 3B) with a nonsignificant trend in comparison to LDLR L5 carriers (P=0.09). We tested for association of the combined L5+H2/H7 haplotype with in vitro LDLR induction following simvastatin exposure (Figure 3C) and found that response was significantly reduced in L5+H2/H7 carriers compared to HMGCR H2/H7 only carriers and noncarriers. No difference in LDLR induction was detected between noncarriers and carriers of either individual haplotype. This indicates that the association of LDLR L5 with attenuated LDLR induction (Figure 2) is due predominantly to the combined L5+H2/H7 carriers.
Figure 3. Combined influence of LDLR L5 and HMGCR H2/H7 haplotypes on in vivo lipid-lowering response and in vitro LDLR protein response to simvastatin exposure.
(A) LDLC and (B) apolipoprotein B were measured before and after simvastatin treatment in 324 African-American CAP participants including 78 noncarriers, 39 LDLR L5 only carriers, 119 HMGCR H2/H7 only carriers, and 89 LDLR L5 + HMGCR H2/H7 carriers. (C)LDLR surface protein was measured following 24 hour exposure with 2mM simvastatin or sham buffer in 193 lymphoblast cell lines derived from African-American CAP participants. Associations adjusted for age, sex, BMI and smoking status. P-values for trends are (A) 0.002, (B) <0.0001, and (C) 0.05.
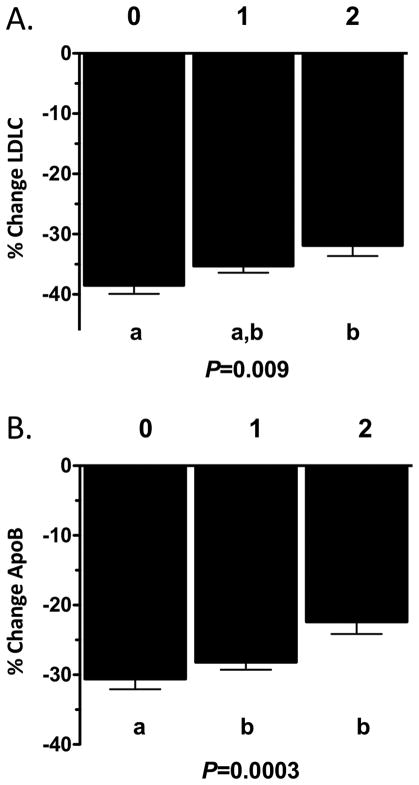
We next assessed whether inclusion of both haplotypes into a combined model would be predictive of statin response in African-Americans. We categorized individuals by lipid-lowering response as carriers of 2 haplotypes (both LDLR L5 and HMGCR H2/H7), 1 haplotype(either the LDLR L5 or HMGCR H2/H7 haplotypes) or 0 haplotypes (neither haplotype). We observed a significant additive association of LDLR/HMGCR haplotypes with statin-induced reduction of LDLC and apoB (Figure 4) as well as TC and nonHDLC (data not shown). In support of these observations, in vitro LDLR protein induction in response to simvastatin was reduced in combined L5+H2/H7 carriers (0.74±0.09A.U. N=57 two haplotype carriers vs. 1.02±0.06 A.U. N=97 one haplotype carriers vs. 0.97±0.10A.U. N=39 noncarriers, P=0.03). These associations were not observed in European-American CAP participants (data not shown), in whom the prevalence of the combined haplotypes was significantly lower (N=89 African-Americans (14.5% prevalence) vs. N=3 European-Americans(<1.0% prevalence), P<0.0001). The previously observed differences between African-Americans and European-Americans in simvastatin-mediated LDLC reduction (−35.7±4.4% AA vs. −38.8±4.4% EA, P=0.001) 7 were reduced to nonsignificance by inclusion of both HMGCR and LDLR haplotypes into the model (−35.6±4.2% AA vs. −36.9±4.2% EA, P=0.23), but not by individual inclusion of either HMGCR or LDLR haplotypes alone.
Figure 4. Association of combined haplotypes on (A) LDLC and (B) apoB change in response to simvastatin treatment.
0, noncarriers (N=78); 1, LDLR L5 only or HMGCR H2/H7 only carriers (N=158); 2, LDLR L5 + HMGCR H2/H7 carriers (N=89). Associations adjusted for age, sex, BMI, and smoking status.
Discussion
In this study, we have found that DNA variation in the 3UTR of LDLR is associated with diminished effects of statin on in vivo lipid reductions and on in vitro LDLR induction. We further demonstrated that there is a combined influence on statin response of the L5 LDLR 3UTR haplotype and the HMGCR haplotypes that we have previously shown to be associated with statin efficacy, such that carriers of both haplotypes had significantly attenuated statin-mediated changes in LDLC and LDLR in comparison to either noncarriers or carriers of individual haplotypes. Furthermore, these effects were more evident in African-Americans than in European-Americans. Although this may be the result of differences between African-Americans and European-Americans in the underlying sequence variation that is tagged by these haplotypes, it is more likely the result of differences in haplotype prevalence. Indeed, these haplotypes were significantly more prevalent in African-Americans than in European-Americans for both HMGCR (32% AA vs. 2% EA, P<0.0001) 9 and LDLR (21% AA vs. 3% EA, P<0.0001). Interestingly, both LDLR and HMGCR haplotypes were specifically represented by the ancestral alleles at all tagSNPs, suggesting that they represent the ancestral haplotypes 9. Furthermore, the combination of both LDLR and HMGCR haplotypes was most strongly associated with attenuated response and this combined genotype was practically absent in the European-Americans within this study population (prevalence <1%).
The LDLR 3UTR haplotypes describe common variation that spans the entire LDLR 3UTR including the proximal AU-rich elements (AREs) and the distal Alu-rich region (ARR). The AREs have been shown to be involved in mediating the LDLC reduction induced by the herbal product, berberine 25. This region promotes mRNA decay and binds to multiple proteins including several involved in mRNA processing and splicing 39–40. In addition to the ARE region, LDLR L5 is also represented by the ancestral alleles of SNPs located within the ARR. This region is responsible for transcriptional stability in response to protein kinase C (PKC) activation 39. The ARR is a rapidly mutagenized genomic region but the influence on LDLR function of SNPs within this region has not been previously studied 41–42. The present study has demonstrated that the LDLR 3UTR ancestral haplotype, which is most prevalent in African-Americans, is associated with attenuated statin response. Because LDLR L5 was represented by ancestral alleles at all tagSNPs, these data suggest that multiple SNPs within the region may modify functionality of this region. Further studies are necessary to determine the functional influence of individual SNPs within the haplotype on statin response.
Previous LDLR 3UTR association analyses focusing on the ARE region have demonstrated that the ancestral haplotype within this region was associated with increased mRNA stability, but contrary to expectation, with increased LDLC in European-American populations 26–27. Association with increased LDLC may reflect linkage disequilibrium between this region and other functional regions of the LDLR gene. Within our extended LDLR 3UTR haplotype analysis, the ancestral tagSNPs within the ARE region are represented by three haplotypes: LDLR L5, for which prevalence is low in European Americans (MAF=0.03), and LDLR L1 and L6, which are more common in European-Americans (MAF=0.22 for each). Both LDLR L1 and L6 are in linkage disequilibrium with an exonic SNP (rs688) that induces LDLR alternate splicing and is associated with increased LDLC 43. This suggests that while the ARE region may influence LDLR mRNA stability, the previously described association of this region with increased LDLC in European-Americans may reflect the functional consequences of variation outside of the 3UTR. In contrast, LDLR L5 was not in linkage disequilibrium with rs688 or any other LDLR SNPs identified by genome-wide association study (GWAS) to be associated with LDL-cholesterol (Supplementary Table II) 16, 21–24, 29, 31, 43–45. Furthermore, we saw no indication that any of these GWAS-identified SNPs were associated with statin-medicated LDLC reduction within our own GWA analysis, although they were all associated with untreated LDLC 46. Studies to assess the influence of LDLR L5 on mRNA stability and processing are ongoing.
Both LDLR L5 and the HMGCR H2/H7 haplotypes were independently associated with reduced simvastatin response specifically in African-American but not European-American CAP participants. The combination of these haplotypes was strongly associated with diminished statin response such that LDLR L5 + HMGCR H2/H7 carriers exhibited significantly attenuated lipid reductions in comparison to carriers of either individual haplotype. Reduced induction of LDLR surface protein was also observed in simvastatin-exposed lymphoblast cell lines derived from carriers of both haplotypes in comparison to cell lines derived from carriers of individual haplotypes. The contribution of these individual haplotypes to the magnitude of variation in statin response was small, with either haplotype explaining 1–4% of the variance in lipid response traits in African-Americans. However, these contributions are additive, with the combined haplotype state accounting for 3–6% of the variation in lipid-lowering simvastatin response in African-Americans. Indeed, presence of both haplotypes diminished LDLC reduction by 17.0% and apoB reduction by 27.4% in African-Americans, suggesting the need for a greater statin dosage requirement in African-American carriers, since a doubling of dose yields a ~6% further reduction in LDLC 47–48. In contrast, the combined haplotypes described less than 1% of the variance in statin response in European Americans, for whom the prevalence of the combined haplotype was much smaller (<1.0%). These data suggest that the effect of racial ancestry on lipid-lowering statin response is caused in part by differential prevalence of these LDLR and HMGCR haplotypes. Indeed, the effect of racial ancestry on lipid-lowering response to simvastatin was reduced to nonsignificance following adjustment for these haplotypes. In conclusion, these data suggest that the dissimilar simvastatin response observed between African-Americans and European-Americans is due in part to the differential prevalence of multiple sites of genetic variation that have independent but compounded effect on statin efficacy.
Supplementary Material
Acknowledgments
a) Acknowledgements – N/A
b) Sources of Funding -This work was funded by grant U01-HL69757 from the National Institutes of Health, National Center for Research Resources (NCRR) grant MO1-RR00425 (GCRC), Diabetes and Endocrinology Research Center (DERC) grant c and the Cedars-Sinai board of Governors’ Chair in Medical Genetics (JIR).
Footnotes
c) Disclosure -LMM, SP, and JIR disclose significant support from the above mentioned NIH grants. RMK discloses modest relationships with ActivX Biosciences (consultant, grant recipient), Celera (consultant), Corcept Therapeutics (consultant), Isis Pharmaceuticals (advisory board), and Merck (advisory board, speakers’ bureau) and a significant relationship with Metabolix (advisory board, grant recipient).
References
- 1.Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation And Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III) Jama. 2001;285:2486–2497. doi: 10.1001/jama.285.19.2486. [DOI] [PubMed] [Google Scholar]
- 2.Grundy SM. Approach to lipoprotein management in 2001 National Cholesterol Guidelines. Am J Cardiol. 2002;90:11i–21i. doi: 10.1016/s0002-9149(02)02631-0. [DOI] [PubMed] [Google Scholar]
- 3.Grundy SM, Cleeman JI, Merz CN, Brewer HB, Jr, Clark LT, Hunninghake DB, Pasternak RC, Smith SC, Jr, Stone NJ. Implications of recent clinical trials for the National Cholesterol Education Program Adult Treatment Panel III guidelines. Circulation. 2004;110:227–239. doi: 10.1161/01.CIR.0000133317.49796.0E. [DOI] [PubMed] [Google Scholar]
- 4.LaRosa JC, He J, Vupputuri S. Effect of statins on risk of coronary disease: a meta-analysis of randomized controlled trials. Jama. 1999;282:2340–2346. doi: 10.1001/jama.282.24.2340. [DOI] [PubMed] [Google Scholar]
- 5.American Heart Association. Heart Disease and Stroke Statistics-2006 Update. Dallas: American Heart Association; 2006. [Google Scholar]
- 6.LaRosa JC. Statins and risk of coronary heart disease. Jama. 2000;283:2935–2936. [PubMed] [Google Scholar]
- 7.Simon JA, Lin F, Hulley SB, Blanche PJ, Waters D, Shiboski S, Rotter JI, Nickerson DA, Yang H, Saad M, Krauss RM. Phenotypic predictors of response to simvastatin therapy among African-Americans and Caucasians: the Cholesterol and Pharmacogenetics (CAP) Study. Am J Cardiol. 2006;97:843–850. doi: 10.1016/j.amjcard.2005.09.134. [DOI] [PubMed] [Google Scholar]
- 8.Chasman DI, Posada D, Subrahmanyan L, Cook NR, Stanton VP, Jr, Ridker PM. Pharmacogenetic study of statin therapy and cholesterol reduction. Jama. 2004;291:2821–2827. doi: 10.1001/jama.291.23.2821. [DOI] [PubMed] [Google Scholar]
- 9.Krauss RM, Mangravite LM, Smith JD, Medina MW, Wang D, Guo X, Rieder MJ, Simon JA, Hulley SB, Waters D, Saad M, Williams PT, Taylor KD, Yang H, Nickerson DA, Rotter JI. Variation in the 3-Hydroxyl-3-Methylglutaryl-Coenzyme A Reductase gene is associated with racial differences in low-density lipoprotein cholesterol response to simvastatin treatment. Circulation. 2008;117:1537–1544. doi: 10.1161/CIRCULATIONAHA.107.708388. [DOI] [PubMed] [Google Scholar]
- 10.Thompson JF, Hyde CL, Wood LS, Paciga SA, Hinds DA, Cox DR, Hovingh GK, Kastelein JJ. Comprehensive whole-genome and candidate gene analysis for response to statin therapy in the Treating to New Targets (TNT) cohort. Circ Cardiovasc Genet. 2009;2:173–181. doi: 10.1161/CIRCGENETICS.108.818062. [DOI] [PubMed] [Google Scholar]
- 11.Mangravite LM, Wilke RA, Zhang J, Krauss RM. Pharmacogenomics of statin response. Curr Opin Mol Ther. 2008;10:555–561. [PubMed] [Google Scholar]
- 12.Thompson JF, Man M, Johnson KJ, Wood LS, Lira ME, Lloyd DB, Banerjee P, Milos PM, Myrand SP, Paulauskis J, Milad MA, Sasiela WJ. An association study of 43 SNPs in 16 candidate genes with atorvastatin response. Pharmacogenomics J. 2005;5:352–358. doi: 10.1038/sj.tpj.6500328. [DOI] [PubMed] [Google Scholar]
- 13.Donnelly LA, Doney AS, Dannfald J, Whitley AL, Lang CC, Morris AD, Donnan PT, Palmer CN. A paucimorphic variant in the HMG-CoA reductase gene is associated with lipid-lowering response to statin treatment in diabetes: a GoDARTS study. Pharmacogenet Genomics. 2008;18:1021–1026. doi: 10.1097/FPC.0b013e3283106071. [DOI] [PubMed] [Google Scholar]
- 14.Medina MW, Gao F, Ruan W, Rotter JI, Krauss RM. Alternative splicing of 3-hydroxy-3-methylglutaryl coenzyme A reductase is associated with plasma low-density lipoprotein cholesterol response to simvastatin. Circulation. 2008;118:355–362. doi: 10.1161/CIRCULATIONAHA.108.773267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reihner E, Rudling M, Stahlberg D, Berglund L, Ewerth S, Bjorkhem I, Einarsson K, Angelin B. Influence of pravastatin, a specific inhibitor of HMG-CoA reductase, on hepatic metabolism of cholesterol. N Engl J Med. 1990;323:224–228. doi: 10.1056/NEJM199007263230403. [DOI] [PubMed] [Google Scholar]
- 16.Linsel-Nitschke P, Gotz A, Erdmann J, Braenne I, Braund P, Hengstenberg C, Stark K, Fischer M, Schreiber S, El Mokhtari NE, Schaefer A, Schrezenmeir J, Rubin D, Hinney A, Reinehr T, Roth C, Ortlepp J, Hanrath P, Hall AS, Mangino M, Lieb W, Lamina C, Heid IM, Doering A, Gieger C, Peters A, Meitinger T, Wichmann HE, Konig IR, Ziegler A, Kronenberg F, Samani NJ, Schunkert H. Lifelong reduction of LDL-cholesterol related to a common variant in the LDL-receptor gene decreases the risk of coronary artery disease--a Mendelian Randomisation study. PLoS One. 2008;3:e2986. doi: 10.1371/journal.pone.0002986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francke U, Brown MS, Goldstein JL. Assignment of the human gene for the low density lipoprotein receptor to chromosome 19: synteny of a receptor, a ligand, and a genetic disease. Proc Natl Acad Sci U S A. 1984;81:2826–2830. doi: 10.1073/pnas.81.9.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown MS, Goldstein JL. Expression of the familial hypercholesterolemia gene in heterozygotes: mechanism for a dominant disorder in man. Science. 1974;185:61–63. doi: 10.1126/science.185.4145.61. [DOI] [PubMed] [Google Scholar]
- 19.Goldstein JL, Dana SE, Brunschede GY, Brown MS. Genetic heterogeneity in familial hypercholesterolemia: evidence for two different mutations affecting functions of low-density lipoprotein receptor. Proc Natl Acad Sci U S A. 1975;72:1092–1096. doi: 10.1073/pnas.72.3.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choumerianou DM, Dedoussis GV. Familial hypercholesterolemia and response to statin therapy according to LDLR genetic background. Clin Chem Lab Med. 2005;43:793–801. doi: 10.1515/CCLM.2005.134. [DOI] [PubMed] [Google Scholar]
- 21.Chasman DI, Pare G, Zee RYL, Parker AN, Cook NR, Buring JE, Kwiatkowski DJ, Rose LM, Smith JD, Williams PT, Rieder MJ, Rotter JI, Nickerson DA, Krauss RM, Miletich JP, Ridker PM. Genetic loci associated with plasma concentration of low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, triglycerides, apolipoprotein A1, and apolipoprotein B among 6382 white women in genome-wide analysis with replication. Circ Cardiovasc Genet. 2008;1:21–30. doi: 10.1161/CIRCGENETICS.108.773168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, Hirschhorn JN, Berglund G, Hedblad B, Groop L, Altshuler DM, Newton-Cheh C, Orho-Melander M. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 24.Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, Voight BF, Bonnycastle LL, Jackson AU, Crawford G, Surti A, Guiducci C, Burtt NP, Parish S, Clarke R, Zelenika D, Kubalanza KA, Morken MA, Scott LJ, Stringham HM, Galan P, Swift AJ, Kuusisto J, Bergman RN, Sundvall J, Laakso M, Ferrucci L, Scheet P, Sanna S, Uda M, Yang Q, Lunetta KL, Dupuis J, de Bakker PI, O’Donnell CJ, Chambers JC, Kooner JS, Hercberg S, Meneton P, Lakatta EG, Scuteri A, Schlessinger D, Tuomilehto J, Collins FS, Groop L, Altshuler D, Collins R, Lathrop GM, Melander O, Salomaa V, Peltonen L, Orho-Melander M, Ordovas JM, Boehnke M, Abecasis GR, Mohlke KL, Cupples LA. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong W, Wei J, Abidi P, Lin M, Inaba S, Li C, Wang Y, Wang Z, Si S, Pan H, Wang S, Wu J, Li Z, Liu J, Jiang JD. Berberine is a novel cholesterol-lowering drug working through a unique mechanism distinct from statins. Nat Med. 2004;10:1344–1351. doi: 10.1038/nm1135. [DOI] [PubMed] [Google Scholar]
- 26.Muallem H, North KE, Kakoki M, Wojczynski MK, Li X, Grove M, Boerwinkle E, Wilhelmsen KC, Heiss G, Maeda N. Quantitative effects of common genetic variations in the 3′UTR of the human LDL-receptor gene and their associations with plasma lipid levels in the Atherosclerosis Risk in Communities study. Hum Genet. 2007;121:421–431. doi: 10.1007/s00439-007-0327-1. [DOI] [PubMed] [Google Scholar]
- 27.Polisecki E, Muallem H, Maeda N, Peter I, Robertson M, McMahon AD, Ford I, Packard C, Shepherd J, Jukema JW, Westendorp RG, de Craen AJ, Buckley BM, Ordovas JM, Schaefer EJ. Genetic variation at the LDL receptor and HMG-CoA reductase gene loci, lipid levels, statin response, and cardiovascular disease incidence in PROSPER. Atherosclerosis. 2008;200:109–114. doi: 10.1016/j.atherosclerosis.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- 29.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howie BN, Carlson CS, Rieder MJ, Nickerson DA. Efficient selection of tagging single-nucleotide polymorphisms in multiple populations. Hum Genet. 2006;120:58–68. doi: 10.1007/s00439-006-0182-5. [DOI] [PubMed] [Google Scholar]
- 31.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 33.Anderson MA, Gusella JF. Use of cyclosporin A in establishing Epstein-Barr virus-transformed human lymphoblastoid cell lines. In Vitro. 1984;20:856–858. doi: 10.1007/BF02619631. [DOI] [PubMed] [Google Scholar]
- 34.Boyum A. Separation of white blood cells. Nature. 1964;204:793–794. doi: 10.1038/204793a0. [DOI] [PubMed] [Google Scholar]
- 35.Pressman S, Rotter JI. Epstein-Barr virus transformation of cryopreserved lymphocytes: prolonged experience with technique. Am J Hum Genet. 1991;49:467. [PMC free article] [PubMed] [Google Scholar]
- 36.Brown MS, Faust JR, Goldstein JL, Kaneko I, Endo A. Induction of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity in human fibroblasts incubated with compactin (ML-236B), a competitive inhibitor of the reductase. J Biol Chem. 1978;253:1121–1128. [PubMed] [Google Scholar]
- 37.Zhao JJ, Xie IH, Yang AY, Roadcap BA, Rogers JD. Quantitation of simvastatin and its beta-hydroxy acid in human plasma by liquid-liquid cartridge extraction and liquid chromatography/tandem mass spectrometry. J Mass Spectrom. 2000;35:1133–1143. doi: 10.1002/1096-9888(200009)35:9<1133::AID-JMS42>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 38.Storey JD. A direct approach to false discovery rates. J R Statist Soc B. 2002;64:479–498. [Google Scholar]
- 39.Wilson GM, Vasa MZ, Deeley RG. Stabilization and cytoskeletal-association of LDL receptor mRNA are mediated by distinct domains in its 3′ untranslated region. J Lipid Res. 1998;39:1025–1032. [PubMed] [Google Scholar]
- 40.Li H, Chen W, Zhou Y, Abidi P, Sharpe O, Robinson WH, Kraemer FB, Liu J. Identification of mRNA binding proteins that regulate thestability of LDL receptor mRNA through AU-rich elements. J Lipid Res. 2009;50:820–831. doi: 10.1194/jlr.M800375-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fagundes NJ, Salzano FM, Batzer MA, Deininger PL, Bonatto SL. Worldwide genetic variation at the 3′-UTR region of the LDLR gene: possible influence of natural selection. Ann Hum Genet. 2005;69:389–400. doi: 10.1046/j.1529-8817.2005.00163.x. [DOI] [PubMed] [Google Scholar]
- 42.Heller AH, Salzano FM, Barrantes R, Krylov M, Benevolenskaya L, Arnett FC, Munkhbat B, Munkhtuvshin N, Tsuji K, Hutz MH, Carnese FR, Goicoechea AS, Freitas LB, Bonatto SL. Intra-and intercontinental molecular variability of an Alu insertion in the 3′ untranslated region of the LDLR gene. Hum Biol. 2004;76:591–604. doi: 10.1353/hub.2004.0056. [DOI] [PubMed] [Google Scholar]
- 43.Zhu H, Tucker HM, Grear KE, Simpson JF, Manning AK, Cupples LA, Estus S. A common polymorphism decreases low-density lipoprotein receptor exon 12 splicing efficiency and associates with increased cholesterol. Hum Mol Genet. 2007;16:1765–1772. doi: 10.1093/hmg/ddm124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, Wahlstrand B, Hedner T, Corella D, Tai ES, Ordovas JM, Berglund G, Vartiainen E, Jousilahti P, Hedblad B, Taskinen MR, Newton-Cheh C, Salomaa V, Peltonen L, Groop L, Altshuler DM, Orho-Melander M. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 46.Barber MJ, Mangravite LM, Hyde CL, Chasman DI, Smith JD, McCarty CA, Li X, Wilke RA, Rieder MJ, Williams PT, Ridker PM, Chatterjee A, Rotter JI, Nickerson DA, Stephens M, Krauss RM. Genome-wide association of lipid-lowering response to statins in combined study populations. PLoS One. 2010;5:e9763. doi: 10.1371/journal.pone.0009763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones P, Kafonek S, Laurora I, Hunninghake D. Comparative dose efficacy study of atorvastatin versus simvastatin, pravastatin, lovastatin, and fluvastatin in patients with hypercholesterolemia (the CURVES study) Am J Cardiol. 1998;81:582–587. doi: 10.1016/s0002-9149(97)00965-x. [DOI] [PubMed] [Google Scholar]
- 48.Roberts WC. The rule of 5 and the rule of 7 in lipid-lowering by statin drugs. Am J Cardiol. 1997;80:106–107. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.