Abstract
Background
Streptococcus dysgalactiae subspecies equisimilis (SDSE) is an emerging global pathogen that can colonize and infect humans. Although most SDSE isolates possess the Lancefield group G carbohydrate, a significant minority have the group C carbohydrate. Isolates are further sub-typed on the basis of differences within the emm gene. To gain a better understanding of their molecular epidemiology and evolutionary relationships, multilocus sequence typing (MLST) analysis was performed on SDSE isolates collected from Australia, Europe and North America.
Methodology/Principal Findings
The 178 SDSE isolates, representing 37 emm types, segregate into 80 distinct sequence types (STs) that form 17 clonal complexes (CCs). Eight STs recovered from all three continents account for >50% of the isolates. Thus, a small number of STs are highly prevalent and have a wide geographic distribution. Both ST and CC strongly correlate with group carbohydrate. In contrast, eleven STs were associated with >1 emm type, suggestive of recombinational replacements involving the emm gene; furthermore, 35% of the emm types are associated with genetically distant STs. Data also reveal a history of extensive inter- and intra-species recombination involving the housekeeping genes used for MLST. Sequence analysis of single locus variants identified through goeBURST indicates that genetic change mediated by recombination occurred ∼4.4 times more frequently than by point mutation.
Conclusions/Significance
A few genetic lineages with an intercontinental distribution dominate among SDSE causing infections in humans. The distinction between group C and G isolates reflects recent evolution, and no long-term genetic isolation between them was found. Lateral gene transfer and recombination involving housekeeping genes and the emm gene are important mechanisms driving genetic variability in the SDSE population.
Introduction
Most streptococci displaying β-hemolysis fall within the pyogenic branch of the 16S rRNA-based taxonomy, and are pathogens or commensals of mammalian hosts [1], [2]. Two species within the pyogenic branch - Streptococcus dysgalactiae subspecies equisimilis (SDSE) and Streptococcus pyogenes (group A streptococcus, GAS) - colonize and/or infect the respiratory tract and skin of the human host [2]. Whereas GAS is an important human pathogen, SDSE is largely considered to be a commensal organism. However, numerous studies report that SDSE can cause disease among otherwise healthy individuals [2], [3], [4], [5]. The disease spectrum of SDSE infection is similar to that of GAS, and includes pharyngitis, post-streptococcal glomerulonephritis, cellulitis, necrotizing fasciitis, septicemia, and streptococcal toxic shock syndrome [6], [7], [8], [9]. Furthermore, in some geographic regions where streptococcal diseases are endemic, surveillance studies report higher rates of throat colonization by SDSE than by GAS [10], [11].
The surface-exposed Lancefield group carbohydrate is an important cell wall antigen that aids in distinguishing between several of the β-hemolytic streptococcal species. S. pyogenes almost exclusively expresses the group A carbohydrate [1]. Although the vast majority of SDSE isolates have group G carbohydrate, and are often referred to as group G streptococci, a significant minority of SDSE isolates have group C carbohydrate; very rarely do SDSE harbor the group A or L carbohydrate [1], [12]. Among the GAS and SDSE populations, differences in the sequences of individual emm genes are widely used for intra-species strain typing. At present, >200 GAS and ∼50 SDSE emm types are recognized (http://www.cdc.gov/ncidod/biotech/strep/strepblast.htm). Although one report finds SDSE isolates expressing emm types stg2078 or stg10 to have enhanced invasive disease potential [5], most studies have failed to uncover disease associations among SDSE emm types [13], [14], [15]. In contrast, associations between emm type and specific diseases are well established for GAS [16], [17], [18], [19].
Comparative genome hybridization studies using a microarray containing probes corresponding to genes encoding virulence factors and putative surface proteins failed to reveal clear cut associations between emm type and gene content among SDSE [13]. Multilocus sequence typing (MLST) is a nucleotide sequence-based method that uses core housekeeping genes to characterize genetic relationships between isolates of the same species. MLST has been used extensively to study the β-hemolytic GAS [20], [21], [22] and S. agalactiae [23], [24] populations, and was recently used to investigate genetic relationships amongst 61 geographically restricted SDSE isolates [25]. In the present study, an intercontinental collection of SDSE isolates is characterized by MLST and emm typing, and the geographic distribution of the identified clones and their genetic relationships are defined.
Results
Molecular typing
MLST was used to characterize 117 SDSE isolates collected from three continents (Table S1). Most of the isolates selected for MLST were derived from large independent collections and were chosen, in part, based on prior knowledge of their emm type and geographic site of isolation, with the goal being the compilation of a genetically diverse data set.
The isolates represent 24 of the ∼50 known SDSE emm types, and include strains bearing the group G or group C carbohydrate. A summary of the epidemiological features of the isolates is provided in Table 1.
Table 1. Characteristics of SDSE isolates included in this study.
| Collection site | No. of isolates | No. of different emm types | Diversity index, D a (95% C.I.)b | No. of different STs | Diversity index, D (95% C.I.) | STs unique to collection site | No. of isolates with group G carbohydrate | No. of isolates with group C carbohydrate | No. of invasive isolates | No. of non-invasive isolates |
| Australia | 55 | 17 | 0.926 (0.901–0.956) | 23 | 0.937 (0.908–0.970) | 14 | 47 | 8 | 24 | 25 |
| Portugal | 36 | 17 | 0.951 (0.930–0.972) | 22 | 0.956 (0.922–0.989) | 11 | 28 | 8 | 10 | 26 |
| USA | 72 | 34 | 0.984 (0.976–0.985) | 45 | 0.975 (0.961–0.988) | 33 | 48 | 23 | 70 | 1 |
| —NYMC | 11 | 8 | 0.927 (0.833–1.020) | 10 | 0.981 (0.936–1.020) | 5 | 6 | 5 | 9 | 0 |
| —CDC | 61 | 33 | 0.983 (0.979–0.988) | 37 | 0.972 (0.957–0.988) | 28 | 42 | 18 | 61 | 1 |
| Other | 15 | 12 | 0.971 (0.951–1.001) | 13 | 0.981 (0.951–1.011) | 10 | 7 | 7 | 1 | 6 |
| Total | 178 | 37 | 0.961 (0.954–0.967) | 80 | 0.966 (0.955–0.976) | n.a. | 131 | 46 | 106 | 59 |
D, Simpsons Index of Diversity.
CI, Confidence Interval.
n.a., not applicable.
With the inclusion of previously published MLST data on 61 invasive SDSE isolates obtained from the USA [25], a total of 37 emm types are represented among a larger set of 178 isolates, which is used for the analyses in this report. The frequencies of the different emm types span a wide range; however, only 38% of the emm types account for the majority (74%) of the isolates.
Among the 178 SDSE isolates, the number of alleles identified for each housekeeping gene locus ranges from 10 for gtr, murI and mutS, to 22 for xpt (Table 2). The gki, recP, and xpt loci exhibit the highest level of nucleotide sequence diversity (π). The percentage of polymorphic nucleotide sites ranges from 2.7 (n = 12) for murI, to 10.1 (n = 50) for gki. A significant portion of the polymorphism observed in gki can be attributed to gki12, a highly divergent allele with greater similarity to GAS gki alleles than to SDSE alleles (Figure S1); when the divergent gki12 allele is removed from the analysis, the nucleotide diversity of gki falls from 0.021 to 0.012, and the percentage of polymorphic sites drops from 10.1 to 5.4 (n = 27). The dn/ds ratio is less than one for each of the seven housekeeping genes, consistent with stabilizing selection.
Table 2. Housekeeping genes used for MLST of SDSE.
| Gene | ORFa | Size of partial gene | No. of alleles | No. of nucleotide variant positions (%) | No. of variant aa positionsb | π | dn | ds | dn/ds |
| Glucose kinase (gki) | SDEG_1515 | 498 | 12 | 50 (10.1) | 7 | 0.021 | 0.0035 | 0.0736 | 0.047 |
| Glutamine transport protein (gtr) | SDEG_1494 | 450 | 10 | 15 (3.3) | 7 | 0.010 | 0.0059 | 0.0248 | 0.240 |
| Glutamate racemase (murI) | SDEG_0413 | 438 | 10 | 12 (2.7) | 1 | 0.012 | 0.0068 | 0.0142 | 0.479 |
| DNA mismatch repair protein (mutS) | SDEG_2091 | 405 | 10 | 27 (6.7) | 7 | 0.016 | 0.0062 | 0.0480 | 0.129 |
| Transketolase (recP) | SDEG_1735 | 459 | 20 | 37 (8.1) | 3 | 0.034 | 0.0008 | 0.1472 | 0.006 |
| Xanthine phosphoribosyl transferase (xpt) | SDEG_0895 | 450 | 22 | 38 (8.4) | 10 | 0.021 | 0.0049 | 0.0736 | 0.0665 |
| Acetoacetyl-coathioloase (atoB) | SDEG_1700 | 434 | 12 | 18 (4.1) | 5 | 0.011 | 0.0035 | 0.0330 | 0.106 |
Based on ORF number in the GGS_124 genome (Genbank number AP010935).
aa, amino acid.
Relationships among STs
The seven housekeeping alleles of each of the 178 isolates yield a total of 80 distinct allelic profiles, referred to as sequence types (STs). Of the 80 STs, 37 are newly identified in this study and 43 were previously defined by Ahmad et al [25]. A minority of STs (10%) account for a disproportionate number (∼50%) of the total SDSE isolates under evaluation. The vast majority of STs (62, or 77%) are represented by only one SDSE isolate. The most prevalent ST (ST15) is represented by 20 isolates. Eight STs, each represented by eight or more isolates, account for 51% of the 178 SDSE isolates characterized by MLST.
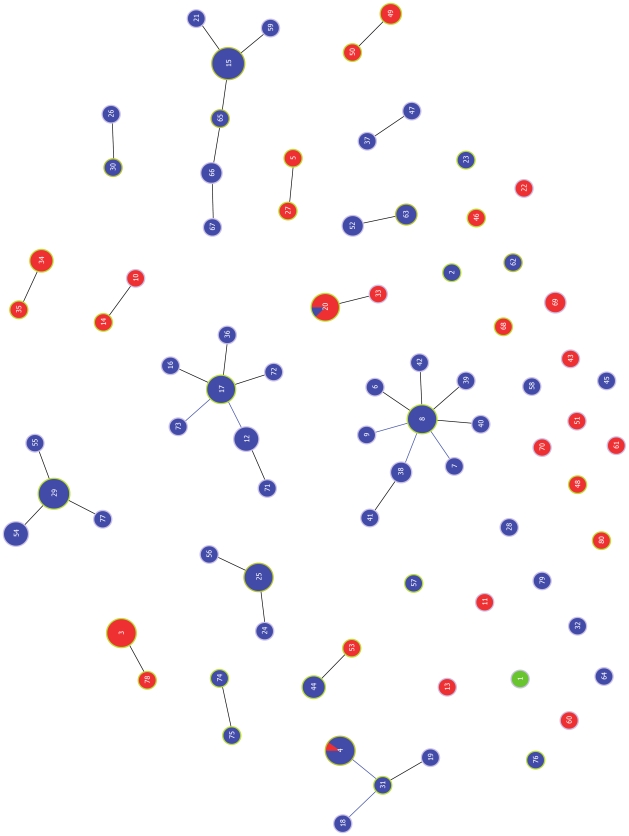
The 80 STs can be grouped into 17 clonal complexes (CCs) by goeBURST, whereby the connected STs are single locus variants (SLVs) of at least one other ST in the group, differing at only one housekeeping gene (Figure 1). However, only six of the 17 CCs contain more than two STs. CC8 contains the highest number of STs (n = 9), whereas CC15 (n = 26) and CC29 (n = 20) contain the most isolates. Twenty-six STs, representing 15% of the 178 isolates, are singletons and differ from all other STs by more than two housekeeping alleles. When clusters are constructed linking STs that are up to triple locus variants (TLVs) of each other, 66 STs are grouped into a single major cluster, whereas only three STs remain ungrouped singletons (Figure S3), indicating that several intermediate genotypes probably exist, but have not yet been sampled.
Figure 1. goeBURST diagram of relationships between 178 global SDSE isolates.
The size of each circle is proportional to the number of isolates with that particular ST in a logarithmic scale. STs assigned to the same CC are linked by straight lines. Blue circles represent isolates that have the group G carbohydrate. Red circles represent isolates expressing the group C carbohydrate. Whenever isolates of the same ST have different group carbohydrates, the number of isolates bearing the same carbohydrate is proportional to the respective color. The green circle represents the single isolate expressing the group L carbohydrate.
Relationships between emm type and ST
The overall correspondence between ST and emm type, as determined by the Wallace Coefficient, is low (ST vs emm type W = 0.473, CI95% 0.332–0.542; emm type vs ST W = 0.384, CI95% 0.311–0.456) reflecting the fact that most emm types are found in multiple STs, and that the same ST can harbor different emm types (Table 3 and Table 4). The correspondence between emm type and CC (W = 0.551, CI95% 0.478 to 0.625) is also weak.
Table 3. Relationship between ST and emm type.
| ST | No. of isolates | Associated emm types | No. of emm types |
| 15 | 20 | stC839, stG10, stG166b, stG2078, stG245, stG6, stG652 | 7 |
| 8 | 10 | stC839, stG11, stG480, stG643, stG7860 | 5 |
| 4 | 10 | stC36, stC5344, stG6792, stG97, stG7882 | 5 |
| 3 | 10 | emm57, stC1400, stC839, stG653 | 4 |
| 25 | 9 | stG166b, stG5420, stG6 | 3 |
| 17 | 9 | stC74a, stG2078, stG485 | 3 |
| 63 | 2 | stG6, stG652 | 2 |
| 52 | 2 | stG6, stG643 | 2 |
| 20 | 8 | stC6979, stG62647 | 2 |
| 34 | 3 | stC1400, stG5063 | 2 |
| 29 | 14 | stC74a, stG485 | 2 |
Table 4. Relationship between emm type and ST.
| emm type | No. of isolates | Associated STs | No. of STs | No. of CCSLV a | No. of CCDLV b | No. of distant STsc |
| stG6 | 13 | 15, 24, 25, 44, 52, 58, 62, 63 | 8 | 4 | 2 | 2 |
| stG480 | 11 | 7, 8, 38, 39, 40, 41, 67 | 7 | 2 | 2 | 2 |
| stC1400 | 8 | 3, 28, 34, 46, 64, 66 | 6 | 3 | 1 | 4 |
| stG485 | 8 | 17, 29, 37, 47, 55, 69 | 6 | 3 | 2 | 2 |
| stG643 | 9 | 8, 12, 22, 48, 52, 73 | 6 | 3 | 4 | 5 |
| stG652 | 7 | 15, 32, 59, 61, 63, 71 | 6 | 3 | 2 | |
| stC6979 | 8 | 9, 19, 20, 54, 80 | 5 | 4 | 4 | 3 |
| stC36 | 6 | 4, 45, 49, 50, 68 | 5 | 2 | 1 | 2 |
| stC74a | 15 | 17, 29, 70, 77 | 4 | 2 | 3 | 3 |
| stC839 | 7 | 3, 8, 15, 78 | 4 | 3 | 2 | 3 |
| stG166b | 5 | 15, 25, 56, 65 | 4 | 2 | 2 | 2 |
| stG11 | 5 | 6, 8, 42 | 3 | 1 | 1 | |
| stG2078 | 9 | 15, 17, 72 | 3 | 2 | 2 | 2 |
| stG245 | 3 | 15, 21, 36 | 3 | 2 | 2 | 2 |
| stG4831 | 3 | 74, 75, 76 | 3 | 1 | 0 | |
| stG62647 | 9 | 20, 33, 60 | 3 | 1 | 1 | |
| stG6792 | 6 | 4, 31, 51 | 3 | 1 | 1 | |
| emm57 | 3 | 3, 57 | 2 | 1 | 0 | 2 |
| stC5344 | 3 | 4, 43 | 2 | 1 | 1 | |
| stC6746 | 2 | 5, 27 | 2 | 1 | 0 | |
| stC9431 | 2 | 13, 14 | 2 | 1 | 1 | |
| stG5063 | 2 | 2, 34 | 2 | 1 | 0 | |
| stG840 | 2 | 26, 30 | 2 | 1 | 1 | |
| stG7882 | 2 | 4, 18 | 2 | 1 | 1 |
CCSLV – Clonal complex based on Single Locus Variant relationships.
CCDLV – Clonal complex based on Double Locus Variant relationships.
Number of STs sharing the same emm type and differing from all other STs harboring that emm type at greater than five housekeeping alleles.
STs associated with multiple emm types most likely arose via recombinational replacement of the emm gene; they are referred to as emm variable STs. Of the 18 STs represented by more than one isolate, 11 STs (61%) are associated with more than one emm type (Table 3). Five STs are associated with two emm types (ST34, 20, 29, 52, 63), two STs (ST17, 25) are associated with three emm types, one ST (ST3) is associated with four emm types, two STs (ST4, ST8) are associated with five emm types and one ST (ST15) is associated with 7 different emm types.
The same emm type is often found in association with multiple STs (Table 4). The association of a given emm type with multiple STs can arise following diversification of housekeeping genes, or by horizontal transfer of the emm gene. An estimate of the horizontal movement of emm is made by enumerating the number of distant STs harboring the same emm type, whereby distant STs are defined as having five or more housekeeping allele differences to any other ST that shares the same emm type; for a given CC, only one representative ST is counted.
Thirteen emm types are associated with distant STs: five emm types are associated with >two distant STs and eight emm types are found among a single pair of distant STs, whereas five emm types are associated with three or more distant STs (Table 4). The most promiscuous emm type is emmstG643, found among five genetically distant strains. A total of 21 horizontal transfer events involving emm genes are evident in the SDSE data set. Taken together, the data provides strong support for the hypothesis that emm genes of SDSE undergo extensive lateral exchange between strains.
Relationships between group carbohydrate and ST
The group specific carbohydrate of the streptococcal cell wall can be used to discriminate among β-hemolytic streptococcal species. The majority of SDSE isolates (74%) in this study express the group G carbohydrate. A sharp distinction between STs associated with strains expressing the group G versus C carbohydrate is observed by goeBURST (Figure 1). Isolates representing 54 STs have group G carbohydrate (group G streptococci, GGS), whereas 27 STs are associated with strains expressing group C carbohydrate (group C streptococci; GCS). A single isolate has the group L carbohydrate.
Only two STs (ST4 and ST20) have isolates associated with both group C and G carbohydrates; for ST4, nine of 10 isolates are GGS and for ST20, seven of eight isolates are GCS. Overall, ST and group carbohydrate, whose biosynthesis locus is unknown, display very strong linkage (W = 0.970, CI95% 0.939 to 1.000). The correspondence between CCs and group carbohydrate is also high, with nine CCs containing only GGS isolates and six CCs restricted to GCS isolates (W = 0.979, CI95% 0.957 to 1.000), indicating that STs belonging to the same genetic lineage almost always also share the same group carbohydrate.
For each of the seven housekeeping loci, the relative distribution of alleles among GCS and GGS isolates was evaluated (Table 5). Overall, 38% of the housekeeping alleles are shared among GCS and GGS isolates. Approximately 36% and 29% of the housekeeping alleles are restricted to GCS and GGS isolates, respectively. This finding shows that there is a common housekeeping gene pool that is shared among numerous GCS and GGS isolates, despite the highly restricted associations that are observed between group carbohydrate and ST.
Table 5. Distribution of housekeeping alleles among GCS and GGS isolates.
| Housekeeping gene locusa | % of alleles shared by GCS and GGS | % of alleles restricted to GCS | % of alleles restricted to GGS |
| murI | 30 | 30 | 40 |
| xpt | 32 | 36 | 32 |
| gtr b | 56 | 33 | 11 |
| gki | 33 | 25 | 43 |
| atoB | 50 | 33 | 17 |
| recP | 35 | 35 | 30 |
| mutS | 40 | 30 | 30 |
| Total for all alleles | 38 | 36 | 29 |
Presented in order of the locus position on the genome of strain GGS_124.
Excludes gtr06 which is restricted to group L.
Relationships between group carbohydrate and emm type
Unlike ST, which is largely restricted to a single group carbohydrate form, 13 (35%) of the 37 emm types are found in association with both GGS and GCS isolates (W = 0.821, CI95% 0.764 to 0.876). Eight of the variable associations between emm type and group carbohydrate likely arose following horizontal transfer of an emm gene to a strain having a different group carbohydrate (data not shown). However, it remains possible that lateral movement of genes encoding carbohydrate biosynthetic enzymes also contributes to the generation of diversity among SDSE, although the frequency of this event is probably low because carbohydrate-variable STs are rare.
Geographic distribution of genetically diverse SDSE isolates
To examine the global distribution of SDSE clones, the ST and emm type of the isolates recovered from Australia, Europe and North America were compared. Clonal diversity based on either ST or emm type, as measured by the Simpson Index of Diversity, was high (>0.9) for isolates collected from each of the three primary locations (Table 1).
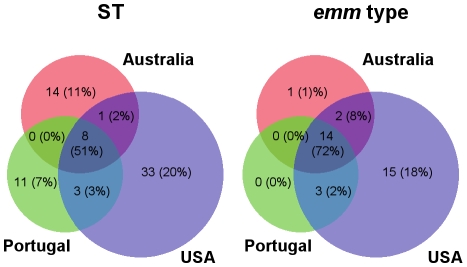
The eight STs that were recovered from all three continents also represent the STs with the highest overall prevalence (ST3, 4, 8, 15, 17, 20, 25, 29) (Table S1; Figure 2). Furthermore, five of the eight highly prevalent STs are predicted to be founders of a CC, wherein the founder ST is defined as that having the highest number of SLVs. The data suggest a wide geographical dispersion of founder STs. Of the next 10 most highly prevalent STs (each of which is represented by two to four isolates), four were associated with isolates from two continents (Table S1). Fourteen emm types were recovered from each of the three continents; 13 of these are represented by the most highly prevalent strains, having greater than 5 SDSE isolates per emm type. Together the results demonstrate that the most highly prevalent strains of SDSE, whereby strain is defined by either ST or emm type, are widely disseminated.
Figure 2. Venn diagram depicting the distribution of ST and emm type across three continents.
Unbracketed numbers represent the total number of STs or emm types. The numbers in brackets indicate the percentage of total isolates in the entire collection.
Among the 178 SDSE isolates, 107 unique combinations of ST, emm type, and group carbohydrate were observed. Isolates corresponding to eight of the 107 unique emm ST-carbohydrate profiles are widespread and were recovered from all three continents (Table 6); another seven were isolated from two continents. Three intercontinental clones belong to CC8, two of which likely arose from a common ancestor by either genetic diversification at a housekeeping gene locus and/or by horizontal transfer of the emm gene. The two intercontinental CC17 clones are SLVs, whereas the CC15 and CC25 sets of intercontinental strains arose via lateral exchange of emm type. The genetic changes are likely to be ancient events that preceded the intercontinental migration of the founders.
Table 6. Intercontinenal clones of SDSE.
| CC | ST | emm type | group carbohydrate | Australia | Europe | North America |
| 3 | 3 | stC839 | C | x | x | x |
| 4 | 4 | stG6792 | G | x | x | |
| 8 | 8 | stG480 | G | x | x | x |
| 8 | 8 | stG11 | G | x | x | |
| 8 | 38 | stG480 | G | x | x | x |
| 15 | 15 | stG10 | G | x | x | x |
| 15 | 15 | stG652 | G | x | x | |
| 15 | 15 | stG166b | G | x | x | |
| 17 | 17 | stG2078 | G | x | x | x |
| 17 | 12 | stG643 | G | x | x | |
| 20 | 20 | stG62647 | C | x | x | x |
| 25 | 25 | stG5420 | G | x | x | x |
| 25 | 25 | stG6 | G | x | x | |
| 29 | 29 | stC74a | G | x | x | x |
| 49 | 49 | stC36 | C | x | x |
Phylogenetic analysis of housekeeping genes
Clonal relationships established via goeBURST are based on the character state of the housekeeping gene allele, and do not take into account the degree of nucleotide sequence heterogeneity. In order to further investigate the relatedness of the housekeeping gene alleles at each locus, phylogenetic trees for each gene were constructed by the neighbor joining method. With the exception of atoB, these trees included the alleles from loci of GAS having the highest percentage nucleotide sequence identity based on BLASTn. Additionally, the housekeeping genes of GAS and SDSE share synteny (Table 2) [20]. In agreement with a previous report [25], several SDSE alleles are more similar to GAS alleles than to other SDSE alleles (Figures S1, S2). Both gki12 and mutS3 form a cluster with GAS alleles, whereas all gtr and murI alleles from SDSE and GAS segregate into distinct species-specific clusters. The relationship between recP and xpt alleles in the two species is more complex, and the phylogenies for the xpt and recP alleles do not resolve into species-specific clusters.
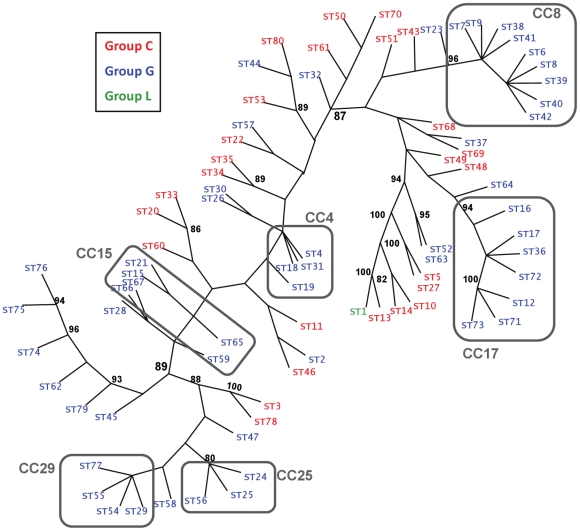
The seven housekeeping alleles were concatenated (3,134 nucleotide sites) for each of the 80 STs of SDSE, and the concatenates used to construct a phylogenetic tree by the maximum parsimony method (Figure 3). The relative distribution of STs along the tree branches is highly concordant with the CCs generated via the goeBURST clustering algorithm that used allele character states (Figure 1). A striking feature of the phylogenetic tree is that GCS and GGS taxa are highly interspersed and fail to form discrete evolutionary lineages, even in portions of the tree having strong bootstrap support. However, the homoplasy index for the phylogenetic tree is high (0.7331, excluding uninformative characters) and strong bootstrap support is absent for many of the deeper branches. Thus, the phylogeny may be less accurate for long term evolutionary events due to a past history of extensive recombination, but nevertheless, it appears to recapitulate the short term evolution detected by goeBURST. The analysis also provides additional support for the horizontal transfer of housekeeping genes between GCS and GGS organisms.
Figure 3. Maximum parsimony tree of concatenated housekeeping alleles.
The housekeeping alleles for each of the 80 STs for SDSE were concatenated (3,134 nt positions), and a maximum parsimony tree was constructed. The radial, unrooted phylogenetic tree is shown. Bootstrap values (500 replicates) showing branch support equal or greater than 80% are indicated; bootstrap analysis used a heuristic search and the 50% majority-rule consensus tree is presented. STs representing GCS and GGS are depicted in red and blue, respectively; the single group L isolate (ST1) is depicted in green. CCs having three or more STs are indicated. Characters: 2937 are constant, 56 variable characters are parsimony-uninformative, 141 are parsimony-informative. Consistency index (CI) = 0.3350; CI excluding uninformative characters = 0.2669; retention index (RI) = 0.7926.
Phylogenetic trees constructed by the minimum evolution (data not shown) or neighbor joining (Figure S4) methods also exhibit high concordance with the CCs generated via goeBURST. The only exception is CC15, which is divided into two or three small subclusters. However, like the maximum parsimony tree (Figure 3), there is little bootstrap support for deep branches in the neighbor joining tree (Figure S4).
Role of recombination in genetic change in SDSE
An analysis of recombination events between the GAS and SDSE housekeeping gene alleles, using the Recombination Detection Program (RDP) suite [26], predicts extensive recombination between GAS and SDSE in the gki gene. The gki75 and gki102 alleles of GAS were identified as having derived from recombination with SDSE alleles (p<0.00001 and p = 0.00037, respectively), whereas the gki12 and gki4 of SDSE appear to result from recombination with GAS alleles (both with p<0.00001). These data account for the high nucleotide percentage diversity observed in the gki locus. Recombination between GAS and SDSE could also be identified using RDP in the recP gene. The GAS recP21, recP40, recP54, recP71, recP85 are presumed to have resulted from recombination with SDSE alleles (p = 0.02469, p = 0.01073, p = 0.00050, p = 0.00900 and p = 0.00900, respectively). In contrast, SDSE alleles recP12 and recP3 seem to result from recombination with GAS alleles (p = 0.00262 and p = 0.00231, respectively). No significant recombination events between the other GAS and SDSE housekeeping alleles (gtr, murI, mutS, xpt) were observed using RDP. The SplitsTrees analysis for networked evolution was also used to assess intragenic recombination involving the MLST genes within the SDSE population. Statistically significant evidence for recombination (PHI test) was observed for murI, recP, xpt and atoB (p<0.01).
It is important to note that the recombination detection algorithms do not detect the complete replacement of the analyzed fragment. As pointed out previously, at least one instance of recombination involving the entire fragment of mutS analyzed is also suggested by phylogenetic analysis (Figure S2). Taken together, these data strongly suggest that intra- and inter-species recombination has occurred for several housekeeping genes.
The largest CC (CC8) contains 11.3% of the total STs, and is within the range of reliable performance of the BURST rules [27]. To estimate the relative role of recombination versus mutation in the short-term evolution of SDSE, the SLVs linked by goeBURST were examined for genetic differences between the variant alleles of each SLV pair. goeBURST identified 38 primary SLVs. Of these, 31 are predicted to have arisen via recombination, and seven SLVs are predicted to arise through point mutation. Thus, recombination occurred 4.4 times more often than mutation. The per site recombination to mutation (r/m) ratio was 20.7. An additional four SLVs, representing alternative ST relationships, are also present in the data set; of these, three SLVs are predicted to arise through recombination. The empirical findings on the relative contribution of recombination versus mutation to the genetic diversification of housekeeping genes are highly consistent with other findings that indicate extensive recombination involving SDSE.
Discussion
S. dysgalactiae subsp equisimilis is increasingly recognized as an important human pathogen that causes disease in many regions of the world. The findings of this report demonstrate that the major genotypes of SDSE have an intercontinental distribution. The recovery from all three continents of the likely founder ST of at least five CCs supports a model whereby a few successful clones have undergone extensive migration, followed by genetic diversification. Several of the descendents are also widely disseminated, indicative of subsequent waves of clonal migration.
SDSE is largely a commensal species, yet the vast majority of isolates evaluated in this study were recovered from cases of human disease. The mode of person-to-person transmission of SDSE has not been well-characterized, and there may be large differences among SDSE strains in terms of their virulence properties and ease of transmission to new hosts. It stands to reason that the most widely disseminated clones are probably among the most readily transmitted. Whether transmission is positively linked to virulence is an important question that remains to be established for SDSE. Molecular typing of SDSE, as provided in this report, provides a framework upon which the question of whether or not subpopulations of SDSE strains have heightened virulence can be addressed.
More than half (60%) of the SDSE isolates studied have a unique combination of emm type, ST and group carbohydrate, indicative of a very high level of genetic diversity within the species. The high level of strain diversity may be a consequence of a high rate of genetic change and/or a very large population size. Nonetheless, molecular typing using only the emm gene versus MLST, yields stratifications that are highly discordant, and neither method by itself is satisfactory for defining strains or clones. This finding provides support for a role of extensive horizontal gene transfer and recombination in promoting random associations of emm and ST.
Recombination following horizontal gene transfer in SDSE is observed at several levels of biological importance, involving core housekeeping genes and the emm gene, and perhaps even the genes encoding the group carbohydrate biosynthetic enzymes, albeit at a much lower frequency. Inter-specific gene transfer between SDSE and GAS is likely for several of the housekeeping genes. Interestingly, ST3 and ST78 isolates possess the SDSE recP6 and xpt2 alleles, which are identical to the GAS derived recP83 and xpt4 alleles respectively. Although it is formally possible that the recP and xpt alleles were transferred in a single genetic exchange event involving a large genome segment, this hypothesis is unlikely because SDSE-like gki and gtr alleles are positioned in between the xpt and recP loci on the SDSE genome.
Transfer of emm genes between SDSE and GAS also seem likely to have occurred, but to a much more limited extent, as evidenced only by the prototypical GAS emm types emm57 [25] and emm12 [28]. In addition to emm and the housekeeping genes, lateral exchange between GAS and SDSE is documented for several other genes, including those encoding a fibronectin-binding protein [29], a DNA gyrase subunit implicated in fluoroquinolone resistance [30], a transcriptional regulator of pilus gene expression [31], and the plasminogen activator streptokinase [32], [33]. Thus, SDSE and GAS share common gene pools for numerous loci.
Intra-species genetic exchange among SDSE organisms, involving either the core housekeeping genes or the emm gene, has been extensive. Data suggest recombination to be the predominant mechanism of genetic diversification among SDSE, occurring four times more often than point mutation in housekeeping loci. The per-site recombination to mutation ratio is also greater than 20. More than half of the STs represented by >one isolate are considered to be emm variable STs, and are found in association with at least two distinct emm types. ST15 is a particularly successful recipient of emm genes originating from multiple SDSE donor strains, as evidenced by its recovery in association with seven different emm types. Mechanisms that might explain the existence of STs having associations with many distinct emm types include possession of genetic machinery that increases their probability for recombinational success, additional accessory genes which facilitate their transmission to new hosts, a high prevalence (eg., ST15 comprises 20 of the 178 isolates) or natural selection favoring the emergence of variants harboring unique emm types.
Numerous emm gene donor-recipient ST pairs are also represented within the SDSE isolate data set, wherein 13 different emm types are associated with two or more genetically distant STs; in total, 21 distinct emm gene horizontal transfer events are suggested by the data. One emm type (stG643) is found in association with five distant STs or CCs. The emm gene of GAS is part of an ancient pathogenicity island [34]. Thus, it will be of interest to determine whether the stG643 gene is harbored by a functional mobile genetic element.
In GAS, the emm gene product (M protein) is a major virulence factor and a primary target of protective immunity by the human host. However, to our knowledge, it remains to be shown that the M protein of SDSE prevents phagocytosis in the absence of M type-specific antibody, which is a hallmark feature of GAS. Thus, the relationship between gene replacements involving the emm gene, and positive selection arising from host immune pressures, remains speculative for SDSE. The M proteins of GAS are also multifunctional proteins, which as a group, exhibits binding for numerous host proteins that include plasminogen, fibrinogen, immunoglobulins and complement regulatory proteins, as recently reviewed by Smeesters et al [35]. Like GAS, M proteins of SDSE that can bind plasminogen have been identified [36]. However, the full extent by which the M proteins of SDSE share functional attributes with the M protein of GAS remains to be established. The acquisition of new, M protein-mediated functional activities by a recipient SDSE strain could conceivably drive selection for the emergence of novel emm-ST combinations among SDSE.
Group carbohydrate is synthesized by biosynthetic enzymes whose genes have yet to be characterized. Among SDSE, the association between group C versus G carbohydrate and ST approaches 100% concordance. The observed linkage between group carbohydrate and ST is probably a large reflection of short term evolution. An evolutionary history of housekeeping gene exchange between GCS and GGS is evident from the phylogenetic tree of concatenated housekeeping gene sequences. While it is likely that GCS and GGS diverged from a common ancestor, subsequent genetic exchange masks that history, making it difficult to ascertain the extent to which GCS and GGS comprise distinct evolutionary lineages. However, MLST data makes it clear that lateral gene transfer leading to a group carbohydrate switch is a rare event.
The goeBURST population snapshot of SDSE (Figure 2) differs from that reported for GAS [16]. For GAS, only 2.8% of the STs are present in the largest CC, as compared to 11.2% of the SDSE STs (i.e., CC8). Based on simulated bacterial populations differing in levels of recombination and diversity generated by mutation [27], the population genetic structure of GAS is best explained by high rates of both recombination and mutation acting on a diverse set of housekeeping genes. For SDSE, the recombination rate and housekeeping gene diversity may be similarly high; however the mutation rate may be somewhat lower than in GAS. This proposed genetic structure for the SDSE population is supported by correspondence to simulated populations [27], combined with findings that show a greater than 4-fold excess of recombinational events in the diversification of SLVs.
SDSE appears to be among the closest extant relatives of GAS. In general terms, a pathogenic species of bacteria can arise from an organism of lower virulence following acquisition of virulence genes (eg., pathogenicity islands). However, evolution can also flow in the opposite direction, as recently evidenced by the loss of virulence genes and descent of the commensal Streptococcus mitis from the pathogen Streptococcus pneumoniae [37]. Determination of whether the most recent common ancestor of GAS and SDSE is more closely related to GAS or to SDSE, will probably require more extensive genomic analyses. The molecular typing and characterization of the population biology of SDSE in this report provides a foundation for future studies that address the evolution and molecular basis for virulence in SDSE.
Materials and Methods
Bacterial Strains
The 117 SDSE isolates analyzed in this study were collected from Australia (n = 55), Portugal (n = 36) and the USA (n = 11). Another 15 SDSE isolates were obtained from other countries or had no associated geographic information (Table S1). All isolates were classified as SDSE on the basis of isolation from a human host, β-hemolysis following growth on sheep blood-containing agar, the presence of group C or G carbohydrate, and the presence of a recognized emm type. emm type was determined by nucleotide sequence typing as described [38], and emm type was assigned using the BLASTn-emm server (http://www.cdc.gov/ncidod/biotech/strep/strepblast.htm). Group carbohydrate was determined using the latex bead agglutination test. Forty-five isolates were obtained from normally sterile tissue or fluid and classified as causing invasive disease; 59 isolates were recovered from other non-sterile sites and classified as causing non-invasive infections (Table 1). Information on the 61 invasive isolates recovered from the USA, and reported in a previous study [25], was integrated into the current analyses to obtain a global overview of the relationships between GGS and GCS causing infections in humans.
MLST
Seven housekeeping genes are used for MLST of SDSE. With the exception of atoB, the housekeeping genes used for MLST of SDSE are the same as those used in the GAS MLST scheme [38]. The seventh allele, atoB, (also called yqiZ) has been described [25]; the allele name has been changed for this report, in order to reduce confusion with the GAS MLST gene, yqiL, which occupies a different locus Primer sequences used for PCR amplification are listed in Table S2. Although our MLST scheme was developed independently from that reported previously [25], the data is directly comparable because the same genes, and regions within genes, are targeted for sequencing. All PCR products were sequenced in both the forward and reverse directions; newly identified alleles and alleles defining new STs were re-sequenced in order to validate the initial findings. Unique alleles at each locus were assigned individual allele numbers. The combination of the seven allele numbers for each isolate was used to define the sequence type (ST). MLST data for SDSE is available at www.mlst.net (pending publication).
Data analysis
goeBURST (http://goeburst.phyloviz.net/) [39], which uses the same clustering rules as eBURST [40] but provides a global optimal solution, was used to determine the relationships between STs. Clonal complexes (CCs) are defined as STs that are linked through single locus variants (SLVs) and are named on the basis of the predicted founder ST, which is the ST having the most SLVs. In cases where a CC contains only two STs, the lower numbered ST was used to define the CC. Isolates that share four of seven alleles (i.e. triple locus variants, TLVs) were used to define larger, more distantly related clonal groups.
The Simpson Index of Diversity (D) and Wallace coefficients (W) were calculated as described previously [41] using www.comparingpartitions.info. A D value equal to one signifies that the typing method distinguishes between all isolates, whereas a D value equal to zero means that all isolates are identical. The W coefficient provides a finer comparison between two typing methods, since the value indicates the probability that two strains classified as the same type by one method are also classified as the same type by the other method. A high value of the W coefficient (values close to 1) indicates that partitions defined by a given method could have been predicted from the results of another method, suggesting that the use of both methodologies could be redundant. Nucleotide diversity (π), nonsynonymous (dn) and synonymous substitution rates (ds) were calculated using DnaSP (version 5) [42].
Phylogenetic analysis
Phylogenetic relationships amongst individual housekeeping alleles was examined using the neighbor joining (NJ) method with Jukes-Cantor substitution algorithm model as implemented in MEGA4 [43]; bootstrapping (1000 replicates) was used to ascertain support for branches. For concatenated housekeeping alleles, evolutionary history was inferred using the NJ method or maximum parsimony method (PAUP 4.0).
Recombination and point mutation
Empirical estimates of the number of mutation and recombination events contributing to the diversification of SLVs were made as previously described [21], [44], with additional modifications. The nucleotide sequence differences in the mismatched allele among the pair of STs that define an SLV are scored as genetic changes that arise due to either mutation or recombination. Nucleotide differences between the two variant alleles at greater than 1 nucleotide site is scored as a probable recombination event. Single nt differences between the two variant alleles that do not occur in other any other alleles among the SDSE set of strains are scored as a change likely due to point mutation. If the single nucleotide polymorphism is present in two or more alleles assigned to different CCs, the genetic change is scored as likely due to recombination. The relative ratio of recombination events versus mutation events was then determined. To calculate the per site recombination/mutation (r/m) ratio, the total number of nucleotide sites that change due to recombination were divided by the total number of nucleotide sites that change due to mutation.
Recombination among SDSE and between the SDSE and GAS at each individual gene was evaluated using RDP [26], which implements a large number of methods (RDP, GENECONV, MaxChi, Chimaera, SiScan, 3Seq) for detecting intragenic recombination. For these analyses the entire collection of GAS alleles was downloaded from the MLST database (http://spyogenes.mlst.net). Since a high number of comparisons were performed, the p values reported are corrected for multiple tests. SplitsTrees4 was also used to assess intragenic recombination [45], excluding parsimony uninformative sites, and using the neighbor net algorithm, uncorrected P distance, and the Phi test for recombination [46].
Supporting Information
Characteristics of SDSE isolates in this study.
(0.25 MB DOC)
PCR primer pairs used for MLST in the study.
(0.03 MB PDF)
Evolutionary history of gki, gtr, murI and atoB alleles from SDSE and GAS. Evolutionary relationships were inferred using the Neighbour-Joining (NJ) method, and evolutionary distances calculated using the the Jukes-Cantor method. Branches with bootstrap support (n = 1000) greater than 80% are shown next to their respective branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Only names of the GGS alleles are shown. Phylogenetic analyses were conducted in MEGA4.
(0.12 MB TIF)
Evolutionary history of mutS, xpt and recP alleles from SDSE and GAS. Evolutionary relationships were inferred using the Neighbour-Joining (NJ) method, and evolutionary distances calculated using the the Jukes-Cantor method. Branches with bootstrap support (n = 1000) greater than 80% are shown next to their respective branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Only names of the GGS alleles are shown. Phylogenetic analyses were conducted in MEGA4.
(0.02 MB PDF)
goeBURST diagram of the relationships between 178 global SDSE isolates grouped up to TLV. The size of each circle is proportional to the number of isolates with that particular ST in a logarithmic scale. STs differing up to three alleles (triple-locus variants - TLVs) are linked by straight lines. Black lines link STs differing at a single gene. Intermediate grey lines link STs differing at a two genes. Light grey lines link STs differing at a three genes. Blue circles represent isolates that have the group G carbohydrate. Red circles represent isolates expressing the group C carbohydrate. Whenever isolates of the same ST have different group carbohydrates, the number of isolates bearing the same carbohydrate is proportional to the respective color. The green circle represents the single isolate expressing the group L carbohydrate. The proposed founders of particular clusters are indicated by a light green outer circle. The sub-founders (defined has having links to three or more STs) are indicated by dark green outer circles.
(0.02 MB PDF)
Neighbor joining tree of concatenated housekeeping alleles. The housekeeping alleles for each of the 80 STs for SDSE were concatenated (3,134 nt positions), and a neighbor joining tree was constructed (MEGA4). Bootstrap values (1,000 replicates) showing branch support equal or greater than 80% are indicated STs representing GCS and GGS are depicted in red and blue, respectively; the single group L isolate (ST1) is depicted in green. CCs having three or more STs are indicated.
(0.06 MB PDF)
Acknowledgments
We thank Awdhesh Kalia for providing genome sequence data that was used in the design of new oligonucleotide primers. We also thank Bernard Beall for providing additional details of previously described isolates [25].
Footnotes
Competing Interests: GlaxoSmithKline does not constitute a competing interest despite their financial support of one of the contributing laboratories. The company was not involved in the study design; collection, analysis and interpretation of data; writing of the paper; or decision to submit for publication. Upon publication, all raw data will be available on a public website (www.mlst.net) that allows all investigators to gain access to the data and perform independent analysis. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: DM is supported by an Australian Government NHMRC Program grant and NHMRC Project grant. DEB received support from the National Institutes of Health (AI-065572; GM-060793). Work conducted by MR was supported in part by Fundao para a Cincia e a Tecnologia (PTDC/SAU-ESA/72321/2006), Fundao Calouste Gulbenkian, Portugal and from an unrestricted grant from Glaxo Smithkline, Portugal. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Facklam R. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin Microbiol Rev. 2002;15:613–630. doi: 10.1128/CMR.15.4.613-630.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chhatwal GS, McMillan DJ, Talay SR. Pathogenicity Factors in Group C and G Streptococci. In: Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI, editors. Gram-positive pathogens. 2 ed. Washington D. C.: ASM Press; 2006. pp. 213–221. [Google Scholar]
- 3.Broyles LN, Van Beneden C, Beall B, Facklam R, Shewmaker PL, et al. Population-based study of invasive disease due to beta-hemolytic streptococci of groups other than A and B. Clin Infect Dis. 2009;48:706–712. doi: 10.1086/597035. [DOI] [PubMed] [Google Scholar]
- 4.Efstratiou A. Pyogenic streptococci of Lancefield groups C and G as pathogens in man. Soc Appl Bacteriol Symp Ser. 1997;26:72S–79S. [PubMed] [Google Scholar]
- 5.Pinho MD, Melo-Cristino J, Ramirez M. Clonal relationships between invasive and noninvasive Lancefield group C and G streptococci and emm-specific differences in invasiveness. J Clin Microbiol. 2006;44:841–846. doi: 10.1128/JCM.44.3.841-846.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Humar D, Datta V, Bast DJ, Beall B, De Azavedo JC, et al. Streptolysin S and necrotising infections produced by group G streptococcus. Lancet. 2002;359:124–129. doi: 10.1016/S0140-6736(02)07371-3. [DOI] [PubMed] [Google Scholar]
- 7.Woo PC, Fung AM, Lau SK, Wong SS, Yuen KY. Group G beta-hemolytic streptococcal bacteremia characterized by 16S ribosomal RNA gene sequencing. J Clin Microbiol. 2001;39:3147–3155. doi: 10.1128/JCM.39.9.3147-3155.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hashikawa S, Iinuma Y, Furushita M, Ohkura T, Nada T, et al. Characterization of group C and G streptococcal strains that cause streptococcal toxic shock syndrome. J Clin Microbiol. 2004;42:186–192. doi: 10.1128/JCM.42.1.186-192.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korman TM, Boers A, Gooding TM, Curtis N, Visvanathan K. Fatal case of toxic shock-like syndrome due to group C streptococcus associated with superantigen exotoxin. J Clin Microbiol. 2004;42:2866–2869. doi: 10.1128/JCM.42.6.2866-2869.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bramhachari PV, Kaul SY, McMillan DJ, Shaila MS, Karmarkar MG, et al. Disease burden due to Streptococcus dysgalactiae subsp. equisimilis (group G and C streptococcus) is higher than that due to Streptococcus pyogenes among Mumbai school children. J Med Microbiol. 2010;59:220–223. doi: 10.1099/jmm.0.015644-0. [DOI] [PubMed] [Google Scholar]
- 11.McDonald M, Towers RJ, Andrews RM, Carapetis JR, Currie B. Epidemiology of Streptococcus dysgalactiae subsp. equisimilis in Tropical Communities, Northern Australia. Emerg Infect Dis. 2007;13:1694–1700. doi: 10.3201/eid1311.061258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka D, Isobe J, Watahiki M, Nagai Y, Katsukawa C, et al. Genetic features of clinical isolates of Streptococcus dysgalactiae subsp. equisimilis possessing Lancefield's group A antigen. J Clin Microbiol. 2008;46:1526–1529. doi: 10.1128/JCM.02188-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies MR, McMillan DJ, Beiko RG, Barroso V, Geffers R, et al. Virulence profiling of Streptococcus dysgalactiae subspecies equisimilis isolated from infected humans reveals 2 distinct genetic lineages that do not segregate with their phenotypes or propensity to cause diseases. Clin Infect Dis. 2007;44:1442–1454. doi: 10.1086/516780. [DOI] [PubMed] [Google Scholar]
- 14.Ikebe T, Murayama S, Saitoh K, Yamai S, Suzuki R, et al. Surveillance of severe invasive group-G streptococcal infections and molecular typing of the isolates in Japan. Epidemiol Infect. 2004;132:145–149. doi: 10.1017/s0950268803001262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Igwe EI, Shewmaker PL, Facklam RR, Farley MM, van Beneden C, et al. Identification of superantigen genes speM, ssa, and smeZ in invasive strains of beta-hemolytic group C and G streptococci recovered from humans. FEMS Microbiol Lett. 2003;229:259–264. doi: 10.1016/S0378-1097(03)00842-5. [DOI] [PubMed] [Google Scholar]
- 16.Bessen DE. Population biology of the human restricted pathogen, Streptococcus pyogenes. Infect Genet Evol. 2009;9:581–593. doi: 10.1016/j.meegid.2009.03.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bessen DE, Lizano S. Tissue tropisms in group A streptococcal infection. Future Microbiology. 2010 doi: 10.2217/fmb.10.28. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cleary PP, Kaplan EL, Handley JP, Wlazlo A, Kim MH, et al. Clonal basis for resurgence of serious Streptococcus pyogenes disease in the 1980s. Lancet. 1992;339:518–521. doi: 10.1016/0140-6736(92)90339-5. [DOI] [PubMed] [Google Scholar]
- 19.Shulman ST, Tanz RR, Dale JB, Beall B, Kabat W, et al. Seven-year surveillance of north american pediatric group a streptococcal pharyngitis isolates. Clin Infect Dis. 2009;49:78–84. doi: 10.1086/599344. [DOI] [PubMed] [Google Scholar]
- 20.Enright MC, Spratt BG, Kalia A, Cross JH, Bessen DE. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect Immun. 2001;69:2416–2427. doi: 10.1128/IAI.69.4.2416-2427.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGregor KF, Spratt BG, Kalia A, Bennett A, Bilek N, et al. Multilocus sequence typing of Streptococcus pyogenes representing most known emm types and distinctions among subpopulation genetic structures. J Bacteriol. 2004;186:4285–4294. doi: 10.1128/JB.186.13.4285-4294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakota V, Fry AM, Lietman TM, Facklam RR, Li Z, et al. Genetically diverse group A streptococci from children in far-western Nepal share high genetic relatedness with isolates from other countries. J Clin Microbiol. 2006;44:2160–2166. doi: 10.1128/JCM.02456-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones N, Bohnsack JF, Takahashi S, Oliver KA, Chan MS, et al. Multilocus sequence typing system for group B streptococcus. J Clin Microbiol. 2003;41:2530–2536. doi: 10.1128/JCM.41.6.2530-2536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Springman AC, Lacher DW, Wu G, Milton N, Whittam TS, et al. Selection, recombination, and virulence gene diversity among group B streptococcal genotypes. J Bacteriol. 2009;191:5419–5427. doi: 10.1128/JB.00369-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmad Y, Gertz RE, Jr, Li Z, Sakota V, Broyles LN, et al. Genetic relationships deduced from emm and multilocus sequence typing of invasive Streptococcus dysgalactiae subsp. equisimilis and S. canis recovered from isolates collected in the United States. J Clin Microbiol. 2009;47:2046–2054. doi: 10.1128/JCM.00246-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin DP. Recombination detection and analysis using RDP3. Methods Mol Biol. 2009;537:185–205. doi: 10.1007/978-1-59745-251-9_9. [DOI] [PubMed] [Google Scholar]
- 27.Turner KM, Hanage WP, Fraser C, Connor TR, Spratt BG. Assessing the reliability of eBURST using simulated populations with known ancestry. BMC Microbiol. 2007;7:30. doi: 10.1186/1471-2180-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simpson WJ, Musser JM, Cleary PP. Evidence consistent with horizontal transfer of the gene (emm12) encoding serotype M12 protein between group A and group G pathogenic streptococci. Infect Immun. 1992;60:1890–1893. doi: 10.1128/iai.60.5.1890-1893.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Towers RJ, Gal D, McMillan D, Sriprakash KS, Currie BJ, et al. Fibronectin-binding protein gene recombination and horizontal transfer between group A and G streptococci. J Clin Microbiol. 2004;42:5357–5361. doi: 10.1128/JCM.42.11.5357-5361.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinho MD, Melo-Cristino J, Ramirez M. Fluoroquinolone resistance in Streptococcus dysgalactiae subsp. equisimilis and evidence for a shared global gene pool with Streptoccocus pyogenes. Antimicrob Agents Chemother. doi: 10.1128/AAC.01377-09. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bessen DE, Manoharan A, Luo F, Wertz JE, Robinson DA. Evolution of transcription regulatory genes is linked to niche specialization in the bacterial pathogen Streptococcus pyogenes. J Bacteriol. 2005;187:4163–4172. doi: 10.1128/JB.187.12.4163-4172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalia A, Bessen DE. Natural selection and evolution of streptococcal virulence genes involved in tissue-specific adaptations. J Bacteriol. 2004;186:110–121. doi: 10.1128/JB.186.1.110-121.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Musser JM, Kapur V, Szeto J, Pan X, Swanson DS, et al. Genetic diversity and relationships among Streptococcus pyogenes strains expressing serotype M1 protein: recent intercontinental spread of a subclone causing episodes of invasive disease. Infect Immun. 1995;63:994–1003. doi: 10.1128/iai.63.3.994-1003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panchaud A, Guy L, Collyn F, Haenni M, Nakata M, et al. M-protein and other intrinsic virulence factors of Streptococcus pyogenes are encoded on an ancient pathogenicity island. BMC Genomics. 2009;10:198. doi: 10.1186/1471-2164-10-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smeesters PR, McMillan DJ, Sriprakash KS. The streptococcal M protein: a highly versatile molecule. Trends Microbiol. 2010 doi: 10.1016/j.tim.2010.02.007. In press. [DOI] [PubMed] [Google Scholar]
- 36.Ben Nasr A, Wistedt A, Ringdahl U, Sjobring U. Streptokinase activates plasminogen bound to human group C and G streptococci through M-like proteins. Eur J Biochem. 1994;222:267–276. doi: 10.1111/j.1432-1033.1994.tb18865.x. [DOI] [PubMed] [Google Scholar]
- 37.Kilian M, Poulsen K, Blomqvist T, Havarstein LS, Bek-Thomsen M, et al. Evolution of Streptococcus pneumoniae and its close commensal relatives. PLoS One. 2008;3:e2683. doi: 10.1371/journal.pone.0002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beall B, Facklam R, Thompson T. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J Clin Microbiol. 1996;34:953–958. doi: 10.1128/jcm.34.4.953-958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Francisco AP, Bugalho M, Ramirez M, Carrico JA. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics. 2009;10:152. doi: 10.1186/1471-2105-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carrico JA, Silva-Costa C, Melo-Cristino J, Pinto FR, de Lencastre H, et al. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J Clin Microbiol. 2006;44:2524–2532. doi: 10.1128/JCM.02536-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 43.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 44.Feil EJ, Enright MC, Spratt BG. Estimating the relative contributions of mutation and recombination to clonal diversification: a comparison between Neisseria meningitidis and Streptococcus pneumoniae. Res Microbiol. 2000;151:465–469. doi: 10.1016/s0923-2508(00)00168-6. [DOI] [PubMed] [Google Scholar]
- 45.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 46.Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics of SDSE isolates in this study.
(0.25 MB DOC)
PCR primer pairs used for MLST in the study.
(0.03 MB PDF)
Evolutionary history of gki, gtr, murI and atoB alleles from SDSE and GAS. Evolutionary relationships were inferred using the Neighbour-Joining (NJ) method, and evolutionary distances calculated using the the Jukes-Cantor method. Branches with bootstrap support (n = 1000) greater than 80% are shown next to their respective branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Only names of the GGS alleles are shown. Phylogenetic analyses were conducted in MEGA4.
(0.12 MB TIF)
Evolutionary history of mutS, xpt and recP alleles from SDSE and GAS. Evolutionary relationships were inferred using the Neighbour-Joining (NJ) method, and evolutionary distances calculated using the the Jukes-Cantor method. Branches with bootstrap support (n = 1000) greater than 80% are shown next to their respective branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Only names of the GGS alleles are shown. Phylogenetic analyses were conducted in MEGA4.
(0.02 MB PDF)
goeBURST diagram of the relationships between 178 global SDSE isolates grouped up to TLV. The size of each circle is proportional to the number of isolates with that particular ST in a logarithmic scale. STs differing up to three alleles (triple-locus variants - TLVs) are linked by straight lines. Black lines link STs differing at a single gene. Intermediate grey lines link STs differing at a two genes. Light grey lines link STs differing at a three genes. Blue circles represent isolates that have the group G carbohydrate. Red circles represent isolates expressing the group C carbohydrate. Whenever isolates of the same ST have different group carbohydrates, the number of isolates bearing the same carbohydrate is proportional to the respective color. The green circle represents the single isolate expressing the group L carbohydrate. The proposed founders of particular clusters are indicated by a light green outer circle. The sub-founders (defined has having links to three or more STs) are indicated by dark green outer circles.
(0.02 MB PDF)
Neighbor joining tree of concatenated housekeeping alleles. The housekeeping alleles for each of the 80 STs for SDSE were concatenated (3,134 nt positions), and a neighbor joining tree was constructed (MEGA4). Bootstrap values (1,000 replicates) showing branch support equal or greater than 80% are indicated STs representing GCS and GGS are depicted in red and blue, respectively; the single group L isolate (ST1) is depicted in green. CCs having three or more STs are indicated.
(0.06 MB PDF)