Summary
We previously showed that FAM29A, a spindle-associated protein, promotes microtubule-dependent microtubule amplification through its interaction with and recruitment of NEDD1, the targeting subunit of the γ-tubulin ring complex. We report here that FAM29A is regulated by Plk1, a kinase essential for spindle assembly and its bipolarity. Plk1, FAM29A and NEDD1 form three separate complexes in vivo, not one single complex. Plk1 recruits FAM29A to spindle microtubules, which, in turn, targets NEDD1 to the spindle. Plk1 also recruits NEDD1 to the centrosomes, probably through a Plk1-NEDD1 interaction, but this interaction does not contribute to targeting NEDD1 to the spindle. Altering intracellular levels of FAM29A changes the distribution of NEDD1 between the centrosomes and the spindle, indicating that FAM29A controls the partition of NEDD1 between these two mitotic structures. Thus, Plk1 promotes microtubule nucleation from the centrosomes through a FAM29A-independent pathway and from the spindle through a FAM29A-dependent pathway. FAM29A controls the relative contributions of these two pathways to microtubule polymerization during mitosis.
Keywords: Plk1, FAM29A, NEDD1, Centrosome, Mitotic spindle, Microtubule amplification
Introduction
Nucleation of microtubules (MTs) initiates the assembly of the mitotic spindle. MT nucleation is also required to maintain the MT density in the spindle (Gadde and Heald, 2004; Wiese and Zheng, 2006). Two well-defined mechanisms of MT nucleation, centrosome-mediated and chromatin-mediated mechanisms, have been extensively characterized previously. Recently, we reported a third mechanism, the MT-dependent MT amplification, mediated by the FAM29A-NEDD1 complex (Goshima et al., 2008; Zhu et al., 2008).
The γ-tubulin ring complex (γTuRC) is a large multi-protein complex required for MT nucleation. The γ-tubulin subunit in the γTuRC promotes the polymerization of the α and β-tubulin subunits into MT polymers (Wiese and Zheng, 2006). During mitosis, γTuRC is localized to both the centrosomes and the spindle MTs and an accessory subunit of the γTuRC, NEDD1, interacts with γTuRC and targets it to these mitotic structures (Haren et al., 2006; Luders et al., 2006). Similarly, during repolymerization of MTs in mitotic cells released from a nocodazole treatment, NEDD1 targets the γTuRC to chromatin foci where chromatin-mediated MT nucleation and polymerization occur (Luders et al., 2006). Thus, the localization of NEDD1 determines the spatial activity of MT nucleation through its recruitment of the γTuRC.
We recently showed that the localization of NEDD1 to spindle MTs requires FAM29A (family with sequence similarity, member 29A), a novel spindle-associated protein (Zhu et al., 2008). FAM29A shares a weak sequence homology to the Dgt6 subunit of the Drosophila Augmin complex (32% amino acid identity over a 100 amino acid region), which is required for centrosome-independent MT generation within the spindle (Goshima et al., 2008). In human cells, knockdown of FAM29A prevents the localization of the NEDD1–γ-tubulin complex to the mitotic spindle, whereas knockdown of NEDD1 does not affect the localization of FAM29A, indicating that FAM29A recruits the NEDD1–γ-tubulin complex to the spindle where γ-tubulin promotes the polymerization of additional MTs independently of the centrosomes and chromatin. This FAM29A-mediated and MT-dependent MT amplification contributes to the spindle assembly and is required for the maturation of kinetochore MT fibers (Zhu et al., 2008). Biochemically, FAM29A interacts with the NEDD1–γ-tubulin complex, but only in mitosis, and this interaction targets NEDD1 to the spindle. However, the mechanism of FAM29A recruitment to the mitotic spindle during mitosis remains unknown.
Polo-like kinase, Plk1, is an essential mitotic kinase (Sunkel and Glover, 1988) that controls mitotic entry, centrosome maturation, bipolar spindle formation, cohesin dissociation, chromosome congression and segregation, as well as cytokinesis (Barr et al., 2004; van de Weerdt and Medema, 2006). It has been reported that Plk1 is involved in the recruitment of γ-tubulin to the centrosomes (Barr et al., 2004; Lane and Nigg, 1996). Although the localization of γ-tubulin to centrosomes requires both NEDD1 and Plk1, whether the recruitment of NEDD1 to the centrosomes is under the control of Plk1 remains unknown. Similarly, although FAM29A is required for targeting NEDD1 to the spindle, whether Plk1 is involved in the recruitment of FAM29A and NEDD1 to the spindle remains to be characterized. Importantly, the molecular mechanism that determines the partition of NEDD1 between the centrosomes and spindle is unclear.
We report here that Plk1, FAM29A and NEDD1 form three separate complexes in mitosis. Plk1 is responsible for recruiting FAM29A, and therefore the NEDDI–γ-tubulin complex to the mitotic spindle. Plk1 also targets NEDD1 to the centrosomes during mitosis, but this recruitment is independent of FAM29A. Thus, FAM29A serves as a bifurcation point to control the localization of NEDD1 to the centrosomes versus the spindle, thereby determining the relative contributions of centrosomal and spindle pathways in MT nucleation and polymerization. Our data identify a novel function of Plk1 in regulation of spindle assembly through targeting FAM29A and NEDD1 to the mitotic spindle, which controls the spindle amplification in mitosis.
Results
FAM29A and NEDD1 interact with Plk1
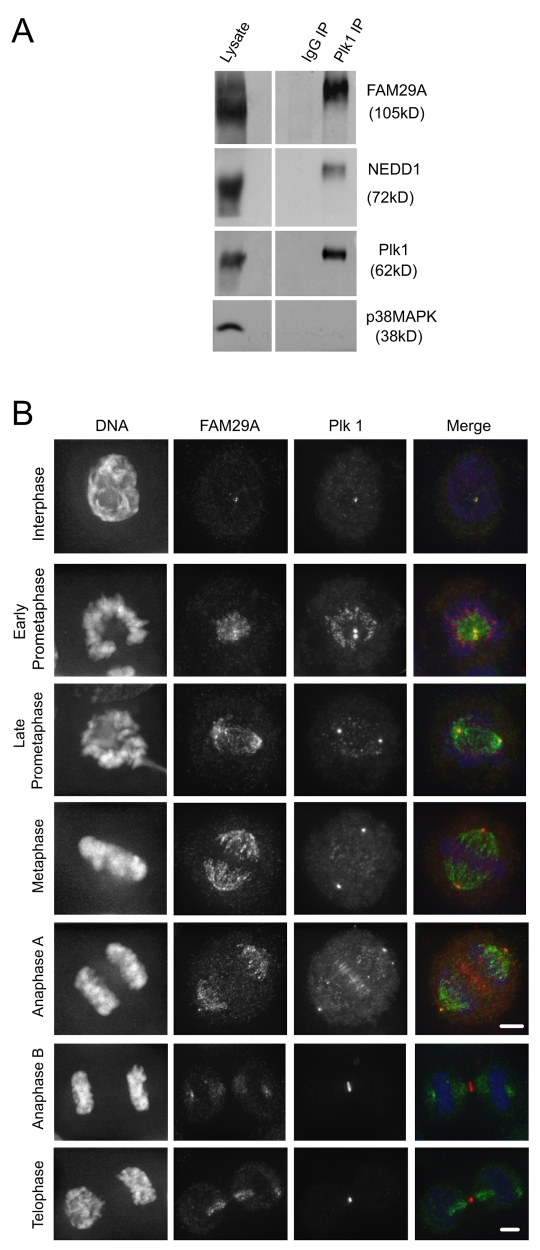
To investigate the function and regulation of Plk1, we previously purified the Plk1 complexes from mitotic HeLa S3 cells and analyzed its associated proteins by mass spectrometry (Seki et al., 2008a; Seki et al., 2008b; Zhu et al., 2008). We identified FAM29A as an interacting protein with high confidence, as reflected in the high XCorr and DeltaCN scores for the FAM29A peptides identified in the Plk1 complex (supplementary material Fig. S1A). Co-immunoprecipitation experiments confirmed the interaction between FAM29A and Plk1 (Fig. 1A). In prometaphase cells synchronized by a thymidine-nocodazole treatment (Fang et al., 1998b), FAM29A was co-precipitated by an anti-Plk1 antibody, but not by a nonspecific antibody. Similarly, NEDD1 also co-precipitated with Plk1 (Fig. 1A).
Fig. 1.
FAM29A and NEDD1 interact with Plk1 during mitosis. (A) HeLa S3 cells were synchronized at prometaphase by a thymidine-nocodazole treatment. Cell lysates were immunoprecipitated (IP) with anti-Plk1 antibodies or with non-specific IgG. The immunoprecipitates were analyzed by western blotting with p38MAPK serving as a control for specificity. (B) Maximum projections from deconvolved z-stacks of representative HeLa cells stained for FAM29A (green), Plk1 (red) and DNA (blue). Scale bars: 5 μm.
Next, we analyzed whether FAM29A and Plk1 co-localized in the cell cycle. As reported previously (Barr et al., 2004), Plk1 was concentrated on centrosomes in interphase cells, and localized to spindle poles and kinetochores in prometaphase and metaphase cells (Fig. 1B). FAM29A was present at the centrosomes in interphase cells and at spindle poles and spindle MTs from prometaphase to metaphase. At anaphase A, FAM29A remained on the spindle, while Plk1 was concentrated at the central spindle and midzone (Fig. 1B). Furthermore, in nocodazole-treated prometaphase cells in which the spindle MTs were depolymerized, FAM29A is localized in centrioles whereas Plk1 is concentrated at pericentrosomal materials (Barr et al., 2004; Zhu et al., 2008). Thus, although FAM29A and Plk1 may interact in centrioles, they also probably form a complex in the cytoplasm during mitosis.
Since Plk1 also interacts with NEDD1 during mitosis (Fig. 1A), we determined the localization of NEDD1 and Plk1 (supplementary material Fig. S1B). Both NEDD1 and Plk1 co-localized on centrosomes in interphase and mitotic cells. However, NEDD1, but not Plk1, was also localized to the mitotic spindle, indicating that these two proteins do not colocalize on the spindle.
Association of Plk1 with FAM29A and NEDD1 is cell-cycle regulated
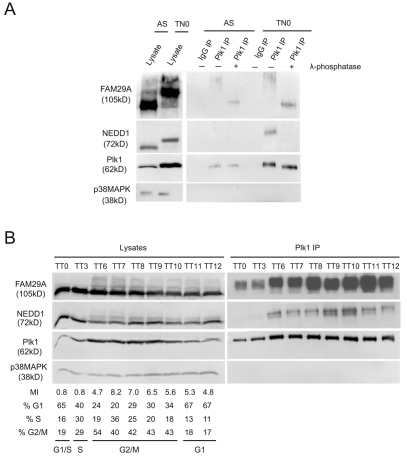
We investigated the association of Plk1 with FAM29A and NEDD1 in the cell cycle. In asynchronous cells, Plk1 was expressed at low levels and FAM29A interacted with Plk1 weakly, but no interaction between NEDD1 and Plk1 was observable within our detection limit, even though NEDD1 was expressed at a level comparable to that in mitotic cells (Fig. 2A). Thus, Plk1, FAM29A and NEDD1 do not form a single complex. The extent of association of Plk1 with both FAM29A and NEDD1 increased substantially in prometaphase cells. Although FAM29A was hyperphosphorylated in mitosis, phosphorylation of FAM29A does not seem to be required for its interaction with Plk1, as incubation of the Plk1-FAM29A complex with λ-phosphatase did not lead to its dissociation (Fig. 2A). However, treatment with λ-phosphatase resulted in the dissociation of the Plk1-NEDD1 complex. We have previously shown that FAM29A and NEDD1 also form a complex that is insensitive to the λ-phosphatase treatment (Zhu et al., 2008). Thus, NEDD1 was not indirectly co-immunoprecipitated with Plk1 through its interaction with FAM29A. We conclude that Plk1, FAM29A and NEDD1 form three separate complexes.
Fig. 2.
Association of Plk1 with FAM29A and NEDD1 is regulated in the cell cycle. (A) HeLa S3 cells were synchronized at prometaphase by a thymidine-nocodazole treatment (TN0). Lysates of asynchronous cells (AS) or prometaphase cells were immunoprecipitated with anti-Plk1 antibodies or with non-specific IgG. The immunoprecipitates (IP) were treated with λ-phosphatase or not treated, washed, and then analyzed by western blotting. (B) HeLa S3 cells were synchronized at the G1-S boundary by a double-thymidine treatment (TT). Cells were released and harvested at the indicated times (TT0-12; hours). The cell cycle profile was analyzed by fluorescence-activated cell sorting with the mitotic index (MI) determined by an antibody (MPM2) specific to mitotic phospho-proteins. Lysates and the anti-Plk1 immunoprecipitates (IP) were analyzed by western blotting.
The association of FAM29A and NEDD1 with Plk1 is cell-cycle regulated. HeLa S3 cells were synchronized at the G1-S boundary by a double-thymidine treatment, and then released (Fang et al., 1998a). The complex between FAM29A and Plk1 was present in G1 and S cells, but increased in its abundance as cells entered G2 and mitosis (Fig. 2B). Interestingly, Plk1 only interacted with the slow-migrating, and therefore hyperphosphorylated FAM29A. The Plk1-associated, hyperphosphorylated FAM29A may correspond to FAM29A in mitotic cells, or simply result from phosphorylation of FAM29A by Plk1 during the immunoprecipitation due to the high local Plk1 concentration on the anti-Plk1 antibody beads.
By contrast, the Plk1-NEDD1 complex was absent in G1 and S cells and the abundance of this complex increased in G2 and in mitosis. Although both hyperphosphorylated and hypophosphorylated NEDD1 were detectable in the cell lysates, as determined by their electrophoretic mobility, only hyperphosphorylated NEDD1 co-precipitated with Plk1 (Fig. 2B), consistent with a requirement of phosphorylation for the complex formation.
Plk1 recruits FAM29A to spindle MTs and NEDD1 to the spindle and centrosomes during mitosis
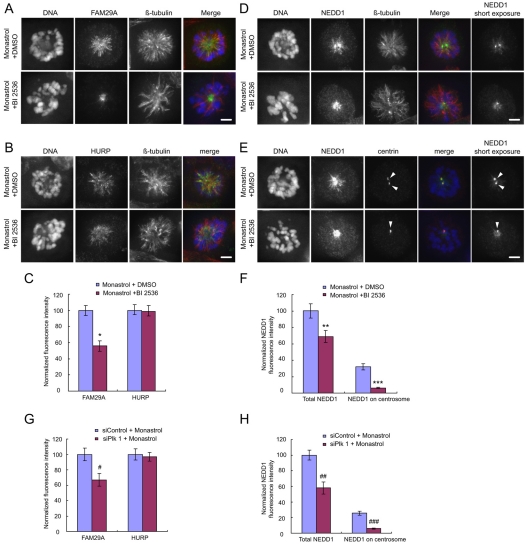
To explore the functional significance of the interaction among Plk1, FAM29A and NEDD1, we analyzed the localization of FAM29A and NEDD1 in the presence of BI 2536, a potent and highly specific inhibitor of Plk1 (Lenart et al., 2007). As inhibition of Plk1 by BI 2536 leads to a monopolar spindle (Lenart et al., 2007), both control and BI 2536-treated cells were arrested with a monopolar spindle by an incubation with monastrol, an inhibitor of the mitotic kinesin Eg5 (Mayer et al., 1999). Inhibition of Plk1 moderately reduced the total amounts of FAM29A (Fig. 3A-C). More strikingly, inhibition of Plk1 prevented the localization of FAM29A to the entire length of the MTs in the monopolar spindle and FAM29A became highly concentrated around the spindle poles in the presence of BI 2536. This effect is specific to FAM29A, but not to other spindle-associated proteins, such as the MT-stabilizing protein HURP (Fig. 3A-C).
Fig. 3.
Inactivation of Plk1 altered the localization of FAM29A and NEDD1. (A-F) HeLa cells were treated with monastrol (100 μM) for 4 hours. DMSO or BI 2536 (300 nM) was also added during the last 2 hours of incubation with monastrol. Images are maximum projections from deconvolved z-stacks of representative cells stained for FAM29A (A), HURP (B), NEDD1 (D,E) (green), β-tubulin (A,B,D), centrin (E; red) and DNA (blue; A,B,D,E). The NEDD1 images with short exposure time are also presented here to better show the NEDD1 signals on centrosomes (D,E). Fluorescence intensities of total FAM29A and total HURP (C), total NEDD1 and centrosomal NEDD1 (F) were quantified, normalized to those of their respective controls, and plotted (n=10 mitotic cells for each quantification). Arrowheads in E indicate centrosomes. *P<0.001; **P<0.01; ***P<1.2×10–5 (two-tailed t-test relative to the DMSO treatment). Centrosomal NEDD1 signals were defined by co-staining with an anti-centrin antibody. (G,H) HeLa cells were transfected with siRNAs, treated with monastrol (100 μM) for 4 hours, fixed and then stained for FAM29A, HURP, NEDD1, β-tubulin, centrin and DNA. Fluorescence intensities of total FAM29A and total HURP (G), total NEDD1 and centrosomal NEDD1 (H) were quantified, normalized to those of their respective control samples, and then plotted (n=10 mitotic cells for each quantification). #P<0.001; ##P<0.0002; ###P<4.3×10–5 (two-tailed t-test relative to siControl cells). Error bars indicate the s.e.m. Scale bars: 5 μm.
We previously reported that FAM29A is required for the recruitment of NEDD1 to the spindle but not for its localization to the centrosomes (Zhu et al., 2008). Consistent with this, inhibition of Plk1 by BI 2536 also moderately reduced the total NEDD1 signal and the degree of reduction was similar to that for FAM29A (Fig. 3D-F). More importantly, the pattern of NEDD1 location altered upon inhibition of Plk1. In control cells, NEDD1 signals spread along the length of MTs in the monopolar spindle, whereas in cells treated with BI 2536, NEDD1 was mostly concentrated at the spindle pole (Fig. 3D,E). Furthermore, NEDD1 signals on centrosomes were also greatly reduced, indicating that Plk1 is required for the recruitment of NEDD1 both to centrosomes and to the spindle. We also noticed that inhibition of Plk1 and kinesin Eg5 prevented the separation of the two centrosomes in the monopolar spindle (arrowheads in Fig. 3E). The effects of Plk1 inhibition on the localization of FAM29A and NEDD1 were independently confirmed in cells depleted of Plk1 (Fig. 3G-H) using a previously characterized specific siRNA (Hansen et al., 2004; Seki et al., 2008a; Seki et al., 2008b). As controls, treatment by BI 2536 or knockdown of Plk1 did not alter the expression levels of FAM29A and NEDD1 (supplementary material Fig. S2).
The kinase activity of Plk1 is required for the interaction between Plk1 and FAM29A, but not for the interaction between FAM29A and NEDD1 or between Plk1 and NEDD1
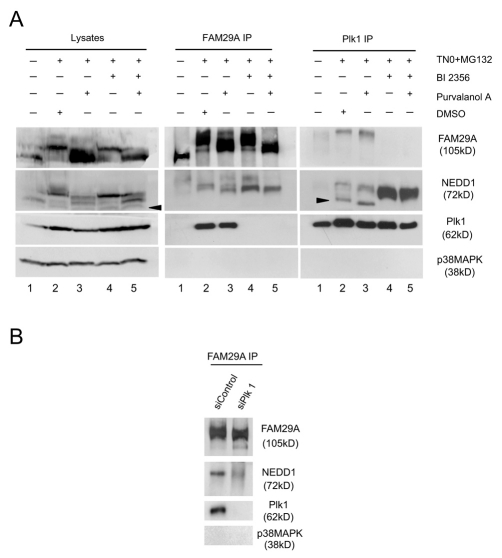
We next investigated the effect of BI 2536 on the formation of various Plk1 complexes in mitosis. Prometaphase cells were arrested by a thymidine-nocodazole treatment and then released into a media containing MG132, an inhibitor of proteasomes, plus BI 2536, purvalanol A, or both (Fig. 4A). Purvalanol A is an inhibitor of Cdk1 (Skoufias et al., 2007) and used as a control here. At the concentration used here (25 μM), purvalanol A effectively inhibits the mitotic Cdk1 in vivo and triggers a premature exit into G1 in nocodazole-arrested mitotic cells (Skoufias et al., 2007). The presence of MG132 prevents mitotic exit upon inhibition of mitotic kinases (Seki et al., 2008b; Skoufias et al., 2007). Inhibition of Plk1 abolished the interaction between Plk1 and FAM29A (Fig. 4A), indicating that the kinase activity of Plk1 is required for the formation of the FAM29A-Plk1 complex, consistent with the requirement of the Plk1 activity for targeting FAM29A to the spindle MTs. Given that the Plk1-FAM29A interaction was not sensitive to λ-phosphatase treatment (Fig. 2A), this experiment suggests either that the phosphorylated residue required for the Plk1-FAM29A interaction is not accessible to the λ-phosphatase, or that the active conformation of Plk1 is required for this interaction.
Fig. 4.
The kinase activity of Plk1 is required for the interaction between Plk1 and FAM29A, but not for the interaction between FAM29A and NEDD1 or between Plk1 and NEDD1. (A) HeLa S3 cells were synchronized at prometaphase by a thymidine-nocodazole arrest, released and incubated for 2 hours with medium containing the proteasome inhibitor MG132 (20 μM) and one of the followings: DMSO (lane 2), purvalanol A (25 μM; lane 3), BI 2536 (1 μM; lane 4), or purvalanol A (25 μM) + BI 2536 (1 μM; lane 5). Asynchronous cells without drug treatment (lane 1) were also included. Cell lysates and the anti-FAM29A and anti-Plk1 immunoprecipitates (IP) were analyzed by western blotting. Arrowheads point to nonspecific cross-reacting proteins. (B) HeLa cells were transfected with siRNAs and synchronized at prometaphase by a thymidine-nocodazole treatment. The anti-FAM29A immunoprecipitates (IP) were analyzed by western blotting.
Inhibition of Plk1 did not, however, affect the association of NEDD1 with either FAM29A or with Plk1 (Fig. 4A), indicating that these interactions are independent of the kinase activity of Plk1. In fact, the extent of association between Plk1 and NEDD1 was substantially increased in the presence of BI 2536. Given that the interaction between Plk1 and NEDD1 was sensitive to the λ-phosphatase treatment (Fig. 2A), we conclude that Plk1 is not responsible for this phospho-dependent regulation of the Plk1-NEDD1 interaction. As Plk1 activity is required for the recruitment of NEDD1 to centrosomes, formation of the Plk1-NEDD1 complex is not sufficient for this recruitment.
Phospho-dependent interaction of Plk1 and NEDD1 suggests that a priming phosphorylation may be required for formation of the complex, as is usually the case for proteins interacting with the Polo-box domain of Plk1 (Barr et al., 2004). We tested whether Cdk1 is the priming kinase for this complex. Inhibition of Cdk1 increased the electrophoretic mobility of FAM28A and NEDD1, suggesting that Cdk1 phosphorylated, either directly or indirectly, both FAM29A and NEDD1 in vivo (Fig. 4A). However, none of the FAM29A-NEDD1 complex, FAM29A-Plk1 complex, or Plk1-NEDD1 complex was altered upon inhibition of Cdk1 (Fig. 4A), suggesting that Cdk1 is not required for any of these interactions. We also noticed that inhibition of Cdk1 increased the levels of FAM29A in mitotic cells (Fig. 4A), suggesting that Cdk1 may downregulate the FAM29A level.
We analyzed whether the presence of the Plk1 protein is required for the FAM29A and NEDD1 interaction. Depletion of Plk1 by a specific siRNA did not abolish the interaction between FAM29A and NEDD1 in prometaphase cells arrested by the thymidine-nocodazole treatment (Fig. 4B), suggesting that formation of this complex is not absolutely dependent on Plk1.
Localization of Plk1 is independent of FAM29A and NEDD1
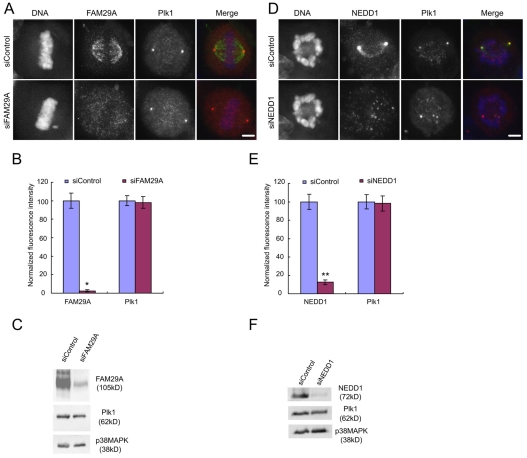
We investigated whether FAM29A and NEDD1 were required for the localization of Plk1. Transfection of previously characterized siRNAs (Luders et al., 2006; Zhu et al., 2008), specifically targeting FAM29A or NEDD1, efficiently depleted endogenous FAM29A or NEDD1, respectively (Fig. 5). However, neither the expression level nor the localization of Plk1 was altered by the depletion of FAM29A or NEDD1 (Fig. 5). Thus, Plk1 acts upstream of FAM29A and NEDD1 to regulate their localization, but not vice versa.
Fig. 5.
Depletion of FAM29A or NEDD1 does not affect the localization of Plk1. (A-F) HeLa cells were transfected with siRNAs and analyzed at 72 hours post-transfection. Maximum projections from deconvolved z-stacks of transfected cells stained for FAM29A (A), NEDD1 (D, green), Plk1 (red; A,D), and DNA (blue; A,D). Fluorescence intensities of total FAM29A (B), total NEDD1 (E), and total Plk1 (B,E) were quantified, normalized to those of their respective control samples, and then plotted (n=10 mitotic cells for each quantification). Lysates of cells transfected with siRNAs were analyzed by western blotting to determine the levels of FAM29A and NEDD1 proteins (C,F). Please note that knockdown of NEDD1 arrests cells at prometaphase (Luders et al., 2006). Thus, prometaphase cells were analyzed in D and E. *P<0.0001; **P<0.0008 (two-tailed t-test relative to siControl cells). Error bars indicate s.e.m. Scale bars: 5 μm.
The cellular activity of FAM29A is under the control of Plk1
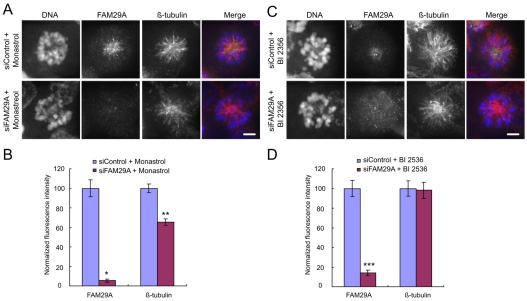
We recently reported that FAM29A promotes MT-dependent MT amplification in mitosis and that depletion of FAM29A reduces MT density in the bipolar spindle (Zhu et al., 2008). Similarly, depletion of FAM29A decreased the MT density by 30% in the monopolar spindle induced by monastrol (Fig. 6A,B). However, upon inhibition of Plk1 by BI 2536, depletion of FAM29A no longer altered the MT density in the monopolar spindle (Fig. 6C,D). Taken together, these data indicate that FAM29A promotes the MT polymerization in the monopolar spindle, but only when Plk1 is active. Although we cannot completely exclude the possibility that inactivation of Plk1 has a dominant effect that bypasses the regulation of spindle MT density by FAM29A, these data also support the conclusion that Plk1 activates FAM29A and that active FAM29A then controls MT density; inhibition of Plk1 abolishes the ability of FAM29A to control the MT density in the spindle. This is consistent with the fact that inactivation of Plk1 prevented the localization of FAM29A along the length of spindle MTs (Fig. 3). Thus, Plk1 is a key regulator of FAM29A, and our data point to a novel function of Plk1 in regulating spindle assembly and MT-dependent MT amplification via the FAM29A–NEDDI–γ-tubulin pathway.
Fig. 6.
Plk1 regulates FAM29A. (A-D) HeLa cells were transfected with siRNAs, treated with monastrol (100 μM) for 4 hours, and then analyzed (A,B). Alternatively, siRNA-transfected cells were treated with monastrol (100 μM) for 2 hours and then with monastrol (100 μM) plus BI 2536 (300 nM) in DMSO for 2 hours (C,D). (A,C) Maximum projections from deconvolved z-stacks of representative cells stained for FAM29A (green), β-tubulin (red) and DNA (blue) are shown. Fluorescence intensities of total FAM29A and total β-tubulin were quantified, normalized to those in their respective control samples, and then plotted (n=10 mitotic cells for each quantification; B,D). *P<0.004; **P<1.5×10–7; ***P<9.9×10–5 (two-tailed t-test relative to siControl cells). Error bars indicate s.e.m. Scale bars: 5 μm.
FAM29A controls the partitioning of NEDD1 between spindle MTs and centrosomes
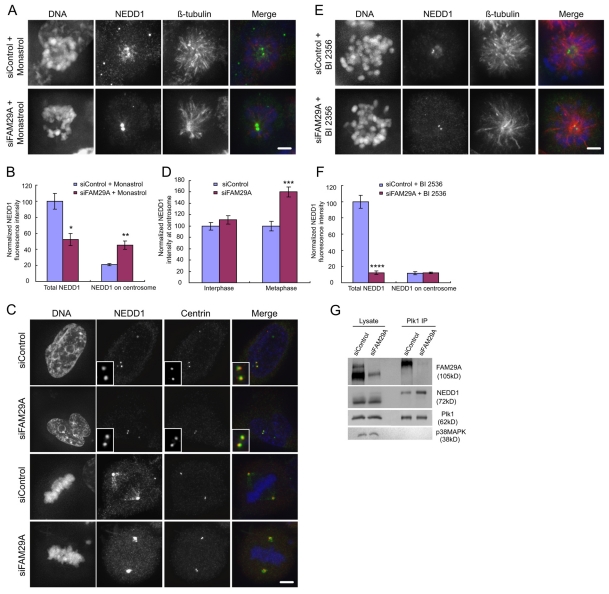
We analyzed the requirement of FAM29A for the recruitment of NEDD1 to the spindle versus centrosomes. Depletion of FAM29A reduced the total NEDD1 signals on the monastrol-induced monopolar spindle, but increased the centrosomal NEDD1 signals twofold (Fig. 7A,B). This finding was also confirmed in cells with bipolar spindles in the absence of monastrol (Fig. 7C,D). This increase in centrosomal NEDD1 only occurred in FAM29A-depleted mitotic cells, not in interphase cells (Fig. 7C,D). Similarly, depletion of FAM29A also increased the centrosomal NEDD1 signals in the presence of nocodazole (data not shown). We conclude that the level of intracellular FAM29A protein influences the distribution of NEDD1 along the spindle MTs versus on centrosomes in mitosis, as the presence of FAM29A directs NEDD1 from centrosomes to the spindle.
Fig. 7.
FAM29A controls the partition of NEDD1 between the spindle and centrosomes. (A-F) HeLa cells were transfected with siRNAs and analyzed at 72 hours post-transfection (C,D). Alternatively, siRNA-transfected cells were treated with monastrol (100 μM) for 2 hours (A,B) or with BI 2536 (300 nM) for 2 hours (E,F). (A,C,E) Maximum projections from deconvolved z-stacks of representative cells stained for NEDD1 (green), β-tubulin (red) and DNA (blue) are shown. Fluorescence intensities of total and centrosomal NEDD1 were quantified and plotted (n=10 mitotic cells for each quantification; B,D,F). Centrosomal NEDD1 signals were defined by co-staining with an anti-centrin antibody (data not shown). *P<0.01; **P<0.0008; ***P<0.001; ****P<0.0005 (two-tailed t-test relative to siControl cells). Error bars indicate s.e.m. Scale bars: 5 μm. (G) Knockdown of FAM29A increases the abundance of the Plk1-NEDD1 complex. HeLa cells were transfected with siRNAs and synchronized at prometaphase by a thymidine-nocodazole treatment. Cell lysates and anti-Plk1 immunoprecipitates (IP) were analyzed by western blotting.
This increase in centrosomal NEDD1 upon depletion of FAM29A requires the kinase activity of Plk1, as inhibition of Plk1 by BI 2536 abolished this increase in siFAM29A cells (Fig. 7E,F). However, NEDD1 signals in BI 2536-treated siControl cells were concentrated around the pole of the monopolar spindle (Fig. 7E,F, Fig. 3D,E). Interestingly, the spindle-pole-associated NEDD1 in BI 2536-treated cells disappeared upon knockdown of FAM29A (Fig. 7E,F). These data suggest that FAM29A is required for targeting NEDD1 to the pole of the monopolar spindle induced by BI 2536, while the kinase activity of Plk1 is essential for distribution of the FAM29A-NEDD1 complex along the entire length of MTs in the spindle.
To understand the biochemical basis of how FAM29A affects the centrosomal localization of NEDD1, we analyzed the Plk1-NEDD1 complex in FAM29A-depelted cells. HeLa cells were transfected with a control or siFAM29A RNA and arrested at prometaphase by a thymidine-nocodazole treatment. The Plk1 complexes were immunopurified using an anti-Plk1 antibody and analyzed by western blotting. Upon depletion of FAM29A, the amount of the Plk1-NEDD1 complex increased several fold (Fig. 7G), probably as a result of an increase in the amounts of free NEDD1 and Plk1 in the cytosol because of the lack of their interacting partner FAM29A. Given that Plk1 is localized to centrosomes and is required for targeting NEDD1 to centrosomes (Fig. 3D-F; Fig. 7B versus 7F), an increase in the abundance of the Plk1-NEDD1 complex could explain the corresponding increase in centrosomal NEDD1 in FAM29A-depleted cells.
Discussion
We report here the regulation of MT nucleation pathways by Plk1 and FAM29A through their recruitment of NEDD1, a subunit of the γ-tubulin ring complex. NEDD1 is localized to both centrosomes and spindle MTs. Through its interaction with the γ-tubulin ring complex, NEDD1 promotes the nucleation of MTs from both centrosomes and the spindle (Haren et al., 2006; Luders et al., 2006). How the two competing processes, nucleation of MTs from centrosomes versus spindle, are coordinately regulated is poorly understood. We show that these two processes are coordinated by the key mitotic kinase, Plk1, through two distinct downstream pathways. Plk1 recruits NEDD1 to centrosomes through their interaction, whereas Plk1 controls the spindle-mediated MT nucleation through its interaction with and regulation of FAM29A.
We previously identified and characterized FAM29A as a key regulator essential for MT-dependent MT polymerization or MT amplification (Goshima et al., 2008; Zhu et al., 2008). Mitotic FAM29A acts upstream of NEDD1. It is localized to the spindle and interacts with the NEDDI–γ-tubulin complex, thereby promoting MT nucleation using existing MTs as templates (Zhu et al., 2008). FAM29A is not required for centrosome-mediated MT nucleation and it promotes MT amplification independently of centrosomes and chromatin. We show here that Plk1 interacts with FAM29A and is required for targeting FAM29A and NEDD1 along the length of spindle MTs. In addition, FAM29A is not only a passive target for Plk1 regulation, but also actively controls the partitioning of NEDD1 between and, by inference, the amounts of MTs polymerized from centrosomes versus the spindle through regulating the abundance of the Plk1-NEDD1 complex.
Plk1 controls nucleation of MTs both from centrosomes and from the spindle
Based on the following observations, we conclude that Plk1 controls the MT nucleation from centrosomes by recruiting NEDD1. First, inhibition of Plk1 activity or depletion of Plk1 protein reduced the amounts of centrosome-associated NEDD1 (Fig. 3). Second, depletion of FAM29A increased the centrosomal NEDD1 signals, but inhibition of Plk1 abolished this increase (Fig. 7). Third, Plk1 and NEDD1 interact (Fig. 1). This interaction is independent of the kinase activity of Plk1 or the presence of FAM29A (Figs 4 and 7). In fact, either knockdown of FAM29A or inhibition of Plk1 enhanced the interaction between NEDD1 and Plk1, possibly resulting from higher concentrations of free NEDD1 and Plk1 in the cytosol, as a consequent of the lack of the FAM29A-NEDD1 or Plk1-FAM29A interaction under these conditions, respectively. The Plk1-NEDD1 interaction is probably dependent on protein phosphorylation (Fig. 2), commonly observed for the association of the Polo-box domain in Plk1 with proteins prime-phosphorylated by another kinase (Barr et al., 2004). However, neither Plk1 nor Cdk1 is responsible for prime-phosphorylation in this case (Fig. 4). We also noted that the interaction between Plk1 and NEDD1 itself is not sufficient to recruit NEDD1 to centrosomes and that the kinase activity of Plk1 is required, because inhibition of Plk1 by BI 2536 substantially reduces the centrosomal NEDD1 signals (Fig. 3), even though this treatment enhanced the Plk1-NEDD1 interaction (Fig. 4).
We also show that Plk1 controls MT amplification by targeting FAM29A and NEDD1 along the spindle MTs (Figs 3, 6 and 7). Plk1 controls the FAM29A-NEDD1 pathway at least at two levels. Inhibition or knockdown of Plk1 abolished the localization of FAM29A and NEDD1 along the spindle MTs (Fig. 3), indicating that the kinase activity is crucial for loading of FAM29A and NEDD1 onto the spindle MTs. In addition, inactivation of Plk1 also altered the distribution patterns of FAM29A and NEDD1 on the spindle, as residual FAM29A and NEDD1 remaining on the spindle were concentrated around the spindle pole, not along the entire length of spindle MTs (Figs 3 and 7). We speculate that spindle localization of FAM29A and NEDD1 in wild-type cells may be mediated by an active transport of the FAM29A-NEDD1 complex toward the plus ends of spindle MTs and this transport is absent upon inhibition of Plk1. Alternatively, spindle localization of FAM29A and NEDD1 may require a Plk1-dependent suppression of the poleward transport of the FAM29A-NEDD1 complex; in the absence of active Plk1, translocation of the FAM29A-NEDD1 complex to the spindle pole concentrates both proteins around the pole.
Biochemically, Plk1 interacts with FAM29A and this interaction requires the kinase activity of Plk1 (Figs 1 and 4), but is independent of the Plk1-NEDD1 interaction (Fig. 2A). FAM29A also interacts with NEDD1 and this interaction does not depend on protein phosphorylation (Zhu et al., 2008) or the activity of the Plk1 kinase (Fig. 4). As FAM29A interacts with both Plk1 and NEDD1, we analyzed whether these three proteins form a single complex. Treatment of the Plk1 immunoprecipitates with λ-phosphatase completely removed associated NEDD1, but retained FAM29A (Fig. 2A); if Plk1, FAM29A and NEDD1 formed a single complex, the phosphorylation-insensitive interactions between Plk1 and FAM29A and between FAM29A and NEDD1 would retain NEDD1 in the Plk1 complex upon phosphatase treatment. Thus, these three proteins are unlikely to form a single complex. We conclude that Plk1, FAM29A and NEDD1 form three separate complexes, although we cannot exclude the possibility that other proteins may also present in each of these complexes.
We investigated the exact biochemical mechanism for Plk1-mediated recruitment of FAM29A and NEDD1 to the spindle. As knockdown of FAM29A abolished the localization of NEDD1 to both bipolar and monopolar spindles, but not vice versa (Fig. 7A,C) (Zhu et al., 2008), and as depletion of FAM29A also removed the spindle pole-associated NEDD1 in BI 2536-treated cells (Fig. 3D, Fig. 7E), we propose that Plk1 targets FAM29A to the spindle, which then serves as a receptor for NEDD1 and recruits NEDD1 to spindle MTs through the FAM29A-NEDD1 interaction. The Plk1-NEDD1 complex does not directly contribute to the recruitment of NEDD1 to the spindle. Given that Plk1 and FAM29A do not colocalize on the mitotic spindle (Fig. 1), it is likely that targeting FAM29A to the spindle by Plk1 is more complex than the simple Plk1-FAM29A interaction.
FAM29A controls the partition of centrosomal versus spindle NEDD1
NEDD1 is localized to both centrosomes and the mitotic spindle and the relative distribution of NEDD1 between these two structures determines the degree of MT nucleation and polymerization from centrosomes versus from the spindle. Coordinated MT nucleation is essential for the proper structure, dynamics and function of the spindle in mitosis. Plk1 is an upstream regulator required for MT nucleation from both structures, and therefore is unlikely to determine the partitioning of NEDD1 between centrosomes and the spindle. Interestingly, we found that cellular levels of the FAM29A protein control the relative distribution of NEDD1. A reduction in FAM29A abundance increased the amounts of the Plk1-NEDD1 complex and the centrosomal NEDD1 signals. Thus, a reduction in MT amplification in the spindle is compensated by an enhanced MT nucleation from centrosomes and this compensatory mechanism ensures proper spindle assembly and function.
Given the requirement of the kinase activity of Plk1 for the cellular recruitment of FAM29A and NEDD1 and for the formation of the Plk1-FAM29A complex, a key unknown is the precise role of phosphorylation. We have extensively analyzed the phospho-regulation of FAM29A by mass spectrometry and identified four sites, Ser406, Ser552, Ser584 and Ser715, that were phosphorylated in endogenous FAM29A during mitosis (H.Z., J. Coppinger, J. Yates and G.F., unpublished data) (Daub et al., 2008; Dephoure et al., 2008; Zhu et al., 2008). However, extensive mutagenesis analyses of these phosphorylation sites, either singularly or in combination, did not reveal any functional requirement for their phosphorylation in mitosis as determined by the ability of the corresponding non-phosphorable mutants to rescue the siFAM29A phenotype (our unpublished results). Elucidation of the exact molecular mechanism underlying this complex regulation by Plk1 awaits future studies.
Materials and Methods
Antibodies
Rabbit anti-FAM29A and anti-HURP antibodies were described previously (Wong and Fang, 2006; Wong et al., 2008; Zhu et al., 2008). The rabbit anti-NEDD1 antibody was kindly provided by Jens Luders (Stanford University, Palo Alto, CA). The following antibodies were from commercial sources: anti-p38MAPK and anti-Plk1 (Santa Cruz Biotechnology, Inc.), mouse anti-NEDD1 (Abnova Corporation), and mouse anti-β-tubulin (Developmental Studies Hybridoma Bank).
Cell culture, siRNAs and transfection
HeLa S3 and HeLa cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and antibiotics (Invitrogen). Cells were synchronized at the G1-S boundary by a double-thymidine treatment (an 18-hour thymidine arrest and then a 6-hour release, followed by an 18-hour thymidine arrest) or at prometaphase by a thymidine-nocodazole treatment (an 18-hour thymidine arrest and then a 6-hour release, followed by a 14-hour nocodazole arrest) (Fang et al., 1998a; Fang et al., 1998b).
OnTargetPlus siRNAs were synthesized by Dharmacon Inc. siRNA sequences targeting FAM29A, NEDD1 and Plk1 were reported and characterized previously (Hansen et al., 2004; Luders et al., 2006; Seki et al., 2008a; Seki et al., 2008b; Zhu et al., 2008). siRNAs were transfected into HeLa cells using DharmaFect 1 (Dharmacon).
Immunoprecipitation
Antibodies against Plk1 and FAM29A were coupled to Affi-Prep protein A beads (Bio-Rad, Richmond, CA) at a concentration of 0.3 mg/ml. HeLa S3 cells were lyzed in the NP-40 lysis buffer (50 mM Hepes, pH 7.4, 200 mM KCl, 0.3% NP-40, 10% glycerol, 1 mM EGTA, 1 mM MgCl2, 0.5 mM DTT, 0.5 μM microcystin, 10 μg/ml each of leupeptin, pepstatin and chymostatin). Lysates were centrifuged, incubated at 4°C for 1 hour with protein A beads coupled with preimmune mouse or rabbit IgG, and then incubated at 4°C overnight with protein A beads coupled with anti-Plk1 or anti-FAM29A antibodies. Antibody beads were recovered by centrifugation, washed four times with the lysis buffer in the presence of 500 mM KCl and once with the lysis buffer, analyzed by SDS-PAGE, and immunoblotted with appropriate antibodies.
For phosphatase treatment (Fig. 2A), anti-Plk1 immunoprecipitates were incubated with 400 units of γ-phosphatase at 30°C for 30 minutes and the immunoprecipitates were then washed extensively to remove dissociated proteins and analyzed by western blotting.
Immunofluorescence
HeLa cells on coverglasses were fixed with –20°C methanol for 5 minutes or with 4% paraformaldehyde for 15 minutes at room temperature. Subsequently, cells were permeabilized and blocked with PBS-BT (1× PBS, 3% BSA, and 0.1% Triton X-100) for 30 minutes at room temperature. Coverslips were then incubated in primary and secondary antibodies diluted in PBS-BT. Images were acquired with Openlab 5.2 (Improvision) under a Zeiss Axiovert 200M microscope using a 1.4 NA Plan-Apo ×100 oil immersion lens. Deconvolved images were obtained using AutoDeblur v9.1 and AutoVisualizer v9.1 (AutoQuant Imaging).
For quantification of immunofluorescence intensity, cells were stained for various spindle, spindle pole or centrosomal antigens and immunofluorescence images of mitotic cells were acquired as z-stacks from the top to the bottom of each cell. For each antigen quantified in different samples, all the samples were stained in parallel and images were acquired under a constant exposure for each antigen. Z-stacks of fluorescence images were deconvolved using AutoDeblur v9.1 and AutoVisualizer v9.1 (AutoQuant Imaging) and the two-dimensional sum-projections of deconvolved z-stacks were used for quantification of the fluorescence intensity of spindle or centrosomal proteins after subtracting background fluorescence measured in areas outside the cells. The total fluorescence intensity of FAM29A (Figs 3, 5 and 6) and HURP (Fig. 3) in the entire cells were measured. In Figs 3, 5 and 7, the fluorescence intensity of total NEDD1 in the entire cells (including the spindle-associated signals, spindle pole-associated signals, and centrosomal signals) as well as of centrosomal NEDD1 were measured.
Plk1 proteomics was performed as described previously (Jang et al., 2008; Seki et al., 2008a; Seki et al., 2008b; Zhao et al., 2008; Zhu et al., 2008).
Supplementary Material
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/15/2750/DC1
We thank Judith A. Coppinger, John R. Yates III (both at The Scripps Research Institute), Chang-Young Jang (Stanford University) for analysis of Plk1-associated proteins by mass spectrometry. We are grateful to members of the Fang lab for stimulating discussions. This work was supported by a Burroughs-Wellcome Career Award in Biomedical Research, by a grant from National Institutes of Health, and by Genentech, Inc (G.F.). Deposited in PMC for release after 12 months.
References
- Barr, F. A., Sillje, H. H. and Nigg, E. A. (2004). Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 5, 429-440. [DOI] [PubMed] [Google Scholar]
- Daub, H., Olsen, J. V., Bairlein, M., Gnad, F., Oppermann, F. S., Korner, R., Greff, Z., Keri, G., Stemmann, O. and Mann, M. (2008). Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell 31, 438-448. [DOI] [PubMed] [Google Scholar]
- Dephoure, N., Zhou, C., Villen, J., Beausoleil, S. A., Bakalarski, C. E., Elledge, S. J. and Gygi, S. P. (2008). A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA 105, 10762-10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, G., Yu, H. and Kirschner, M. W. (1998a). Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol. Cell 2, 163-171. [DOI] [PubMed] [Google Scholar]
- Fang, G., Yu, H. and Kirschner, M. W. (1998b). The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 12, 1871-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadde, S. and Heald, R. (2004). Mechanisms and molecules of the mitotic spindle. Curr. Biol. 14, R797-R805. [DOI] [PubMed] [Google Scholar]
- Goshima, G., Mayer, M., Zhang, N., Stuurman, N. and Vale, R. D. (2008). Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 181, 421-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, D. V., Loktev, A. V., Ban, K. H. and Jackson, P. K. (2004). Plk1 regulates activation of the anaphase promoting complex by phosphorylating and triggering SCFbetaTrCP-dependent destruction of the APC Inhibitor Emi1. Mol. Biol. Cell 15, 5623-5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren, L., Remy, M. H., Bazin, I., Callebaut, I., Wright, M. and Merdes, A. (2006). NEDD1-dependent recruitment of the gamma-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J. Cell Biol. 172, 505-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, C. Y., Wong, J., Coppinger, J. A., Seki, A., Yates, J. R., 3rd and Fang, G. (2008). DDA3 recruits microtubule depolymerase Kif2a to spindle poles and controls spindle dynamics and mitotic chromosome movement. J. Cell Biol. 181, 255-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, H. A. and Nigg, E. A. (1996). Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J. Cell Biol. 135, 1701-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenart, P., Petronczki, M., Steegmaier, M., Di Fiore, B., Lipp, J. J., Hoffmann, M., Rettig, W. J., Kraut, N. and Peters, J. M. (2007). The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 17, 304-315. [DOI] [PubMed] [Google Scholar]
- Luders, J., Patel, U. K. and Stearns, T. (2006). GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 8, 137-147. [DOI] [PubMed] [Google Scholar]
- Mayer, T. U., Kapoor, T. M., Haggarty, S. J., King, R. W., Schreiber, S. L. and Mitchison, T. J. (1999). Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science 286, 971-974. [DOI] [PubMed] [Google Scholar]
- Seki, A., Coppinger, J. A., Du, H., Jang, C. Y., Yates, J. R., 3rd and Fang, G. (2008a). Plk1- and beta-TrCP-dependent degradation of Bora controls mitotic progression. J. Cell Biol. 181, 65-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, A., Coppinger, J. A., Jang, C. Y., Yates, J. R. and Fang, G. (2008b). Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science 320, 1655-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias, D. A., Indorato, R. L., Lacroix, F., Panopoulos, A. and Margolis, R. L. (2007). Mitosis persists in the absence of Cdk1 activity when proteolysis or protein phosphatase activity is suppressed. J. Cell Biol. 179, 671-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkel, C. E. and Glover, D. M. (1988). polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J. Cell Sci. 89, 25-38. [DOI] [PubMed] [Google Scholar]
- van de Weerdt, B. C. and Medema, R. H. (2006). Polo-like kinases: a team in control of the division. Cell Cycle 5, 853-864. [DOI] [PubMed] [Google Scholar]
- Wiese, C. and Zheng, Y. (2006). Microtubule nucleation: gamma-tubulin and beyond. J. Cell Sci. 119, 4143-4153. [DOI] [PubMed] [Google Scholar]
- Wong, J. and Fang, G. (2006). HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J. Cell Biol. 173, 879-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, J., Lerrigo, R., Jang, C. Y. and Fang, G. (2008). Aurora A regulates the activity of HURP by controlling the accessibility of its microtubule-binding domain. Mol. Biol. Cell 19, 2083-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, W. M., Coppinger, J. A., Seki, A., Cheng, X. L., Yates, J. R., 3rd and Fang, G. (2008). RCS1, a substrate of APC/C, controls the metaphase to anaphase transition. Proc. Natl. Acad. Sci. USA 105, 13415-13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H., Coppinger, J. A., Jang, C. Y., Yates, J. R., 3rd and Fang, G. (2008). FAM29A promotes microtubule amplification via recruitment of the NEDD1-{gamma}-tubulin complex to the mitotic spindle. J. Cell Biol. 183, 835-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.