Abstract
Interactions between genetic and∕or environmental factors are ubiquitous, affecting the phenotypes of organisms in complex ways. Knowledge about such interactions is becoming rate-limiting for our understanding of human disease and other biological phenomena. Phenomics refers to the integrative analysis of how all genes contribute to phenotype variation, entailing genome and organism level information. A systems biology view of gene interactions is critical for phenomics. Unfortunately the problem is intractable in humans; however, it can be addressed in simpler genetic model systems. Our research group has focused on the concept of genetic buffering of phenotypic variation, in studies employing the single-cell eukaryotic organism, S. cerevisiae. We have developed a methodology, quantitative high throughput cellular phenotyping (Q-HTCP), for high-resolution measurements of gene-gene and gene-environment interactions on a genome-wide scale. Q-HTCP is being applied to the complete set of S. cerevisiae gene deletion strains, a unique resource for systematically mapping gene interactions. Genetic buffering is the idea that comprehensive and quantitative knowledge about how genes interact with respect to phenotypes will lead to an appreciation of how genes and pathways are functionally connected at a systems level to maintain homeostasis. However, extracting biologically useful information from Q-HTCP data is challenging, due to the multidimensional and nonlinear nature of gene interactions, together with a relative lack of prior biological information. Here we describe a new approach for mining quantitative genetic interaction data called recursive expectation-maximization clustering (REMc). We developed REMc to help discover phenomic modules, defined as sets of genes with similar patterns of interaction across a series of genetic or environmental perturbations. Such modules are reflective of buffering mechanisms, i.e., genes that play a related role in the maintenance of physiological homeostasis. To develop the method, 297 gene deletion strains were selected based on gene-drug interactions with hydroxyurea, an inhibitor of ribonucleotide reductase enzyme activity, which is critical for DNA synthesis. To partition the gene functions, these 297 deletion strains were challenged with growth inhibitory drugs known to target different genes and cellular pathways. Q-HTCP-derived growth curves were used to quantify all gene interactions, and the data were used to test the performance of REMc. Fundamental advantages of REMc include objective assessment of total number of clusters and assignment to each cluster a log-likelihood value, which can be considered an indicator of statistical quality of clusters. To assess the biological quality of clusters, we developed a method called gene ontology information divergence z-score (GOid_z). GOid_z summarizes total enrichment of GO attributes within individual clusters. Using these and other criteria, we compared the performance of REMc to hierarchical and K-means clustering. The main conclusion is that REMc provides distinct efficiencies for mining Q-HTCP data. It facilitates identification of phenomic modules, which contribute to buffering mechanisms that underlie cellular homeostasis and the regulation of phenotypic expression.
A phenotype, or “trait,” is a physical manifestation of an organism. Perhaps the most fundamental phenotype among all organisms is survival and proliferation of a cell. This phenotype has been and continues to be extensively analyzed for the budding yeast, Saccharomyces cerevisiae, which has proven to be a valuable model for genetic analysis and of high relevance to cancer and other human diseases.1 The power of yeast genetics stems from its extreme tractability regarding genotype-phenotype interplay; however, only recently have tools been developed for systematic analysis of gene-gene and gene-environment interactions.2 Moreover, since the arrival of genome sequencing, there has been increased appreciation for the evolutionary conservation of genes across different life forms,3 creating new opportunities, from a systems biology perspective, to achieve an integrative understanding of how genetic and environmental factors interact with respect to phenotypes.4 Inherent limitations with systematic studies of gene interaction include (1) involvement of combinatorial tests (combinatorial explosion), (2) the abundance of natural genetic and phenotypic variation (intractability), and (3) an emphasis of biological research in human and mammalian model systems where gene interaction effects cannot be tested in a controlled manner, and where acquisition of data is more difficult and expensive. Genetically tractable model systems, although recognized for a role in biomedical research, remain underutilized. Research employing quantitative high throughput cellular phenotyping (Q-HTCP) analysis of the genomic collection of deletion strains has the potential to address many current limitations for understanding gene interaction networks. In addition to functional conservation between the S. cerevisiae and human genomes, S. cerevisiae is easy and inexpensive to culture, its generation time is less than a tenth that of human cells, and its genes are much easier to manipulate. Moreover it is a single celled organism, and thus exists experimentally in a more natural state than cells cultured in vitro from multicellular organisms. Evolutionary constraints placed on biological systems naturally result in conservation of cellular processes, and thus S. cerevisiae can provide initial insight regarding biological principles of gene interaction that underlie the genetic complexity of human disease.4 For all of these reasons, we are utilizing this model system to assess the effects of gene-gene and gene-drug interaction on phenotypes. In this report we revisit some data from previously published experiments.5 In the earlier analysis of the data, we perceived strengths and weaknesses in the use of hierarchical clustering (Hc) for mining high throughput quantitative gene interaction data. Here we describe our efforts to address limitations of Hc while preserving useful features, an approach called recursive expectation-maximization clustering (REMc).
INTRODUCTION
A genomic set of gene deletion, or “knockout,” strains is currently the most advanced resource for studying genetic interaction networks.6, 7 Using this collection of 5000 strains one can systematically test the effects of single gene deletion effects in combination with drug treatments5, 8, 9 or introduction of a particular gene mutation of interest into the entire collection.10, 11, 12 Prior to creation of the yeast gene deletion strain library resource, yeast cell proliferation phenotypes were traditionally screened in a qualitative manner,13 and only genes of special interest quantitatively analyzed using kinetic growth curves. Since creation of the library, we have worked to develop methodologies to measure tens of thousands of growth curves in a single experiment in order to resolve gene interactions quantitatively.14 We have found that quantitative resolution of gene interactions can be a critical factor for identifying phenomic “modules.”5, 15, 16 Other research groups have also shown that clustering quantitative gene interaction data is useful for identifying pathways and protein complexes.17
An unnecessary distinction is often drawn between gene-gene and gene-environment interactions. For practical and biological reasons, we consider all gene interactions to be fundamentally similar and mutually informative, because gene mutations, environmental exposures, and drug treatments in a broader context are simply different types of “perturbations.” Interactions can be quantified the same way regardless of which perturbation types are combined.5 There is plenty of support from the literature for this integrated systems view of gene interaction, since the genomic interaction profiles of a drug treatment versus mutation of the corresponding drug target in theory should (and in reality do) share high similarity.9, 18 Another note about the term gene interaction (also called epistasis) is it has been defined various ways in genetic research.19, 20 We think of genetic interactions in a mathematical sense; meaning that the observed phenotype resulting from a combination of perturbations departs from an assumed neutral (noninteractive) phenotype; the expected phenotype being based on the phenotypes observed in the setting of the respective single perturbations. The strength of interaction reflects the degree of “surprise,” or departure from expectation. Thus, genetic interactions derive from two essential components: a neutrality function and quantitative phenotypic measures.21 Synergistic, or “enhancing,” interactions reflect an accelerated effect on the phenotype in the same direction; antagonistic, or “suppressing,” interactions indicate alleviation or counteracting effects on the phenotype by different perturbations when combined. We are interested in how the neutrality function and quantitative phenotypic measure affect the interpretation of Q-HTCP data; however, the work here is not focused on these issues directly, but rather on development of data mining tools to assess genetic interactions, however defined. One possibility is that improvement in the quality of Q-HTCP data5, 14, 22 together with development of flexible and robust data mining tools will help advance our understanding of the biological relevance of different neutrality models in large-scale studies of gene interaction.21, 23
To assist in development and testing REMc, we used a previously published data set of gene-drug interactions consisting of a 297 gene by 14 drug perturbation matrix of gene-drug interactions in which interactions were quantified as a z-score, called the Growth Index (see below and supplementary material).5, 24 As seen in the equation below, the neutrality function assumed that reduction in the area under the growth curve induced by growth inhibitory drug treatment would be proportional for any deletion mutant compared to the wild type control strain. Noise in the assay was accounted for by replicate assays of the wild type strain (no gene deletion). This definition is a form of the “multiplicative” neutrality function,21, 25
We screened the entire collection of 5000 knockout strains at different concentrations of hydroxyurea (HU), an inhibitor of ribonucleotide reductase (RNR). To further understand biological differences between 297 putative RNR-interactive genes, they were tested for phenotypic interaction with drugs having different cellular effects including (1) cisplatin, which like HU, induces DNA damage, but by a different mechanism (intercalating in DNA). (2) Miconazole, which is an inhibitor of the ERG11 gene and essential enzyme in ergosterol biosynthesis. (3) T-butyl hydrogen peroxide (TBHP), which induces oxidative damage, stressing many cellular processes, including DNA and protein metabolism. (4) Cycloheximide, which is an inhibitor of a gene RPL28, a component of the large ribosomal subunit essential for translation, making protein synthesis rate-limiting for cell proliferation. Genes having function(s) important for phenotypic stability in the presence of one perturbation often have different importance in another context, hence the rationale of clustering matrices of quantitative gene interaction data to identify genetic pathways that buffer (e.g., compensation by alternative pathways) loss of RNR function.5
Our primary goal for development of a new clustering approach was to achieve objective results that could be easily interpreted—interpretation including the total number of clusters as well as the statistical quality and biological meaning. These objectives were born from our experience with Hc, for which the flexibility and scalability seemed limited by subjectivity, making it labor intensive and nonquantitative. K-means clustering (KMc), also commonly used, requires a priori knowledge of the number of clusters, and like Hc, employs metrics such as Euclidean distance (Euc) or Pearson correlation (Pc), introducing another subjective parameter that impacts the result. Other methods, such as biclustering, offer an advantage over KMc or Hc, in that the multifunctional aspects of genes may be better accounted for by allowing genes to appear in multiple clusters.8, 26 However, with biclustering there are also numerous different algorithms and data visualization is a challenge.27, 28 REMc resulted from taking a fresh look at developing a flexible, quantitative, and visually intuitive clustering tool for discovering phenomic modules from Q-HTCP data. In previous work we had demonstrated that such modules can contain information about novel buffering mechanisms that regulate phenotypic expression.15 Recently, others have shown independently that such mechanisms are evolutionarily conserved.29 We wondered if, by using REMc, we could arrive at similar conclusions, but in a more objective, efficient, and potentially automated way than with other clustering approaches.
REMc utilizes a probabilistic framework, enabling determination of cluster likelihood, and objective estimation of the total number and rank order of clusters. Advantages of REMc thus include (1) direct analysis of data, avoiding use of gene similarity metrics, such as Euclidean distance or Pc coefficient; (2) objective determination of the total number of clusters; (3) ranking of clusters according to their quality; and (4) a view of hierarchical relationships between clusters. To assess potential advantages of REMc, we compared properties of REMc clusters with those obtained from Hc and KMc.
In addition to the “statistical quality” of clusters provided by REMc, we desired a tool to assess gene interaction clusters with regard to biological function. Gene ontology (GO) is a computational resource for systematic assessment of genomic data for biological functions.30 Although many computational tools have been developed for use with GO, we did not find one for summarizing, in a single quantitative score, the enrichment over all GO terms within a single list of genes. Thus we devised a method called gene ontology information divergence z-score (GOid_z). GOid_z can be thought of as a score that summarizes overall enrichment of biological functions within a gene list. GOid_z is useful for assessing the relative enrichment of all biological functions between different gene clusters. In addition, we usedGO TERMFINDER (GTF),31 available at the SGD website,32 to identify specific annotation terms and the genes that compose each term.
RESULTS
REMc offers theoretical and practical advantages over popular clustering methods
Hc and KMc are commonly used to mine biological data. Each entails the use of metrics, such as Euclidean distance or Pc coefficient, and Hc additionally employs different linkage methods, namely, average or complete. Clustering results thus vary according to the particular combination of algorithm and similarity metric used.33 The rationale for choosing a particular algorithm and∕or metric to evaluate a data set is difficult to establish, and there is no statistical basis upon which to assess the number or quality of clusters. By contrast, expectation-maximization clustering (EMc) is a model-based clustering method, implemented by fitting the raw data matrix to Gaussian distributions to calculate the most probable number of distinct groupings [Gaussian mixture model (GMM) optimized by expectation-maximization (EM) algorithm]. EMc eliminates the need to choose among distance metrics for clustering analysis, while objectively specifying the number and quantifying the likelihood of clusters. For EMc, we used the freely available Waikato Environment for Knowledge Analysis (WEKA) software.34 We found that by recursively applying EMc (REMc), additional clusters could be found, a log-likelihood (LL) value could be obtained for every cluster, and hierarchical relationships were established in the process. Thus, REMc avoids subjectively cutting a dendrogram to define clusters, as is often done with Hc, or guessing, a priori, the number of clusters, as required for KMc. To investigate these advantages, we compared REMc results with (1) two KMc methods, using either Euclidean distance or Pc as a similarity metric, and (2) four Hc methods, combining Pc or Euclidean distance with either average or complete linkage.
REMc provides an objective assessment of cluster number and quality
An overview of the REMc clustering algorithm is depicted in Fig. 1a. A 297×14 matrix of gene-drug interactions was analyzed, and characteristics of the clusters resulting from REMc are further detailed in Table 1. In the first round of clustering, there were four clusters with a LL of −41.1. In the second round of clustering, each of the first round clusters was further subdivided; cluster 0 giving rise to two additional clusters with LL=−30.9, cluster 1 giving rise to five clusters with LL of −37.8, and so on. By recursively applying the algorithm to each new cluster, until subclusters are no longer obtained, a LL value can be obtained for “terminal” clusters (larger LL indicates a more significant cluster). In addition to cluster quality values, REMc establishes hierarchical relationships through the generation of subclusters with iterations of the method (e.g., clusters 0_0 and 0_1 comprise branches of cluster 0). To validate the number of clusters predicted by REMc, we specified the number of clusters incrementally (an optional parameter in WEKA), generating a plot of LL versus number of clusters. We observed a steep increase in LL versus cluster number followed by a leveling off of the LL when the number of clusters reached 17, equal to the number of clusters predicted by REMc [Fig. 1b]. Our interpretation of Fig. 1b is that any increase in LL associated with greater than 17 clusters represents overfitting. Recognizing that every gene interaction profile is unique, incremental linear increases in LL obtained by increasing cluster number are probably noninformative and thus not worth attention.
Figure 1.
Algorithm for REMc. (a) REM clustering is performed on the unclustered data and then repeatedly on each new cluster until no additional subcluster is obtained. The first round of clustering yielded four clusters. Each cluster was subdivided in a second round of clustering (thick lines). Only two clusters were divided further in the third round of clustering (thin lines). No new clusters were found in the fourth round of REMc. A LL score is obtained for each round of clustering, thus when a single cluster is no longer subclassified, the LL provides a quantitative indication of the probability that the cluster represents a uniform class of data. (b) To evaluate the number of clusters obtained by REMc, EMc was performed by fixing the number of clusters between 1 and 40 (instead of determining the optimal number of clusters). The arrow indicates that the number of clusters determined by REMc, 17, was at an inflection point of the plot of LL vs cluster number.
Table 1.
Results from REMc are indicated. For each round of clustering, the LL is given along with the name of corresponding clusters and their number of genes. “1” indicates that the gene list submitted for REMc did not reveal additional subclusters. See also Fig. 1 and supplementary material.
| First Rd LL | First Rd | No. of genes | Second Rd LL | Second Rd | No. of genes | Third Rd LL | Third Rd | No. of genes | Fourth Rd LL | Fourth Rd | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 44 | −30.88 | 0_0 | 18 | −32.64 | 1 | |||||
| 0_1 | 26 | −28.52 | 1 | ||||||||
| 1 | 63 | −37.58 | 1_0 | 16 | −40.06 | 1 | |||||
| 1_1 | 11 | −36.21 | 1 | ||||||||
| 1_2 | 23 | −35.33 | 1 | ||||||||
| 1_3 | 10 | −33.12 | 1 | ||||||||
| 1_4 | 3 | −30.98 | 1 | ||||||||
| 297 ORFs | −41.13 | 2 | 96 | −40.22 | 2_0 | 47 | −34.07 | 2_0_0 | 15 | −31.13 | 1 |
| 2_0_1 | 12 | −35.20 | 1 | ||||||||
| 2_0_2 | 20 | −33.08 | 1 | ||||||||
| 2_1 | 21 | −44.71 | 1 | ||||||||
| 2_2 | 28 | −36.03 | 2_2_0 2 0 | 9 | −35.32 | 1 | |||||
| 2_2_1 2 1 | 19 | −35.44 | 1 | ||||||||
| 3 | 94 | −33.93 | 3_0 | 9 | −32.20 | 1 | |||||
| 3_1 | 34 | −32.02 | 1 | ||||||||
| 3_2 | 24 | −33.94 | 1 | ||||||||
| 3_3 | 27 | −32.46 | 1 |
REMc surpasses other clustering methods for biological discovery
CLUSTERJUDGE, which assesses enrichment of GO terms across all clusters, was used to assess the overall quality of each clustering method.35 The basic idea is genes that function together biologically (i.e., they are coannotated with GO terms) will cluster together, and CLUSTERJUDGE assesses this partitioning of genetic data with respect to biological information. The CLUSTERJUDGE score is calculated from the sum of mutual information (MI) correlation scores for all clusters based on the biological attribute (GO category) vector and the vector of cluster assignments for all genes. A z-score is created by comparing the global MI score for clusters derived by a particular method using as a benchmark a distribution of random clusterings. CLUSTERJUDGE is run multiple times for each cluster to achieve a robust comparison. CLUSTERJUDGE results were obtained (15 replicate runs for each clustering method with average and standard error) using the online tool provided by the Roth laboratory.35 Since other methods do not determine the number of clusters, we employed 17 clusters, the number determined by REMc (Fig. 1), for comparison of other methods. Hc tended to yield clusters containing only one gene [Fig. 2a], perhaps contributing to lower CLUSTERJUDGE scores, since single attributes by definition do not exhibit MI [Fig. 2b]. Thus, REMc and KMc yielded more information than Hc regarding biological enrichment in gene clusters. Since Hc_Pc with complete linkage performed best among Hc methods, it was carried forward in the additional comparisons between REMc and KMc described below.
Figure 2.
Comparison of cluster distributions and yield of biological information by REMc, Hc, and KMc. (a) Cluster size distributions from each of seven different clustering methods. With the cluster number fixed at 17, Hc results in a wider range of cluster sizes relative to other methods. (b) The output from 15 runs of CLUSTERJUDGE (CJ) using the entire result of each indicated clustering method as an input. 17 clusters, the number predicted by REMc, were assumed for each method. The p-value refers to t-test results comparing distributions of CJ scores between REMc and each other method. Clustering method abbreviations are REMc (recursive expectation-maximization), KMc_Euc (K-means with Euclidean distance), KMc_Pc (K-means with Pc), Hc_Pc_comp (hierarchical with Pc and complete linkage), Hc_Euc_comp (hierarchical with Euclidian distance and complete linkage), Hc_Pc_avg (hierarchical with Euclidian distance and average linkage), and Hc_Euc_avg (hierarchical with Pc and average linkage).
Log-likelihood and GOid_z discriminate REMc cluster quality
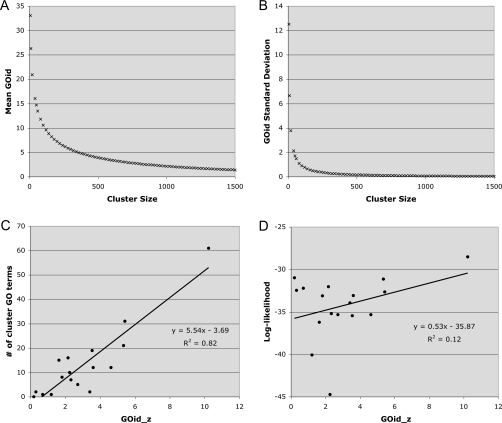
As described above, a LL measure is obtained for each individual cluster by recursively clustering until there are no significant subclasses (Fig. 1 and Table 1). Independent of this statistical value assigned to each cluster by REMc, we sought a convenient biological measure for cluster quality. For this purpose, we developed a method, GOid, to assess functional enrichment within a gene cluster with respect to all GO terms. GOid is converted to a z-score (GOid_z) to correct for the effect of gene cluster size, which correlates negatively with the GOid mean and standard deviation of randomly chosen gene sets [Figs. 3a, 3b]. The GOid_z is a quantitative measure summarizing the enrichment of biological functions in a gene cluster. As expected, GOid_z correlates positively with the total number of GO terms enriched within a cluster [Fig. 3c]. Note that GOid_z estimates enrichment of biological functions in a single cluster (e.g., for comparing relative quality of clusters obtained by a single method), in contrast to CLUSTERJUDGE, which compares different clustering methods with respect to the entire result (all clusters).
Figure 3.
The GOid_z score as a summary of functional enrichment of clustered genes. For random clusters, the mean GOid and standard deviation were inversely correlated with the number of genes per cluster. Thus, the GOid mean (a) and standard deviation (b) were calculated for 1000 random clusters and used to determine a z-score for the GOid for each REMc cluster. (c) For REMc clusters, a positive correlation was found between GOid_z and the number of enriched GO terms for each cluster, as calculated by GTF. (d) To investigate the complementary nature of the REMc LL and GOid_z score for mining gene interaction data, LL and GOid_z were plotted for all clusters; see text for discussion.
Whereas the GOid_z provides an assessment of biological enrichment, the LL provides an indicator of statistical quality, which can be thought of as the uniformity of the gene profiles in the cluster. These are complementary measures to objectively assess cluster quality and identify phenomic modules. From a scatter plot of LL versus GOid_z, we observed a positive, although weak, correlation between LL and GOid_z [Fig. 3d]. Considering four groups of clusters, corresponding to the four quadrants of this plot: group 1 consisted of clusters with high LL and high GOid_z values. These represent gene clusters where the experimental signature (LL) is strongly detected, and the associated biology (GOid_z) is well described in the literature. Cluster 0_1 is the representative cluster in this group, containing DNA damage response genes that have a strong and uniform profile of response to HU and cisplatin, and are highly annotated due to extensive study of these genes, which are of high cancer-relevance. Group 2 clusters for which the LL was high, but the GOid_z was relatively low, indicated a set of genes whose functions affect phenotype of the organism in a similar manner, however for which the biological relationships of the genes with respect to one another are less well characterized in the literature. Group 3 held clusters with relatively low LL and low GOid_z scores, probably representing heterogeneous data with low biological information quality. Notably, we did not find any clusters in the potential group 4, with low LL and high GOid_z, consistent with the thought that sets of genes that do not have good statistical cluster quality (i.e., the gene interaction profiles are heterogeneous) are less likely to contain biologically related genes.
Partitioning biological information by different clustering methods: A case study
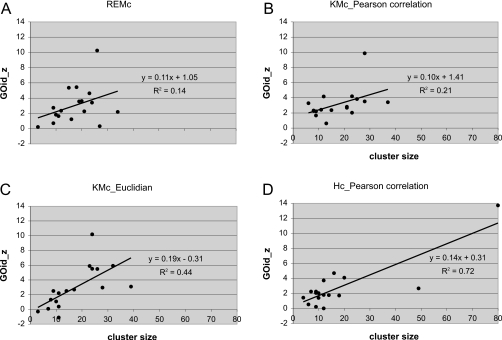
When plots of GOid_z versus cluster size were compared between REMc, KMc, and Hc_Pc (Fig. 4), two differences were apparent: first, Hc tended to yield clusters of more extreme size, less than 20 or greater than 50 [Fig. 4d], whereas the other three methods yielded similar size distributions. The extreme size of some Hc clusters was consistent with the fact that three out of the four Hc methods yielded multiple clusters containing only one gene [Fig. 2a]. This is partially a consequence of constraining the cluster number to 17, but highlights the difficulty in objectively determining the absolute number of clusters with Hc. The range of cluster GOid_z values was notably different for KMc using Pc [Fig. 4b] than it was for REMc and KMc using the Euclidean distance metric [Figs. 4a, 4c]. Most KMc_Pc clusters had GOid_z between the range of 2 and 4, lacking discrimination between clusters. In contrast, the distributions of GOid_z observed for KMc_Euc and REMc suggested greater discrimination between different clusters. The differences above can also be appreciated in Fig. 5, in which the data in Fig. 4 were ranked and viewed together in separate plots of cluster size and GOid_z. A biological explanation for the difference in the range of GOid_z values between Pc and Euclidean distance metric-derived cluster is that Euclidean distance takes more into account the strength of gene interactions. In contrast, Pc is more sensitive to the pattern, and less so to the magnitude of effects across an effect profile. Thus we reason that KMc_Euc may partition discrete biological functions more precisely than KMc_Pc, because it better incorporates information about the strength of gene interactions. Importantly, REMc shares more the characteristic of KMc_Euc, which is desirable for biological discrimination between phenomic modules. Accordingly the background size for GO terms resulting from REMc and KMc_Euc tended to be smaller than for KMc_Pc, indicating discovery of more specific biological functions by REMc and KMc_Euc. On the other hand, Pc has been particularly useful in genome-wide analysis of gene expression where identification of functionally related genes hinges on detection of the direction of change, perhaps more so than the absolute amount of change.35 Taken together, we conclude that the strength of gene interaction is a key component in identifying phenomic modules, and that REMc and nonmodel based methods using Euclidian distance are better at detecting this than methods using Pc.
Figure 4.
Differential partitioning of biological information by REMc and other clustering methods. Cluster size is plotted against GOid_z for four clustering methods. REMc (a) shared similarity with KMc_Euc (c), consistent with assessment of overlap in genes per cluster (see Table 2). By comparison with REMc and KMc_Euc, KMc_Pc (b) exhibited different GOid_z range, while Hc_Pc (d) differed with respect to cluster size distribution.
Figure 5.
Comparison of cluster size and GOid_z for clusters from each method. The histograms represent a different view of the data presented in Fig. 4, showing similarity between REMc and KMc, and depicting that Pc tends to result in the even distribution of biological information across all clusters (a), while Hc tends to yield extreme cluster sizes (b).
We next mapped REMc clusters to those obtained using Hc_Pc_complete, KMc_Pc, and KMc_Euc by comparing the overlap of respective gene clusters. We hypothesized that tightly correlated gene interaction profiles and∕or those containing genes with highly related functions would be identified in common by different clustering methods. REMc clusters were considered to match those obtained by another method if there was a 10% overlap of genes in both directions and at least a 25% overlap in either direction, thus a REM cluster could map to multiple clusters from another method. For each REM cluster, the best match was determined by the largest two-way overlap with a cluster from another method. For example, there were 26 genes in REMc cluster 0_1. The total number of overlapping genes appearing in other matching clusters—22 (Hc̱Pc=85%), 23 (KMc̱Euc=88%), and 23 (KMc̱Pc=88%)—was comparable for each of the methods; however, the sizes of those clusters and thus the percentage matches were different between Hc_PC (22∕80), KMc_Euc (23∕24), and KM_PC (23∕28), and thus KMc_Euc was considered the best match to REMc for cluster 0_1. The result of cluster mapping is summarized in Table 2. As suggested by the GOid_z versus cluster size plots (Fig. 4), the cluster mapping exercise indicated greatest similarity between REMc and KMc_Euc. Although not an entirely simple relationship, high quality REMc clusters, i.e., clusters with high LL and∕or high GOid_z, tended to overlap between methods (Table 2).
Table 2.
An overview of mappings between REMc clusters and clusters obtained by other methods. Based on the total number of overlapping genes and the relative size of each cluster, REMc clusters were matched to Hc (Euclidian distance and complete linkage) and KMc (Euclidian distance or Pc) clusters. A match was defined as at least 0.10 overlap in both directions and 0.25 overlap in one direction. The best matches are in bold.
| GOid_z rank | LL rank | REMc ID | No. of genes | Matches | Best match | He_rank | No. of genes | No. of match | EM (%) | Hc (%) | KMc_Eu_rank | No. of genes | No. of match | EM (%) | K_EU (%) | KMc_Pc_rank | No. of genes | No. of match | EM (%) | K_Pc (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 0_1 | 26 | All | K_Euc | 1 | 80 | 22 | 0.85 | 0.28 | 1 | 24 | 23 | 0.88 | 0.96 | 1 | 28 | 23 | 0.88 | 0.82 |
| 2 | 7 | 0_0 | 18 | All | K_Euc | 3 | 20 | 11 | 0.61 | 0.55 | 5 | 26 | 16 | 0.89 | 0.62 | 7 | 6 | 6 | 0.33 | 1.00 |
| 2 | 7 | 0_0 | 18 | 2 | 16 | 4 | 0.22 | 0.25 | 4 | 25 | 9 | 0.50 | 0.36 | |||||||
| 3 | 3 | 2_0_0 | 15 | All | N∕A | 4 | 12 | 10 | 0.67 | 0.83 | 2 | 32 | 14 | 0.93 | 0.44 | 3 | 12 | 9 | 0.60 | 0.75 |
| 3 | 6 | 9 | 3 | 0.20 | 0.33 | 13 | 8 | 4 | 0.27 | 0.50 | ||||||||||
| 4 | 13 | 1_2 | 23 | N∕A | N∕A | 1 | 80 | 10 | 0.43 | 0.13 | 10 | 14 | 6 | 0.26 | 0.43 | 6 | 37 | 8 | 0.35 | 0.22 |
| 4 | 13 | 1_3 | 24 | 5 | 49 | 7 | 0.30 | 0.14 | 7 | 28 | 8 | 0.35 | 0.29 | 2 | 23 | 6 | 0.26 | 0.26 | ||
| 5 | 8 | 2_0_2 | 20 | Hc; K_Euc | K_Euc | 1 | 80 | 15 | 0.75 | 0.19 | 3 | 23 | 12 | 0.60 | 0.52 | 5 | 28 | 8 | 0.40 | 0.29 |
| 5 | 8 | 2_0_2 | 20 | 13 | 4 | 2 | 0.10 | 0.50 | 4 | 25 | 5 | 0.25 | 0.20 | |||||||
| 6 | 14 | 2_2_1 | 19 | All | K_Euc | 10 | 12 | 7 | 0.37 | 0.58 | 9 | 9 | 7 | 0.37 | 0.78 | 17 | 13 | 7 | 0.37 | 0.54 |
| 6 | 14 | 2_2_1 | 19 | 14 | 12 | 6 | 0.32 | 0.50 | 8 | 21 | 5 | 0.26 | 0.24 | |||||||
| 6 | 14 | 2_2_1 | 19 | 9 | 10 | 4 | 0.21 | 0.40 | 3 | 12 | 3 | 0.16 | 0.25 | |||||||
| 7 | 10 | 3_2 | 24 | K_Euc | K_Euc | 8 | 9 | 5 | 0.21 | 0.56 | 6 | 39 | 13 | 0.54 | 0.33 | 14 | 9 | 5 | 0.21 | 0.56 |
| 7 | 10 | 3_2 | 24 | 5 | 49 | 6 | 0.25 | 0.12 | 11 | 11 | 3 | 0.13 | 0.27 | 12 | 8 | 3 | 0.13 | 0.38 | ||
| 7 | 10 | 3_2 | 24 | 2 | 16 | 4 | 0.17 | 0.25 | 16 | 9 | 3 | 0.13 | 0.33 | |||||||
| 8 | 12 | 2_2_0 | 9 | All | Hc | 7 | 7 | 5 | 0.56 | 0.71 | 11 | 11 | 4 | 0.44 | 0.36 | 10 | 11 | 4 | 0.44 | 0.36 |
| 8 | 12 | 2_2_0 | 9 | 8 | 9 | 3 | 0.33 | 0.33 | 4 | 24 | 3 | 0.33 | 0.13 | 14 | 9 | 3 | 0.33 | 0.33 | ||
| 9 | 11 | 2_0_1 | 12 | Hc: K_Euc | Hc | 12 | 18 | 6 | 0.50 | 0.33 | 8 | 17 | 5 | 0.42 | 0.29 | 9 | 21 | 5 | 0.42 | 0.24 |
| 9 | 11 | 2_0_1 | 12 | 3 | 20 | 4 | 0.33 | 0.20 | 4 | 25 | 4 | 0.33 | 0.16 | |||||||
| 10 | 17 | 2_1 | 21 | K_Euc; K_Pc | K_Euc | 16 | 9 | 6 | 0.29 | 0.67 | 15 | 7 | 7 | 0.33 | 1.00 | 8 | 21 | 8 | 0.38 | 0.38 |
| 10 | 17 | 2_1 | 21 | 16 | 3 | 2 | 0.10 | 0.67 | 5 | 28 | 6 | 0.29 | 0.21 | |||||||
| 10 | 17 | 2_1 | 21 | 12 | 8 | 4 | 0.19 | 0.50 | ||||||||||||
| 10 | 17 | 2_1 | 21 | 3 | 23 | 6 | 0.29 | 0.26 | ||||||||||||
| 11 | 4 | 3_1 | 34 | K_Euc | K_Euc | 1 | 80 | 17 | 0.50 | 0.21 | 7 | 28 | 15 | 0.44 | 0.54 | 5 | 28 | 9 | 0.26 | 0.32 |
| 11 | 4 | 3_1 | 34 | 5 | 49 | 10 | 0.29 | 0.20 | 6 | 39 | 11 | 0.32 | 0.28 | 6 | 37 | 10 | 0.29 | 0.27 | ||
| 11 | 4 | 3_1 | 34 | |||||||||||||||||
| 12 | 9 | 1_3 | 10 | K_Euc | K_Euc | 3 | 20 | 4 | 0.40 | 0.20 | 5 | 26 | 10 | 1.00 | 0.38 | 4 | 25 | 4 | 0.40 | 0.16 |
| 12 | 9 | 1_3 | 10 | 2 | 16 | 3 | 0.30 | 0.19 | 2 | 23 | 3 | 0.30 | 0.13 | |||||||
| 12 | 9 | 1_3 | 10 | 12 | 8 | 2 | 0.20 | 0.25 | ||||||||||||
| 13 | 15 | 1_1 | 11 | Hc | Hc | 15 | 6 | 4 | 0.36 | 0.67 | 4 | 24 | 6 | 0.55 | 0.25 | 6 | 37 | 5 | 0.45 | 0.14 |
| 13 | 15 | 1_1 | 11 | 9 | 10 | 3 | 0.27 | 0.30 | 8 | 17 | 4 | 0.36 | 0.24 | |||||||
| 14 | 16 | 1_0 | 16 | K_Euc | K_Euc | 5 | 49 | 11 | 0.69 | 0.22 | 13 | 10 | 7 | 0.44 | 0.70 | 15 | 23 | 5 | 0.31 | 0.22 |
| 14 | 16 | 1_0 | 16 | 10 | 14 | 6 | 0.38 | 0.43 | 1 | 28 | 4 | 0.25 | 0.14 | |||||||
| 15 | 5 | 3_0 | 9 | K_Euc; K_Pc | K_Euc | 14 | 11 | 7 | 0.78 | 0.64 | 15 | 23 | 7 | 0.78 | 0.30 | |||||
| 16 | 6 | 3_3 | 27 | K_Euc | K_Euc | 11 | 14 | 6 | 0.22 | 0.43 | 17 | 11 | 10 | 0.37 | 0.91 | 11 | 15 | 6 | 0.22 | 0.40 |
| 16 | 6 | 3_3 | 27 | 14 | 10 | 4 | 0.15 | 0.40 | 6 | 39 | 15 | 0.56 | 0.38 | 17 | 13 | 4 | 0.15 | 0.31 | ||
| 16 | 6 | 3_3 | 27 | 17 | 12 | 4 | 0.15 | 0.33 | 10 | 11 | 3 | 0.11 | 0.27 | |||||||
| 17 | 2 | 1_4 | 3 | N∕A |
REMc reveals a hierarchical aspect of quantitative phenomic information
A useful feature of Hc, which is lacking from KMc, is the representation of hierarchical relationships between genes and gene clusters.36 In contrast to KMc, hierarchical relationships are an emergent aspect of REMc, as illustrated by the heat maps representing the intermediate and final clusters of REMc (Fig. 6). By combining knowledge associated with (1) the molecular effect of the perturbations (e.g., drugs with known targets), (2) statistical and biological cluster quality, and (3) visual data such as heat map images (see Fig. 6), one can mine the biological relevance of each cluster.
Figure 6.
Heat maps of REMc clusters illustrate hierarchical relationships and cluster quality. Heat maps represent each round of REMc. Each REM cluster was subjected to Hc to produce a gene dendrogram and heat map. Green shading indicates synergistic interaction, and blue shading indicates antagonistic interaction (see scale). The interaction values (each shaded box) correspond to Growth Index scores (described in Sec. 1 and reproduced in supplementary material) (Ref. 5). The cluster names are given to left of heat map. GOid_z and LL are displayed to the right of clusters (rank indicated in parentheses). Gene names are given along the right side of each heat map. Drug treatment conditions are labeled A-N at the bottom and the same order is maintained for each cluster, as detailed for cluster 0. See text and supplemental material for additional detail about each perturbation (Ref. 24).
With respect to Fig. 6, recall that the unclustered set of 297 genes is in fact a highly select subset of genes from the genomic set of 4850 knockout strains that exhibit interactions with HU, an inhibitor of RNR and DNA synthesis.5 To better understand biological differences between these genes, they were tested for phenotypic interaction with drugs having different cellular effects. Cisplatin, like HU, induces DNA damage, but does so by a different mechanism (intercalating in DNA). Miconazole is an inhibitor of the ERG11 gene and essential enzyme in ergosterol biosynthesis. TBHP induces oxidative damage, which stresses many cellular processes, including DNA and protein metabolism. Finally cycloheximide is an inhibitor of a gene RPL28, a component of the large ribosomal subunit essential for translation and thus makes protein synthesis rate-limiting for cell proliferation.
Since HU and cisplatin are most related among the perturbations (both perturb DNA metabolism), and the genes were selected originally based on interaction with HU, cisplatin is the next most “gene interactive” drug that drives clustering among these selected deletion strains. This is evidenced in the first round cluster heat maps, which can largely be described in terms of HU and cisplatin gene-drug interactions: (1) cluster 0 shows strong interactions with both HU and cisplatin; (2) cluster 1 shows strong interaction with HU, but intermediate strength interaction with cisplatin; (3) cluster 2 shows intermediate strength interaction with HU and cisplatin and more assorted interactions with other drugs; and (4) cluster 3 shows weak-to-intermediate strength interaction with HU and cisplatin, with fewer pleiotorpic interactions.
A biological expectation of REMc is increasing enrichment in the sharing of annotation terms among genes with successive rounds of clustering. In general, this was what we observed. For example, cluster 0 displays genes with an interaction profile indicative of a strong requirement for buffering DNA damage, which is induced by treatment with HU (Fig. 6, columns a–c in each heat map) or cisplatin (columns g–i in each heat map); i.e., there is a synergistic growth inhibition effect from deleting any one of these genes and thus sensitivity (i.e., synergistic interaction) to either of these drugs is greatly increased when any of the genes in the cluster are knocked out of a cell’s genome. Interestingly, cluster 0 breaks down into two subclusters; cluster 0_1 has high statistical quality and contains more highly annotated genes (reflected by high GOid_z) than cluster 0_0. Cluster 0_0 has lower statistical quality and differs by exhibiting strong sensitivity to cycloheximide (columns l–n in each heat map) and weak sensitivity to both miconazole (columns d–f in each heat map) and hydrogen peroxide (columns j and k in each heat map). The high GOid_z and LL of cluster 0_1 are consistent with the fact that genes in that subcluster function relatively specifically in DNA damage repair, as reflected by the uniformity of their gene interaction profiles relative to those in cluster 0_0, representing more pleiotropic phenotypes. To add finer grain to assessment of relationships within E-M clusters, we employed Hc using Euclidean distance and complete linkage (Fig. 6). As can be seen (gene names are provided beside the heat maps), genes with the most similar interaction profiles are often genes in a common pathway.5 Another example is seen in cluster 2, where genes are grouped in the subcluster 2_1 when they confer increased resistance (blue shading) to miconazole and hydrogen peroxide treatment; however, subcluster 2_0 contains genes conferring weak resistance or no phenotype in response to miconazole or hydrogen peroxide and cluster 2_0_2 contains genes that, although they exhibit synergistic interaction with HU, actually have stronger interactions (darker green shading) with cisplatin, a drug used in treatment of many human cancers.
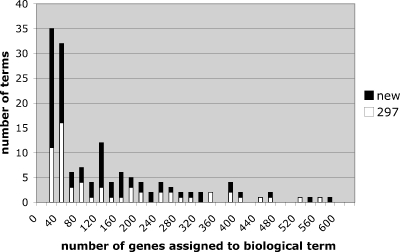
The overall enrichment of biological functions (measured by the number of GO terms) attributable to clustering is summarized in Table 3, and was obtained using GO TERMFINDER.31 Our first hypothesis was simply that clustering of gene interaction data would increase the discovery of GO terms. REMc increased the total number of GO terms by about threefold over unclustered data (which was already enriched based on selection for HU gene-drug interactions) (Table 3). We next traced the segregation of GO terms following successive rounds of REMc. Occasionally terms would fall out, being present in an intermediate cluster, but not in subsequent clusters. These were sometimes terms comprised of very large gene sets (over 100 genes per term), meaning they were biologically nonspecific, in which case genes annotated to a disappearing term might be associated with different, smaller terms in the clustered data. On the other hand, genes that were annotated to the same term in an intermediate cluster did not exhibit similar enough gene interaction profiles to stay together in subsequent rounds of REMc, thus distinguishing between genes that function as tight modules from those that are (although co-annotated) more heterogeneous, or pleiotropic in their phenotypic effects. We further observed from the GO TERMFINDER analysis that terms represented in the unclustered data segregated into more than one cluster, meaning that functional subsets of genes assigned to the same cellular process can be differentiated by unique aspects of their phenotypic profiles. The majority of new terms, which were not enriched in the unclustered data but emerged during REMc, were specific to a single cluster. In general, new GO terms emerging during REMc represented more specific biological functions involving smaller groups of genes. A frequency histogram, plotting together the number of GO terms identified versus the total number of genes annotated to a given term, revealed that the new GO terms discovered by REMc primarily reflected cellular processes annotated to 60 or fewer genes (Fig. 7).
Table 3.
Summary of total GO terms emerging during REMc. Unclustered data were enriched for 71 GO terms. The first round of clustering yielded four clusters and a total of 166 GO terms, indicating an increase in the detection of functionally related genes by clustering. However, 42 of the GO terms across the four clusters were redundant (reducing the total number of unique GO terms to 124, given in parentheses), and 15 terms (“missed previous”) associated with the unclustered data were not associated with one of the four clusters. The cluster ID name indicates the parent-child relationships for each clustering. Clustering was performed recursively until no new clusters were obtained. See supplemental material for list of GO terms for each cluster.
| Unclustered | Cluster ID | Rd 1 GT | Cluster ID | Rd 2 GT | Cluster ID | Rd 3 GT | |
|---|---|---|---|---|---|---|---|
| 71 | 1.0 | 69 | 2.0-0 | 31 | |||
| 2.0-1 | 61 | ||||||
| 1.1 | 20 | 2.1-0 | 1 | ||||
| 2.1-1 | 15 | ||||||
| 2.1-2 | 12 | ||||||
| 2.1-3 | 8 | ||||||
| 2.1-4 | 0 | ||||||
| 1.2 | 56 | 2.2-0 | 41 | 3.2-0-0 | 21 | ||
| 3.2-0-1 | 7 | ||||||
| 3.2-0-2 | 12 | ||||||
| 2.2-1 | 10 | ||||||
| 2.2-2 | 17 | 3.2-2-0 | 5 | ||||
| 3.2-2-1 | 19 | ||||||
| 1.3 | 21 | 2.3-0 | 0 | ||||
| 2.3-1 | 16 | ||||||
| 2.3-2 | 0 | ||||||
| 2.3-3 | 2 | ||||||
| Unique GT (total) | 71(71) | 124(166) | 144(214) | 146(220) | |||
| New in round | 68 | 39 | 11 | ||||
| Missed previous | 15 | 19 | 9 |
Figure 7.
Increase in GO terms associated with REMc. The frequency of GO terms obtained from the unclustered data (white) and following REMc clustering (black) is plotted against bins representing total number of genes annotated to the corresponding terms.
We note that GO comprises an acyclic graph structure,30 thus with GO TERMFINDER analysis, an increased number of GO terms overrepresents the increase in biological processes, because the same gene set often accounts for multiple related terms (supplementary material).24 We can see this in the example of clusters 0_0 and 0_1 resulting from the second round of E-M clustering, where cluster 0_1 resulted in 24 GO terms not enriched in the unclustered data; however, only three completely distinct (i.e., nonoverlapping genes representing the terms) gene modules accounted for all of them. Similarly, cluster 0_0, which contained 18 new GO terms, shared two genes in common between all of the terms, although some of the terms were broader and contained as many as nine of the 18 genes in the cluster. Interestingly, however, there was no overlap in the terms associated with clusters 0_1 and 0_0. Thus, in the example of these two clusters, there were five nonoverlapping gene sets contributing to most of the annotations, they were functionally distinct, and segregated distinctly.
REMc highlights phenomic modules that cooperate to buffer cellular perturbations
REMc determined about the same number of clusters as we previously described using Hc. Reassuringly, REMc cluster heat maps and associated enrichment in gene functions tracked those surmised from our previously published analysis using Hc.5, 24 Cluster 0 involves genes strongly required for tolerating DNA damage. Cluster 0_1 is highly specific in this regard and contains genes involved in double strand break repair (RAD57, RAD55, XRS2, RAD51, RAD54, RAD5, MMS22, RAD52, MRE11, RAD50), post replication repair (RAD18, RAD6, POL32, RAD52), and DNA replication (RNR4, POL32, TOP3, SGS1, CTF4). Cluster 0_0 also buffers DNA damage, but is less specific, being comprised of genes functioning in regulation of mRNA stability (CCR4, DHH1, RPB4, POP2), a cellular process that is of broader utility. Nevertheless, these genes have been recently been confirmed as particularly important in regulating RNR, providing a mechanistic molecular explanation for the genetic interactions.37 Cluster 1 highlights genes strongly required to buffer HU perturbation, but only have a moderate role in buffering the DNA damaging effects of cisplatin. Cluster 1_1 reveals genes involved in threonine (HOM2, HOM3, THR1) and sterol (ERG3, CYB5) metabolism, while cluster 1_0 contains genes cofunctioning in meiotic recombination (MMS4,UME6, MUS81, RAD17), and cluster 1_2 indicates that telomere maintenance (PTC1, RPB9, SRB5, BUD32, RPB4) could partially underlie this interaction profile. Cluster 2 contains genes that on average are more strongly required to buffer cisplatin damage than genes from other clusters. Cluster 2_1, like cluster 1_2, involves a significant number of genes cofunctioning in telomere maintenance (HPR1, KEM1, CAX4, CHO2, ADO1, EST1, VPS9, NAT3).38 Cluster 2_0_0 reveals components of the vacuolar H+∕ATPase (VMA2, TFP1, CUP5, VMA8, VMA10, VMA5, VMA6) that functions in vacuolar acidification and regulation of cellular pH; cluster 2_0_2 shows functional enrichment in sister chromatic cohesion (DCC1, CTF8, RSC2) related to chromosome segregation (DCC1, CTF8, RSC2, CSE2), chromosome localization (NUP84, NUP133), and protein sumoylation (SLX8, WSS1). Cluster 3 consists of genes with relatively weak drug-gene interactions; nevertheless, the subclusters represent modules that buffer HU and cisplatin (3_1), those more specifically involved in buffering only HU (3_3), or those exhibiting a heterogeneous pattern of interactions (3_2). Consistent with the subtler phenotypes of cluster 3, the genes were less well annotated. However, the weaker phenotypes were informative, cluster 3_0 revealing involvement of the tubulin complex assembly (YKE2, GIM5), cluster 3_1 highlighting mitochondrial signaling (RTG3, RTG2, MKS1, RTG1) and cell cycle checkpoint (MRC1, RAD9, BIM1, RAD24, ELM1, DDC1), and cluster 3_3 pointing to genes functioning in mitochondrial organization and biogenesis (MGM101, MDM35, MDM30, SML1).
Phenomic modules enable hypothesis generation regarding buffering networks
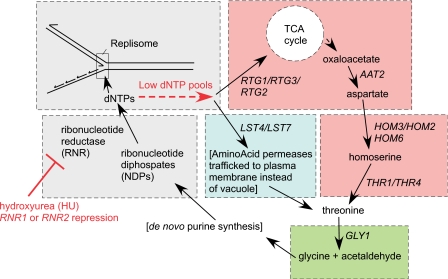
The overall goal of this data set was to gain insight into how cells buffer replication stress.4, 5 Replication stress was induced by HU, which depletes dNTP pools through inhibition of the rate-limiting enzyme for dNTP biosynthesis, RNR. A model attempting to connect as many of the modules as possible was generated. Some were straightforward such as DNA repair pathways, which were easily identified (cluster 1 and cluster 3). Other modules were not known to be involved in dNTP metabolism, having been studied previously in other context. With discovery of their co-occurrence as phenomic modules buffering replication stress, it was possible to create a model hypothesizing their cooperation in a buffering mechanism (Fig. 8), involving rerouting of metabolic fluxes.15 It was surprising that genes involved in threonine biosynthesis (AAT2, HOM3, HOM2, HOM6, THR1, and THR4) would be HU sensitive, and moreover that successive genes in the pathway were progressively more sensitive. The threonine biosynthesis pathway is evidenced by HOM3, HOM2, and THR1 in cluster 1_1 (Fig. 6). Aspartate is a precursor in the biosynthesis of threonine and is produced from the substrate oxaloacetate by aspartate amino transferase (AAT2). Oxaloacetate is a product of the TCA cycle, so it was intriguing that RTG1∕RTG2∕RTG3, known to transmit “retrograde” (nuclear-to-mitochondrial) signals that regulate TCA cycle activity,39, 40 represented another module (cluster 3_1). These two modules suggested de novo threonine biosynthesis to be important for buffering dNTP pool depletion. Additionally, LST4 (cluster 1_1) and LST7 (cluster 3_2), two genes that cofunction with LST8 and SEC13 in regulating amino acid permease trafficking, had suggestive gene interaction profiles,41 and furthermore LST8 is a known regulator of the RTG pathway.42 We validated this model by knocking down the RNR gene activity directly (reducing gene expression) to deplete dNTP pools and induce replication stress in these deletion strains (rather than using HU, which inhibits RNR activity, but could potentially have other “off-target” effects). The gene-drug interactions discovered with HU were validated as gene-gene interactions, and furthermore two parallel (“extrinsic”) buffering paths of this homeostatic circuit (threonine biosynthesis and uptake) were found to be synthetic lethal. The model was further validated by the finding that the unexplained slow growth phenotype of a strain with a deletion mutation in the gene GLY1, encoding threonine aldolase, was associated with a low basal level of dNTP pools and a slow homeostatic response following induction of RNR deficiency.15
Figure 8.
A model for buffering of dNTP pool homeostasis by threonine metabolism. Please see text and references for further explanation (Refs. 15, 29).
In summary, the work above illustrates how identification of phenomic modules can lead to discovery of buffering mechanisms, as described above for maintenance of dNTP pools via regulation of threonine metabolism. The extent to which buffering mechanisms are evolutionarily conserved remains unknown. However, evolutionary conservation of threonine catabolism for maintenance of normal dNTP pools has been recently reported in mice. Furthermore, the mechanism by which threonine metabolism regulates dNTP pool homeostasis and cell proliferation was specific to embryonic stem cells, suggesting that yeast may be a good model for this cell type.29 At some level of granularity, the yeast and ES cell models for buffering DNA synthesis by regulation of threonine metabolism will be different. Nevertheless, it provides an example that buffering mechanisms, like genes, are conserved over long evolutionary distances.4
DISCUSSION
Living organisms are dynamic, nonlinear systems with modular and hierarchical designs having been engineered by natural selection over evolutionary time in response to environmental pressures. Living systems are also robust;43 however, in contrast to man-made systems, the generation of diversity is a fundamental characteristic.44 High throughput analysis of genetic interactions reveals aspects of robustness and diversity in biological systems representing a sort of reverse engineering approach to dissect the complexity of cellular design.45 Genome-scale, systematic analysis of gene interactions is relatively new, but from it we already know that genes are highly interactive,46, 47 and that quantitative assessment of interactions aids the effort to resolve their complexity.5, 11, 15, 48 Having developed tools with enhanced capacity for collecting Q-HTCP data, we sought streamlined and efficient ways to mine the resulting large-scale quantitative genetic interaction data, which lead to the development of REMc. The goal during development of REMc was to establish objectivity regarding where clusters exist, and which ones are of the highest quality. We sought to test whether comparable interpretation would be reached for a large data set that had been previously mined using Hc.5 In general it appears REMc provides useful features of other clustering methods (identification of genes with similar profiles), but with increased objectivity and efficiency. There are theoretical and technical issues that remain to be addressed to deploy REMc on a larger scale, as well as questions regarding further development of REMc. These considerations overlook for the moment matters regarding how phenotypic data are collected and∕or how genetic interactions are calculated.
An unrealistic aspect of gene clustering is that genes are typically assigned a single cluster. Just as the Beadle–Tatum hypothesis of one gene-one enzyme no longer fits with our appreciation of biological complexity, neither does the notion of a gene interacting within one or even a small number of phenomic modules.49, 50 Instead, genes interact, and phenomic modules function, dynamically within a changing cellular context. Although the REMc outputs we focused on in this initial study were the most probable ones (i.e., genes were constrained to one cluster), it is possible to relax the model and to consider individual genes as part of multiple clusters. Considering genes to interact in multiple modules adds dimensions to the data mining problem. Like biclustering,8, 26, 28, 51 REMc should enable investigation of this question as a future direction. Other factors affecting REMc, such as selecting the most mutually informative features for clustering, should also be examined.
GOid_z could be improved by creation of additional algorithms that perform tasks a biologist would typically undertake, such as correcting for increases in the GOid_z score due to the same set of genes being associated with multiple related terms. Similarly the GTF tool could be integrated within the REMc framework for streamlining related data mining tasks.
In summary, from the outset our benchmark for development of REMc was to recapitulate biological insights described in detail previously where the same data were analyzed by Hc and without the use of GO tools.5 The earlier effort demonstrated that a relatively small amount of quantitative gene-drug interaction data revealed many of the same functional modules as a much larger set of qualitative gene-gene and gene-drug interaction data.5, 9 As described below, REMc recapitulated the biological findings that stood out from the earlier more subjective analysis, thus achieving the objective. We refer to that paper for more detailed discussions, emphasizing here that the fundamental advantages of REMc are objective determination of the absolute cluster number and hierarchy together with quantification of cluster quality. These advantages reduce laborious and subjective scrutiny of clustering results, increase reproducibility, providing a scalable clustering approach.
As tools for studying gene interaction networks improve, the challenge of data visualization increases. Given the diversity of living organisms and universal requirements for homeostatic mechanisms there would seem to be nearly infinite ways that genes and pathways can interact. However, evolutionary constraints and modularization of biological processes may make it possible to understand and extrapolate gene interactions to buffering mechanisms across species. REMc may help by making clustering more flexible, objective, and quantitative, allowing more attention to focus on utilization of cluster information for data integration. Tools and interdisciplinary approaches for systems biology are under rapid development, and hopefully REMc can assist the impending phenomics effort by providing a useful data mining tool for large-scale quantitative analysis of gene interactions.
METHODS
Quantification of genetic interactions
We used previously published genetic interaction data obtained from 297 yeast gene deletion strains, selected from a screen of over 4800 deletion strains for chemical-genetic interactions with HU (an inhibitor of RNR activity that arrests the cell cycle by rendering dNTP biosynthesis rate-limiting for cell proliferation). These strains were further tested for interactions with four additional chemicals, each at multiple concentrations, to aid discrimination of gene functions with respect to a range of cellular perturbations, as previously described.5
Clustering analysis
EM-optimized Gaussian mixture model clustering
REMc was developed using WEKA 3.5,34 which provides an EM clustering module that was incorporated into JAVA code to perform the clustering recursively (for help incorporating WEKA in JAVA code see http://weka.wikispaces.com/Use+Weka+in+your+Java+code). WEKA accepts comma-delimited files containing a data matrix, which can be optionally converted to ARFF files (see http://weka.wikispaces.com/Creating+an+ARFF+file) prior to clustering. Parameters that can be selected are the number of clusters and the degree of cross-validation. We used the default settings, which include that the algorithm optimizes the number of clusters and with tenfold cross-validation.
In GMM clustering, a finite mixture of Gaussian densities is fit to the data. Each of the N genes is represented as a vector with components corresponding to the 15 perturbations. The likelihood function for finding gene i in cluster j is , where is the vector of Gaussian distribution parameters. A function proportional to the posterior probability for gene i being generated by the collection of cluster classes is a mixture or linear combination of these Gaussians: , where πj is the prior probability of a gene coming from cluster j. The goal of GMM clustering is to maximize the LL,
| (1) |
The EM algorithm (maximum likelihood method) is applied to yield the class membership and fit the mixture components.52 The algorithm alternates between E and M steps. In an E step, the probability of each observation belonging to each cluster is estimated conditionally on the current parameter set (cluster means and standard deviations). In an M step, the model parameters are estimated given the current class membership probabilities. It is likely an oversimplification of biology to force each gene to be in only one cluster because we expect genes to have more than one function. Thus, unlike K-means, which assigns each gene to a single cluster, GMM clustering does not assign a gene to any single cluster but rather gives the gene’s probability of being in all clusters. However, to simplify interpretation, once the EM algorithm converges, genes are assigned to the class with the maximum conditional probability.
Training the GMM models on the entire data set increases overfitting and decreases the generality of the model. To achieve more robust clusters and more reliable LL estimates, tenfold cross-validation is used with the EM algorithm. The data set is divided into ten equal partitions, and the EM algorithm is trained on each partition and tested on the remaining data. The LLs are averaged over all ten folds. The number of clusters is determined iteratively by incrementing the number of clusters until the average LL does not increase appreciably from the previous number of clusters. To perform the maximum likelihood GMM clustering, we used the WEKA open source data mining software written in JAVA. We modified the WEKA algorithm to include the recursive approach described above.34
Hierarchical and K-means clustering
Hc and KMc were performed using the MATLAB Bioinformatics Toolbox. Function “pdist” was used to calculate the distance for either Euclidean or Pc distance (http://www.mathworks.com/access/helpdesk/help/toolbox/stats/pdist.html). Function “linkage” was used to establish the hierarchical cluster tree (http://www.mathworks.com/access/helpdesk/help/toolbox/stats/linkage.html). Function “cluster” creates clusters from linkages (http://www.mathworks.com/access/helpdesk/help/toolbox/stats/cluster.html). Function “kmeans” was used for KMc (http://www.mathworks.com/access/helpdesk/help/toolbox/stats/kmeans.html). Function “clustergram” was used to generate all heat maps with one-dimensional clustering (perturbation axis fixed), Euclidean distance metric, and complete linkage provided http://www.mathworks.com/access/helpdesk/help/toolbox/bioinfo/ref/clustergram.html).
Gene ontology methods
GO provides biological knowledge about genes from curated literature information using a controlled vocabulary and systematic annotation of genes. It provides a computational way to assess for biological functions within a list of genes relative to a background set.53
CLUSTERJUDGE
A GO-based method used in this study compares the relative quality of different clustering methods based on MI regarding enrichment of gene functions across all clusters. The online tool provided by the Roth laboratory was used.35
GO TERMFINDER
GTF is an online tool available from the Saccharomyces Genome Database website.31, 54 It takes input files consisting of a gene list (cluster) and a background set (deletion strains).
Gene ontology information divergence z-score
We developed a new tool for this study, GOid, to summarize the overall enrichment of gene functions (across all biological processes) in a single cluster. GOid was converted to a GOid_z score, as indicated in Fig. 3; GOid_z was in turn used to compare REMc, Hc, and KMc results. Whereas the LL from REMc provides a data-driven quality of classes predicted by model-based clustering, knowledge-driven cluster quality methods assign biological function to clusters. Most clustering evaluation tools provide a global score for the collection of clusters produced by a method. In order to assess the quality of individual clusters, we introduced an information theoretic score to quantify the GO enrichment of each class. Specifically, we use the Kullback–Leibler divergence (KLD) between a cluster of genes C and a background list of genes B for a given GO term t,
| (2) |
The divergence D measures the degree of dissimilarity between the discrete posterior distribution C with probability spectra ck and a background or prior distribution B with probability spectra bk. In this application, each gene can be in binary state k=1 or 0, corresponding to the probability of genes being associated with the GO term or not. Although not a true metric, it satisfies many important mathematical properties such as being non-negative and equaling zero only if ck=bk. In other areas of bioinformatics KLD (referred to as relative entropy in this context) has been used to quantify sequence alignments and visualized by sequence logos.55 In the alignment application, the posterior was the observed probability of a residue at an alignment column and the prior∕background was the expected probability of the residue to occur at random. To understand the output of the GO term divergence for a cluster, it is instructive to compare with the output of GTF. Consider five highly enriched hypothetical GO terms and the results for a hypothetical cluster with 28 genes compared with 7292 background genes.
| GO term |
Cluster frequency |
Background frequency |
GTF p-value |
Divergence |
| 1 | 28∕28 | 104∕7292 | 7.84×10−52 | 5.99 |
| 2 | 28∕28 | 108∕7292 | 2.65×10−51 | 5.93 |
| 3 | 28∕28 | 215∕7292 | 4.6×10−42 | 4.95 |
| 4 | 23∕28 | 98∕7292 | 1.01×10−37 | 4.43 |
| 5 | 23∕28 | 200∕7292 | 5.63×10−30 | 3.59 |
The output of GTF is a p-value, while the output of KLD is a raw score or strength, but the table shows the same trend down a column: less significant p-values and decreasing divergence scores. It is also possible to assign an approximate p-value to the divergence score by either assuming its asymptotic distribution of chi-square or by generating bootstrap samples.56
To estimate the biological quality of C with respect to GO, we compute the GOid as the sum of the divergence between C and B across all t,
| (3) |
Similar to CLUSTERJUDGE,35 we filter extremely sparse attributes to avoid division by zero in the divergence calculation, but we do not otherwise filter attributes. For a given attribute, a large information divergence suggests that the enrichment of genes in C for this GO category diverges from the fraction of genes associated with this term for the entire genome (background enrichment), signifying significant enrichment of this classified set of genes. If the class and background probabilities are equal, then GOid=0, consistent with a cluster providing no significant GO enrichment. The output of GTF would be less suitable for estimating cluster quality than KLD because it is less statistically sound to average p-values, whereas it makes sense to average a strength like KLD. The divergence from Eq. 2 can be used to rank the contribution of individual GO terms to the enrichment of a cluster class, and the total divergence in Eq. 3 adds these contributions to give a quality score for a cluster class.
For GOid_z, we incorporated the mean and standard deviation from a set of 1000 random clusters R, generated with the same number of genes as the corresponding real cluster C (see Fig. 2). The random clusters were generated from the same background set of genes, corresponding to those represented in the gene deletion strain library (gene lists available from Open Biosystems). The GOid_z was calculated as
| (4) |
To reduce computing requirements for calculation of GOid for R and GOid_z for C, we fit the data shown in Figs. 3a, 3b, and used the resulting functions (data not shown) to determine the mean and standard deviation of GOid scores from 1000 random clusters of any size.
ACKNOWLEDGMENTS
The authors are grateful for the following grants which supported this work: HHMI Physician-Scientist Early Career Award (J.L.H.), NIH K08-CA-90637 (J.L.H.), American Cancer Society Research Scholar Grant (J.L.H.), and Cystic Fibrosis Foundation Graduate Student Fellowship awarded to J.G. by the UAB CF Center (P.I. Eric Sorscher). The authors also thank Jennifer Bryan and Fritz Roth for helpful comments on the manuscript.
NOMENCLATURE
- CJ
CLUSTERJUDGE
- Euc
Euclidean distance
- EMc
Expectation-maximization clustering
- GO
Gene ontology
- GOid_z
Gene ontology information divergence z-score
- Hc
Hierarchical clustering
- HU
Hydroxyurea
- GMM
Gaussian mixture model
- GTF
GO TERMFINDER
- KMc
K-means clustering
- LL
Log-likelihood
- Pc
Pearson correlation
- Q-HTCP
Quantitative high throughput cellular phenotyping
- REMc
Recursive expectation-maximization clustering
- RNR
Ribonucleotide reductase
- TCA
Tricarboxylic acid
References
- Hartwell L. H., Biosci Rep. 22, 373 (2002). 10.1023/A:1020918107706 [DOI] [PubMed] [Google Scholar]
- Dixon S. J., Costanzo M., Baryshnikova A., Andrews B., and Boone C., Annu. Rev. Genet. 43, 601 (2009). 10.1146/annurev.genet.39.073003.114751 [DOI] [PubMed] [Google Scholar]
- Rubin G. M., Yandell M. D., Wortman J. R., Gabor Miklos G. L., Nelson C. R., Hariharan I. K., Fortini M. E., Li P. W., Apweiler R., Fleischmann W., Michael Cherry J., Henikoff S., Skupski M. P., Misra S., Ashburner M., Birney E., Boguski M. S., Brody T., Brokstein P., Celniker S. E., Chervitz S. A., Coates D., Cravchik A., Gabrielian A., Galle R. F., Gelbart W. M., George R. A., Goldstein L. S. B., Gong F., Guan P., Harris N. L., Hay B. A., Hoskins R. A., Li J., Li Z., Hynes R. O., Jones S. J. M., Kuehl P. M., Lemaitre B., Troy Littleton J., Morrison D. K., Mungall C., O’Farrell P. H., Pickeral O. K., Shue C., Vosshall L. B., Zhang J., Zhao Q., Zheng X. H., Zhong F., Zhong W., Gibbs R., Craig Venter J., Adams M. D., and Lewis S., Science 287, 2204 (2000). 10.1126/science.287.5461.2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- J. L.HartmanIV, Garvik B., and Hartwell L., Science 291, 1001 (2001). 10.1126/science.291.5506.1001 [DOI] [PubMed] [Google Scholar]
- J. L.HartmanIV and Tippery N. P., Genome Biol. 5, R49 (2004). 10.1186/gb-2004-5-7-r49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., Chu A. M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., Arkin A. P., Astromoff A., El Bakkoury M., Bangham R., Benito R., Brachat S., Campanaro S., Curtiss M., Davis K., Deutschbauer A., Entian K. -D., Flaherty P., Foury F., Garfinkel D. J., Gerstein M., Gotte D., Güldener U., Hegemann J. H., Hempel S., Herman Z., Jaramillo D. F., Kelly D. E., Kelly S. L., Kötter P., LaBonte D., Lamb D. C., Lan N., Liang H., Liao H., Liu L., Luo C., Lussier M., Mao R., Menard P., Loon Ooi S., Revuelta J. L., Roberts C. J., Rose M., Ross-Macdonald P., Scherens B., Schimmack G., Shafer B., Shoemaker D. D., Sookhai-Mahadeo S., Storms R. K., Strathern J. N., Valle G., Voet M., Volckaert G., Wang C., Ward T. R., Wilhelmy J., Winzeler E. A., Yang Y., Yen G., Youngman E., Yu K., Bussey H., Boeke J. D., Snyder M., Philippsen P., Davis R. W., and Johnston M., Nature (London) 418, 387 (2002). 10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J. D., Bussey H., Chu A. M., Connelly C., Davis K., Dietrich F., Whelen Dow S., El Bakkoury M., Foury F., Friend S. H., Gentalen E., Giaever G., Hegemann J. H., Jones T., Laub M., Liao H., Liebundguth N., Lockhart D. J., Lucau-Danila A., Lussier M., M’Rabet N., Menard P., Mittmann M., Pai C., Rebischung C., Revuelta J. L., Riles L., Roberts C. J., Ross-MacDonald P., Scherens B., Snyder M., Sookhai-Mahadeo S., Storms R. K., Véronneau S., Voet M., Volckaert G., Ward T. R., Wysocki R., Yen G. S., Yu K., Zimmermann K., Philippsen P., Johnston M., and Davis R. W., Science 285, 901 (1999). 10.1126/science.285.5429.901 [DOI] [PubMed] [Google Scholar]
- Dudley A. M., Janse D. M., Tanay A., Shamir R., and Church G. M., Mol. Syst. Biol. 1, 2005:0001 (2005). 10.1038/msb4100004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons A. B., Brost R. L., Ding H., Li Z., Zhang C., Sheikh B., Brown G. W., Kane P. M., Hughes T. R., and Boone C., Nat. Biotechnol. 22, 62 (2003). 10.1038/nbt919 [DOI] [PubMed] [Google Scholar]
- Pan X., Ye P., Yuan D. S., Wang X., Bader J. S., and Boeke J. D., Cell 124, 1069 (2006). 10.1016/j.cell.2005.12.036 [DOI] [PubMed] [Google Scholar]
- Schuldiner M., Collins S. R., Thompson N. J., Denic V., Bhamidipati A., Punna T., Ihmels J., Andrews B., Boone C., Greenblatt J. F., Weissman J. S., and Krogan N. J., Cell 123, 507 (2005). 10.1016/j.cell.2005.08.031 [DOI] [PubMed] [Google Scholar]
- Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., Pagé N., Robinson M., Raghibizadeh S., Hogue C. W. V., Bussey H., Andrews B., Tyers M., and Boone C., Science 294, 2364 (2001). 10.1126/science.1065810 [DOI] [PubMed] [Google Scholar]
- Koning A. J., Larson L. L., Cadera E. J., Parrish M. L., and Wright R. L., Genetics 160, 1335 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N. A., Laws R. J., Wardman B., Zhao L. P., and J. L.HartmanIV, BMC Syst. Biol. 1, 3 (2007). 10.1186/1752-0509-1-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- J. L.HartmanIV, Proc. Natl. Acad. Sci. U.S.A. 104, 11700 (2007). 10.1073/pnas.0705212104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Hopfield J. J., Leibler S., and Murray A. W., Nature (London) 402, C47 (1999). 10.1038/35011540 [DOI] [PubMed] [Google Scholar]
- Collins S. R., Schuldiner M., Krogan N. J., and Weissman J. S., Genome Biol. 7, R63 (2006). 10.1186/gb-2006-7-7-r63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T. R., Marton M. J., Jones A. R., Roberts C. J., Stoughton R., Armour C. D., Bennett H. A., Coffey E., Dai H., He Y. D., Kidd M. J., King A. M., Meyer M. R., Slade D., Lum P. Y., Stepaniants S. B., Shoemaker D. D., Gachotte D., Chakraburtty K., Simon J., Bard M., and Friend S. H., Cell 102, 109 (2000). 10.1016/S0092-8674(00)00015-5 [DOI] [PubMed] [Google Scholar]
- Phillips P. C., Genetics 149, 1167 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips P. C., Nat. Rev. Genet. 9, 855 (2008). 10.1038/nrg2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani R., St Onge R. P., J. L.HartmanIV, Giaever G., and Roth F. P., Proc. Natl. Acad. Sci. U.S.A. 105, 3461 (2008). 10.1073/pnas.0712255105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh I., Pass R., Togay S. O., Rodgers J. W., and J. L.HartmanIV, Genetics 181, 289 (2009). 10.1534/genetics.108.092981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Granka J. M., and Feldman M. W., Genetics 184, 827 (2010). 10.1534/genetics.109.111120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- See supplementary material at http://dx.doi.org/10.1063/1.3455188 for supplemental data file 1 with clustering results and supplemental data file 2 with GO TERMFINDER results for REMc clusters.
- Segre D., Deluna A., Church G. M., and Kishony R., Nat. Genet. 37, 77 (2005). [DOI] [PubMed] [Google Scholar]
- Cheng Y. and Church G. M., Proc. Int. Conf. Intell. Syst. Mol. Biol. 8, 93 (2000). [PubMed] [Google Scholar]
- Santamaría R., Therón R., and Quintales L., BMC Bioinf. 9, 247 (2008). 10.1186/1471-2105-9-247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria R., Theron R., and Quintales L., Bioinformatics 24, 1212 (2008). 10.1093/bioinformatics/btn076 [DOI] [PubMed] [Google Scholar]
- Wang J., Alexander P., Wu L., Hammer R., Cleaver O., and McKnight S. L., Science 325, 435 (2009). 10.1126/science.1173288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M. A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Richter J., Rubin G. M., Blake J. A., Bult C., Dolan M., Drabkin H., Eppig J. T., Hill D. P., Ni L., Ringwald M., Balakrishnan R., Cherry J. M., Christie K. R., Costanzo M. C., Dwight S. S., Engel S., Fisk D. G., Hirschman J. E., Hong E. L., Nash R. S., Sethuraman A., Theesfeld C. L., Botstein D., Dolinski K., Feierbach B., Berardini T., Mundodi S., Rhee S. Y., Apweiler R., Barrell D., Camon E., Dimmer E., Lee V., Chisholm R., Gaudet P., Kibbe W., Kishore R., Schwarz E. M., Sternberg P., Gwinn M., Hannick L., Wortman J., Berriman M., Wood V., de la Cruz N., Tonellato P., Jaiswal P., Seigfried T., and White R., Nucleic Acids Res. 32, D258 (2004). 10.1093/nar/gkh066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle E. I., Weng S., Gollub J., Jin H., Botstein D., Michael Cherry J., and Sherlock G., Bioinformatics 20, 3710 (2004). 10.1093/bioinformatics/bth456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk D. G., Ball C. A., Dolinski K., Engel S. R., Hong E. L., Issel-Tarver L., Schwartz K., Sethuraman A., Botstein D., Michael Cherry J., and The Saccharomyces Genome Database Project, Yeast 23, 857 (2006). 10.1002/yea.1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’haeseleer P., Nat. Biotechnol. 23, 1499 (2005). 10.1038/nbt1205-1499 [DOI] [PubMed] [Google Scholar]
- Hall M., Frank E., Holmes G., Pfahringer B., Reutemann P., and Witten I. H., SIGKDD Explor. 11, 10 (2009). 10.1145/1656274.1656278 [DOI] [Google Scholar]
- Gibbons F. D. and Roth F. P., Genome Res. 12, 1574 (2002). 10.1101/gr.397002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen M. B., Spellman P. T., Brown P. O., and Botstein D., Proc. Natl. Acad. Sci. U.S.A. 95, 14863 (1998). 10.1073/pnas.95.25.14863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolstencroft R. N., Beilharz T. H., Cook M. A., Preiss T., Durocher D., and Tyers M., J. Cell. Sci. 119, 5178 (2006). 10.1242/jcs.03221 [DOI] [PubMed] [Google Scholar]
- Gatbonton T., Imbesi M., Nelson M., Akey J. M., Ruderfer D. M., Kruglyak L., Simon J. A., and Bedalov A., PLoS Genet. 2, e35 (2006). 10.1371/journal.pgen.0020035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X. and Butow R. A., Cell 72, 61 (1993). 10.1016/0092-8674(93)90050-Z [DOI] [PubMed] [Google Scholar]
- Butow R. A. and Avadhani N. G., Mol. Cell 14, 1 (2004). 10.1016/S1097-2765(04)00179-0 [DOI] [PubMed] [Google Scholar]
- Roberg K. J., Bickel S., Rowley N., and Kaiser C. A., Genetics 147, 1569 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Sekito T., Epstein C. B., and Butow R. A., EMBO J. 20, 7209 (2001). 10.1093/emboj/20.24.7209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U., Surette M. G., Barkai N., and Leibler S., Nature (London) 397, 168 (1999). 10.1038/16483 [DOI] [PubMed] [Google Scholar]
- Kirschner M. and Gerhart J., Proc. Natl. Acad. Sci. U.S.A. 95, 8420 (1998). 10.1073/pnas.95.15.8420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csete M. E. and Doyle J. C., Science 295, 1664 (2002). 10.1126/science.1069981 [DOI] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., Sevier C. S., Ding H., Koh J. L. Y., Toufighi K., Mostafavi S., Prinz J., St. Onge R. P., VanderSluis B., Makhnevych T., Vizeacoumar F. J., Alizadeh S., Bahr S., Brost R. L., Chen Y., Cokol M., Deshpande R., Li Z., Lin Z. -Y., Liang W., Marback M., Paw J., San Luis B. -J., Shuteriqi E., Hin Yan Tong A., van Dyk N., Wallace I. M., Whitney J. A., Weirauch M. T., Zhong G., Zhu H., Houry W. A., Brudno M., Ragibizadeh S., Papp B., Pál C., Roth F. P., Giaever G., Nislow C., Troyanskaya O. G., Bussey H., Bader G. D., Gingras A. -C., Morris Q. D., Kim P. M., Kaiser C. A., Myers C. L., Andrews B. J., and Boone C., Science 327, 425 (2010). 10.1126/science.1180823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A. H., Lesage G., Bader G. D., Ding H., Xu H., Xin X., Young J., Berriz G. F., Brost R. L., Chang M., Chen Y., Cheng X., Chua G., Friesen H., Goldberg D. S., Haynes J., Humphries C., He G., Hussein S., Ke L., Krogan N., Li Z., Levinson J. N., Lu H., Ménard P., Munyana C., Parsons A. B., Ryan O., Tonikian R., Roberts T., Sdicu A. -M., Shapiro J., Sheikh B., Suter B., Wong S. L., Zhang L. V., Zhu H., Burd C. G., Munro S., Sander C., Rine J., Greenblatt J., Peter M., Bretscher A., Bell G., Roth F. P., Brown G. W., Andrews B., Bussey H., and Boone C., Science 303, 808 (2004). 10.1126/science.1091317 [DOI] [PubMed] [Google Scholar]
- Ihmels J., Collins S. R., Schuldiner M., Krogan N. J., and Weissman J. S., Mol. Syst. Biol. 3, 86 (2007). 10.1038/msb4100127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon S. J., Fedyshyn Y., Koh J. L., Keshava Prasad T. S., Chahwan C., Chua G., Toufighi K., Baryshnikova A., Hayles J., Hoe K. -L., Kim D. -U., Park H. -O., Myers C. L., Pandey A., Durocher D., Andrews B. J., and Boone C., Proc. Natl. Acad. Sci. U.S.A. 105, 16653 (2008). 10.1073/pnas.0806261105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A., Bandyopadhyay S., Zofall M., Zhang K., Fischer T., Collins S. R., Qu H., Shales M., Park H. O., Hayles J., Hoe K. L., Kim J. U., Ideker T., Grewal S. I., Weissman J. S., and Krogan N. J., Science 322, 405 (2008). 10.1126/science.1162609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. J. and Kasif S., Nucleic Acids Res. 33, W596 (2005). 10.1093/nar/gki469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X. -L. and Rubin D. B., Biometrika 80, 267 (1993). 10.1093/biomet/80.2.267 [DOI] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Michael Cherry J., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., and Sherlock G., Nat. Genet. 25, 25 (2000). 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E. L., Balakrishnan R., Dong Q., Christie K. R., Park J., Binkley G., Costanzo M. C., Dwight S. S., Engel S. R., Fisk D. G., Hirschman J. E., Hitz B. C., Krieger C. J., Livstone M. S., Miyasato S. R., Nash R. S., Oughtred R., Skrzypek M. S., Weng S., Wong E. D., Zhu K. K., Dolinski K., Botstein D., and Cherry J. M., Nucleic Acids Res. 36, D577 (2007). 10.1093/nar/gkm909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorodkin J., Heyer L. J., Brunak S., and Stormo G. D., Nucleic Acids Res. 25, 3724 (1997). 10.1093/nar/25.18.3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleks J. and Ivan B., Proceedings of the 21st International Conference on Machine Learning, Banff, Alberta, Canada, 2004.