Abstract
Background
Translation termination is mediated through an interaction between the release factors eRF1 and eRF3 and the stop codon within its nucleotide context. Although it is well known that the nucleotide contexts both upstream and downstream of the stop codon, can modulate readthrough, little is known about the mechanisms involved.
Results
We have performed an in vivo analysis of translational readthrough in mouse cells in culture using a reporter system that allows the measurement of readthrough levels as low as 10-4. We first quantified readthrough frequencies obtained with constructs carrying different codons (two Gln, two His and four Gly) immediately upstream of the stop codon. There was no effect of amino acid identity or codon frequency. However, an adenine in the -1 position was always associated with the highest readthrough levels while an uracil was always associated with the lowest readthrough levels. This could be due to an effect mediated either by the nucleotide itself or by the P-site tRNA. We then examined the importance of the downstream context using eight other constructs. No direct correlation between the +6 nucleotide and readthrough efficiency was observed.
Conclusions
We conclude that, in mouse cells, the upstream and downstream stop codon contexts affect readthrough via different mechanisms, suggesting that complex interactions take place between the mRNA and the various components of the translation termination machinery. Comparison of our results with those previously obtained in plant cells and in yeast, strongly suggests that the mechanisms involved in stop codon recognition are conserved among eukaryotes.
Background
Translation termination is a crucial step in the process of information decoding. Its accuracy reaches about 10-4 and ensures that only very few abnormal products are synthesised under normal conditions [1]. Conversely, translation termination is widely used by both animal and plant viruses as a mean of controling expression, through recoding events [2, 3]. Readthrough is the process by which a stop codon is misread as sense by the translational apparatus, allowing the synthesis of an extended polypeptide which carries novel activities [2]. Up to now, no specific gene products have been implicated in the control of translational readthrough, strongly suggesting that only normal interactions between the mRNA and components of the translational machinery are involved. This differs from the mechanism of selenocysteine incorporation, which requires a whole set of specific partners, including a specific tRNA and a particular elongation factor [4]. Readthrough can therefore be seen as "programmed translational errors" occurring at specific sequences on the mRNA.
Several studies, either in vivo or "in silico", have analyzed the termination context in mammalian cells [5,6,7]. In particular, it has been demonstrated that the nucleotide following the stop codon (defined as +4) is non random, with purines over-represented for the three stop codons. This + 4 nucleotide in fact plays a key role in termination efficiency, leading to the proposition that termination is directed by a "four base signal" [8]. Clearly, some programmed readthrough events are based on alteration of these interactions. This is the case in Sindbis virus where the UGAC sequence, which is a very poorly used stop context in mammals, is sufficient to drive an efficient readthrough (2-5%) [5]. More generally, it has been demonstrated in numerous experimental systems that the nucleotide at position +4 plays an important role in suppression efficiency [5, 6, 9,10,11,12,13,14]. Other nucleotide biases are found around stop signals, non-randomness being observed at up to eight positions downstream of the stop codon and three positions upstream [15]. Such biases possibly reflect the existence of interactions between the mRNA and other components of the ribosomal machinery, which are important for termination efficiency.
Elucidating the mechanisms at play during readthrough may help understand normal translation termination. To achieve this goal, we have analyzed the rules governing readthrough in mouse cells in culture. We focused our studies on the sequence CAA UAG CAA UUA, derived from the plant Tobbaco Mosaic Virus (TMV) [16]. This sequence was previously shown to drive a high level readthrough both in in vitro systems [12] and in vivo in plant cells [9, 17] and yeast [10, 18]. This sequence is also functional in mammalian cells in culture [19]. We used a highly sensitive luciferase reporter to study the role of amino acid identity, codon frequency and nucleotide context, at the 5' and 3' triplets flanking the stop codon.
Results
To study the effect of the context on readthrough efficiency, we used a leaky termination signal derived from the TMV: CAA UAG CAA UUA. Different mutations were introduced into the triplets immediately preceeding or following the stop codon (Figure 1). The constructs were transfected in mouse cells and the readthrough level was evaluated by quantifying the luciferase activity obtained (see Materials and Methods). The results are summarized in the Tables and represented as histograms in the Figures.
Figure 1.
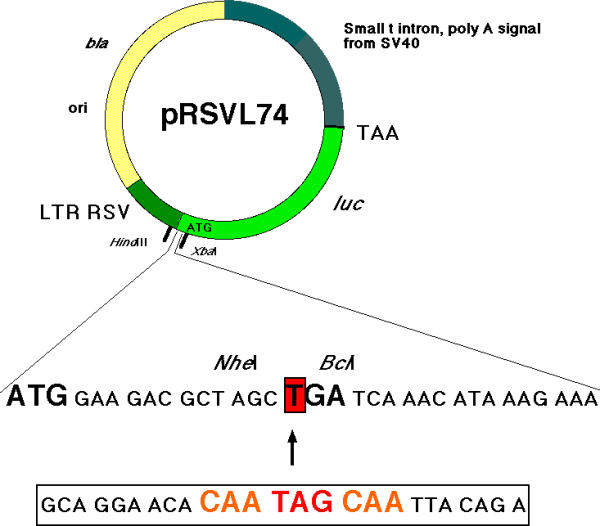
Description of the pRSVL74 vector and of the cloning strategy. The sequence of the 5' end of the luc gene coding sequence is shown with the common sequence of the inserted oligonucleotide containing the stop with its surrounding context. This sequence is inserted in place of the boxed "T" in the luc gene sequence. For each test construct, a control with the same sequence where a CAG replaced the TAG stop codon was used to evaluate the luciferase specific molecular activity.
Effect of upstream mutations
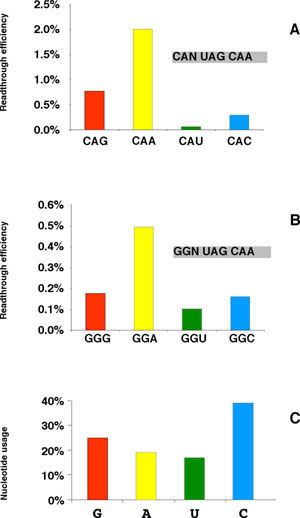
An important methodological problem is that changing a nucleotide could modify several parameters in addition to nucleotide identity: mRNA secondary structure, codon frequency, amino acid identity and luciferase molecular specific activity. To overcome the problem of possible changes in molecular specific activity of the modified luciferase, control constructs were designed in which the stop codon has been replaced by a CAG glutamine codon. As shown in Table 1, the activities of the modified luciferase proteins vary from 10% to 40% compared to the wild type pRSVL reporter, emphasizing the importance of this type of control. To differentiate at least some of the various structural parameters that may be involved in termination efficiency, we generated series of comparable constructs, harboring synonymous codons. One can thus analyze the effect of amino acid identity on readthrough and assess the effect of codon frequency for a given amino acid; finally, the effect of nucleotide identity at a given position can be studied in different contexts. This last point is crucial, since the occurrence of complex interactions between nucleotides located at different positions, would prevent a consensus sequence to emerge when analysing the stop codon context at a genomic level. A series of mutations were introduced in the CAA codon preceeding the stop. CAA was changed for the other glutamine codon CAG, for the two histidine codons CAU and CAC and for the four glycine (GGN) codons. Results are shown in Figure 2A-B. We first examined whether amino acid identity could interfere with readthrough. In this case, one would expect to see only a weak variation between constructs harboring the same amino acid. Conversely, variation should be higher for different amino acids. Comparing the results within a series and between different series, showed that variation between synonymous codons of the GLY family was greater than variation from one amino acid to another. These observations ruled out that the identity of the last amino acid was the main determinant of readthrough level.
Table 1.
Luciferase activity from in frame controls
| Control | Activity compared to | Standard | |
| Sequences | wild type construct | Deviation | |
| CAA CAG CAA | 13,8% | 6,4% | |
| CAU CAG CAA | 31,8% | 6,0% | |
| GGC CAG CAA | 38,2% | 13,4% | |
| CAA CAG CAU | 27,5% | 3,8% | |
| CAA CAG GGC | 11,1% | 4,1% | |
Figure 2.
Readthrough efficiencies of upstream mutants. The different stop codon context mutations were inserted at the beginning of the luc gene, as illustrated in Figure 1. Each construct was used to transfect NIH3T3 mouse cells and luciferase activity was determined. Each value is the mean of at least 3 independent experiments. Readthrough is expressed as percentage of the corresponding control where the stop codon has been replaced by a sense CAG glutamine codon. A: CAN UAG CAA mutants B: GGN UAG CAA mutants C: Nucleotide usage at the -1 position relative to the stop.
Another parameter that might affect readthrough is the availability of the tRNA decoding the last sense codon, for example through a different ribosome pausing kinetics. Since rare tRNAs generally correspond to rare codons, we checked whether a bias in codon frequency was correlated with readthrough efficiency. Results shown in Table 2 demonstrate that codon frequency and readthrough efficiency were not correlated, either in the whole set of data or within a given set of synonymous codons [20].
Table 2.
Readthrough efficiencies from the mutant constructs
| Test | Codon | Readthrough | Standard |
| Sequnces | Frequency | Efficiency | Deviation |
| CAGGLN UAG CAA | 3.41% | 0.77% | 0.08% |
| CAAGLN UAG CAA | 1.18% | 2.01% | 0.80% |
| CAUHIS UAG CAA | 0.97% | 0.06% | 0.03% |
| CACHIS UAG CAA | 1.50% | 0.30% | 0.18% |
| GGGGLY UAG CAA | 1.59% | 0.18% | 0.08% |
| GGAGLY UAG CAA | 1.80% | 0.49% | 0.23% |
| GGUGLY UAG CAA | 1.21% | 0.10% | 0.04% |
| GGCGLY UAG CAA | 2.31% | 0.16% | 0.08% |
| CAA UAG CAG | 0.43% | 0.05% | |
| CAA UAG CAA | 2.01% | 0.80% | |
| CAA UAG CAU | 0.07% | 0.04% | |
| CAA UAG CAC | 0.066% | 0.05% | |
| CAA UAG GGG | 0.022% | 0.01% | |
| CAA UAG GGA | 0.028% | 0.01% | |
| CAA UAG GGU | 0.033% | 0.01% | |
| CAA UAG GGC | 0.053% | 0.03% |
(a) The last amino acid inserted before the stop is indicated together with the codon frequency compiled from Mus musculus [20]).
As shown by Tate and coworkers, nucleotides surrounding the stop codons are not randomly distributed [8, 21, 22]. In particular, a strong bias is observed at the -1 position (see Figure 2C). Comparing Figure 2A-B to Figure 2C shows that, although the infrequent A was associated with the highest readthrough, no correlation was observed with the other nucleotides.
Finally, the only parameter that could be correlated with the readthrough efficiency was the nucleotide identity at the -1 position. Adenine was associated with the highest readthrough and uracil with the lowest. Guanine and cytosine gave intermediate values, guanine being in both cases higher than cytosine, although this difference is only significant in the CAN series.
Effect of dowstream mutations
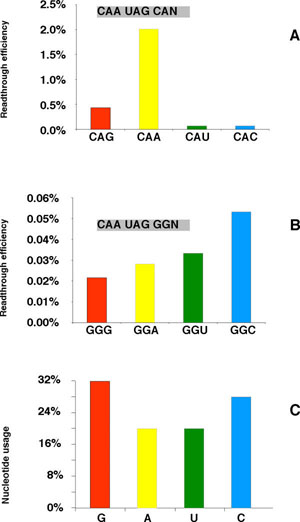
The same two series of constructs were analyzed for the CAA following the stop: CAN and GGN. Figure 3A-B illustrates that, in this case, the pattern was strikingly different between the two series. For the CAN series the readthrough efficiency followed the hierarchy: A>G>U=C, with A showing a 30 fold higher readthrough level than C. By contrast, for the GGN series, the hierarchy was C>U≥A≥G with C showing a 2.5 fold higher readthrough than G. As above, we examined the correlation between nucleotide usage at the variable position and readthrough efficiency. The histogram shown in Figure 3C demonstrates no correlation between these two values.
Figure 3.
Readthrough efficiencies of downstream mutants Same legend as Figure 2. A: CAA UAG CAN mutants B: CAA UAG GGN mutants C: Nucleotide usage at the +6 position relative to the stop.
Are these results related to readthrough efficiency ?
In this last set of results, the readthrough activities measured for many constructs were very low (around 10-4). This raised the general point of whether the measured enzymatic activities might reflect other translational events such as sliding, internal ribosome entry or reinitiation. To determine whether the observed patterns were relevant to leaky translation termination, we repeated the GGN upstream and downstream series of experiments, which gave the lowest readthrough values, in the presence of an amber suppressor tRNA [23]. The experimental scheme was exactly the same, except that a construct allowing expression of the suppressor tRNA was cotransfected along with the test constructs. The results are shown in Table 3. A 50 to 100 fold increase in readthrough activities was observed, demonstrating that very high levels of suppression were reached. Strikingly, Figure 4 illustrates that the same pattern was obtained either in the presence or in the absence of UAG suppressor tRNA. This indicates that the results observed under basal conditions actually corresponded to bona fide translational readthrough.
Table 3.
Effect of an amber supressor tRNA
| Readthrough Efficiency | ||
| Basal | + Sup | |
| GGG UAG CAA | 0.18% | 16.2% |
| GGA UAG CAA | 0.49% | 33.9% |
| GGU UAG CAA | 0.10% | 11.9% |
| GGC UAG CAA | 0.16% | 13.0% |
| CAA UAG GGG | 0.022% | 12.1% |
| CAA UAG GGA | 0.028% | 16.3% |
| CAA UAG GGU | 0.033% | 30.6% |
| CAA UAG GGC | 0.053% | 34.4% |
Figure 4.
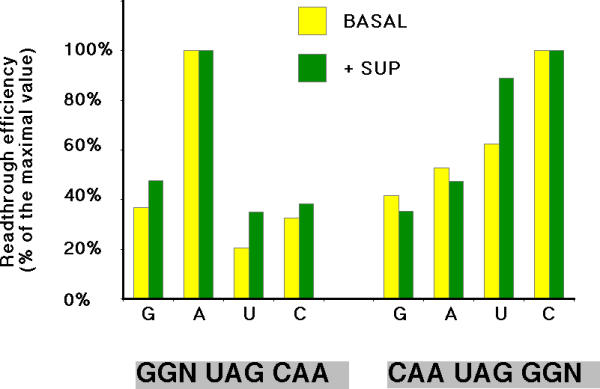
Readthrough efficiencies in the presence of a suppressor tRNA. Readthrough efficiencies were determined as described in Figure 1. To compare the values that differed by several orders of magnitude between the two sets of experiments, results are given as percentage of the highest readthrough obtained in each condition.
Discussion
The mechanisms involved in translation termination are still not completely understood, partly because more interactions than those required for codon-anticodon recognition are involved. In particular, protein-RNA and protein-protein interactions are other potential targets of translation termination control. Since in vitro translation may not always accurately reflect the in vivo conditions, we chosed to perform such analyses in living mouse cells. This requires the use of a system that allows the detection of very low readthrough levels. Thus, we used a reporter system bearing: i) the highly sensitive luciferase reporter gene; ii) the powerful RSV LTR promoter [24, 25].
We used the sequence CAA UAG CAA UUA, that drives a high level of translational readthrough in mouse cells in culture [19], and introduced targeted mutations in the sequences surrounding the stop codon. We found that single changes in this sequence were sufficient to decrease readthrough by up to 30 fold in vivo. We constructed a set of mutants where changes corresponded to synonymous codons. In this case, one can analyze separately at least some of the different parameters - amino acid identity, codon frequency, nucleotide identity - that may be involved in termination efficiency. We were also able to analyze nucleotide effects in different contexts, i.e. determine whether a given change would give the same effect in different sequence environments. This last point is crucial to assess the physiological significance of nucleotide usage found in stop signal regions.
We focused our analysis on two positions which have already been shown to be involved in termination or readthrough in eukaryotes, but which have not been studied in mammalian cells so far: the nucleotide preceeding the stop (-1) and the third nucleotide following the stop (+6) [15].
Two sets of modifications were introduced in the CAA triplet preceeding the stop: CAN and GGN. We found that the same amino acid can drive very different readthrough efficiencies (5 fold differences between GGA and GGU), suggesting that the amino acid itself does not play a major role in readthrough efficiency. Availability of the tRNA preceeding the stop is another parameter that could modulate ribosome pausing [26] and thus may affect readthrough. Since rare tRNAs generally correspond to rare codons [27], we checked whether a bias in codon usage was correlated with readthrough efficiency. This seemed to be the case for some of the mutants presented here. For example, the rare CAA glutamine codon gave a higher readthrough than the frequent CAG codon. However, when we systematically examined codon frequency in parallel with readthrough efficiency, no correlation could be found (see Table 2). Although we cannot rule out a marginal effect of codon frequency, it is unlikely that it represents the main determinant of readthrough efficiency.
Readthrough efficiency is clearly correlated with the nucleotide immediately preceeding the stop codon. Hence, an adenine in the -1 position is associated with a high readthrough while a pyrimidine is associated with a low readthrough. Taken together, these results show that there is a gradation in the observed effect on readthrough efficiency by which A>G>Y. The fact that a rule could be drawn suggests that relatively simple interactions are involved, possibly between the tRNA in the P site, the incomming potential suppressor tRNA and the message, as it has been shown in yeast [28]. However, since the information on the Glycine-, Histidine- or Glutamine-isoacceptor tRNAs in mouse is not yet available (see M. Sprinzl, K.S. Vassilenko, J. Emmerich, F. Bauer "Compilation of tRNA sequences and sequences of tRNA genes". http://www.uni-bayreuth.de/departments/biochemie/trna/), this possibility cannot be explored further for the time being.
In a second series of experiments, two sets of mutations were introduced in the triplet following the stop codon: CAA was changed for CAN and for GGN. A drastic effect on readthrough, up to 100 fold, was observed. In this case however, no uniform rule emerged, the hierarchy being A>G>U=C in the CAN series, but C>U≥A≥G for the GGN series. These contrasted results imply that complex interactions are taking place downstream of the stop codon. Although the nature of these interactions cannot be deduced from our results, one possibility would be that mRNA structure is involved in this phenomenon, as it is in numerous translational events [29]. Another obvious candidate potentially interacting with the stop context is the release factor eRF1 [30]. If this is the case, one would predict to be able to obtain mutant forms of eRF1 showing a better recognition of poor codon context. The three dimensional structure of eRF1 has been published recently and could help to design mutants affected in stop codon context interaction [31].
It is striking that the CAA UAG CAA UUA sequence directs a high readthrough in several very different eukaryotes. In plants, the readthrough efficiency reaches 4-5% [9], a level similar to what is observed in mouse cells (2%). In the yeast S. cerevisiae, it is even more efficient, driving a readthrough of 15-25% in a [psi-] strain [18] and up to 55% in a [PSI+] strain (Olivier Namy and Jean-Pierre Rousset, unpublished results). Furthermore, when our results are compared with those obtained in plant cells and yeast, a similar effect of the nucleotide context is observed [9, 10, 28]. By contrast, this sequence is unable to drive a significant readthrough activity (4,5 10-4) in E. coli (JPR, MC, unpublished results, see also [9]).
Conclusions
Altogether, our results indicate that different signals are involved in the interaction between the translational machinery and the upstream and downstream stop codon contexts. In addition, they also suggest that the mechanisms involved in readthrough are common to eukaryotes, pointing to either an ancient origin of the translation termination machinery, or to very strong structural constraints at the level of the eukaryotic ribosome. Another implication of these observations is that the readthrough sequence analyzed here, although first described in a plant virus, might be used as a recoding signal also in mammals and yeast. Analyzing DNA sequence data bases should help in identifying such putative physiological recoding events.
Materials and Methods
Plasmids
The pRSVL74 cloning vector is shown in Figure 1 and the procedures used to construct the mutant derivatives have been described previously [25]. Briefly, 25 nt synthetic double stranded oligonucleotides were inserted in the oriented NheI-BclI cloning site of the vector. In the pRSVL74 derived plasmids, the luc gene encoding the very sensitive luciferase reporter is under the control of the strong Rous Sarcoma Virus promoter [24]. Mutations were introduced in the sequence GCA GGA ACA CAA TAG CAA TTA CAG A. All constructs were verified by sequencing 200 nt around the mutation. pCH110 carries a lacZ gene under the control of the SV40 promoter-enhancer region [32].
ptRNAam is a gift of Olivier Jean-Jean; it carries the entire coding sequence and promoter region of a human tRNASer gene, in which the anticodon has been replaced by CUA, therefore recognizing the UAG stop codon [23].
Cells
NIH 3T3 cells were cultured in Ham's F12 or DMEM medium supplemented with 5 % fetal calf serum, at 37°C in humidity saturated 7% CO2 in air.
Transfection
One million cells were transfected with 10 μg of each test vector and 5 μg of the pCH110 vector, using DNA/phosphate coprecipitation. Cells were rinsed after overnight incubation with the DNA, and the medium was changed the next day. Cells were harvested on day 3 and crude extracts were prepared as described [25]. To check the effect of suppression, the same protocol was used except that 5 μg of the ptRNAam vector was added just before the step of DNA/phosphate coprecipitation.
Enzyme assays
β-galactosidase and luciferase were assayed using standard methods [33, 34].
Readthrough efficiency quantification
To evaluate readthrough efficiency, the luciferase activity obtained from a test construct was compared to the activity of the wild type pRSVL vector, which was used as an internal control of each individual transfection experiment. The β-galactosidase activities driven by the cotransfected pCH110 plasmid reflected transfection efficiencies and were used to standardized luciferase activities. The readthrough level was expressed as the ratio of standardized activity from the test construct to the standardized activity from the wild type pRSVL construct. Since luciferase molecular specific activity may vary depending on the amino acid sequence of the protein, control vectors in which the UAG stop codon has been replaced by a CAG glutamine codon were similarly transfected and their activities standardized for transfection efficiency.
Acknowledgments
Acknowledgments
We are grateful to Jonathan Gallant for numerous suggestions concerning the manuscript and to Olivier Jean-Jean for the ptRNAam vector. Part of this work was supported by the << Association pour la Recherche contre le Cancer >> (contract 9873 to JPR) and by the Association Française contre les Myopathies (contract 7623 to JPR).
Contributor Information
Michel Cassan, Email: Michel.Cassan@cgm.cnrs-gif.fr.
Jean-Pierre Rousset, Email: rousset@igmors.u-psud.fr.
References
- Kirkwood TBL, Rosenberger RF, Galas DJ. Accuracy in Molecular Processes. London: Chapman and Hall; 1986.
- Atkins JF, Weiss RB, Gesteland RF. Ribosome gymnastics-Degree of difficulty 9.5, style 10.0. Cell. 1990;62:413–423. doi: 10.1016/0092-8674(90)90007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland RF, Weiss RB, Atkins JF. Recoding: reprogrammed genetic decoding. Science. 1992;257:1640–1641. doi: 10.1126/science.1529352. [DOI] [PubMed] [Google Scholar]
- Böck A, Forchhammer K, Heider J, Leinfelder W, Sawers G, Veprek B, Zinoni F. Selenocysteine: the 21st amino acid. Mol Microbiol. 1991;5:515–520. doi: 10.1111/j.1365-2958.1991.tb00722.x. [DOI] [PubMed] [Google Scholar]
- Li G, Rice CM. The signal for translational readthrough of a UGA codon in Sindbis virus RNA involves a single cytidine residue immediately downstream of the termination codon. J Virol. 1993;67:5062–5067. doi: 10.1128/jvi.67.8.5062-5067.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. On the relationship between preferred termination codon contexts and nonsense suppression in human cells. Nucleic Acids Res. 1994;22:15–19. doi: 10.1093/nar/22.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Jones MK, Watson FJ, Martin R. The 3' codon context effect on UAG suppressor tRNA is different in Escherichia coli and human cells. J Mol Biol. 1993;233:1–6. doi: 10.1006/jmbi.1993.1479. [DOI] [PubMed] [Google Scholar]
- Brown CM, Stockwell PA, Trotman CN, Tate WP. Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Res. 1990;18:6339–6345. doi: 10.1093/nar/18.21.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuzeski JM, Nichols LM, Gesteland RF, Atkins JF. The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J Mol Biol. 1991;218:365–373. doi: 10.1016/0022-2836(91)90718-l. [DOI] [PubMed] [Google Scholar]
- Bonetti B, Fu LW, Moon J, Bedwell DM. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J Mol Biol. 1995;251:334–345. doi: 10.1006/jmbi.1995.0438. [DOI] [PubMed] [Google Scholar]
- McCaughan KK, Brown CM, Dalphin ME, Berry MJ, Tate WP. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc Natl Acad Sci USA. 1995;92:5431–5435. doi: 10.1073/pnas.92.12.5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle RP, Drugeon G, Devignes MM, Legocki AB, Haenni AL. Codon context effect in virus translational readthrough. A study in vitro of the determinants of TMV and Mo-MuLV amber suppression. Febs Lett. 1992;306:133–139. doi: 10.1016/0014-5793(92)80984-O. [DOI] [PubMed] [Google Scholar]
- Howard MT, Shirts BH, Petros LM, Flanigan KM, Gesteland RF, Atkins JF. Sequence specificity of aminoglycoside-induced stop condon readthrough: potential implications for treatment of Duchenne muscular dystrophy. Ann Neurol. 2000;48:164–169. doi: 10.1002/1531-8249(200008)48:2<164::AID-ANA5>3.3.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Manuvakhova M, Keeling K, Bedwell DM. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. Rna. 2000;6:1044–1055. doi: 10.1017/S1355838200000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate WP, Poole ES, Mannering SA. Hidden infidelities of the translational stop signal. Prog Nucleic Acid Res Mol Biol. 1996;52:293–335. doi: 10.1016/s0079-6603(08)60970-8. [DOI] [PubMed] [Google Scholar]
- Pelham HRB. Leaky UAG termination codon in tobacco mosaic virus RNA. Nature. 1978;272:469–471. doi: 10.1038/272469a0. [DOI] [PubMed] [Google Scholar]
- Skuzeski JM, Nichols LM, Gesteland RF. Analysis of leaky viral translation termination codons in vivo by transient expression of improved beta-glucuronidase vectors. Plant Mol Biol. 1990;15:65–79. doi: 10.1007/BF00017725. [DOI] [PubMed] [Google Scholar]
- Stahl G, Bidou L, Rousset JP, Cassan M. Versatile vectors to study recoding: conservation of rules between yeast and mammalian cells. Nucleic Acids Res. 1995;23:1557–1560. doi: 10.1093/nar/23.9.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berteaux V, Rousset JP, Cassan M. UAG readthrough is not increased in vivo by Moloney murine leukemia virus infection. Biochimie. 1991;73:1291–1293. doi: 10.1016/0300-9084(91)90091-E. [DOI] [PubMed] [Google Scholar]
- Jacobs GH, Stockwell PA, Schrieber MJ, Tate WP, Brown CM. Transterm: a database of messenger RNA components and signals. Nucleic Acids Res. 2000;28:293–295. doi: 10.1093/nar/28.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CM, Stockwell PA, Trotman CN, Tate WP. The signal for the termination of protein synthesis in procaryotes. Nucleic Acids Res. 1990;18:2079–2086. doi: 10.1093/nar/18.8.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate WP, Poole ES, Dalphin ME, Major LL, Crawford DJ, Mannering SA. The translational stop signal: codon with a context, or extended factor recognition element? Biochimie. 1996;78:945–952. doi: 10.1016/S0300-9084(97)86716-8. [DOI] [PubMed] [Google Scholar]
- Capone JP, Sharp PA, RajBhandary UL. Amber, ochre and opal suppressor tRNA genes derived from a human serine tRNA gene. Embo J. 1985;4:213–221. doi: 10.1002/j.1460-2075.1985.tb02338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wet JR, Wood KV, DeLuca M, Helinski DR, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassan M, Delaunay N, Vaquero C, Rousset JP. Translational frameshifting at the gag-pol junction of human immunodeficiency virus type 1 is not increased in infected T-lymphoid cells. J Virol. 1994;68:1501–1508. doi: 10.1128/jvi.68.3.1501-1508.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindhout P, Neeleman L, Van Tol H, Van Vloten-Doting L. Ribosomes are stalled during in vitro translation of alfalfa mosaic virus RNA 1. Eur J Biochem. 1985;152:625–631. doi: 10.1111/j.1432-1033.1985.tb09241.x. [DOI] [PubMed] [Google Scholar]
- Bulmer M. Coevolution of codon usage and transfer RNA abundance. Nature. 1987;325:728–730. doi: 10.1038/325728a0. [DOI] [PubMed] [Google Scholar]
- Mottagui-Tabar S, Tuite MF, Isaksson LA. The influence of 5' codon context on translation termination in Saccharomyces cerevisiae. Eur J Biochem. 1998;257:249–254. doi: 10.1046/j.1432-1327.1998.2570249.x. [DOI] [PubMed] [Google Scholar]
- McCarthy JEG. Posttranscriptional control of gene expression in yeast. Microbiol Mol Biol Rev. 1998;62:1492–1553. doi: 10.1128/mmbr.62.4.1492-1553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni AL, Celis JE, Philippe M, et al. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor [see comments]. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D. The crystal structure of human eukaryotic release factor eRF1-mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- Hall C, Jacob E, Ringold G, Lee F. Expression and regulation of Escherichia coli lacZ gene fusions in mammalian cells. J Mol Appl Genet. 1983;2:101–109. [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor laboratory; 1972.
- Nguyen VT, Morange M, Bensaude O. Firefly luminescence assays using scintillation counters for quantitation in transfected mammalian cells. Ann Biochem. 1988;171:404–408. doi: 10.1016/0003-2697(88)90505-2. [DOI] [PubMed] [Google Scholar]