Abstract
The ability to monitor health status, disease onset and progression, and treatment outcome through non-invasive means is a most desirable goal in the health care promotion and delivery. There are three prerequisites to materialize this goal: specific biomarkers associated with a health or disease state; a non-invasive approach to detect and monitor the biomarkers; and the technologies to discriminate the biomarkers. A national initiative catalyzed by the National Institute of Dental & Craniofacial Research (NIDCR) has created a roadmap to achieve these goals through the use of oral fluids as the diagnostic medium to scrutinize the health and/or disease status of individuals. Progress has shown this is an ideal opportunity to bridge state of the art saliva-based biosensors, optimized to disease discriminatory salivary biomarkers, for diagnostic applications. Oral fluid being the ‘mirror of body’ is a perfect medium to be explored for health and disease surveillance. The translational applications and opportunities are enormous.
Keywords: genomics, nanotechnology, oral cancer, proteomics, saliva
Introduction
High impact human diseases, including cancer, cardiovascular, metabolic and neurological diseases, are challenging to diagnose without supplementing clinical evaluation with laboratory testing. Even with laboratory tools, definitive diagnosis often remains elusive. Three roadblocks hold back the realization of clinical diagnostics’ potential: 1) definitive disease-associated protein and genetic markers; 2) easy and inexpensive sampling methods that effect minimal subject discomfort; and 3) an accurate, portable, and easy to use diagnostic platform. Saliva, a biofluid that is totally non-invasive and readily available, has long been recognized to address the second road block (1). Until recently and with the visionary investment by the National Institute of Dental & Craniofacial Research (NIDCR), saliva biomarker discovery and salivary diagnostic technologies are currently in development that are addressing the first and third roadblocks. It is safe to predict that the use of saliva for disease diagnostics and normal health surveillance is a few years away. This is an exciting time as we are seeing the applications of saliva diagnostics for oral diseases, which will soon to be followed by systemic diseases. This will allow the bridging of oral health research into systemic diseases via the biofluid that filters and processes itself from the vasculature that nourishes the salivary glands into the oral cavity (Figs 1 and 2). Oral fluid being the ‘mirror of body’ is a perfect medium to be explored for health and disease surveillance. The translational applications and opportunities are enormous.
Fig. 1.
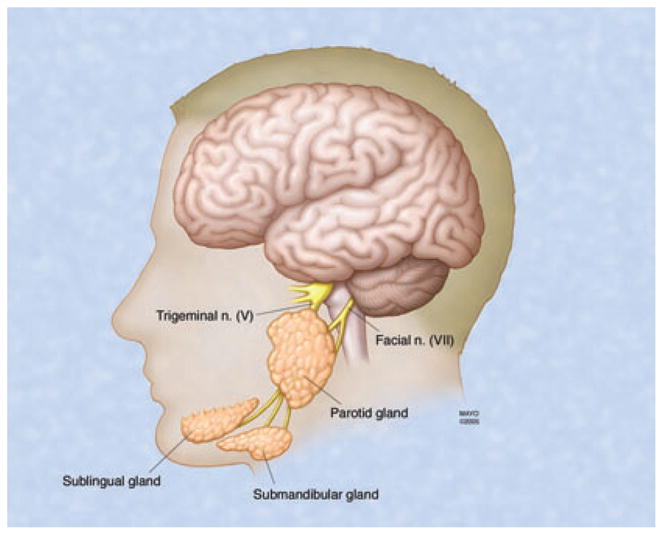
Anatomical locations of the three major salivary glands: parotid, submandibular and sublingual.
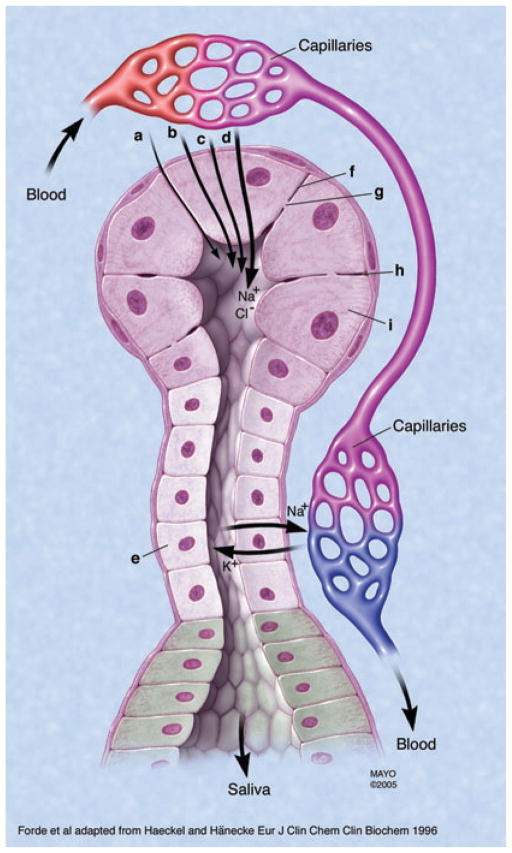
Fig. 2.
Mechanisms of transport of proteins and ions from serum into salivary gland ducts.
A growing number of proof-of-principle examples have been established for using saliva to monitor systemic diseases and conditions. The barriers to widespread implementation of saliva diagnostics derive from technological problems such as sensitivity, miniaturization, high-throughput, automation, portability, low cost, high functionality, and speed to enable detection and measurements of multiple disease markers in saliva have largely been overcome. Techniques are emerging from a combination of miniaturization technologies and discoveries in many different fields of biology, chemistry, physics, and engineering are leading to high-throughput, automated, portable, low cost, more efficient, and rapid biochemical analyzes. Miniaturized diagnostic technologies will be able, with minute amounts of body fluids, to yield critical patient information reflecting health and disease status. These ‘lab-on-a-chip’ platforms will be able to perform multiple operations in parallel in non-laboratory settings such as the field, factory, hospital clinic or home. It is envisioned that such technologies will allow the simultaneous assessment of multiple conditions of health and disease and provide clinicians with prevention and therapeutic strategies to meet patient needs.
Vision and challenges
The post-genomic era provides opportunities for high-throughput approaches to genomics and proteomics. The novel technologies of miniaturization coupled with the highly parallel detection create the possibility of radically new ways to detect and diagnose health and disease states in an individual, even in remote or impoverished settings. These discoveries and technological advances in conjunction with the ability for disease diagnostics in a non-invasive biofluid would offer a revolutionary change in medicine.
There is a great need for convenient and accurate point of care disease diagnostic tools in a non-invasive manner. This is of particular relevance in the developing world where many health risks and illnesses remain poorly defined and receive inappropriate treatment. In addition, little information about the burden of disease is available to guide population health decisions.
The challenge to making saliva diagnostic a clinical reality is to establish the scientific foundation and clinical validations necessary to position salivary diagnostics to be a novel, highly accurate and feasible technologies to achieve definitive point of care assessment of individuals’ health and disease status. Inherent in this vision is to establish the science and diagnostic targets in saliva and the development of robust, simple-to-use biosensor technologies for reliable and valid clinical applications.
Saliva as a diagnostic fluid
Saliva is a mirror of the body. The ability to utilize saliva to monitor the health and disease state of an individual is a highly desirable goal for health promotion and health care research. However, saliva diagnostics is a later bloomer, as only recently has there been a growing appreciation of saliva as a mirror of the body which can reflect virtually the entire spectrum of normal and disease states (1). These include tissue levels of natural substances and a large variety of molecules introduced for therapeutic, dependency or recreational purposes, emotional status, hormonal status, immunological status, neurological effects, and nutritional and metabolic influences. A major drawback to use saliva as a diagnostic fluid has been the notion that informative analytes are generally present in lower amounts in saliva than in serum (2). With new and very sensitive techniques, the lower level of analytes in saliva is no longer a limitation. Almost anything one can measure in blood, one can measure in saliva. Saliva has been reliably used to detect HIV 1 and 2, and viral hepatitis A, B and C. It can also be used to monitor a variety of drugs including marijuana, cocaine and alcohol (1).
There are compelling reasons to use saliva as a diagnostic fluid to monitor health and diseases. It meets the demands for inexpensive, non-invasive and easy to use diagnostic methods. As a clinical tool, saliva has many advantages over serum. Saliva is easy to collect, store and ship and can be obtained at low cost in sufficient quantities for analysis. For patients, the non-invasive collecting techniques dramatically reduce anxiety and discomfort and simplify procurement of repeated samples for longitudinal monitoring over time. For professionals, saliva collection is safer than blood tests, which could expose health care providers to HIV or hepatitis virus. Saliva is also easier to handle for diagnostic procedures since it does not clot, lessening the manipulations required. Saliva-based diagnostics are therefore more accessible, accurate, less expensive and present less risk to the patient than current methodologies.
Development of technologies for saliva-based diagnostics
Six years ago, in 2002, the NIDCR initiated a concerted research effort in the area of saliva diagnostics and progress is currently being advanced towards technologically viable systems moving towards commercialization. NIDCR funded seven UO1 awards to develop microfluidics and microelectricomechanical systems (MEMS) for saliva diagnostics. MEMS are integrating systems consisting of mechanical elements, sensors, actuators, and electronics on a common silicon substrate developed through microfabrication technology. These systems use small sample and reagent volumes coupled with integrated detection methods to perform analysis. The seven NIDCR-supported U01 awards focused on the development of microfluidic and MEMS technologies for measuring DNA, gene transcripts (mRNA), proteins, electrolytes and small molecules in saliva as well as overall profile correlates of a particular disease state, such as cardiovascular disease.
However, it is clear that none of the new technologies will become a practical and clinical reality without strong partnerships with industry early in the development stage. The reasons include the many challenges that such technologies face reaching the stage of fabrication, integration of individual components, validation, regulatory approval and finally commercialization. This has sparked a new initiative for the ‘Development and Validation of Technologies for Saliva-Based Diagnostics’ in order for the currently developed academic saliva diagnostic technologies to team up with industrial partners to further develop functional prototypes and test their robustness for clinical applications for saliva-based detection of clinical diseases. The outcome of this initiative is the anticipated commercialization of saliva-based diagnostic technologies optimized for the detection of highly sensitive and specific detection of salivary biomarkers for human diseases. Four of the initial seven groups were recently renewed for 5 years for the second round technology optimization and commercialization goal. The mandate is that by year 2011, these federally funded, point of care salivary diagnostic technologies would have been prototyped, clinically validated and ready for commercialization.
Diagnostic molecular targets in saliva
In year 2003, NIDCR funded three UO1 awards aiming to comprehensively identify and catalog human salivary proteins from the three major salivary glands. It was envisioned that the Human Salivary Proteome will be a resource to help elucidate disease pathogenesis and evaluate the influence of medications on the structure, composition and secretion of all salivary secretory constituents.
The envisioned end product of the NIDCR-program will be a single Human Salivary Proteome with input from all three groups. Each of the groups will contribute their bioinformatics specialists to team together to develop computational methods for maintenance of a stable, comprehensive, fully classified, and accurately annotated protein sequence knowledgebase. This database, with extensive cross-references and querying interfaces of structural and functional proteomics projects will be important in order to understand the genetic and biological mechanisms causing human disease. This first comprehensive list of the 1166 salivary secretory proteins was recently published (3). The entire content of the salivary proteome in healthy subjects can be viewed at the UCLA data centralization site (http://www.hspp.ucla.edu). The human salivary proteome creates the ‘periodic table’ of the parotid, submandibular and sublingual secretory components to help elucidate disease pathogenesis and evaluate the influence of medications on the structure, composition and secretion of all salivary secretory constituents. Two major translational outcomes have recently emerged based on the salivary proteome. First is that we have worked with the leadership at the human plasma proteome project and did a systematic and comprehensive comparison of the saliva and plasma proteomes (4). A second outcome is that we have identified a panel of highly discriminatory salivary proteomic biomarkers for oral cancer detection with sensitivities and specificities at 89% respectively (5).
The UCLA collaborative oral fluid diagnostic research center
The UCLA School of Dentistry is engaged in both the technology development and the salivary proteome initiatives for saliva diagnostics. During the past 6 years, we have established the ‘UCLA Collaborative Oral Fluid Diagnostic Research Center’ to develop the platform of using nano/micro technology to detect salivary protein and genomic biomarkers for point of care applications of high impact human diseases.
For salivary diagnostic technology development, we have partnered with engineers at the UCLA School of Engineering who are pioneers in the development of micro- and nano-electrical-mechanical systems (MEMS & NEMS) biosensors that exhibit exquisite sensitivity and specificity for analyte detection, down to single molecule level (6, 7). Our research consortium has established a firm and committed collaboration toward the development of MEMS/NEMS biosensors for the real time, ultrasensitive and ultraspecific detection of salivary diagnostic analytes. This is a robust forum of interactions between engineers and biologists/clinicians towards the development of MEMS/NEMS biosensors for saliva-based disease diagnostics. Our prediction is that in ~3 years there will be ‘lab-on-a-chip’ prototypes available for research as well as patient applications (8). The envisioned product is the ‘Oral Fluidic NanoSensor Test (OFNASET)’. The OFNASET is a handheld, automated, easy to use integrated system that will enable simultaneous and rapid detection of multiple salivary protein and nucleic acid targets (Fig. 3; http://www.skb.ucla.edu/). This saliva biomarker detector can be used in dentist or health care provider’s office for point of care disease screening and detection.
Fig. 3.

UCLA’s Oral Fluid NanoSensor Test (OFNASET). A point of care biosensor optimized for saliva detection of multiplex biomarkers, self-contained and being able to detect nucleic acids and proteins without thermal cyclers and ELISA readers nor trained personnel.
Of interest and in a very serendipitous manner, our laboratory has recently made the discovery that discriminatory and diagnostic human mRNAs are present in saliva of normal and disease individuals. The salivary transcriptome presents an additional valuable resource for disease diagnostics. Our first report of the salivary transcriptome, demonstrating that the normal salivary transcriptome consists of ~3000 mRNAs (9). Of particular value is that of the 3000 mRNAs, 180 are common between different normal subjects, constituting the normal salivary transcriptome core (NSTC). To demonstrate the diagnostic and translation potential of the salivary transcriptome, saliva from head & neck cancer patients were profiled and analyzed. Four genes from the NSTC (IL8, OAZ1, SAT and IL1B) were able to discriminate and predict if a saliva sample is from a cancer or normal subject with a sensitivity and specificity of 91% respectively (ROC = 0.95). While head & neck cancer was used as the first proof-of-principle disease for salivary transcriptome diagnostics, data will soon be available for systemic diseases. These data, while early and exploratory, provide sufficient rationale and demonstrate the urgent need to fully explore ‘Salivary Transcriptome Diagnostics’ for major human disease translational applications. Adding to this urgency is our recent finding that the serum transcriptome from the same patients we examined for their salivary transcriptome yield 4 RNA biomarkers that have a sensitivity and specificity of 91% and 71% respectively (ROC = 0.88), demonstrating clearly that for oral cancer detection, saliva transcriptome diagnostics has a slight edge over serum (10). We have recently advanced the technology to profile the salivary transcriptome at the exon level, providing a sixfold increase in diagnostic resolution and well position saliva for pharmacogenomic applications (11) (Fig. 4).
Fig. 4.

Saliva exon expression profiling.
There are advantages of utilizing transcriptome markers for diseases diagnostics. The marker discovery process is high-throughput using genome-wide microarray platforms. As a biomarker, RNA is as robust, as informative as any other analyte. Thus salivary transcriptome offers the combined advantages of high-throughput marker discovery in a non-invasive biofluid with very high patient compliance. Highly diagnostic RNA signatures have been identified for head & neck cancer and Sjögren’s syndrome (5, 12).
Conclusions and future perspectives
While it is clear that there is a national agenda to turn saliva diagnostics into a clinical and commercial reality, much work needs to be done before this vision can be realized. There remains the need to identify definitive disease-associated salivary biomarkers (proteins and genetic) that can be use in conjunction with the technology platforms for saliva diagnostics. The scientific community is poised to develop and validate saliva-based tests as a point of care chair side, portable and multiplexible devices to be used for diagnostic applications. Collectively, technology platform advancements and the identification and validation of robust and discriminatory suites of salivary biomarkers for disease diagnostics represent the necessary marriage to propel saliva diagnostics into a clinical and commercial reality.
Clinical Relevance
Saliva is an ideal translational research tool and diagnostic medium and is being used in novel ways to provide molecular biomarkers for a variety of oral and systemic diseases and conditions. The ability to analyze saliva to monitor health and disease is a highly desirable goal. Saliva has been used to detect caries risk, periodontitis, oral cancer, breast cancer, salivary gland diseases, and systemic disorders such as hepatitis, HIV and HCV. Technology advancement has allowed high-throughput studies to be performed at a scale unrealized previously and is serving to advance the discovery and validation of salivary disease biomarkers.
Acknowledgments
Supported by PHS grants UO1 DE-15018, UO1 DE-16275, UO1 DE-17790, RO1 CA-017170 and RO1 DE-17593 and the Jonsson Comprehensive Cancer Center.
Footnotes
Disclosure: DT Wong is the co-founder of RNAmeTRIX.
Contributor Information
J.M. Lee, UCLA Lung Cancer Research Program, Division of Cardiothoracic Surgery, University of California Los Angeles, Jonsson Comprehensive Cancer Center, University of California Los Angeles, Los Angeles, CA, USA
E. Garon, Division of Hematology-Oncology, David Geffen School of Medicine at UCLA, University of California Los Angeles, Jonsson Comprehensive Cancer Center, Los Angeles, CA, USA
D.T. Wong, School of Dentistry and Dental Research Institute, University of California Los Angeles, School of Engineering, University of California Los Angeles, Division of Head & Neck Surgery/Otolaryngology, University of California Los Angeles, Jonsson Comprehensive Cancer Center, University of California Los Angeles, Molecular Biology Institute, University of California Los Angeles, Los Angeles, CA, USA
References
- 1.Mandel ID. Salivary diagnosis: more than a lick and a promise. J Am Dent Assoc. 1993;124:85–7. doi: 10.14219/jada.archive.1993.0007. [DOI] [PubMed] [Google Scholar]
- 2.Miller SM. Saliva testing – a nontraditional diagnostic tool. Clin Lab Sci. 1994;7:39–44. [PubMed] [Google Scholar]
- 3.Denny P, Hagen FK, Hardt M, Liao L, Yan W, Arellanno M, et al. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J Proteome Res. 2008;7:1994–2006. doi: 10.1021/pr700764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan W, Apweiler R, Balgley BM, Boontheung P, Bundy JL, Cargile BJ, et al. Systematic comparison of the human saliva and plasma proteomes. Proteomics. 2009;3:116–34. doi: 10.1002/prca.200800140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu S, Arellano M, Boontheung P, Wang J, Zhou H, Elashoff D, et al. Salivary proteomics for oral cancer biomarker discovery. Clin Cancer Res. 2008;14:6246–52. doi: 10.1158/1078-0432.CCR-07-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang TJ, Shalci A, Ho D, Zhang Y-H, McCabe ERB, Ho C-M. Rapid Bacterial Diagnosis: MEMS-Based DNA Detection. Pediatric Academic Societies’ Annual Meeting; 2002; Baltimore MD. 2002. [Google Scholar]
- 7.Soong RK, Bachand GD, Neves HP, Olkhovets AG, Craighead HG, Montemagno CD. Powering an inorganic nanodevice with a biomolecular motor. Science. 2000;290:1555–8. doi: 10.1126/science.290.5496.1555. [DOI] [PubMed] [Google Scholar]
- 8.St John M, Li Y, Zhou X, Denny P, Ho C-M, Montemagno CD, et al. IL-6 and IL-8: potential biomarkers for oral cavity and oropharyngeal SCCA. Arch Otolaryngol Head Neck Surg. 2004;130:929–35. doi: 10.1001/archotol.130.8.929. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Zhou X, St John MA, Wong DT. RNA profiling of cell-free saliva using microarray technology. J Dent Res. 2004;83:199–203. doi: 10.1177/154405910408300303. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Elashoff D, Oh M, Sinha U, St John MA, Zhou X, et al. Serum circulating human mRNA profiling and its utility for oral cancer detection. J Clin Oncol. 2006;24:1754–60. doi: 10.1200/JCO.2005.03.7598. [DOI] [PubMed] [Google Scholar]
- 11.Hu Z, Zimmermann BG, Zhou H, Wang J, Henson BS, Yu W, et al. Exon-level expression profiling: a comprehensive transcriptome analysis for oral fluids. Clin Chem. 2008;54:824–32. doi: 10.1373/clinchem.2007.096164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu S, Wang J, Meijer J, Ieong S, Xie Y, Yu T, et al. Salivary proteomic and genomic biomarkers for primary Sjogren’s syndrome. Arthritis Rheum. 2007;56:3588–600. doi: 10.1002/art.22954. [DOI] [PMC free article] [PubMed] [Google Scholar]