Abstract
Current strategies for deploying synthetic nanocarriers involve the creation of agents that incorporate targeting ligands, imaging agents, and/or therapeutic drugs into particles as an integral part of the formulation process. Here we report the development of an amphipathic peptide linker that enables postformulation editing of payloads without the need for reformulation to achieve multiplexing capability for lipidic nanocarriers. To exemplify the flexibility of this peptide linker strategy, 3 applications were demonstrated: converting nontargeted nanoparticles into targeting vehicles; adding cargo to preformulated targeted nanoparticles for in vivo site-specific delivery; and labeling living cells for in vivo tracking. This strategy is expected to enhance the clinical application of molecular imaging and/or targeted therapeutic agents by offering extended flexibility for multiplexing targeting ligands and/or drug payloads that can be selected after base nanocarrier formulation.—Pan, H., Myerson, J. W., Ivashyna, O., Soman, N. R., Marsh, J. N., Hood, J. L., Lanza, G. M., Schlesinger, P. H., Wickline, S. A.. Lipid membrane editing with peptide cargo linkers in cells and synthetic nanostructures.
Keywords: molecular imaging, targeted drug delivery, biomarker, nanoparticles, liposome
The optimal design and deployment of nanotechnologies for diagnosis and treatment of complex diseases will require the ready incorporation of multiplexed targeting ligands and drug combinations to improve cell-specific delivery and therapeutic efficacy (1,2,3). In view of the considerable overlap in gene expression between physiological and pathological processes during the evolution of a disease, the use of a unique signature for a pathological condition would be enhanced by the simultaneous detection of diverse molecular biomarkers (4,5,6,7). To date, typical cargos such as targeting ligands, imaging agents, and/or drugs are incorporated into the nanocarriers during the formulation process, which requires dedicated particle design and formulation for each individual application. However, the potential of targeted nanocarriers to achieve improved selectivity and combinatorial therapy against complex diseases may not be fully exploited if flexible multiply targeted and/or drug loadings are not developed and deployed.
In contrast, a strategy designed to accommodate swapping and/or combining multiple cargos in generic base nanocarriers would enable flexible and personalized customization of the nanocarriers for application to specific pathologic lesions and unique disease stages. Despite the apparent regulatory challenges to the clinical adoption of such a paradigm, the design of flexible nanocarriers with polyvalent and rapidly swappable targeting and drug delivery cargos represents a necessary first step. Among numerous existing nanoparticles, lipidic nanoparticles either have U.S. Food and Drug Administration approval or are in clinical trials. The potential of rapid clinical translation continues to inspire researchers and scientists to generate novel applications for lipidic nanoparticles (8). To fully exploit and extend existing lipidic nanoparticle carriers, we now report the design, characterization, and evaluation of a peptide linker that can quickly load cargoes into lipidic nanoparticles after they are formulated and stored. The source for this linker derives from the bee venom peptide component, melittin, which has been shown previously to insert into lipid membranes and cause disruption (9, 10). We have demonstrated that this modified peptide linker can insert into both perfluorocarbon (PFC) nanoparticles and liposomes without affecting the integrity of these nanocarriers. By using this linker strategy, VCAM-1-targeted PFC nanoparticles were generated, and their specific molecular targeting was quantified by magnetic resonance spectroscopy; the VCAM-1-targeted drug Doxil (doxorubicin HCl) presents enhanced cellular drug delivery. Moreover, by loading cargo into ανβ3-integrin-targeted PFC nanoparticles, we achieved in vivo cargo delivery using this linker approach. This approach preserves the integrity of both the manufacturing process and the product for the base nanocarrier, while enabling the addition of myriad cargos after formulation. Furthermore, we showed that this strategy could also load cargos onto living cells for in vivo tracking or cell-based Trojan horse therapeutic applications.
MATERIALS AND METHODS
Nanoparticle preparation
PFC nanoparticle emulsions were formulated using methods described previously (11). In brief, a lipid/surfactant comixture of 99 mol% egg lecithin and 1 mol% dipalmitoyl-phosphatidylethanolamine (Avanti Polar Lipids, Piscataway, NJ, USA) was dissolved in methanol-chloroform(1:3, v/v). The solvent was evaporated under reduced pressure to produce a lipid film, which was dried in a 50°C vacuum oven overnight to obtain the surfactant. Then the surfactant (2.0%, w/v), PFC (Gateway Specialty Chemicals, St. Peters, MO, USA) (20%, w/v), and distilled, deionized water were blended and emulsified at 20,000 psi for 4 min in an ice bath (S110 Microfluidics emulsifier; Microfluidics, Newton, MA, USA). For fluorescent nanoparticles, Alexa Fluor 488 was incorporated into the surfactant layer. For ανβ3-integrin-targeted nanoparticles, 0.1 mol% of a peptidomimetic vitronectin antagonist (U.S. patent 6,322,770) linked to phosphatidylethanolamine was incorporated into the surfactant layer.
Giant unilamellar vesicle (GUV) preparation
GUVs were prepared by the electroformation method (12) from a lipid mixture containing 99.9 mol% of 1,2-dioleoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids) and 0.1 mol% of the fluorescent dye DiD (Invitrogen, Carlsbad, CA, USA). In brief, a chloroform mixture of lipids and dye at 2 mg/ml total lipid concentration was dried on a surface of two parallel platinum electrodes, resulting in the creation of a thin lipid film on each electrode. Next, platinum electrodes were immersed in a chamber containing 300 mM sucrose solution and connected to a power generator. Electroformation of GUVs attached to the platinum electrodes was performed at 2.3 V and 10 Hz for 1 h at room temperature followed by the detachment of GUVs from the platinum electrodes at 2.3 V and 2 Hz for 30 min.
Peptide synthesis
All peptides were purchased from GenScript USA Inc. (Piscataway, NJ, USA). D1-7 and TCP1 were also synthesized automatically by Fmoc solid-phase peptide synthesis in our laboratory. The product identities were assessed by mass spectrometry (Washington University Proteomics Center, St. Louis, MO, USA), and the purity (>95%) was confirmed by analytical HPLC.
Incorporation of peptides onto PFC nanoparticles and Doxil
Incorporation procedures were described previously (13). By controlling the amount of peptide added, the lipid/peptide ratios of peptide-loaded nanoparticles ranged from 300 to 30. The peptide-liposome complex was prepared by mixing Doxil (obtained from Barnes-Jewish Hospital Pharmacy, St. Louis, MO, USA) with 10 mM TCP1 at 30:1 (v/v).
Cell culture
All of the cells were maintained in a humidified atmosphere of 95% air and 5% CO2. The murine 2F2B endothelial cells (American Type Culture Collection, Manassas, VA, USA) were maintained in DMEM with 10% (v/v) heart-inactivated FBS (Sigma-Aldrich), and 1% (v/v) sodium pyruvate (Washington University Tissue Culture Support Center, St. Louis, MO, USA). The human C32 melanoma cells (Washington University Tissue Culture Support Center) were maintained in modified Eagle’s medium containing Earle’s salts and 10% (v/v) FBS. The murine monocytic RAW 246.7 cells (Washington University Tissue Culture Support Center) were maintained in DMEM with 10% (v/v) heart-inactivated FBS.
Cell viability assay
An XTT cell viability assay kit (Biotium, Hayward, CA, USA) was used to assess the cytotoxicity of mutated melittin peptides at different concentrations and the cell-killing effect of Doxil with or without editing at indicated volumes in 1 ml of final treatment volume. For testing peptides, 2F2B cells were seeded in the 96-well plates at a seeding density of 20,000 cells/well. After 24-h incubations, cells were treated with different peptides at selected concentrations for 3 h. For testing Doxil with or without editing, 2F2B cells were seeded in the 24-well plates at a seeding density of 53,333 cells/well. Cells were treated with targeted or nontargeted Doxil for 48 h. The XTT cell viability assay was then performed according to the manufacturer’s instructions.
Surface plasmon resonance (SPR)
The kinetics of D1-7, TCP1, and TCP2 insertion in lipid monolayers of PFC nanoparticles was studied by SPR. SPR detects change in the reflective index of a surface (Biacore X100 and carboxymethylated dextran chip L1; Biacore Inc., Piscataway, NJ, USA). A uniform lipid monolayer on an L1 chip was created by injecting PFC nanoparticles (3 μl/min) for 30 min. Loosely deposited nanoparticles were removed by performing extra washing steps after immobilization to ensure a stable baseline. Complete coverage was confirmed by injecting BSA (1 mg/ml in PBS) at 15 μl/min for 2 min. Different peptides in selected concentrations were injected at a flow rate of 30 μl/min for 1 min. At the end of each experiment the chip was regenerated by two consecutive injections of 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (50 μl; 100 μl/min). The data were analyzed with BIAevaluation software (Biacore Inc.). A two-state model was used for curve fitting (Eq. 1):
 |
1 |
where P and L represent the peptide and lipid membrane of PFC nanoparticles, respectively; PL and PL* represent the electrostatic and the hydrophobic interaction between peptide and lipid membrane, respectively; the unit of the rate constant ka1 is 1/M · s (second-order reaction); and the unit of the rate constants kd1, ka2, and kd2 is 1/s (first-order reactions).
Size distribution and ζ potential of nanoparticles
The size distributions of the nanoparticles with or without cargo incorporated were analyzed by dynamic light scattering (Brookhaven Instruments Corp., Holtsville, NY, USA). The size distribution was plotted by particle number. ζ-Potential values were determined with a PALS Zeta Potential Analyzer (Brookhaven Instruments Corp.). Data were acquired in the phase-analysis light-scattering (PALS) mode after solution equilibration at 25°C.
Electron microscopy
The effect of D1-7 on PFC nanoparticles was examined by transmission electron microscopy. Procedures have been described in detail previously (14).
Circular dichroism (CD) spectroscopy
A J-810 spectropolarimeter (Jasco Inc., Easton, MD, USA) was used for CD spectra measurements of free D1-7 and lipid-bound D1-7 (lipid/peptide molar ratio 8:1). Spectra were scanned in a 1-mm path length quartz cuvette in the far-UV range from 190 to 260 nm at a scan rate of 100 nm/min, and all spectra were collected under argon. An average of 20 scans was used for all spectra. Buffer used was 10 mM potassium phosphate buffer, pH 7.0.
Immunofluorescence
Expression of VCAM-1 on 2F2B cells was determined by using antibody to VCAM-1 (clone 429, MVCAM.A; BD Pharmingen, San Jose, CA, USA) at a dilution of 1:50, along with FITC-conjugated secondary antibody (BD Pharmingen) at dilution of 1:50.
Real-time RT-PCR
RNA was extracted from 2F2B cells with or without TNF-α stimulation by using an RNeasy Mini Kit (Qiagen, Valencia, CA, USA). Mouse endothelial cell biology RT2 PCR Array, RT2 First Strand Kit, and RT2 First SYBR Green/ROX PCR Master Mix were purchased from SABiosciences (Frederick, MD, USA). Real-time RT-PCR was performed on an ABI 7300 system (Applied Biosystems, Foster City, CA, USA).
Confocal microscopy
VCAM-targeted or nontargeted Alexa Fluor 488 colabel PFC nanoparticles were incubated with 4-h TNF-α (10 ng/ml)-stimulated 2F2B cells for 1 h at 37°C. Fluorescein-D1-7-loaded PFC nanoparticles with or without targeting ligands (a lipid-linked peptidomimetic vitronectin antagonist to the ανβ3-integrin receptor) were incubated with C-32 melanoma cells for 1 h at 37°C. Unbound nanoparticles were removed by washing 5 times with PBS. VCAM-targeted or nontargeted Doxil was incubated with 4-h TNF-α (10 ng/ml) -stimulated 2F2B cells for 2.5 h at 37°C. Unbound Doxil was removed by washing 5 times with PBS. Then cells were fixed in paraformaldehyde (4% in PBS) for 5 min at 37°C and visualized by using a confocal microscope (Meta 510; Carl Zeiss, Thornwood, NY, USA). Fluorescence imaging of cells was performed with a confocal microscope using standard filter sets. Concomitant differential interference contrast imaging was used to determine the location of nanoparticles in the cells. For GUV observation, 50 μl of solution containing GUVs was transferred into a Lab-Tek observation chamber (Fisher Scientific, Pittsburgh, PA, USA) containing 450 μl of 10 mM HEPES, pH 7.2, 100 mM KCl, and 20 μM Alexa Fluor 546 (Invitrogen). Alexa Fluor 546 dye was used to assess the permeabilization of GUVs. Observation of GUVs and confocal microscopy were done on an LSM 510 microscope (Carl Zeiss).
19F magnetic resonance spectroscopy (MRS) and magnetic resonance imaging (MRI) at 11.7 T
19F MRS and MRI of 2F2B cells were performed on a Varian 11.7-T scanner using a custom-designed 0.5-cm 4-turn solenoid radiofrequency (RF) coil per published procedures (15). Cells were contained within a centrifuge tube and analyzed together with an internal standard of 3, 0.3, or 0.03 μl of PFC nanoparticle. 19F MRS (number of averages=256, 90° flip angle, acquisition time: 6 min) of cells was performed for quantitative evaluation of intracellular labeling of nanoparticles using a spin-echo sequence. Proton and 19F spin-echo images were exactly coregistered. The proton images were acquired over two signal averages with a 128 × 256 matrix and a 1.5 × 2 cm field of view with a 1.5-mm depth. A 1-s repetition time and 0.017-s echo time were used. Fluorine images used 16 signal averages, a 64 × 128 matrix, and an identical field of view with a 1-cm depth. A 1.25-s repetition time and 0.01-s echo time were used.
Animal experimental protocol
The experimental and humane animal care protocols were approved by the Animal Care Committee of the Washington University School of Medicine. Tumor xenografts for in vivo tumor site delivery (male C57BL/6 mice; National Cancer Institute, Bethesda, MD, USA) at the age of 4–6 wk were subcutaneously inoculated into the inguinal region with 1 × 106 B16F10 cells in 50 μl of sterile saline. Ten days after tumor implantation, the tumor-bearing mice were injected intravenously with ανβ3-targeted nanoparticles with or without FITC loading (1 ml/kg), and nanoparticles were allowed to circulate and bind to the neovasculature for 2 h before the tumors were excised, snap-frozen, and sliced for fluorescence visualization using a BX61 microscope (Olympus America Inc., Center Valley, PA, USA). For in vivo optical imaging of circulating red blood cells, 1 ml of whole blood was withdrawn from the left ventricle of a male donor athymic nude mouse (nu/nu) (National Cancer Institute) and RBCs were isolated after centrifugation. After cell labeling with FITC-D1-7, RBCs suspended in 50 μl of sterile saline were intravenously injected into athymic nude mice (nu/nu) for in vivo fluorescence imaging.
In vivo ultrasound tumor imaging
Tumor growth was confirmed by in vivo transcutaneous ultrasound imaging with an animal ultrasound imager. Procedures and image processing have been described in detail previously (13). In brief, a high-frequency ultrasound imaging system (Vevo 660; VisualSonics, Toronto, ON, Canada) was used to acquire backscatter data from the mouse tumors. The system was modified to output analog RF data and associated trigger signals to permit digitization of the raw RF waveforms. Waveforms were digitized at 500 MHz with an 8-bit digitizer (CS82G; Gage Applied Technologies, Lockport, IL, USA) and stored for offline analysis. The transducer probe (40-MHz wobbler, 6-mm focal length, and 20-Hz frame rate) was affixed to a motorized gantry under computer control to enable automated scanning of the probe across the length of the tumor. Each anesthetized animal was placed on its back on a platform beneath the probe, and a small amount of ultrasound coupling gel was applied to the area proximal to the tumor. The probe was positioned so that the central area of the tumor was situated in the focal region of the transducer. RF data corresponding to cross-sectional views of the tumor were acquired at multiple sites along the length of the tumor, so that the entire tumor volume was interrogated. The probe was translated laterally (perpendicularly to the swing of the wobbler) in 100-μm steps between each scan plane acquisition.
Tumor histology
Frozen acetone-fixed tumor sections were stained with hematoxylin and eosin. Expression of the endothelium-specific marker platelet/endothelial cell adhesion molecule 1 (PECAM-1) were determined by using monoclonal antibody to PECAM-1 (clone MEC13.3; BD Biosciences) at dilutions of 1:50. The antibody was developed using an ABC Kit and VIP Substrate Kit (Vector Laboratories, Burlingame, CA, USA) with methyl green nuclear staining.
Cell labeling
RAW 246.7 cells were incubated with FITC-linker peptide (10 μM) for 1 h at 4°C. Subsequently, cells were washed and analyzed by using flow cytometry (CyAn ADP with Summit Software; Dako, Carpinteria, CA, USA). RBCs were incubated with FITC-linker peptide (10 μM) for 1 h at 4°C. After incubation, the RBCs were washed and visualized with a BX61 microscope (Olympus America).
In vivo optical mice imaging
In vivo fluorescence images were acquired and analyzed with a Xenogen IVIS Spectrum imaging system (Caliper LifeSciences, Hopkinton, MA, USA). During the image acquisition, the mice were maintained under isoflurane inhalation anesthesia. The settings (excitation, 500 nm; emission, 540 nm; exposure time, 0.5 s; binning factor, 8; f value, 2; and field of view, 12.9) were used for imaging acquisitions from the animals intravenously injected with fluorescence-labeled RBCs. For the control animals, all of the settings were the same except that exposure time was 1 s. The resultant pseudocolor “efficiency image” was used to illustrate the systemic distribution of the labeled cells.
RESULTS
Modification of melittin attenuates its pore-forming and lytic activity
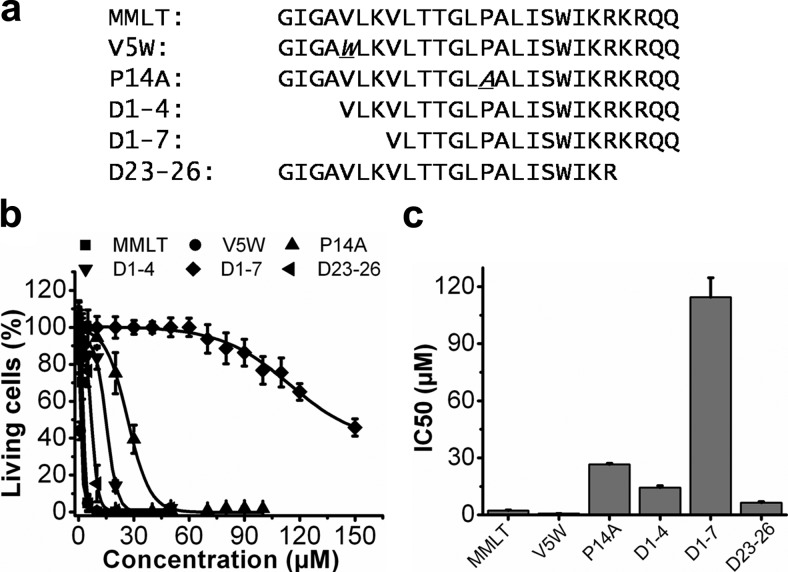
Previously we demonstrated that the amphipathic host-defense peptide melittin rapidly inserts and remains stably bound in the lipid monolayer surrounding the PFC nanoparticles for in vivo application (13, 14). We hypothesized that melittin could be engineered to attenuate its lytic activity while retaining its property of stable insertion into the lipidic nanocarriers. Melittin was altered by point mutations and truncations (Fig. 1a). For each peptide, cytotoxicity was assessed in the murine 2F2B endothelial cells by the XTT cell viability assay (Fig. 1b). Among all the peptide mutations tested, the cytotoxicity index of D1-7 (114.4±10.2 μM) was reduced most compared with melittin modified with N-terminal acetylation and C-terminal amidation (MMLT) (2.362±0.228 μM), representing an ∼50-fold decrease in cytotoxic effect due to the reduced lytic activity of the modified peptide compared with MMLT (Fig. 1c).
Figure 1.
Peptide mutations designed to alter melittin lytic activity. a) Sequences of melittin peptide mutations. Sequences are aligned; point mutations are marked in italic with underscore. Point mutations chosen were V5W and P14A. For V5W, the 5th amino acid, valine, was substituted by tryptophan. For P14A, the 14th amino acid, proline, was substituted by alanine. The 3 truncations were D1-4, D1-7, and D23-26. D1-4 and D1-7 had the first 4 and 7 aa on the N terminus deleted, and D23-26 had the last 4 aa on the C terminus deleted. The MMLT retained the native melittin sequence. All of the peptides contained N-terminal acetylation and C-terminal amidation to mimic the situation of cargo attachment that would apply when these peptides would be used as a linker on the nanocarriers. b) Cell viabilities after treatment with melittin mutants at selected concentrations. Shown is 2F2B cell viability induced by 3 h incubation with MMLT (▪), V5W (•), P14A (▴), D1-4 (▾), D1-7 (♦), or D23-26 (◂). Data were fitted with a Boltzmann equation. c) IC50 values (μM) of MMLT, V5W, P14A, D1-4, D1-7, and D23-26. All data points represent means ± sd (n=4).
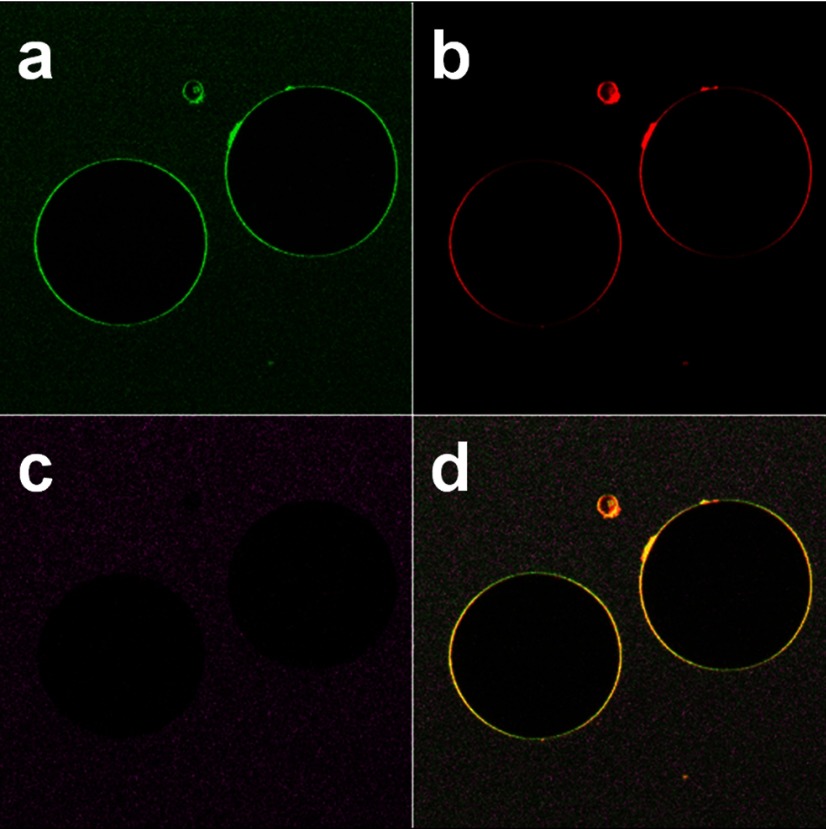
As demonstrated in our previous study, melittin destroys liposomes at a peptide to lipid molar ratio of 1:40 (13). To further demonstrate the attenuated lytic activity of D1-7 in a lipid bilayer system, we conjugated FITC onto the N terminus of D1-7 and visualized the integrity of the DiD-labeled GUV membrane by confocal microscopy at a peptide/lipid molar ratio of 1:40. The fluorescence from the FITC-labeled linker peptides (Fig. 2a, green rings) and that from GUV (Fig. 2b, red rings) were colocalized (Fig. 2d, yellow rings). In addition, we added a soluble dye, Alexa Fluor 456, outside the GUV to test the permeability of the GUV membrane after the membrane insertion of the D1-7. As shown in Fig. 2c, the dye fluorescence signal could only be detected outside of the GUV. This result indicates that D1-7 was inserted into the lipid membrane of GUVs without affecting the membrane permeability. These observations indicate that D1-7 exhibits dramatically attenuated lytic activity compared with that of melittin. Therefore, D1-7 was selected as a candidate peptide, and its utility was assessed as a linker for membrane insertion into PFC nanoparticles.
Figure 2.
Lipid integrity remains intact after the D1-7 incorporation. Confocal microscope images show linker peptide incorporated onto the lipid membrane GUVs. a) Confocal image of FITC-labeled linker peptide. b) Confocal image of GUV membranes labeled with the lipophilic dye DiD. c) Confocal image of soluble dye, Alexa Fluor 456, added outside the GUV. d) Colocalization of FITC-labeled linker peptides and lipid membrane GUV.
Stable insertion of D1-7 peptide into PFC nanoparticles
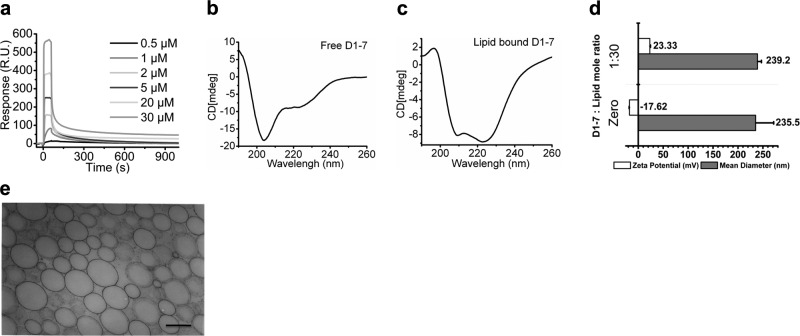
To investigate the kinetics of D1-7 insertion into the lipid membrane of the PFC nanoparticles, we used SPR to evaluate the dynamics of the 2-state interaction between the D1-7 and PFC nanoparticles. PFC nanoparticles were immobilized on an L1 sensor chip as described previously (14). D1-7 was injected over the PFC nanoparticle surface in selected concentrations. The sensorgram responses directly mapped the association and dissociation kinetics of the D1-7 with the PFC nanoparticles (Fig. 3a). A 2-state model was applied as shown in Eq. 1 (see Materials and Methods). In the first state, D1-7 rapidly associates with the lipid membrane of PFC nanoparticles, where ka1 is 2619 (1/M · s) and kd1 is 2.424 × 10−3 (1/s). In the second state, D1-7 inserts into the lipid membrane and undergoes a structural change, where ka2 is 6.725 × 10−3 (1/s), and kd2 is 6.05 × 10−4 (1/s). The dissociation constant of D1-7 from PFC nanoparticles is 7.64 × 10−8 M.
Figure 3.
Stable insertion of D1-7 into nanocarriers. a) Insertion of D1-7 into PFC nanoparticles. Sensorgram, acquired by Biacore X100, depicts the kinetics of the D1-7 insertion into the PFC nanoparticles immobilized on the surface of a L1 sensor chip. D1-7 concentrations were, from bottom to top, 0.5, 1, 2, 5, 20, and 30 μM. R.U., response units. b, c) Secondary structural change of D1-7 after lipid insertion. b) Free D1-7 presented unordered structure. c) Lipid-bounded D1-7 adopted α-helical structure. d) Mean hydrodynamic diameter and ζ potential of nanoparticles with or without insertion of D1-7, respectively. e) Transmission electron micrograph of PFC nanoparticles after insertion of D1-7. Scale bar = 200 nm.
After melittin inserts into the lipid membrane, it undergoes a secondary structural change from a random unordered structure to an α-helical conformation (14, 16,17,18). Therefore, we measured CD spectra for free D1-7 and PFC nanoparticle-bound D1-7 to determine whether D1-7 undergoes a similar structural change as does melittin on membrane binding. Free D1-7 exhibited a very strong negative peak near 200 nm and a relatively weak peak between 220 and 230 nm, which represented the unordered structure (Fig. 3b). However, the CD spectrum of lipid membrane-bound D1-7 exhibits two negative peaks: one at 209.2 nm and the other at 222.8 nm (Fig. 3c), which demonstrate the α-helical secondary structure formation of D1-7 after incorporation into the lipid membrane.
The ζ potential was measured to determine the surface charge of the PFC nanoparticles and the extent of peptide incorporation. The ζ potential of native PFC nanoparticles is −17.62 mV. After incorporating positively charged D1-7 peptides, PFC nanoparticles exhibited a ζ-potential shift to +23.33 mV (Fig. 3d). This change is consistent with the insertion of the positively charged D1-7 into the PFC nanoparticles. On the other hand, the average hydrodynamic diameter of the PFC nanoparticles was not affected by D1-7 insertion (Fig. 3d). Furthermore, the structural integrity of D1-7-inserted PFC nanoparticles was confirmed by transmission electron microscopy (Fig. 3e), for which the lipid membrane of the PFC nanoparticles appears intact. These observations taken together indicate that D1-7 stably inserts into the PFC nanoparticles to form a stable peptide-nanoparticle complex.
After demonstrating the stable D1-7 peptide-nanoparticle complex formation, we investigated the cytotoxicity of this new nanoconstruct, by extending our standard cytotoxicity study to D1-7 incorporated into αvβ3 integrin-targeted nanoparticles applied against B16F10 melanoma cells, which express ανβ3-integrin on the cell surface in vitro. At a concentration of 250 μM, PFC nanoparticles incorporating D1-7 only induce ∼10% of the cell death (Supplemental Fig. 1). These results further illustrate the safety of D1-7 as a linker in particle formulations.
Cargo attachment does not disrupt the linker properties of D1-7
So far, we have demonstrated that D1-7 functions as a linker for PFC nanoparticles and can be added after the formulation of the base nanoparticles. Next, we sought to demonstrate that the insertion of D1-7 into PFC nanoparticles would not be affected by conjugation with a targeting cargo. As a test cargo, we conjugated a 7-aa peptide-targeting ligand against the adhesion molecule VCAM-1 that was previously reported by Kelly et al.(19). VCAM-1, an inflammatory biomarker, was selected for practical purposes because it plays a pivotal role in the evolution of atherosclerosis and other inflammatory diseases and therefore is an excellent candidate for nanoparticle-based molecular imaging and targeted drug delivery in these pathological conditions (1, 20,21,22). The anti-VCAM-1 peptide was conjugated onto either the C terminus or N terminus of D1-7 with two glycines in between as a spacer to form two types of VCAM-1 targeting combination peptides called TCP1 and TCP2, respectively. The sequences of TCP1 and TCP2 are shown in Supplemental Fig. 1a.
The insertion kinetics of TCP1 and TCP2 into PFC nanoparticles were examined with SPR (Supplemental Fig. 2b, c). Values for ka1 were 257.8 and 1792 (1/M · s), for kd1 were 8.74e−4 and 1.438e−3 (1/s); for ka2 were 1.536e−3 and 3.454e−3 (1/s), and for kd2 were 1.57e−4 and 2.36e−4 (1/s) for TCP1 and TCP2, respectively. The dissociation constants of TCP1 and TCP2 were 3.15 × 10−7 and 5.13 × 10−8 M, respectively. These data suggest that the insertion of D1-7 into PFC nanoparticles is not appreciably affected by the addition of a targeting ligand cargo. Similar to the behavior of D1-7 insertion, TCP1 and TCP2 altered the ζ potential of PCF nanoparticles from −17.67 to +13.47 and +12.43 mV, respectively (Supplemental Fig. 2d). The mean hydrodynamic diameter of the PFC nanoparticles was not affected by the insertion of either TCP1 or TCP2 (Supplemental Fig. 2d). These results indicate that D1-7 is capable of stably and rapidly linking a small targeting ligand into the preformulated PFC nanoparticles.
Linker strategy enables specific molecular targeting, imaging, and quantification
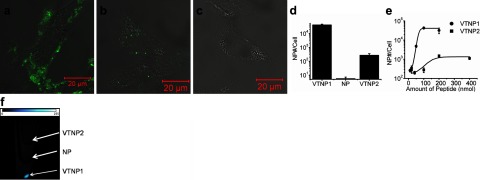
Validation of specific binding of the VCAM-1-targeted nanoparticles was studied in cultured murine endothelial cells (2F2B) in the presence of serum. Two formulations of VCAM-1-targeted nanoparticles (VTNP1 and VTNP2) were produced by loading preformulated PFC nanoparticles with TCP1 or TCP2, respectively. Specific targeting to 2F2B cells was investigated by confocal microscopy, 19F MRS, and MRI. In each case, lipid-linked Alexa Fluor 488 also was included in the membrane of these PFC nanoparticles to allow confocal fluorescence visualization. Up-regulation of VCAM-1 expression by TNF-α stimulation on the 2F2B cells was verified on both mRNA and protein expression (Supplemental Fig. 3a–c). Therefore, TNF-α-stimulated 2F2B cells that have enhanced VCAM-1 expression on their surface were treated with Alexa Fluor 488-colabeled VTNP1, VTNP2, or nontargeted nanoparticles. Manifestly stronger fluorescent signals emanated from the cells treated with VCAM-targeted nanoparticles (Fig. 4a, b) than from cells treated with nontargeted nanoparticles (Fig. 4c), which demonstrates the specific binding of targeted nanoparticles to cellular VCAM-1.
Figure 4.
Specific binding of targeted nanoparticles to 2F2B endothelial cells. a–c) Confocal microscopic images of 2F2B endothelial cells treated with VTNP1, VTNP2, or nontargeted Alexa Fluor 488 colabeled PFC nanoparticles, respectively. d) Quantification of specific delivery of VTNP1, VTNP2, and nontargeted nanoparticles to 2F2B cells calculated from 19F magnetic resonance spectra. VTNP1 or VTNP2 were generated with addition of 100 nmol of TCP1 or TCP2 on 20 μl of PFC nanoparticles, respectively. Data are means ± sd (n=6). e) Dose-dependent comparison between VTNP1 and VTNP2 at selected peptide loadings into 20 μl of PFC nanoparticles in 10 ml of final treatment volume. The x axis represents the amount of peptide in nanoparticles; the y axis represents the number of nanoparticles delivered to one 2F2B endothelial cell. A log scale was used. f) Fluorine magnetic resonance image overlaid onto proton images of cell pellets in test tubes, acquired from cells treated with VTNP2, nanoparticles (NP), or VTNP1, from top to bottom. The 19F signal (bright pellet) is not apparent in the cells treated with nontargeted nanoparticles.
In addition, the 19F PFC core of PFC nanoparticles enables these nanoparticles to serve as unique magnetic resonance spectroscopic and imaging agents with no background signal in vivo(23). Because of the known linear relation between 19F signal intensity and the concentration of 19F molecules (15, 24), the amount of nanoparticles delivered to the cell can be estimated. Using this approach, 19F magnetic resonance spectra of TNF-α stimulated 2F2B endothelial cells treated with VTNP1, VTNP2, or nontargeted nanoparticles were acquired. We normalized the number of bound nanoparticles by the total number of measured cells to calculate the number of the bound nanoparticles per cell (Fig. 4d). These data confirmed that VTNP1 manifests better targeting efficiency than does VTNP2 and further support the specific binding of targeted nanoparticles.
We also evaluated the dose dependencies of VTNP1 and VTNP2 targeting. Sigmoidal dose-dependent responses (Fig. 4e) again confirm the better targeting efficiency of VTNP1, while also demonstrating that the binding of VCAM-targeted nanoparticles is dependent on the amount of targeting peptide incorporated onto the nanoparticle, which provides the guidance for optimal and efficient cargo loading. As expected, the 19F image of the cells treated with VTNP1, VTNP2, or nontargeted nanoparticles showed the same preference for C terminus addition of the targeting sequence (Fig. 4f). A stronger 19F signal was detected from the cells treated with VTNP1 than with VTNP2, whereas no 19F signal was detected from the cells treated with nontargeted nanoparticles. It is also clear that the exposure of VCAM-1-targeting peptides fused to the C terminus of the linker generates better binding than the VCAM-1-targeting peptide attached to the N terminus.
We also investigated the cytotoxicity of VTNP1 and VTNP2 against TNF-α-stimulated 2F2B cells, which exhibit up-regulated VCAM-1 expression on cell membranes. No significant cell toxicity was observed even at the concentration of 200 μM (Supplemental Fig. 4). The VCAM-1-targeted nanoparticles reach their targeting saturation at a concentration of 20 μM (Fig. 4e), which illustrates the safety of these targeted cargo carrying nanoparticles.
Linker strategy adds cargo to a conventional targeted nanosystem for in vivo applications
To illustrate the potential for therapeutic cargo delivery into cells and the utility of the system for in vivo application, we next loaded FITC conjugated on D1-7, as a mock cargo, into integrin-targeted PFC nanoparticles that were preformulated with a lipid-linked ανβ3-integrin-binding peptidomimetic moiety (25). First, in vitro confocal imaging revealed marked cytoplasmic uptake of the FITC from ανβ3-integrin-targeted nanoparticles by C-32 cells (Supplemental Fig. 5a). In contrast, after nontargeted FITC-loaded nanoparticles were incubated with C-32 cells for the same time period, cellular internalization of FITC was not apparent (Supplemental Fig. 5b).
To demonstrate in vivo application, we injected FITC-loaded ανβ3-integrin-targeted nanoparticles intravenously into mice harboring 10-d-old B16F10 melanoma tumors (Fig. 5a, b). Previous reports indicated that ανβ3-integrin expression is intensely up-regulated on neovasculature at tumor sites, and specific vascular targeting of ανβ3-targeted nanoparticles can be achieved with lipid-conjugated targeting ligands prepared in the usual fashion (25). Here, we observed a significant fluorescent signal at the tumor site in the mice injected with FITC-loaded ανβ3-integrin-targeted nanoparticles after 2 h of circulation (Fig. 5c) but not in the mouse treated with the ανβ3-targeted nanoparticles without the FITC-linker cargo, which provided tissue autofluorescence control (Fig. 5d). This result confirms the stability of linker-nanoparticle complexes for in vivo application.
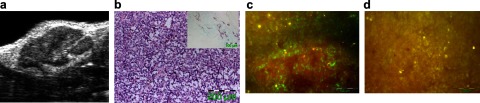
Figure 5.
In vivo cellular cargo deliveries by the linker strategy. a) Representative ultrasound in vivo visualization of B16F10 subcutaneous melanoma tumor in mouse inguinal region illustrating complex internal architecture of growing tumor mass. b) Histological assessment of B16F10 melanoma tumors excised after 10 d of implantation. Hematoxylin and eosin shows dense proliferation at tumor site. Inset: PECAM-1 staining illustrates tumor neovascularization prominently located around the tumor capsule. c) In vivo fluorophore delivery into the tumor site by FITC-loaded αγβ3-integrin-targeted nanoparticles (green). A representative fluorescence image was acquired from the tumor tissue, after 2 h circulation of intravenously injected FITC-loaded αγβ3-integrin-targeted nanoparticles. d) Representative fluorescence image from control shows only autofluorescence (yellow).
Linker strategy converts Doxil into a targeted delivery system and enhances drug delivery
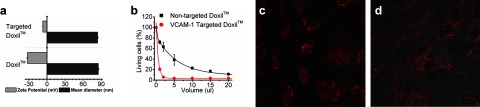
To demonstrate the direct translational potential of the system, we loaded TCP1 onto Doxil, which is a lipid bilayer drug delivery nanosystem, to generate VCAM-1-targeted Doxil. Consistent with our findings in the lipid monolayer nanosystem, the loading of the targeting ligand onto Doxil did not affect the size of this liposomal agent (Fig. 6a) but did shift the ζ potential of the Doxil to a less negative voltage range as expected from effective conjugation (Fig. 6a). When the VCAM-1-targeted Doxil was applied to cells with surface VCAM-1 expression in vitro, only 10% of the typical nontargeted Doxil dosage was able to achieve the equivalent cell-killing effects (Fig. 6b). To differentiate the cellular uptake of targeted and nontargeted Doxil, confocal visualization demonstrated significantly greater doxorubicin, both cytoplasmic and nuclear uptake in the treatments, with targeted Doxil (Fig. 6c) than with nontargeted Doxil (Fig. 6d).
Figure 6.
Targeting feature introduced by linker strategy enhanced Doxil effect. a) Mean hydrodynamic diameter and ζ potential of Doxil with or without insertion of TCP1, respectively. b) Cell killing induced after treatments with VCAM-1-targeted or nontargeted Doxil at various volumes in a 1-ml final treatment volume. All data points represent means ± sd (n=3). c, d) Confocal microscopic images of 2F2B endothelial cells treated with VCAM-1-targeted (c) or nontargeted (d) Doxil.
Linker strategy enables live cell labeling and tracking
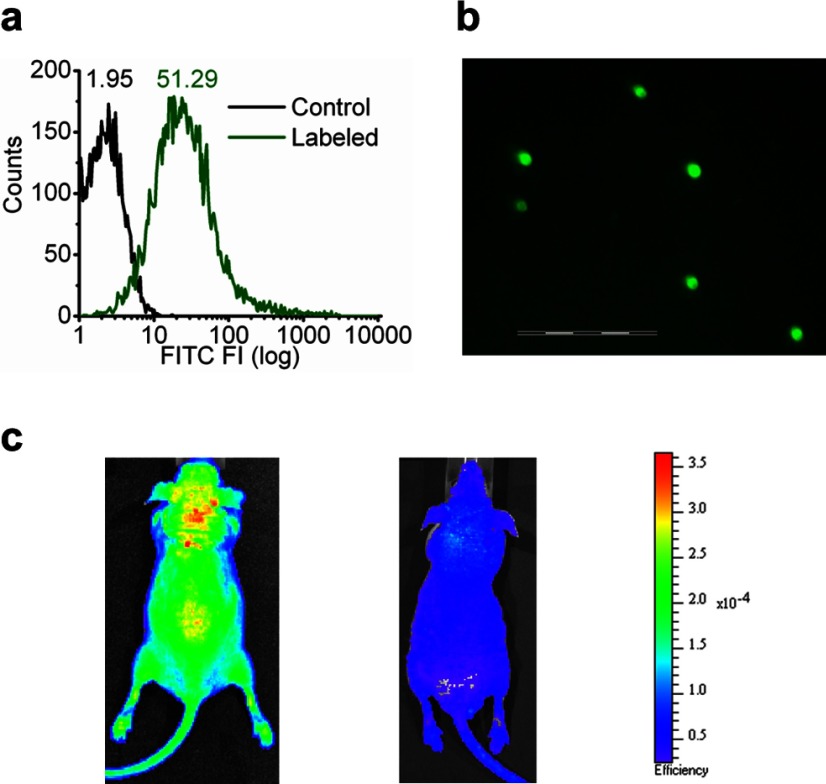
For investigating potential cell labeling and imaging applications, FITC loading onto cultured monocytes (RAW264.7) by this linker strategy was demonstrated by flow cytometry. Significant rightward shifts of mean florescence intensity (Fig. 7a) confirmed the loading of the FITC onto the monocytes. Also, this linker was capable of loading FITC onto isolated RBCs, which were then visualized intact under fluorescence microscopy (Fig. 7b). The labeled RBCs then were injected into mice to produce a prominent and distributed fluorescent signal by whole-body fluorescence imaging (Fig. 7c, left) within minutes after intravenous injection, which distinguished from the autofluorescence (Fig. 7c, right). These results illustrate the cell tracking potential of the linker labeling system.
Figure 7.
Linker strategy for cell labeling and in vivo application. a) Fluorescence labeling of macrophages through linker strategy. Monocytes (RAW264.7) were labeled with FTIC-conjugated linker peptide. Histograms were from one of 4 sets of independent experiments. Numbers represented mean fluorescence intensity. b) Fluorescence labeling of RBCs through linker strategy. A representative fluorescence image was acquired after RBCs were labeled with FITC-conjugated linker peptide. Scale bar = 100 μm. c) Immediate systemic distribution of labeled cells after intravenous injection. Fluorescence images were acquired from athymic nude mouse (nu/nu) (left) after FITC-labeled RBCs were intravenously injected, and from athymic nude mouse without injection (right) with the use of a Xenogen IVIS Spectrum imaging system. Resultant pseudocolor “efficiency image” illustrates systemic distribution of the labeled cells.
DISCUSSION
We have demonstrated recently that native melittin can stably insert into PFC nanoparticles for use as anticancer therapy (13, 14). Here, for the first time, we show that modified melittin, D1-7, can be converted into a cargo linker for lipid membrane editing of nanostructures and living cells. Melittin is a 26-aa peptide that comprises more than half of the dry weight of the venom of the honeybee Apis mellifera(26). The residues 1–20 of the melittin form two amphipathic α-helixes with a proline hinge (27), whereas the residues 21–26 of the melittin are positively charged. Both hydrophobic and hydrophilic segments of melittins are essential for its lytic activity (9, 28,29,30,31,32,33). Sequence modifications of melittin alter its lytic activity (28,29,30, 32,33,34,35). However, the membrane binding of those modified melittins has not been investigated previously. Consistent with previous reports (30, 33, 35), D1-7 with a shortened hydrophobic segment exhibited dramatically attenuated cytolytic and pore-forming activity on the lipid membrane. Furthermore, the stable membrane insertion of D1-7 was illustrated in 3 independent tests: SPR to study the kinetics of D1-7 lipid insertion; CD spectroscopy to define the secondary structural change after D1-7 insertion into lipid membrane; and ζ potential to confirm the surface charge change after the insertion of positively charged D1-7 into the lipid membrane.
Inspection of the kinetic data suggests that the off rates of integrated D1-7 and native melittin are similar. The major difference between D1-7 and melittin resides in the first state of integration, which suggests that by extending the loading period, similar PFC nanoparticle loading can be achieved compared with that for native melittin. The truncated melittin analog, D1-7, exhibited an interaction with the PFC nanoparticles that is consistent with prior observations (14). The CD spectra demonstrated that D1-7 formed an α-helical structure after insertion into the lipid membrane similar in extent to that of native melittin. According to Klocek et al.(36), the hydrophobic interaction of the α-helical segment of the amphipathic peptide contributes to the negative free energy change that is generated by the hydrophobic integration (PL*), which is characterized by ka2 and kd2. We observed that the kd2/ka2 of D1-7 (0.0899) was comparable to that of melittin (0.0673) (14), thus confirming that the structural accommodation that insertion of D1-7 provides would be as stable in the lipid membrane for in vivo application as is that of melittin (13). The stable insertion of D1-7 into PFC nanoparticles for in vivo applications was also confirmed in Fig. 5. FITC, a fluorophore loaded into ανβ3-integrin-targeted nanoparticles by D1-7, was delivered into tumor sites after 2 h of in vivo circulation. These results indicate that this linker can postedit a variety of membrane-functionalized nanocarriers, suggesting the possibility of converting existing functionalized nanocarriers into a multiplexing nanosystem for enhanced molecular imaging and therapy.
In the data presented so far, we have demonstrated the ability to load a cargo into nanocarriers with negative ζ potential by using the positively charged D1-7. However, the loading capability of D1-7 is not limited to the lipidic nanosystems with negative ζ potential, as it also can insert into the cationic nanoparticles. In Supplemental Fig. 6, we demonstrate that D1-7 incorporates into cationic nanoparticles and that the average hydrodynamic diameter of the PFC nanoparticles was not affected, whereas the ζ potential of the cationic nanoparticles shifts from +45.85 ± 1.9 to +76.03 ± 2.01 mV. These results further demonstrate the pivotal role of hydrophobic interaction in the stable insertion of D1-7 into the lipid membrane, and the insertion is membrane charge-independent, which is consistent with previous findings (9, 28).
We also observed that exposure of VCAM-1-targeting peptides fused to the C-terminus of the linker generates better targeting than did the VCAM-1 targeting peptide attached to the N terminus. This observation was consistent with the known orientation of melittin in lipid membranes where the N terminus situates more deeply into the lipid membrane and the C terminus remains exposed on the membrane surface (37, 38). Because the N terminus of the linker peptide presumably situates more deeply into the particle lipid monolayer, the orientation of the targeting ligands is undoubtedly more complex than that of the C-terminal-loaded peptides, which would not be optimal for interacting with surface biomarkers. Therefore, VTNP1 exhibits better targeting efficiency than does VTNP2. The exact configuration of the N- and C-terminal-loaded peptides in the monolayer is a subject of further investigation in an attempt to achieve optimally performing structures. Moreover, we demonstrated that these targeted nanoparticles responded to the level of VCAM-1 expression, such that the more VCAM-1 that was expressed, the more targeted nanoparticles were delivered (Supplemental Fig. 3d). Therefore, we conclude that we succeeded in converting nontargeted PFC nanoparticles into targeted nanovehicles with the use of this linker strategy and that the fundamental interaction of the targeted nanoparticles with the cells was specific and occurred independently of the generalized cell penetration mechanism that might be expected of other cationic peptides, such as TAT (39). Moreover, the specific targeting of the Doxil liposomal carrier markedly enhances the deposition of doxorubicin into the cytoplasm, which then presumably leads to better nuclear delivery through conventional doxorubicin trafficking. The Doxil treatment results demonstrated that targeting dramatically augmented the cell killing, which is of definite interest for nanomedical applications in cancer therapy.
In summary, we have designed, characterized, and verified a novel linker strategy to generate biocompatible peptide nanostructures for lipidic nanocarriers including PFC nanoemulsions, liposomes, and cells for combined molecular imaging and cell-targeted therapeutic agents. This linker offers flexible and rapid switching and multiplexing of targeting ligands and/or therapeutic payloads as dictated by specific medical needs. In this approach, the targeting ligand and/or therapeutic ingredients can be added to an existing nanosystem without having to reformulate the base nanocarriers. Such a strategy should prove useful in biomarker discovery by speeding adoption of new targets and/or drugs into the nanoparticle formulary. Moreover, the extension of this paradigm to rapid cellular or organelle labeling also appears promising for in vivo cell tracking and molecular imaging. In short, the concept of “nanostructure editing” with the use of the novel linker strategy presented here introduces a novel paradigm for achieving the rapid deployment of multiple targeting ligands and/or therapeutic cargos in a large class of clinically acceptable nanomedicinal agents.
Supplementary Material
Acknowledgments
The authors thank Stacy J. Allen, Michael J. Scott, and Xiaoxia Yang for critical support with the animal study, Eric T. Christenson for helpful discussions, Ralph Fuhrhop for the PFC nanoparticle formulation, and Huiying Zhang for assisting with the histological analysis. This work was supported by National Institutes of Health grants HL073646 and CA119342 to S.A.W.
References
- Blankenberg S., Barbaux S., Tiret L. Adhesion molecules and atherosclerosis. Atherosclerosis. 2003;170:191–203. doi: 10.1016/s0021-9150(03)00097-2. [DOI] [PubMed] [Google Scholar]
- Ferrari M. Cancer nanotechnology: opportunities and challenges. Nat Rev. 2005;5:161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- Cai W., Chen X. Multimodality molecular imaging of tumor angiogenesis. J Nucl Med. 2008;49(Suppl. 2):113S–128S. doi: 10.2967/jnumed.107.045922. [DOI] [PubMed] [Google Scholar]
- Saul J. M., Annapragada A. V., Bellamkonda R. V. A dual-ligand approach for enhancing targeting selectivity of therapeutic nanocarriers. J Control Release. 2006;114:277–287. doi: 10.1016/j.jconrel.2006.05.028. [DOI] [PubMed] [Google Scholar]
- Willmann J. K., Lutz A. M., Paulmurugan R., Patel M. R., Chu P., Rosenberg J., Gambhir S. S. Dual-targeted contrast agent for US assessment of tumor angiogenesis in vivo. Radiology. 2008;248:936–944. doi: 10.1148/radiol.2483072231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAteer M. A., Schneider J. E., Ali Z. A., Warrick N., Bursill C. A., von zur Muhlen C., Greaves D. R., Neubauer S., Channon K. M., Choudhury R. P. Magnetic resonance imaging of endothelial adhesion molecules in mouse atherosclerosis using dual-targeted microparticles of iron oxide. Arterioscler Thromb Vasc Biol. 2008;28:77–83. doi: 10.1161/ATVBAHA.107.145466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Tang G. P., Li J. Z., Kong Y., Huang H. L., Min L. J., Zhou J., Shen F. P., Wang Q. Q., Yu H. Dual-targeting non-viral vector based on polyethylenimine improves gene transfer efficiency. J Biomater Sci. 2007;18:545–560. doi: 10.1163/156856207780852532. [DOI] [PubMed] [Google Scholar]
- Namiki Y., Namiki T., Yoshida H., Ishii Y., Tsubota A., Koido S., Nariai K., Mitsunaga M., Yanagisawa S., Kashiwagi H., Mabashi Y., Yumoto Y., Hoshina S., Fujise K., Tada N. A novel magnetic crystal-lipid nanostructure for magnetically guided in vivo gene delivery. Nat Nanotechnol. 2009;4:598–606. doi: 10.1038/nnano.2009.202. [DOI] [PubMed] [Google Scholar]
- Sessa G., Freer J. H., Colacicco G., Weissmann G. Interaction of alytic polypeptide, melittin, with lipid membrane systems. J Biol Chem. 1969;244:3575–3582. [PubMed] [Google Scholar]
- Weissmann G., Hirschhorn R., Krakauer K. Effect of melittin upon cellular and lysosomal membranes. Biochem Pharmacol. 1969;18:1771–1775. doi: 10.1016/0006-2952(69)90167-1. [DOI] [PubMed] [Google Scholar]
- Winter P. M., Neubauer A. M., Caruthers S. D., Harris T. D., Robertson J. D., Williams T. A., Schmieder A. H., Hu G., Allen J. S., Lacy E. K., Zhang H., Wickline S. A., Lanza G. M. Endothelial αvβ3 integrin-targeted fumagillin nanoparticles inhibit angiogenesis in atherosclerosis. Arterioscler Thromb Vasc Biol. 2006;26:2103–2109. doi: 10.1161/01.ATV.0000235724.11299.76. [DOI] [PubMed] [Google Scholar]
- Angelova M. I., Dimitrov S. D. Liposome electroformation. Faraday Discuss Chem Soc. 1986;81:303–311. [Google Scholar]
- Soman N. R., Baldwin S. L., Hu G., Marsh J. N., Lanza G. M., Heuser J. E., Arbeit J. M., Wickline S. A., Schlesinger P. H. Molecularly targeted nanocarriers deliver the cytolytic peptide melittin specifically to tumor cells in mice, reducing tumor growth. J Clin Invest. 2009;119:2830–2842. doi: 10.1172/JCI38842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soman N. R., Lanza G. M., Heuser J. M., Schlesinger P. H., Wickline S. A. Synthesis and characterization of stable fluorocarbon nanostructures as drug delivery vehicles for cytolytic peptides. Nano Lett. 2008;8:1131–1136. doi: 10.1021/nl073290r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters E. A., Chen J., Yang X., Zhang H., Neumann R., Santeford A., Arbeit J., Lanza G. M., Wickline S. A. Detection of targeted perfluorocarbon nanoparticle binding using 19F diffusion weighted MR spectroscopy. Magn Reson Med. 2008;60:1232–1236. doi: 10.1002/mrm.21794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey C. E. The actions of melittin on membranes. Biochim Biophys Acta. 1990;1031:143–161. doi: 10.1016/0304-4157(90)90006-x. [DOI] [PubMed] [Google Scholar]
- Toraya S., Nishimura K., Naito A. Dynamic structure of vesicle-bound melittin in a variety of lipid chain lengths by solid-state NMR. Biophys J. 2004;87:3323–3335. doi: 10.1529/biophysj.104.046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom R., Crifo C., Viti V., Guidoni L., Podo F. Variations in circular dichroism and proton-NMR relaxation properties of melittin upon interaction with phospholipids. FEBS Lett. 1978;96:45–50. doi: 10.1016/0014-5793(78)81059-x. [DOI] [PubMed] [Google Scholar]
- Kelly K. A., Nahrendorf M., Yu A. M., Reynolds F., Weissleder R. In vivo phage display selection yields atherosclerotic plaque targeted peptides for imaging. Mol Imaging Biol. 2006;8:201–207. doi: 10.1007/s11307-006-0043-6. [DOI] [PubMed] [Google Scholar]
- Hartge M. M., Unger T., Kintscher U. The endothelium and vascular inflammation in diabetes. Diab Vasc Dis Res. 2007;4:84–88. doi: 10.3132/dvdr.2007.025. [DOI] [PubMed] [Google Scholar]
- Wu T. C. The role of vascular cell adhesion molecule-1 in tumor immune evasion. Cancer Res. 2007;67:6003–6006. doi: 10.1158/0008-5472.CAN-07-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAteer M. A., Sibson N. R., von Zur Muhlen C., Schneider J. E., Lowe A. S., Warrick N., Channon K. M., Anthony D. C., Choudhury R. P. In vivo magnetic resonance imaging of acute brain inflammation using microparticles of iron oxide. Nat Med. 2007;13:1253–1258. doi: 10.1038/nm1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza G. M., Winter P. M., Neubauer A. M., Caruthers S. D., Hockett F. D., Wickline S. A. 1H/19F magnetic resonance molecular imaging with perfluorocarbon nanoparticles. Curr Top Dev Biol. 2005;70:57–76. doi: 10.1016/S0070-2153(05)70003-X. [DOI] [PubMed] [Google Scholar]
- Morawski A. M., Winter P. M., Yu X., Fuhrhop R. W., Scott M. J., Hockett F., Robertson J. D., Gaffney P. J., Lanza G. M., Wickline S. A. Quantitative “magnetic resonance immunohistochemistry” with ligand-targeted 19F nanoparticles. Magn Reson Med. 2004;52:1255–1262. doi: 10.1002/mrm.20287. [DOI] [PubMed] [Google Scholar]
- Schmieder A. H., Winter P. M., Caruthers S. D., Harris T. D., Williams T. A., Allen J. S., Lacy E. K., Zhang H., Scott M. J., Hu G., Robertson J. D., Wickline S. A., Lanza G. M. Molecular MR imaging of melanoma angiogenesis with ανβ3-targeted paramagnetic nanoparticles. Magn Reson Med. 2005;53:621–627. doi: 10.1002/mrm.20391. [DOI] [PubMed] [Google Scholar]
- Tosteson M. T., Tosteson D. C. The sting Melittin forms channels in lipid bilayers. Biophys J. 1981;36:109–116. doi: 10.1016/S0006-3495(81)84719-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzo R., Tappin M. J., Pastore A., Harvey T. S., Carver J. A., Campbell I. D. The structure of melittin A 1H-NMR study in methanol. Eur J Biochem. 1988;173:139–146. doi: 10.1111/j.1432-1033.1988.tb13977.x. [DOI] [PubMed] [Google Scholar]
- DeGrado W. F., Musso G. F., Lieber M., Kaiser E. T., Kezdy F. J. Kinetics and mechanism of hemolysis induced by melittin and by a synthetic melittin analogue. Biophys J. 1982;37:329–338. doi: 10.1016/S0006-3495(82)84681-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey C. E., Bazzo R., Harvey T. S., Syperek I., Boheim G., Campbell I. D. Contribution of proline-14 to the structure and actions of melittin. FEBS Lett. 1991;281:240–244. doi: 10.1016/0014-5793(91)80402-o. [DOI] [PubMed] [Google Scholar]
- Gevod V. S., Birdi K. S. Melittin and the 8–26 fragment Differences in ionophoric properties as measured by monolayer method. Biophys J. 1984;45:1079–1083. doi: 10.1016/S0006-3495(84)84255-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex S. A Pro → Ala substitution in melittin affects self-association, membrane binding and pore-formation kinetics due to changes in structural and electrostatic properties. Biophys Chem. 2000;85:209–228. doi: 10.1016/s0301-4622(00)00121-6. [DOI] [PubMed] [Google Scholar]
- Rivett D. E., Kirkpatrick A., Hewish D. R., Reilly W., Werkmeister J. A. Dimerization of truncated melittin analogues results in cytolytic peptides. Biochem J. 1996;316(Pt. 2):525–529. doi: 10.1042/bj3160525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werkmeister J. A., Kirkpatrick A., McKenzie J. A., Rivett D. E. The effect of sequence variations and structure on the cytolytic activity of melittin peptides. Biochim Biophys Acta. 1993;1157:50–54. doi: 10.1016/0304-4165(93)90077-l. [DOI] [PubMed] [Google Scholar]
- Blondelle S. E., Simpkins L. R., Perez-Paya E., Houghten R. A. Influence of tryptophan residues on melittin’s hemolytic activity. Biochim Biophys Acta. 1993;1202:331–336. doi: 10.1016/0167-4838(93)90024-l. [DOI] [PubMed] [Google Scholar]
- Dawson C. R., Drake A. F., Helliwell J., Hider R. C. The interaction of bee melittin with lipid bilayer membranes. Biochim Biophys Acta. 1978;510:75–86. doi: 10.1016/0005-2736(78)90131-1. [DOI] [PubMed] [Google Scholar]
- Klocek G., Schulthess T., Shai Y., Seelig J. Thermodynamics of melittin binding to lipid bilayers aggregation/pore formation. Biochemistry. 2009;49:2586–2596. doi: 10.1021/bi802127h. [DOI] [PubMed] [Google Scholar]
- Dufourcq J., Faucon J. F. Intrinsic fluorescence study of lipid-protein interactions in membrane models Binding of melittin, an amphipathic peptide, to phospholipid vesicles. Biochim Biophys Acta. 1977;467:1–11. doi: 10.1016/0005-2736(77)90236-x. [DOI] [PubMed] [Google Scholar]
- Mollay C., Kreil G. Fluorometric measurements on the interaction of melittin with lecithin. Biochim Biophys Acta. 1973;316:196–203. doi: 10.1016/0005-2760(73)90009-x. [DOI] [PubMed] [Google Scholar]
- Rapoport M., Lorberboum-Galski H. TAT-based drug delivery system—new directions in protein delivery for new hopes? Expert Opin Drug Deliv. 2009;6:453–463. doi: 10.1517/17425240902887029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.