Abstract
Tumor-driving mutations in the TP53 gene occur frequently in human cancers. These inactivating mutations arise predominantly from a single-point mutation in the DNA-binding domain of this tumor suppressor gene (i.e., exons 4–9). The human p53 knock-in (Hupki) mouse model was constructed using gene-targeting technology to create a mouse strain that harbors human wild-type TP53 DNA sequences in both copies of the mouse TP53 gene. Replacement of exons 4–9 of the endogenous mouse TP53 alleles in the Hupki mouse with the homologous normal human TP53 gene sequences has offered a humanized replica of the TP53 gene in a murine genetic environment. The Hupki mouse model system has proven to be an invaluable research tool for studying the underlying mechanisms of human TP53 mutagenesis. The utility of the Hupki mouse model system for exploring carcinogen-induced TP53 mutagenesis has been demonstrated in both in vivo animal experiments and in vitro cell culture experiments. Here, we highlight applications of the Hupki mouse model system for investigating mutagenesis induced by a variety of environmental carcinogens, including sunlight ultraviolet radiation, benzo[a]pyrene (a tobacco smoke-derived carcinogen), 3-nitrobenzanthrone (an urban air pollutant), aristolochic acid (a component of Chinese herbal medicine), and aflatoxin B1 (a food contaminant). We summarize the salient findings of the respective studies and discuss their relevance to human cancer etiology.—Besaratinia, A., Pfeifer, G. P. Applications of the human p53 knock-in (Hupki) mouse model for human carcinogen testing.
Keywords: cancer, etiology, mutation, tumor suppressor gene
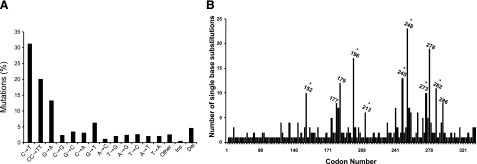
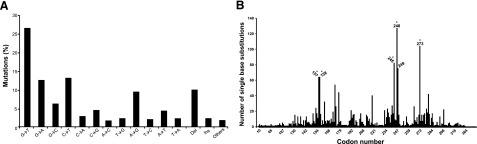
The pattern of somatic alterations in a human cancer genome is shaped by a number of factors, of which mutagen exposure holds great importance (1,2,3). Numerous mutagenic agents are present in the air we breathe, in the food we eat, and in the water we drink (4, 5). The complexity of human exposure to environmental mutagens can variably influence the compendium of somatic mutations occurring in human cancers (2, 3, 6, 7). For example, human nonmelanoma skin- and lung cancer genomes bear unique mutational signatures, which are attributable to exposure to sunlight ultraviolet (UV) radiation and tobacco smoke, respectively (8). Tumor genomes from nonmelanoma skin- and lung cancer patients, respectively, show a characteristic preponderance of C→T or CC→TT transitions at dipyrimidine sites (Fig. 1), and G→T transversions at methylated CpG dinucleotides (Fig. 2), which occur preferentially on the nontranscribed DNA strand (9,10,11).
Figure 1.
Mutation spectrum (A) and codon distribution (B) of the TP53 tumor suppressor gene in human nonmelanoma skin tumors (basal cell and squamous cell carcinomas; n=541). Data were obtained from the TP53 mutation database of the International Agency for Research on Cancer (http://www-p53.iarc.fr/p53DataBase.htm; R12 version) (79). Codons containing methylated CpG sequences are indicated by asterisks. Ins, insertions; Del, deletions.
Figure 2.
Mutation spectrum (A) and codon distribution (B) of the TP53 tumor suppressor gene in tobacco smoke-associated lung cancer (n=2340). Data were obtained from the TP53 mutation database of the International Agency for Research on Cancer (http://www-p53.iarc.fr/p53DataBase.htm; R12 version) (79). Entries with confounding exposure to asbestos, mustard gas, or radon were excluded. Codons containing methylated CpG sequences are indicated by asterisks. Ins, insertions; Del, deletions.
The recent advent of next-generation sequencing platforms has massively reduced the cost and effort of cataloguing human cancer mutations (12, 13). Currently, high throughput next-generation sequencing projects are interrogating a variety of cancer genomes, including various types of human tumors and cancer cell lines (14,15,16,17,18). These projects are poised to identify unique somatic changes in the genome of cancer patients and/or cell lines with a history of exposure to specific mutagens. To address the issue of cancer etiology, however, the occurrence of these genetic mutations does need to be recapitulated experimentally. Demonstration of a link between exposure to certain mutagens and induction of specific mutations in genes that promote tumorigenesis when mutated should be done in validated experimental model systems and under well-defined and controlled exposure conditions (8). By helping draw causality inference, this approach can greatly improve our understanding of the etiology of human cancer.
Inactivating mutations in the TP53 tumor suppressor gene are frequent events in human cancers (19,20,21,22), and TP53 generally stands at the top of the list of the most frequently mutated genes even when all coding sequences of the human genome are analyzed (11). The vast majority of TP53 mutations arise from a single point mutation in the segment encoding the DNA-binding domain of the TP53 protein (19, 20). These inactivating mutations render the mutant TP53 protein unable to carry out its normal functions, i.e., transcriptional transactivation of downstream target genes that regulate the cell cycle and apoptosis (21, 22). Scrutiny of the available databases of TP53 gene mutations in human cancers [e.g., IRAC TP53 database (http://www-p53.iarc.fr/p53DataBase.htm), 27,132 entries; and UMD TP53 mutation database (http://p53.free.fr/Database/ p53_database.html), 28,513 entries] has revealed significant links between exposure to mutagenic compounds and TP53 genetic alterations specific for certain types of human cancers (20, 23, 24). Attempts have been made to reproduce these putative links in experimental settings using a wide range of model systems, including bacterial Ames test (25, 26), functional analysis of separated alleles in yeast (FASAY; refs. 27, 28), reporter gene-based transgenic rodents, e.g., BigBlue® system (29), or analysis of endogenous noncancer-related genes, e.g., the house-keeping hypoxanthine phosphoribosyltransferase (HPRT) gene in mammalian or human tissues/cells (30). Although these model systems have all provided invaluable information on many aspects of mutagenesis-derived carcinogenesis, they all lack, in one way or another, crucially important factors that contribute to TP53 mutations and human cancers (8). For instance, in these model systems, the known determinants of mutagenesis, e.g., DNA-sequence contexts, DNA repair efficiency, and fidelity of replicative DNA polymerases, which are species/cell-type dependent, may not represent the respective parameters in the human TP53 gene (8).
More recently, a novel model system has been developed to investigate experimentally induced mutations in the human TP53 gene, in its natural mammalian context. The human p53 knock-in (Hupki) mouse model has addressed the issue of DNA-sequence context by replacing exons 4–9 of the endogenous mouse TP53 allele with the homologous normal human TP53 gene sequence (31). In the following sections, we will discuss the Hupki mouse model system, which has the utility to detect both spontaneously arisen- and carcinogen-induced mutations in the human TP53 gene in vitro(32,33,34,35,36,37,38) or in vivo(31, 39, 40).
Hupki MOUSE STRAIN
The Hupki mouse model system was constructed using gene-targeting technology to create a mouse strain that harbors human wild-type TP53 DNA sequences from exons 4 to 9 in place of the homologous murine DNA sequences in both copies of the mouse TP53 gene (31). The substituted segment encodes the polyproline domain and DNA-binding domain of wild-type human TP53, and the chimeric TP53 gene remains under normal transcription regulation at the mouse locus. The Hupki mice develop normally, exhibit no apparent physiological defects, remain fertile, and show no susceptibility to spontaneous lymphomas, sarcomas, or other neoplasms, which are common in TP53-deficient or null mice (31). The Hupki mice retain a variety of normal TP53 functions and characteristics, including nuclear accumulation of TP53 protein after exposure to DNA-damaging agents, transcriptional activation of known TP53 downstream targets, and induction of apoptosis in thymocytes after γ-irradiation, an outcome modulated by a functional TP53 gene (31, 35, 41).
In addition to its application for in vivo animal experiments (31, 39, 40), the Hupki model system is also amenable to in vitro cell culture experiments (32,33,34,35,36,37,38). Ideally, the origin of TP53 mutations in human cancers should be determined using a mutagenesis assay in which suspected endogenous metabolites or exogenous carcinogens can target the DNA sequences of this tumor suppressor gene, thereby leading to mutant TP53 proteins that confer a selectable phenotype, preferably one that resembles the aberrant functions that are typical for human tumor TP53 mutants. Theoretically, normal human cells would be the ideal cell type for such assay, were they not resistant to undergo immortalization or transformation in vitro. In practice, proliferative nonsenescent cultures are required for amplification and subsequently detection of the few phenotypically expressed TP53 mutant cells, which are usually outnumbered by an overwhelming pool of wild-type nonmutant cells (even in cultures that have undergone numerous rounds of passaging) (8). Murine fibroblasts, in contrast to human cells, spontaneously undergo immortalization during in vitro culturing, and require only one key genetic defect, such as loss of TP53 function, thus, allowing the selection of TP53 mutant cells in vitro(42,43,44). Likewise, primary embryonic fibroblasts from the Hupki mice readily undergo immortalization during in vitro passaging, which allows for the selection of dysfunctional TP53 point mutations that are characteristic of human tumors (32,33,34,35,36,37,38).
Given the importance of DNA sequence context in human TP53 mutagenesis and carcinogenesis (8, 31), the murine TP53 gene, however, cannot optimally represent its human counterpart gene due to the 15% discrepancy in base sequence in the DNA-binding domain and amino acid differences between these two species (35, 44, 45). This incomparability might explain, for example, the observation that whereas codon 248 (CGG) of the human TP53 gene is the most prominent mutation hotspot in nonmelanoma skin cancers (Fig. 1B), the mouse equivalent codon (CGC) (45) hardly harbors any mutations in UVB-induced tumors (46). The Hupki model system has resolved the above issue of incomparability by genetically modifying the murine TP53, and creating a humanized replica of the TP53 gene in the mouse genome (35). The system offers a promising venue for assaying spontaneous or experimentally induced human TP53 gene mutations both in vitro(32,33,34,35,36,37,38) and in vivo(31, 39, 40). In the following paragraphs, we will highlight applications of the Hupki mouse model system for mutagenicity analysis of various physical or chemical carcinogens and discuss the advantages and disadvantages of this model system in vitro and in vivo.
Hupki MUTAGENESIS ASSAY IN VITRO
The Hupki mouse embryonic fibroblasts treated with benzo[a]pyrene (B[a]P), a prominent tobacco-derived carcinogen, harbored TP53 mutations consisting of predominantly single base substitutions in the DNA-binding domain of this gene [29 of 36 (∼81%) of all mutations] (32, 34, 36). G→T transversion mutations constituted half of all B[a]P-induced mutations, of which all but one (17 of 18) occurred at sites where the permutated guanines were positioned on the nontranscribed strand of the TP53 gene. Distribution of the 29 B[a]P-induced mutations in the DNA-binding domain of the TP53 gene revealed codons 157, 158, and 273 as the most frequently mutated sites. The overall pattern and distribution of B[a]P-induced mutations in the Hupki mouse model system (32, 34, 36) mirror the characteristic features of TP53 mutations in lung tumors of smokers (Figs. 2A and 3A) (47,48,49).
Figure 3.
Induced mutation spectra of the TP53 tumor suppressor gene in the Hupki mouse model system. A) B[a]P-treated Hupki mouse embryonic fibroblasts (n=36) (data compiled from refs. 32, 34, 36). B) AA-treated Hupki mouse embryonic fibroblasts (n=37) (data compiled from refs. 32, 33, 38, 52). C) 3-NBA-treated Hupki mouse embryonic fibroblasts (n=29) (data obtained from ref. 37).
The established cultures of Hupki mouse embryonic fibroblasts irradiated with UVC gave rise to point mutations in the DNA-binding domain of the TP53 gene (33). Two of the seven induced mutations recovered from the UVC-irradiated cultures were at codons 248 and 273 (33), two major TP53 mutational hotspots in human nonmelanoma skin cancers (Fig. 1B) (50). Of all UVC-induced mutations, three were C→T transitions on the nontranscribed strand of the TP53 gene (33), a common feature of mutant TP53 gene in sun-exposed human skin tumors (Fig. 1A) (10, 50).
The Hupki mouse embryonic fibroblasts were treated with aristolochic acid (AA) (32, 33, 38), a plant extract potentially involved in Chinese herbs nephropathy and possibly leading to urothelial cancer development (51). Twenty of the 37 AA-induced TP53 mutations (54%) were A→T transversion mutations (52) (Fig. 3B), reflecting the hallmark mutation detected in the urothelial tumor cells of patients with documented AA exposure (53, 54). The induced A→T transversion mutations were presumably due to the permutated adenines located almost exclusively on the nontranscribed strand of the TP53 gene (32, 33, 38). This finding is consistent with the preferential formation of AA-adenine adducts found in the DNA of AA-treated Hupki cells and nephropathy patients (51, 54,55,56), as well as in the DNA from target organs of AA-exposed rats (57, 58). Recently, Grollman et al.(53) have also identified AA-adenine and -guanine adducts in the renal cortex of nephropathy patients and in the transitional cell tumors of patients with upper urinary tract malignancies who were residents of the endemic (Balkan) villages in which chronic dietary exposure to AA is prevalent. Mutation analysis of the TP53 gene in this population showed a predominance of mutations at A:T base pairs accounting for 89% of all detected mutations, with the vast majority (15 of 19) being A:T→T:A transversions (53).
The Hupki mouse embryonic fibroblasts were treated with 3-nitrobenzanthrone (3-NBA) (37), a member of the class of nitropolycyclic aromatic hydrocarbons, present in particulate fraction of diesel exhaust (59), and a ubiquitous urban air pollutant (60, 61). The established cultures of 3-NBA-treated cells harbored TP53 mutations in the DNA-binding domain of this gene, which consisted mainly of base substitutions (22 of 29, ∼76%) (37). Of these, G:C→T:A transversions were the major type of mutations (10 of 22, ∼46%) followed by A:T→T:A transversions (3 of 22, ∼14%) (Fig. 3C). This ratio of G:C→T:A to A:T→T:A transversions (3:1) perfectly mirrored the ratio of dG/dA adduct formation (75:25%) determined in similarly treated cells with 3-NBA or its reactive metabolite, N-hydroxy-3-aminobenzanthrone (N-OH-3-ABA) (37). A similar correlation in ratios of 3-NBA-derived purine adducts to transversion mutations was previously found in liver tissues of the MutaMouse, where the proportion of induced dG to dA adducts was 6 to 1 and that of corresponding G:C→T:A and A:T→T:A mutations was 5 to 1 (61). G:C→T:A transversions were also the predominant type of mutations found in N-OH-3-ABA-treated shuttle vector plasmids propagated in human cells (62), as well as in livers of 3-NBA-treated MutaMice (61) and in the lungs of gpt-Δ transgenic mice following inhalation of diesel exhaust (63).
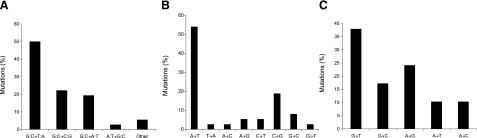
Whibley et al. (64) have recently shown that primary murine embryonic fibroblasts from wild-type and Hupki mice alike undergo in vitro spontaneous immoratalization consequent to successive passaging in culture. The authors demonstrated that basic features of oxidative-stress-induced senescence and subsequent immortalization of wild-type mouse embryonic fibroblasts are preserved in the counterpart cells from the Hupki mice. While wild-type mouse embryonic fibroblasts entered and exited senescence slightly later than their counterpart Hupki cells, reactive oxygen species (ROS) levels and the extent of DNA damage, determined by staining with a ROS-labile dye and the alkaline comet assay, respectively, were similar in the two cell lines at corresponding passages. TP53 mutations and p19 deletions occurred in a significant proportion of spontaneously immortalized cells in both wild-type and Hupki embryonic fibroblasts, and the frequency of these events did not differ significantly between these two genotypes (64). Notably, TP53 mutations arising during spontaneous immortalization of the Hupki fibroblast cultures correspond to human cancer TP53 mutations (64) that are known to be deficient in transcriptional activity (65). Although a limited number of TP53 mutation data was available from the immortalized wild-type mouse embryonic fibroblasts as compared to that from the counterpart Hupki cells (12 vs. 64 entries, respectively), the spectra of TP53 mutations found in the respective genotypes were similar in that G:C→C:G transversions constituted the most common type of base substitutions (Fig. 4) (64). This type of base alteration occurs at high frequency (up to ∼30%) in human breast cancer (1, 17, 66), although the driving force behind this mutagenic event is currently unknown (11). The prevalence of G:C→C:G transversions in the spectra of TP53 mutations in both wild-type and Hupki immortalized mouse embryonic fibroblasts remains a matter of further investigation (M. Hollstein, University of Leeds, Leeds, UK; personal communication, February 2, 2010).
Figure 4.
Spontaneous mutation spectra of the TP53 tumor suppressor gene in immortalized Hupki (A) and wild-type (B) mouse embryonic fibroblasts. Total numbers of TP53 mutations in wild-type and Hupki immortalized cells were 12 and 64, respectively. Data were obtained from ref. 64.
Hupki MUTAGENESIS ASSAY IN VIVO
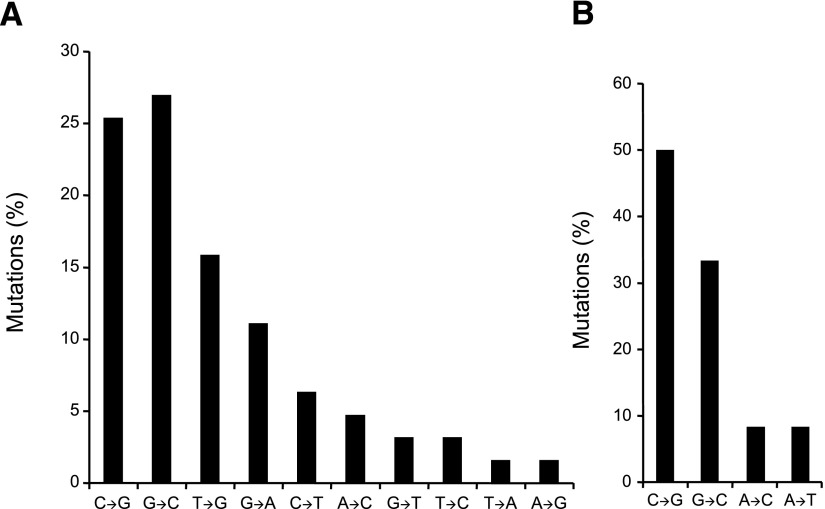
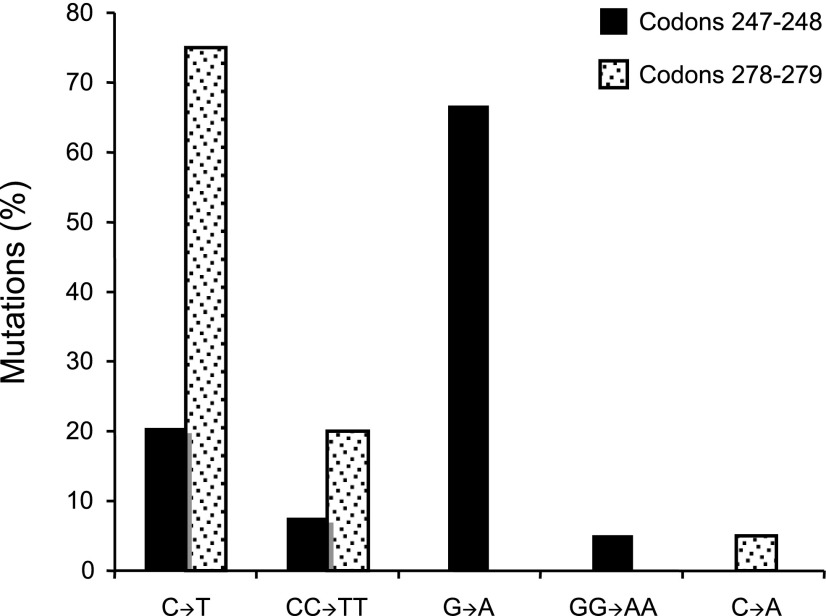
Luo et al. (35) have demonstrated that UVB-irradiated Hupki mice exhibit characteristic molecular pathology features of sunlight-associated human skin cancers (67, 68), including development of clones of epidermal cell patches with TP53-immunoreactive nuclei; formation of UV-induced cyclobutane pyrimidine dimers at skin cancer mutational hotspots in the TP53 gene, which colocalize with the respective lesions induced in UVB-exposed human keratinocytes; and induction of signature C→T transition mutations in the respective TP53 mutational hotspots (35). Screening of the UVB-irradiated Hupki mouse epidermal cells for mutations at codons 247–248 and 278–279, two major skin cancer TP53 mutational hotspots (Fig. 1B), showed that 58 of the 59 mutant clones (98%) harbored a transition mutation, including 9 (15%) with tandem CC→TT mutations (35). All of the UVB-induced mutations in codons 278–279 and ∼28% of mutations in codons 247–248 occurred at sites where the premutated pyrimidine dinucleotides were on the nontranscribed strand of the TP53 gene (Fig. 5) (35). The overall pattern of TP53 mutations found in UVB-irradiated Hupki mice (35) corresponds to the unique features of TP53 mutations in sunlight-associated human skin tumors (69,70,71).
Figure 5.
Mutation spectrum of the TP53 tumor suppressor gene in UVB-irradiated Hupki mice. Restriction enzyme digestion-coupled to PCR amplification of codons 247–248 and codons 278–279 followed by DNA sequencing were performed to establish the spectrum of mutations in the respective codons, two major TP53 mutational hotspots in human nonmelanoma skin cancers (see Fig. 1B) (50). Total numbers of mutant clones at codons 247–248 and 278–279 were 39 and 20, respectively.
Tong et al. (40) have used the Hupki mice to investigate the effect of local DNA sequence on TP53 codon 249 mutation, a prevalent occurrence in human hepatocellular carcinoma associated with synergistic exposure to aflatoxin B1 (AFB1) and hepatitis B virus (HBV) infection (72). A single intraperitoneal injection of AFB1 to the Hupki mice and counterpart wild-type animals showed that the mice expressing the humanized TP53 were more prone to hepatocellular carcinoma development and death, compared to mice expressing the murine TP53, without acquiring any mutations in the TP53 gene (40). These findings support the notion that the specificity of TP53 codon 249 mutation in human hepatocellular carcinoma is not solely dependent on DNA sequence context of this gene, and other determining factors, e.g., concomitant HBV infection, may synergistically be involved in this process (73). Currently, work is underway to address this question in a more relevant model system, i.e., the Hupki × Chisari’s HBV cross-strain mice (M. Hollstein, personal communication, November 18, 2008).
ADVANTAGES AND DISADVANTAGES OF THE Hupki MOUSE MODEL SYSTEM
The Hupki mouse embryonic fibroblasts have proven to be an invaluable in vitro model system for mutagenicity testing of various physical or chemical carcinogens (32,33,34,35,36,37,38). The system has appeared metabolically competent by converting all the tested chemicals to their reactive metabolites (32,33,34,35,36,37,38), and expressing a number of phase I and II metabolic enzymes, including Cyp 1b1, Cyp 1a2, microsomal epoxide hydrolase, NAD(P)H:quinone oxidoreductase, and nitroreductase (33). However, the Hupki system has yet to resolve its inherent challenges, including time- and labor-intensiveness, two common drawbacks of the currently available in vitro mutagenesis assays (8). The latter hinders generation of a sizable mutation database. Another limitation of the Hupki model system, both in vitro and in vivo, is the intrinsic possibility that the Hupki TP53 protein may perform suboptimally in the genetic environment of the murine cells, either due to the absence of other human-specific proteins that interact with TP53, or because of blocking of its function by mouse-specific proteins. Also, uncertainties remain about the ability of the Hupki TP53 protein to function exactly as expected in a genetically engineered environment. For example, species-specific differences in phosphorylation sites within the substituted polyproline domain of the Hupki model system may lead to a differential post-translational regulation of TP53 function in this model system (39, 74, 75). Moreover, the ability of the Hupki TP53 protein to participate in processes of DNA repair and recombination (76, 77) remains a matter of uncertainty. Despite the overall conservation in evolution of DNA repair mechanisms, differences exist between humans and mice, such as the efficiency of the global genomic repair subpathway of nucleotide excision repair (78). Such discrepancies may prove problematic because promutagenic lesions in the Hupki TP53 gene are subject to the murine DNA repair machinery (33). Altogether, although the Hupki TP53 model system has recapitulated many aspects of TP53 mutagenesis and human cancer (31,32,33,34,35,36,37,38,39,40), future studies will determine the extent to which this model system can precisely delineate human TP53 mutagenesis and carcinogenesis.
CONCLUSIONS
Inactivating mutations in the TP53 tumor suppressor gene occur frequently in a variety of human cancers (19,20,21,22). The pattern (spectrum) and frequency distribution (hotspots) of mutations in the TP53 gene are tumor-specific and reflective of past mutagen exposure (11). Thus, investigating human TP53 mutagenesis in relation to exposure to mutagenic agents can provide information on the underlying etiology of human cancers. The Hupki mouse model system was constructed using gene-targeting technology to create a humanized replica of the TP53 gene in mouse, thus allowing for experimental recapitulation of human TP53 mutagenesis (31). The utility of the Hupki mouse model system for studying the etiologic involvement of suspect environmental mutagens in human TP53 mutagenesis has been demonstrated in a number of in vivo animal experiments (35, 40) and/or in vitro cell culture experiments (32,33,34, 36,37,38, 52). For the most part, these investigations have reproduced the respective human TP53 mutagenesis data obtained from populations with documented exposure to mutagens of interest. A recent study has also shown the validity of the Hupki mouse model system for portraying upstream events leading to, and downstream events caused by, human TP53 mutagenesis (64). Further studies of such design must determine the accuracy of the Hupki mouse model system for representing the events preceding, and those following, human TP53 mutagenesis in vivo. Another important area of research, which awaits further exploration, is establishing the status of CpG methylation in the substituted segment of the Hupki mouse genome. The methylated CpGs (mCpGs) in this segment in the human genome constitute the single most significant mutational target in the TP53 gene (11). The importance of mCpGs in human TP53 mutagenesis is borne out by the observation that TP53 mutational hotspots in certain types of human cancers localize almost exclusively to mCpG-containing codons (e.g., the majority of both lung and colon cancer mutational hotspots have mCpGs in their sequence contexts, or nonmelanoma skin cancer mutational hotspots cluster at pyrimidine-mCpG sequence contexts) (11).
Acknowledgments
This work was supported by grants from the University of California Tobacco Related Disease Research Program (18KT-0040 to A. B.) and the National Cancer Institute (R01CA084469 to G. P. P.).
References
- Greenman C., Stephens P., Smith R., Dalgliesh G. L., Hunter C., Bignell G., Davies H., Teague J., Butler A., Stevens C., Edkins S., O'Meara S., Vastrik I., Schmidt E. E., Avis T., Barthorpe S., Bhamra G., Buck G., Choudhury B., Clements J., Cole J., Dicks E., Forbes S., Gray K., Halliday K., Harrison R., Hills K., Hinton J., Jenkinson A., Jones D., Menzies A., Mironenko T., Perry J., Raine K., Richardson D., Shepherd R., Small A., Tofts C., Varian J., Webb T., West S., Widaa S., Yates A., Cahill D. P., Louis D. N., Goldstraw P., Nicholson A. G., Brasseur F., Looijenga L., Weber B. L., Chiew Y. E., DeFazio A., Greaves M. F., Green A. R., Campbell P., Birney E., Easton D. F., Chenevix-Trench G., Tan M. H., Khoo S. K., Teh B. T., Yuen S. T., Leung S. Y., Wooster R., Futreal P. A., Stratton M. R. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance E. D., Cheetham R. K., Stephens P. J., McBride D. J., Humphray S. J., Greenman C. D., Varela I., Lin M. L., Ordonez G. R., Bignell G. R., Ye K., Alipaz J., Bauer M. J., Beare D., Butler A., Carter R. J., Chen L., Cox A. J., Edkins S., Kokko-Gonzales P. I., Gormley N. A., Grocock R. J., Haudenschild C. D., Hims M. M., James T., Jia M., Kingsbury Z., Leroy C., Marshall J., Menzies A., Mudie L. J., Ning Z., Royce T., Schulz-Trieglaff O. B., Spiridou A., Stebbings L. A., Szajkowski L., Teague J., Williamson D., Chin L., Ross M. T., Campbell P. J., Bentley D. R., Futreal P. A., Stratton M. R. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance E. D., Stephens P. J., O'Meara S., McBride D. J., Meynert A., Jones D., Lin M. L., Beare D., Lau K. W., Greenman C., Varela I., Nik-Zainal S., Davies H. R., Ordonez G. R., Mudie L. J., Latimer C., Edkins S., Stebbings L., Chen L., Jia M., Leroy C., Marshall J., Menzies A., Butler A., Teague J. W., Mangion J., Sun Y. A., McLaughlin S. F., Peckham H. E., Tsung E. F., Costa G. L., Lee C. C., Minna J. D., Gazdar A., Birney E., Rhodes M. D., McKernan K. J., Stratton M. R., Futreal P. A., Campbell P. J. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wogan G. N., Hecht S. S., Felton J. S., Conney A. H., Loeb L. A. Environmental and chemical carcinogenesis. Semin Cancer Biol. 2004;14:473–486. doi: 10.1016/j.semcancer.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Carbone M., Klein G., Gruber J., Wong M. Modern criteria to establish human cancer etiology. Cancer Res. 2004;64:5518–5524. doi: 10.1158/0008-5472.CAN-04-0255. [DOI] [PubMed] [Google Scholar]
- Hussain S. P., Harris C. C. Molecular epidemiology of human cancer: contribution of mutation spectra studies of tumor suppressor genes. Cancer Res. 1998;58:4023–4037. [PubMed] [Google Scholar]
- Forbes S. A., Bhamra G., Bamford S., Dawson E., Kok C., Clements J., Menzies A., Teague J. W., Futreal P. A., Stratton M. R. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr Protoc Hum Genet. 2008;10:11. doi: 10.1002/0471142905.hg1011s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besaratinia A., Pfeifer G. P. Investigating human cancer etiology by DNA lesion footprinting and mutagenicity analysis. Carcinogenesis. 2006;27:1526–1537. doi: 10.1093/carcin/bgi311. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Denissenko M. F., Olivier M., Tretyakova N., Hecht S. S., Hainaut P. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene. 2002;21:7435–7451. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., You Y. H., Besaratinia A. Mutations induced by ultraviolet light. Mutat Res. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Besaratinia A. Mutational spectra of human Cancer Hum. Genet. 2009;125:493–506. doi: 10.1007/s00439-009-0657-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguerat S., Wilhelm B. T., Bahler J. Next-generation sequencing: applications beyond genomes. Biochem Soc Trans. 2008;36:1091–1096. doi: 10.1042/BST0361091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova O., Marra M. A. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Mardis E. R., Wilson R. K. Cancer genome sequencing: a review. Hum Mol Genet. 2009;18:R163–168. doi: 10.1093/hmg/ddp396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Getz G., Wheeler D. A., Mardis E. R., McLellan M. D., Cibulskis K., Sougnez C., Greulich H., Muzny D. M., Morgan M. B., Fulton L., Fulton R. S., Zhang Q., Wendl M. C., Lawrence M. S., Larson D. E., Chen K., Dooling D. J., Sabo A., Hawes A. C., Shen H., Jhangiani S. N., Lewis L. R., Hall O., Zhu Y., Mathew T., Ren Y., Yao J., Scherer S. E., Clerc K., Metcalf G. A., Ng B., Milosavljevic A., Gonzalez-Garay M. L., Osborne J. R., Meyer R., Shi X., Tang Y., Koboldt D. C., Lin L., Abbott R., Miner T. L., Pohl C., Fewell G., Haipek C., Schmidt H., Dunford-Shore B. H., Kraja A., Crosby S. D., Sawyer C. S., Vickery T., Sander S., Robinson J., Winckler W., Baldwin J., Chirieac L. R., Dutt A., Fennell T., Hanna M., Johnson B. E., Onofrio R. C., Thomas R. K., Tonon G., Weir B. A., Zhao X., Ziaugra L., Zody M. C., Giordano T., Orringer M. B., Roth J. A., Spitz M. R., Wistuba I. I., Ozenberger B., Good P. J., Chang A. C., Beer D. G., Watson M. A., Ladanyi M., Broderick S., Yoshizawa A., Travis W. D., Pao W., Province M. A., Weinstock G. M., Varmus H. E., Gabriel S. B., Lander E. S., Gibbs R. A., Meyerson M., Wilson R. K. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons D. W., Jones S., Zhang X., Lin J. C., Leary R. J., Angenendt P., Mankoo P., Carter H., Siu I. M., Gallia G. L., Olivi A., McLendon R., Rasheed B. A., Keir S., Nikolskaya T., Nikolsky Y., Busam D. A., Tekleab H., Diaz L. A., Jr, Hartigan J., Smith D. R., Strausberg R. L., Marie S. K., Shinjo S. M., Yan H., Riggins G. J., Bigner D. D., Karchin R., Papadopoulos N., Parmigiani G., Vogelstein B., Velculescu V. E., Kinzler K. W. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T., Jones S., Wood L. D., Parsons D. W., Lin J., Barber T. D., Mandelker D., Leary R. J., Ptak J., Silliman N., Szabo S., Buckhaults P., Farrell C., Meeh P., Markowitz S. D., Willis J., Dawson D., Willson J. K., Gazdar A. F., Hartigan J., Wu L., Liu C., Parmigiani G., Park B. H., Bachman K. E., Papadopoulos N., Vogelstein B., Kinzler K. W., Velculescu V. E. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Wood L. D., Parsons D. W., Jones S., Lin J., Sjoblom T., Leary R. J., Shen D., Boca S. M., Barber T., Ptak J., Silliman N., Szabo S., Dezso Z., Ustyanksky V., Nikolskaya T., Nikolsky Y., Karchin R., Wilson P. A., Kaminker J. S., Zhang Z., Croshaw R., Willis J., Dawson D., Shipitsin M., Willson J. K., Sukumar S., Polyak K., Park B. H., Pethiyagoda C. L., Pant P. V., Ballinger D. G., Sparks A. B., Hartigan J., Smith D. R., Suh E., Papadopoulos N., Buckhaults P., Markowitz S. D., Parmigiani G., Kinzler K. W., Velculescu V. E., Vogelstein B. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Hofseth L. J., Hussain S. P., Harris C. C. p53: 25 years after its discovery. Trends Pharmacol Sci. 2004;25:177–181. doi: 10.1016/j.tips.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Olivier M., Hussain S. P., Caron de Fromentel C., Hainaut P., Harris C. C. TP53 mutation spectra and load: a tool for generating hypotheses on the etiology of cancer. IARC Sci Publ. 2004;157:247–270. [PubMed] [Google Scholar]
- Petitjean A., Achatz M. I., Borresen-Dale A. L., Hainaut P., Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26:2157–2165. doi: 10.1038/sj.onc.1210302. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Lane D., Levine A. J. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Hussain S. P., Hollstein M. H., Harris C. C. p53 tumor suppressor gene: at the crossroads of molecular carcinogenesis, molecular epidemiology, and human risk assessment. Ann N Y Acad Sci. 2000;919:79–85. doi: 10.1111/j.1749-6632.2000.tb06870.x. [DOI] [PubMed] [Google Scholar]
- Thilly W. G. Have environmental mutagens caused oncomutations in people? Nat Genet. 2003;34:255–259. doi: 10.1038/ng1205. [DOI] [PubMed] [Google Scholar]
- Gee P., Maron D. M., Ames B. N. Detection and classification of mutagens: a set of base-specific Salmonella tester strains. Proc Natl Acad Sci U S A. 1994;91:11606–11610. doi: 10.1073/pnas.91.24.11606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortelmans K., Zeiger E. The Ames Salmonella/microsome mutagenicity assay. Mutat Res. 2000;455:29–60. doi: 10.1016/s0027-5107(00)00064-6. [DOI] [PubMed] [Google Scholar]
- Fronza G., Inga A., Monti P., Scott G., Campomenosi P., Menichini P., Ottaggio L., Viaggi S., Burns P. A., Gold B., Abbondandolo A. The yeast p53 functional assay: a new tool for molecular epidemiology Hopes and facts. Mutat Res. 2000;462:293–301. doi: 10.1016/s1383-5742(00)00011-9. [DOI] [PubMed] [Google Scholar]
- Smardova J., Smarda J., Koptikova J. Functional analysis of p53 tumor suppressor in yeast. Differentiation. 2005;73:261–277. doi: 10.1111/j.1432-0436.2005.00028.x. [DOI] [PubMed] [Google Scholar]
- Lambert I. B., Singer T. M., Boucher S. E., Douglas G. R. Detailed review of transgenic rodent mutation assays. Mutat Res. 2005;590:1–280. doi: 10.1016/j.mrrev.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Albertini R. J. HPRT mutations in humans: biomarkers for mechanistic studies. Mutat Res. 2001;489:1–16. doi: 10.1016/s1383-5742(01)00064-3. [DOI] [PubMed] [Google Scholar]
- Luo J. L., Yang Q., Tong W. M., Hergenhahn M., Wang Z. Q., Hollstein M. Knock-in mice with a chimeric human/murine p53 gene develop normally and show wild-type p53 responses to DNA damaging agents: a new biomedical research tool. Oncogene. 2001;20:320–328. doi: 10.1038/sj.onc.1204080. [DOI] [PubMed] [Google Scholar]
- Feldmeyer N., Schmeiser H. H., Muehlbauer K. R., Belharazem D., Knyazev Y., Nedelko T., Hollstein M. Further studies with a cell immortalization assay to investigate the mutation signature of aristolochic acid in human p53 sequences. Mutat Res. 2006;608:163–168. doi: 10.1016/j.mrgentox.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Liu Z., Hergenhahn M., Schmeiser H. H., Wogan G. N., Hong A., Hollstein M. Human tumor p53 mutations are selected for in mouse embryonic fibroblasts harboring a humanized p53 gene. Proc Natl Acad Sci U S A. 2004;101:2963–2968. doi: 10.1073/pnas.0308607101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Muehlbauer K. R., Schmeiser H. H., Hergenhahn M., Belharazem D., Hollstein M. C. p53 mutations in benzo(a)pyrene-exposed human p53 knock-in murine fibroblasts correlate with p53 mutations in human lung tumors. Cancer Res. 2005;65:2583–2587. doi: 10.1158/0008-5472.CAN-04-3675. [DOI] [PubMed] [Google Scholar]
- Luo J. L., Tong W. M., Yoon J. H., Hergenhahn M., Koomagi R., Yang Q., Galendo D., Pfeifer G. P., Wang Z. Q., Hollstein M. UV-induced DNA damage and mutations in Hupki (human p53 knock-in) mice recapitulate p53 hotspot alterations in sun-exposed human skin. Cancer Res. 2001;61:8158–8163. [PubMed] [Google Scholar]
- Reinbold M., Luo J. L., Nedelko T., Jerchow B., Murphy M. E., Whibley C., Wei Q., Hollstein M. Common tumour p53 mutations in immortalized cells from Hupki mice heterozygous at codon 72. Oncogene. 2008;27:2788–2794. doi: 10.1038/sj.onc.1210932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vom Brocke J., Krais A., Whibley C., Hollstein M. C., Schmeiser H. H. The carcinogenic air pollutant 3-nitrobenzanthrone induces GC to TA transversion mutations in human p53 sequences. Mutagenesis. 2009;24:17–23. doi: 10.1093/mutage/gen049. [DOI] [PubMed] [Google Scholar]
- Vom Brocke J., Schmeiser H. H., Reinbold M., Hollstein M. MEF immortalization to investigate the ins and outs of mutagenesis. Carcinogenesis. 2006;27:2141–2147. doi: 10.1093/carcin/bgl101. [DOI] [PubMed] [Google Scholar]
- Jaworski M., Hailfinger S., Buchmann A., Hergenhahn M., Hollstein M., Ittrich C., Schwarz M. Human p53 knock-in (hupki) mice do not differ in liver tumor response from their counterparts with murine p53. Carcinogenesis. 2005;26:1829–1834. doi: 10.1093/carcin/bgi142. [DOI] [PubMed] [Google Scholar]
- Tong W. M., Lee M. K., Galendo D., Wang Z. Q., Sabapathy K. Aflatoxin-B exposure does not lead to p53 mutations but results in enhanced liver cancer of Hupki (human p53 knock-in) mice. Int J Cancer. 2006;119:745–749. doi: 10.1002/ijc.21890. [DOI] [PubMed] [Google Scholar]
- Chao C., Hergenhahn M., Kaeser M. D., Wu Z., Saito S., Iggo R., Hollstein M., Appella E., Xu Y. Cell type- and promoter-specific roles of Ser18 phosphorylation in regulating p53 responses. J Biol Chem. 2003;278:41028–41033. doi: 10.1074/jbc.M306938200. [DOI] [PubMed] [Google Scholar]
- Harvey D. M., Levine A. J. p53 alteration is a common event in the spontaneous immortalization of primary BALB/c murine embryo fibroblasts. Genes Dev. 1991;5:2375–2385. doi: 10.1101/gad.5.12b.2375. [DOI] [PubMed] [Google Scholar]
- Hahn W. C., Weinberg R. A. Modelling the molecular circuitry of cancer. Nat Rev Cancer. 2002;2:331–341. doi: 10.1038/nrc795. [DOI] [PubMed] [Google Scholar]
- Hergenhahn M., Luo J. L., Hollstein M. p53 designer genes for the modern mouse. Cell Cycle. 2004;3:738–741. [PubMed] [Google Scholar]
- Bienz B., Zakut-Houri R., Givol D., Oren M. Analysis of the gene coding for the murine cellular tumour antigen p53. EMBO J. 1984;3:2179–2183. doi: 10.1002/j.1460-2075.1984.tb02110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumaz N., van Kranen H. J., de Vries A., Berg R. J., Wester P. W., van Kreijl C. F., Sarasin A., Daya-Grosjean L., de Gruijl F. R. The role of UV-B light in skin carcinogenesis through the analysis of p53 mutations in squamous cell carcinomas of hairless mice. Carcinogenesis. 1997;18:897–904. doi: 10.1093/carcin/18.5.897. [DOI] [PubMed] [Google Scholar]
- Hainaut P., Pfeifer G. P. Patterns of p53 G–>T transversions in lung cancers reflect the primary mutagenic signature of DNA-damage by tobacco smoke. Carcinogenesis. 2001;22:367–374. doi: 10.1093/carcin/22.3.367. [DOI] [PubMed] [Google Scholar]
- Toyooka S., Tsuda T., Gazdar A. F. The TP53 gene, tobacco exposure, and lung cancer. Hum Mutat. 2003;21:229–239. doi: 10.1002/humu.10177. [DOI] [PubMed] [Google Scholar]
- Besaratinia A., Pfeifer G. P. Second-hand smoke and human lung cancer. Lancet Oncol. 2008;9:657–666. doi: 10.1016/S1470-2045(08)70172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besaratinia A., Pfeifer G. P. Sunlight ultraviolet irradiation and BRAF V600 mutagenesis in human melanoma. Hum Mutat. 2008;29:983–991. doi: 10.1002/humu.20802. [DOI] [PubMed] [Google Scholar]
- Nortier J. L., Martinez M. C., Schmeiser H. H., Arlt V. M., Bieler C. A., Petein M., Depierreux M. F., De Pauw L., Abramowicz D., Vereerstraeten P., Vanherweghem J. L. Urothelial carcinoma associated with the use of a Chinese herb (Aristolochia fangchi) N Engl J Med. 2000;342:1686–1692. doi: 10.1056/NEJM200006083422301. [DOI] [PubMed] [Google Scholar]
- Nedelko T., Arlt V. M., Phillips D. H., Hollstein M. TP53 mutation signature supports involvement of aristolochic acid in the aetiology of endemic nephropathy-associated tumours. Int J Cancer. 2009;124:987–990. doi: 10.1002/ijc.24006. [DOI] [PubMed] [Google Scholar]
- Grollman A. P., Shibutani S., Moriya M., Miller F., Wu L., Moll U., Suzuki N., Fernandes A., Rosenquist T., Medverec Z., Jakovina K., Brdar B., Slade N., Turesky R. J., Goodenough A. K., Rieger R., Vukelic M., Jelakovic B. Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc Natl Acad Sci U S A. 2007;104:12129–12134. doi: 10.1073/pnas.0701248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord G. M., Hollstein M., Arlt V. M., Roufosse C., Pusey C. D., Cook T., Schmeiser H. H. DNA adducts and p53 mutations in a patient with aristolochic acid-associated nephropathy. Am J Kidney Dis. 2004;43:e11–17. doi: 10.1053/j.ajkd.2003.11.024. [DOI] [PubMed] [Google Scholar]
- Lord G. M., Cook T., Arlt V. M., Schmeiser H. H., Williams G., Pusey C. D. Urothelial malignant disease and Chinese herbal nephropathy. Lancet. 2001;358:1515–1516. doi: 10.1016/s0140-6736(01)06576-x. [DOI] [PubMed] [Google Scholar]
- Arlt V. M., Stiborova M., Schmeiser H. H. Aristolochic acid as a probable human cancer hazard in herbal remedies: a review. Mutagenesis. 2002;17:265–277. doi: 10.1093/mutage/17.4.265. [DOI] [PubMed] [Google Scholar]
- Pfau W., Schmeiser H. H., Wiessler M. Aristolochic acid binds covalently to the exocyclic amino group of purine nucleotides in DNA. Carcinogenesis. 1990;11:313–319. doi: 10.1093/carcin/11.2.313. [DOI] [PubMed] [Google Scholar]
- Kohara A., Suzuki T., Honma M., Ohwada T., Hayashi M. Mutagenicity of aristolochic acid in the lambda/lacZ transgenic mouse (MutaMouse) Mutat Res. 2002;515:63–72. doi: 10.1016/s1383-5718(01)00350-3. [DOI] [PubMed] [Google Scholar]
- Gilman P. U.S. Environmental Protection Agency, National Center for Environmental Assessment; Washington, DC: Health Assessment Document for Diesel Engine Exhaust EPA/600/8–90/057F. 2002 [Google Scholar]
- Arlt V. M. 3-Nitrobenzanthrone, a potential human cancer hazard in diesel exhaust and urban air pollution: a review of the evidence. Mutagenesis. 2005;20:399–410. doi: 10.1093/mutage/gei057. [DOI] [PubMed] [Google Scholar]
- Arlt V. M., Zhan L., Schmeiser H. H., Honma M., Hayashi M., Phillips D. H., Suzuki T. DNA adducts and mutagenic specificity of the ubiquitous environmental pollutant 3-nitrobenzanthrone in Muta Mouse. Environ Mol Mutagen. 2004;43:186–195. doi: 10.1002/em.20014. [DOI] [PubMed] [Google Scholar]
- Nishida H., Kawanishi M., Takamura-Enya T., Yagi T. Mutagenic specificity of N-acetoxy-3-aminobenzanthrone, a major metabolically activated form of 3-nitrobenzanthrone, in shuttle vector plasmids propagated in human cells. Mutat Res. 2008;654:82–87. doi: 10.1016/j.mrgentox.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Hashimoto A. H., Amanuma K., Hiyoshi K., Sugawara Y., Goto S., Yanagisawa R., Takano H., Masumura K., Nohmi T., Aoki Y. Mutations in the lungs of gpt delta transgenic mice following inhalation of diesel exhaust. Environ Mol Mutagen. 2007;48:682–693. doi: 10.1002/em.20335. [DOI] [PubMed] [Google Scholar]
- Whibley C., Odell A. F., Nedelko T., Balaburski G., Murphy M., Liu Z., Stevens L., Walker J. H., Routledge M., Hollstein M. Wild-type and HUPKI (human P53 knock-in)murine embryonic fibroblasts: P53/ARF pathway disruption in spontaneous escape from senescence [E-pub ahead of print] J Biol Chem. 2010 doi: 10.1074/jbc.M109.064444. doi: 10.1074/jbc.M109.064444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S., Han S. Y., Liu W., Otsuka K., Shibata H., Kanamaru R., Ishioka C. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci U S A. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S., Zhang X., Parsons D. W., Lin J. C., Leary R. J., Angenendt P., Mankoo P., Carter H., Kamiyama H., Jimeno A., Hong S. M., Fu B., Lin M. T., Calhoun E. S., Kamiyama M., Walter K., Nikolskaya T., Nikolsky Y., Hartigan J., Smith D. R., Hidalgo M., Leach S. D., Klein A. P., Jaffee E. M., Goggins M., Maitra A., Iacobuzio-Donahue C., Eshleman J. R., Kern S. E., Hruban R. H., Karchin R., Papadopoulos N., Parmigiani G., Vogelstein B., Velculescu V. E., Kinzler K. W. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brash D. E. Sunlight and the onset of skin cancer. Trends Genet. 1997;13:410–414. doi: 10.1016/s0168-9525(97)01246-8. [DOI] [PubMed] [Google Scholar]
- Jonason A. S., Kunala S., Price G. J., Restifo R. J., Spinelli H. M., Persing J. A., Leffell D. J., Tarone R. E., Brash D. E. Frequent clones of p53-mutated keratinocytes in normal human skin. Proc Natl Acad Sci U S A. 1996;93:14025–14029. doi: 10.1073/pnas.93.24.14025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brash D. E., Rudolph J. A., Simon J. A., Lin A., McKenna G. J., Baden H. P., Halperin A. J., Ponten J. A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc Natl Acad Sci U S A. 1991;88:10124–10128. doi: 10.1073/pnas.88.22.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A., Leffell D. J., Kunala S., Sharma H. W., Gailani M., Simon J. A., Halperin A. J., Baden H. P., Shapiro P. E., Bale A. E., Brash D. E. Mutation hotspots due to sunlight in the p53 gene of nonmelanoma skin cancers. Proc Natl Acad Sci U S A. 1993;90:4216–4220. doi: 10.1073/pnas.90.9.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A., Jonason A. S., Leffell D. J., Simon J. A., Sharma H. W., Kimmelman J., Remington L., Jacks T., Brash D. E. Sunburn and p53 in the onset of skin cancer. Nature. 1994;372:773–776. doi: 10.1038/372773a0. [DOI] [PubMed] [Google Scholar]
- Montesano R., Hainaut P., Wild C. P. Hepatocellular carcinoma: from gene to public health. J Natl Cancer Inst. 1997;89:1844–1851. doi: 10.1093/jnci/89.24.1844. [DOI] [PubMed] [Google Scholar]
- Hussain S. P., Schwank J., Staib F., Wang X. W., Harris C. C. TP53 mutations and hepatocellular carcinoma: insights into the etiology and pathogenesis of liver cancer. Oncogene. 2007;26:2166–2176. doi: 10.1038/sj.onc.1210279. [DOI] [PubMed] [Google Scholar]
- Oda K., Arakawa H., Tanaka T., Matsuda K., Tanikawa C., Mori T., Nishimori H., Tamai K., Tokino T., Nakamura Y., Taya Y. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell. 2000;102:849–862. doi: 10.1016/s0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- Dohoney K. M., Guillerm C., Whiteford C., Elbi C., Lambert P. F., Hager G. L., Brady J. N. Phosphorylation of p53 at serine 37 is important for transcriptional activity and regulation in response to DNA damage. Oncogene. 2004;23:49–57. doi: 10.1038/sj.onc.1207005. [DOI] [PubMed] [Google Scholar]
- Dudenhoffer C., Kurth M., Janus F., Deppert W., Wiesmuller L. Dissociation of the recombination control and the sequence-specific transactivation function of P53. Oncogene. 1999;18:5773–5784. doi: 10.1038/sj.onc.1202964. [DOI] [PubMed] [Google Scholar]
- Albrechtsen N., Dornreiter I., Grosse F., Kim E., Wiesmuller L., Deppert W. Maintenance of genomic integrity by p53: complementary roles for activated and non-activated p53. Oncogene. 1999;18:7706–7717. doi: 10.1038/sj.onc.1202952. [DOI] [PubMed] [Google Scholar]
- Hanawalt P. C. Subpathways of nucleotide excision repair and their regulation. Oncogene. 2002;21:8949–8956. doi: 10.1038/sj.onc.1206096. [DOI] [PubMed] [Google Scholar]
- Olivier M., Eeles R., Hollstein M., Khan M. A., Harris C. C., Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat. 2002;19:607–614. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]