Abstract
The enormous diversity of extant animal forms is a testament to the power of evolution, and much of this diversity has been achieved through the emergence of novel morphological traits. The origin of novel morphological traits is an extremely important issue in biology, and a frequent source of this novelty is co-option of pre-existing genetic systems for new purposes (Carroll et al., 2008). Appendages, such as limbs, fins and antennae, are structures common to many animal body plans which must have arisen at least once, and probably multiple times, in lineages which lacked appendages. We provide evidence that appendage proximodistal patterning genes are expressed in similar registers in the anterior embryonic neurectoderm of Drosophila melanogaster and Saccoglossus kowalevskii (a hemichordate). These results, in concert with existing expression data from a variety of other animals suggest that a pre-existing genetic system for anteroposterior head patterning was co-opted for patterning of the proximodistal axis of appendages of bilaterian animals.
Keywords: Proximodistal appendage patterning, Anteroposterior neurectoderm patterning, Genetic co-option, btd/Sp8, Dll/Dlx, dac/Dach, hth/Meis
Introduction
Since the advent of molecular biology, many morphological traits that are shared between disparate animal clades have been found to be controlled by conserved underlying genetic systems (McGinnis and Krumlauf, 1992; Holley et al., 1995; Bier, 1997; Silver and Rebay, 2005; Olson, 2006). Morphological novelty, on the other hand, involves the evolution of new traits that are often patterned by co-opted genes or genetic systems that originally performed other developmental functions. For example, eye spots on butterfly wings are patterned through the re-deployment, in small foci corresponding to the eye spots, of genes that also control the growth and patterning of the entire insect wing (Keys et al., 1999). Another example is seen in the redeployment of a few Hox genes to pattern the paired appendages of vertebrates; these genes having been co-opted from an ancestral role in patterning posterior structures on the main body axis of chordates (McGinnis and Krumlauf, 1992; Zakany and Duboule, 2007). In this study we wished to explore the origins of the proximodistal appendage patterning system.
The patterning of Drosophila appendages is a well studied system of proximodistal axis specification. Although numerous genes participate in patterning of Drosophila appendages, there is a core group of genes which is responsible for establishing the gross morphological divisions. The gene pair buttonhead (btd):D-Sp1 [despite its name, D-Sp1 gene is not an ortholog of vertebrate Sp1, being a member of the Sp8 family (Beermann et al., 2004), and it will hereafter be referred to as D-Sp8], and the genes Distal-less (Dll), dachshund (dac), and homothorax (hth), are expressed in and regulate the growth and boundaries of the distal, medial, and proximal appendage domains (Kojima, 2004). All of these genes encode DNA binding transcription factors, and we refer to them as the core proximodistal appendage patterning system.
In Drosophila embryos, hth and btd are both expressed at very early stages in the appendage primordia. btd and D-Sp8 have overlapping functions in activating Dll transcription in embryonic thoracic appendage primordia (Estella et al., 2003). As the domains of Dll expressing cells expand, hth becomes excluded from a subset of these cells in response to repression by Dll (Bolinger and Boekhoff-Falk, 2005). Cells from these primordia go on to form the larval Keilin’s organs and leg imaginal discs.
Early in leg imaginal disc development, as in the embryonic appendage primordia, cells are divided into two major domains by a central cluster of Dll expressing cells surrounded by hth expressing cells (Abu-Shaar and Mann, 1998; Wu and Cohen, 1999). As development progresses, dac expression comes on in a medial region of leg discs (Abu-Shaar and Mann, 1998; Wu and Cohen, 1999), as well as in antennal discs (Dong et al., 2001). The expression domains of Dll, dac, and hth overlap at later stages of appendage disc development, although the genes also exhibit mutually repressive interactions in some cells of the leg discs (Abu-Shaar and Mann, 1998; Wu and Cohen, 1999; Dong et al., 2001). Although the regulatory relationships between btd:D-Sp8 and other core appendage patterning genes in the larval imaginal discs are unknown, the btd gene is expressed in a disc domain that overlaps with hth, dac and Dll, but is excluded from the most distal and most proximal cells (Estella et al., 2003). The developing Drosophila antenna has a slightly different imaginal disc expression profile than that of the leg, with the medial dac domain being smaller and hth expression overlapping that of both dac and Dll (Dong et al., 2001). However, expression domains of these genes still roughly correspond to the same proximodistal fates in both developing leg and antennae (Dong et al., 2001), and similarly ordered and overlapping expression domains of the core genes are conserved in many developing arthropod appendages (Angelini and Kaufman, 2005; Beermann et al., 2004; Schaeper et al., 2009).
Investigation of genes underlying proximodistal development of vertebrate appendages has revealed that, despite structural dissimilarity to arthropod appendages, they develop under the control of a genetic patterning system that includes orthologs of Drosophila btd:D-Sp8, Dll, dac, and hth genes (Pueyo and Couso, 2005). Vertebrate Sp8 genes are expressed in evolutionarily conserved patterns in distal ectoderm of limb buds, and knockdown of Sp8 function in chick results in defects of limb outgrowth and patterning (Kawakami et al., 2004). Dlx family genes (Dlx1, 2, 5 & 6; Dll orthologs) are also expressed in distal ectoderm of mouse limb buds, and Dlx5:Dlx6 double mutants have distal limb defects (Panganiban and Rubenstein, 2002; Kraus and Lufkin, 2006). Dach1 (a dac ortholog) is expressed in a complex pattern in developing mouse limb buds, with a transient stage when expression is limited to anterior-medial limb bud cells (Hammond et al., 1998; Davis et al., 1999). Meis1 (a vertebrate hth ortholog) is expressed in the proximal regions of vertebrate limb buds, and required for the normal development of the proximal domain of chick appendages (Mercader et al., 1999).
Available fossil data from the Pre-Cambrian does not allow us to be sure of the body plan of the last common ancestor of vertebrates and arthropods (Valentine, 2004). However, a synthesis of comparative morphology suggests that it either existed with rudimentary appendages or lacked them entirely (Shubin et al., 1997). The appendages of disparate extant bilaterian groups almost certainly evolved independently in multiple lineages subsequent to their divergence from a common ancestor which lacked appendages (Shubin et al., 1997). If animal appendages are not derived from a common ancestral appendage, the involvement of a common genetic system in proximodistal patterning could be due to random convergence of the same set of genes to pattern non-homologous appendages, or independent co-option of the same genetic system that functioned to pattern an ancestral structure shared by both vertebrates and arthropods (Panganiban et al., 1997; Davidson and Erwin, 2006; Tabin et al., 1999). Involvement of a genetic system in essential developmental roles (e.g. insect wing patterning) may make the regulatory interactions within the system resistant to change (Davidson and Erwin, 2006). This does not, however, preclude redeployment of such a patterning system using different genetic inputs and outputs, which could then contribute to novel morphological structures (Davidson and Erwin, 2006), such as butterfly wing eye spots (Keys et al., 1999).
It has been previously proposed that lateral appendages might have originated through the co-option of a pre-existing group of genes, including Dll, that controlled a rudimentary appendage-like outgrowth in the ancestor of vertebrates and arthropods (Tabin et al., 1999). It has also been proposed that the appendages of vertebrates and arthropods might be modified duplicates of the entire anteroposterior body axis (Minelli, 2000). This proposal is based in part on an ancestral role of Hox genes in patterning the main body axis, and the involvement of a subset of Hox genes in patterning the proximal-distal axis of vertebrate appendages (Zakany and Duboule, 2007). Hox genes do not have similar expression patterns in vertebrate and arthropod appendages, so for this and other reasons the model that the entire anteroposterior body axis patterning system is redeployed in most animal appendages (Minelli, 2000) is not well supported in our opinion. Our proposition relates an ancient conserved genetic system for patterning the anterior neurectoderm of animals to the proximodistal patterning of bilateral animal appendages.
A survey of previous research provides data from a few different animal groups on expression patterns of the core proximodistal appendage patterning genes in various tissues. We noticed that these genes, as well as other genes that are part of the proximodistal appendage patterning system in Drosophila, such as aristaless (al), apterous (ap), and BarH1, are expressed in discrete domains in the anterior embryonic neurectoderm of many chordates and arthropods (Supplementary material). We considered the hypothesis that a shared anteroposterior expression regimen of these genes in head neurectoderm might be common in bilateral animals. We wished to evaluate this hypothesis by testing the relative expression patterns of core appendage patterning genes in the anterior neurectoderm of Drosophila embryos, as well as in embryos of Saccoglossus, a basal deuterostome that lacks bilateral appendages.
Results and Discussion
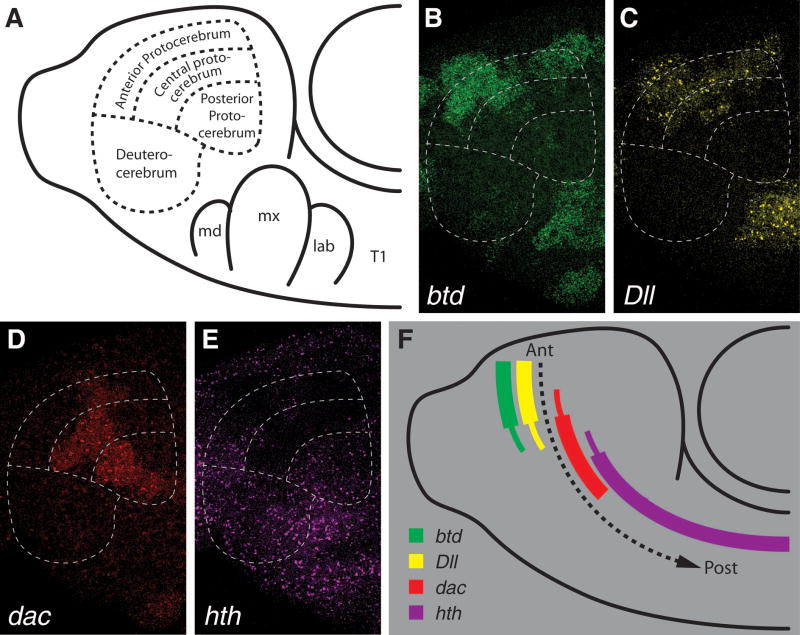
Genes of the core proximodistal appendage patterning system are expressed in a spatially and temporally complex manner during Drosophila development. However, in the anterior neurectoderm of Drosophila embryos, these genes are expressed in a clear anteroposterior order. We determined the relative expression patterns of Dll, dac, hth, and btd:D-Sp8 using combinatorial in situ hybridizations, and analyzed their relative expression patterns in germband extended (stage 11) embryos (Fig. 1B–E). At this stage the procephalic neurectoderm can be divided into anterior, central, and posterior protocerebral areas, and a more posterior deuterocerebral area, with the most anterior cells being those flanking the dorsal midline of the procephalon and posterior cells located more ventrolaterally (Younossi-Hartenstein et al., 1996) (Fig. 1A). btd:D-Sp8 and Dll are transcribed in overlapping patches covering most of the anterior protocerebral neurectoderm (Fig. 1B–C). The domain of dac transcription is mainly in central and posterior protocerebral neurectoderm, with small regions of overlap with Dll and btd:D-Sp8 (Fig. 1D). hth transcripts are largely absent in anterior neurectoderm (Fig. 1E), and are completely excluded from domains which transcribe Dll and btd:D-Sp8 (compare to Fig. 1C). The posterior protocerebral region contains cells which transcribe both hth and dac, but the majority of hth transcription is found in the deuterocerebrum and more posterior neurectoderm (compare Fig. 1D–E). Taken together, these data reveal an expression order of btd:D-Sp8 and Dll in the most anterior neurectodermal cells, dac in medial cells, and hth in posterior cells, with small zones of overlap at the borders of the three major domains (Fig. 1F).
Figure 1. Expression of core appendage patterning genes in the head neurectoderm of Drosophila embryos.
Data are presented as maximum projections of confocal sections through embryonic procephalic neurectoderm of a stage 11 Drosophila embryo. (A) Schematic of the procephalic region of a stage 11 Drosophila embryo displaying subdivisions of the neurectoderm. Indicated are anterior, central, and posterior protocerebral and deuterocerebral regions of the head neurectoderm (adapted from (Younossi-Hartenstein et al., 1996)), as well as the mandibular (md), maxillary (mx), labial (lab), and first thoracic (T1) segments. (B) btd is transcribed mainly in the anterior protocerebral neurectoderm with a small region of expression in central regions. Also seen are antennal and maxillary segment expression outside the neurectoderm. The D-Sp8 transcription pattern is the same as btd in this region (data not shown). (C) Dll is transcribed in nearly the same neurectodermal pattern as btd at this stage. (D) dac is transcribed mainly in central and posterior protocerebral neurectodermal cells and overlaps with the posterior expression of Dll and btd. (E) hth is expressed in posterior protocerebral cells and deuterocerebral cells, overlapping with dac expression in its posterior expression domain. (F) Diagram indicating relative expression domains of Dll, btd, dac, and hth in the procephalic neurectoderm. Dashed arrow indicates general anterior to posterior orientation of the head neurectoderm.
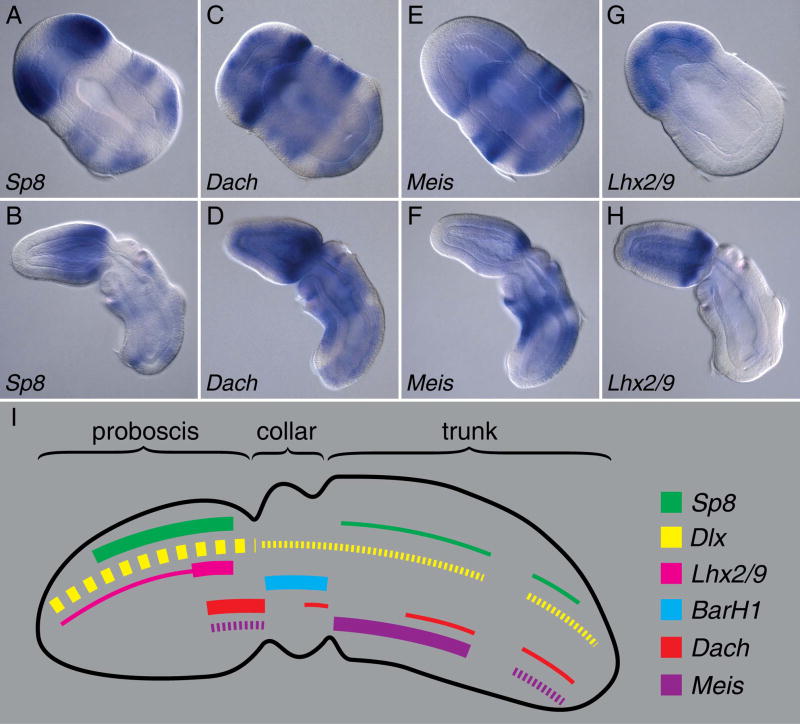
Using in situ hybridization, we also tested the expression patterns of the orthologous genes in Saccoglossus embryos ranging from gastrula to early gill slit stages. In post gastrulae, Sp8 is transcribed at high levels in the anterior third of the embryonic neurectoderm, and to a lesser degree in multiple medial to posterior stripes (Fig. 2A). In embryos at the one gill slit stage, expression is similar with the major expression confined to the proboscis (Fig. 2B). The Saccoglossus dac ortholog (Dach) is transcribed at high levels in a neurectodermal stripe just anterior to the collar, and at low levels throughout most of the rest of the embryo, both in post gastrula (Fig. 2C) and one gill slit stage embryos (Fig. 2D). Meis, the ortholog of hth, is transcribed at high levels in a broad band in the trunk neurectoderm, as well as in two dorsal patches - one just anterior to the collar; the other in the posterior trunk (Fig. 2E–F). The Saccoglossus Lhx2/9 gene, orthologous to apterous, is transcribed throughout the proboscis neurectoderm in late gastrulae (Fig. 2G) and then becomes restricted mainly to a strong stripe just anterior to the collar (Fig. 2H). Along with previously documented expression patterns for the Saccoglossus BarH1 and Dlx orthologs (Lowe et al., 2003) we provide an expression model (Fig. 2I) summarizing the transcription domains of all of these genes.
Figure 2. Expression of core appendage patterning genes in Saccoglossus embryos.
Data are presented as saggital optical sections of in situs with anterior to the upper right of each panel of post gastrula (A,C,E, and G) and one gill slit stage embryos (B,D,F, and H). (A, B) High levels of Sp8 transcripts could be detected in two broad lateral patches in the proboscis. Expression is absent from both the dorsal and ventral midlines and the most anterior region of the proboscis. Sp8 transcripts are additionally detected at low levels in two broad lateral stripes in the trunk ectoderm with both dorsal and ventral midlines and ciliated band free of expression. (C, D) Dach is expressed at high levels in an ectodermal stripe just anterior to the collar. Additional low level ectodermal expression is detected throughout much of the embryo. (E, F) Meis is expressed strongly in the trunk ectoderm excluding the ciliated band and at the base of the proboscis at early developmental stages, which then subsequently refines to a strong dorsal domain. (G, H) At post gastrula stage Lhx2/9 is expressed throughout the proboscis ectoderm. By one gill slit stage expression becomes restricted mainly to a strong stripe at the base of the proboscis. (I) A schematic combining these data with previously published expression data (Lowe et al., 2003) for Dlx and BarH1 indicating the relative levels and anteroposterior extents of neurectodermal expression of the appendage patterning genes. Dashed lines indicate expression in a subset of cells for the indicated anteroposterior domain.
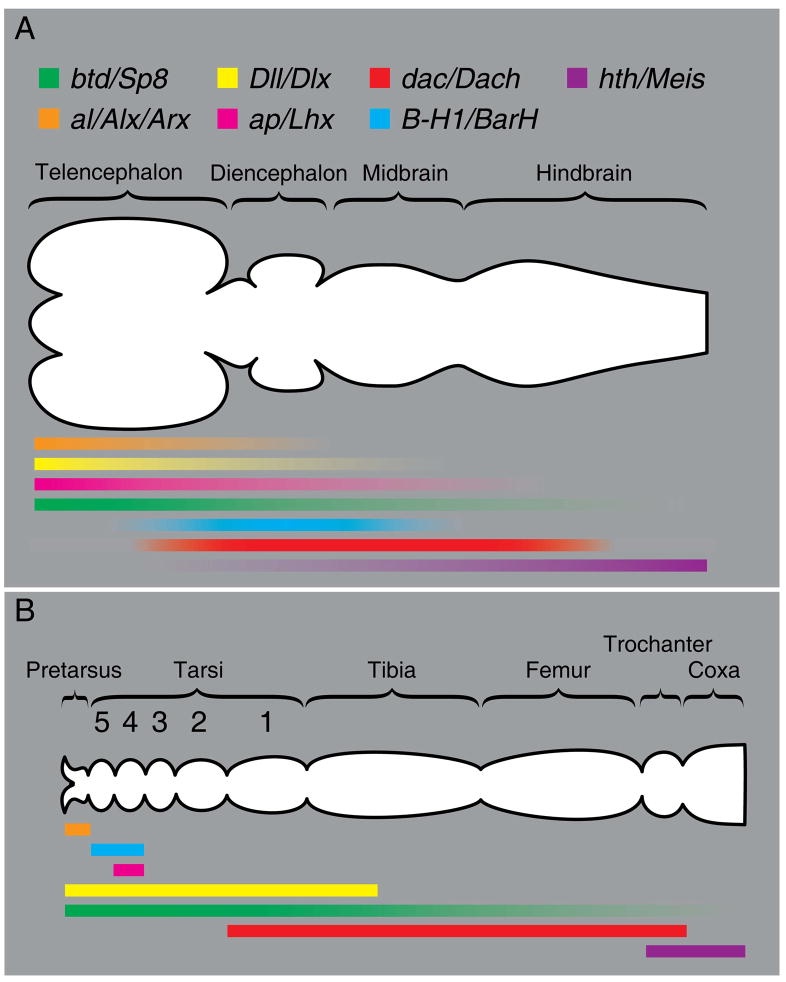
Based on the above data and previously published work (Supplementary material), we estimated the ancestral expression domains of the anteroposterior head patterning system (Fig. 3A), and compared them to the approximate domains of the proximodistal appendage patterning system in the developing Drosophila leg (Fig. 3B). Our proposal posits that a “head-appendage” genetic patterning system, consisting of the btd/Sp8, Dll/Dlx, dac/Dach, hth/Meis genes (and likely other genes, some of which are shown in Fig. 3), was present in a bilaterian ancestor that lacked appendages, where the system functioned to pattern the anteroposterior head axis. The evolution of this system may even have contributed to the process of cephalization in early animals. Subsequently, this system was co-opted to pattern the proximodistal axis of bilateral appendages through modification of input and output connections.
Figure 3. Schematic diagram comparing expression of core appendage patterning genes in limbs and anterior neurectoderm.
(A) Estimated ancestral expression patterns in embryonic anterior neurectoderm (based on conserved domains of expression) is displayed on a generalized diagram of a chordate brain. (B) Expression of proximodistal appendage patterning genes is displayed on a diagram of an adult Drosophila leg (adapted from (Kojima, 2004)).
After a system for anteroposterior head patterning had been co-opted for proximodistal appendage patterning, it could be used specify and diversify the pattern of many body wall outgrowths (e.g. sensory structures, locomotory appendages, external genitalia, feeding appendages, etc.) through changes in the system. These could include variations in the regulatory relationships and expression patterns of the core appendage patterning genes, as well as further modifications of input and output connections (Dong et al., 2001). Consistent with this theory, inputs into this system during Drosophila appendage formation, such as Dpp and wg, are not conserved in this role among insects (Angelini and Kaufman, 2005). At least part of the same system has apparently been coopted for the development of beetle horns (Moczek et al., 2006; Moczek and Rose, 2009), an appendage-like body wall outgrowth, long after the evolutionary advent of bilateral appendages. It will be interesting to study the expression pattern of the head-appendage patterning genes in other branches of the evolutionary tree, especially in cnidarians, acoel flatworms, and lophotrochozoans.
Materials and methods
Drosophila in situ hybridizations were performed as in Kosman et al (Kosman et al., 2004). btd antisense probes were made from a 2.6 kb genomic fragment starting 49 bp 5′ of the coding region. Dll antisense probes were made from a 1.4 kb EcoRI cDNA fragment (Cohen et al., 1989). dac antisense probes were made from a genomic PCR fragment cloned into pCRII (Invitrogen), the primers for the dac fragment were: 5′ AAGCAAAGTATAGAACGGATTAGCA 3′; 5′ TCCAACGAATCTTTCACTTCG 3′. Saccoglossus in situ hybridizations were performed as in Lowe et al (Lowe et al., 2004). Antisense probes for Sp8, Lhx2/9, Dach, and Meis were made from Saccoglossus kowalevskii cDNAs (Freeman et al., 2008), accession numbers NM_001168189, NM_001164971, NM_001164944, and GU384871 respectively.
Supplementary Material
Acknowledgments
We thank D. Kosman for advice on in situ hybridization and synthesis of some of the Drosophila probes; A. Pare and E. Tour for comments on the manuscript. This research was supported by NIH grant R37HD28315 to WM., NIH grant T32GM007240 to DL, NIH grant HD42724 to JG.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abu-Shaar M, Mann RS. Generation of multiple antagonistic domains along the proximodistal axis during Drosophila leg development. Development. 1998;125:3821–30. doi: 10.1242/dev.125.19.3821. [DOI] [PubMed] [Google Scholar]
- Angelini DR, Kaufman TC. Insect appendages and comparative ontogenetics. Dev Biol. 2005;286:57–77. doi: 10.1016/j.ydbio.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Beermann A, Aranda M, Schroder R. The Sp8 zinc-finger transcription factor is involved in allometric growth of the limbs in the beetle Tribolium castaneum. Development. 2004;131:733–42. doi: 10.1242/dev.00974. [DOI] [PubMed] [Google Scholar]
- Bier E. Anti-neural-inhibition: a conserved mechanism for neural induction. Cell. 1997;89:681–4. doi: 10.1016/s0092-8674(00)80250-0. [DOI] [PubMed] [Google Scholar]
- Bolinger RA, Boekhoff-Falk G. Distal-less functions in subdividing the Drosophila thoracic limb primordium. Dev Dyn. 2005;232:801–16. doi: 10.1002/dvdy.20329. [DOI] [PubMed] [Google Scholar]
- Carroll SB, Grenier JK, Weatherbee SD. From DNA to diversity: molecular genetics and the evolution of animal design. Blackwell; Malden, Mass. u.a: 2008. [Google Scholar]
- Cohen SM, Bronner G, Kuttner F, Jurgens G, Jackle H. Distal-less encodes a homoeodomain protein required for limb development in Drosophila. Nature. 1989;338:432–4. doi: 10.1038/338432a0. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- Davis RJ, Shen W, Heanue TA, Mardon G. Mouse Dach, a homologue of Drosophila dachshund, is expressed in the developing retina, brain and limbs. Dev Genes Evol. 1999;209:526–36. doi: 10.1007/s004270050285. [DOI] [PubMed] [Google Scholar]
- Dong PD, Chu J, Panganiban G. Proximodistal domain specification and interactions in developing Drosophila appendages. Development. 2001;128:2365–72. doi: 10.1242/dev.128.12.2365. [DOI] [PubMed] [Google Scholar]
- Estella C, Rieckhof G, Calleja M, Morata G. The role of buttonhead and Sp1 in the development of the ventral imaginal discs of Drosophila. Development. 2003;130:5929–41. doi: 10.1242/dev.00832. [DOI] [PubMed] [Google Scholar]
- Freeman RM, Jr, Wu M, Cordonnier-Pratt MM, Pratt LH, Gruber CE, Smith M, Lander ES, Stange-Thomann N, Lowe CJ, Gerhart J, Kirschner M. cDNA sequences for transcription factors and signaling proteins of the hemichordate Saccoglossus kowalevskii: efficacy of the expressed sequence tag (EST) approach for evolutionary and developmental studies of a new organism. Biol Bull. 2008;214:284–302. doi: 10.2307/25470670. [DOI] [PubMed] [Google Scholar]
- Hammond KL, Hanson IM, Brown AG, Lettice LA, Hill RE. Mammalian and Drosophila dachshund genes are related to the Ski proto-oncogene and are expressed in eye and limb. Mech Dev. 1998;74:121–31. doi: 10.1016/s0925-4773(98)00071-9. [DOI] [PubMed] [Google Scholar]
- Holley SA, Jackson PD, Sasai Y, Lu B, De Robertis EM, Hoffmann FM, Ferguson EL. A conserved system for dorsal-ventral patterning in insects and vertebrates involving sog and chordin. Nature. 1995;376:249–53. doi: 10.1038/376249a0. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Esteban CR, Matsui T, Rodriguez-Leon J, Kato S, Izpisua Belmonte JC. Sp8 and Sp9, two closely related buttonhead-like transcription factors, regulate Fgf8 expression and limb outgrowth in vertebrate embryos. Development. 2004;131:4763–74. doi: 10.1242/dev.01331. [DOI] [PubMed] [Google Scholar]
- Keys DN, Lewis DL, Selegue JE, Pearson BJ, Goodrich LV, Johnson RL, Gates J, Scott MP, Carroll SB. Recruitment of a hedgehog regulatory circuit in butterfly eyespot evolution. Science. 1999;283:532–4. doi: 10.1126/science.283.5401.532. [DOI] [PubMed] [Google Scholar]
- Kojima T. The mechanism of Drosophila leg development along the proximodistal axis. Dev Growth Differ. 2004;46:115–29. doi: 10.1111/j.1440-169X.2004.00735.x. [DOI] [PubMed] [Google Scholar]
- Kosman D, Mizutani CM, Lemons D, Cox WG, McGinnis W, Bier E. Multiplex detection of RNA expression in Drosophila embryos. Science. 2004;305:846. doi: 10.1126/science.1099247. [DOI] [PubMed] [Google Scholar]
- Kraus P, Lufkin T. Dlx homeobox gene control of mammalian limb and craniofacial development. Am J Med Genet A. 2006;140:1366–74. doi: 10.1002/ajmg.a.31252. [DOI] [PubMed] [Google Scholar]
- Lowe CJ, Tagawa K, Humphreys T, Kirschner M, Gerhart J. Hemichordate embryos: procurement, culture, and basic methods. Methods Cell Biol. 2004;74:171–94. doi: 10.1016/s0091-679x(04)74008-x. [DOI] [PubMed] [Google Scholar]
- Lowe CJ, Wu M, Salic A, Evans L, Lander E, Stange-Thomann N, Gruber CE, Gerhart J, Kirschner M. Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell. 2003;113:853–65. doi: 10.1016/s0092-8674(03)00469-0. [DOI] [PubMed] [Google Scholar]
- McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- Mercader N, Leonardo E, Azpiazu N, Serrano A, Morata G, Martinez C, Torres M. Conserved regulation of proximodistal limb axis development by Meis1/Hth. Nature. 1999;402:425–9. doi: 10.1038/46580. [DOI] [PubMed] [Google Scholar]
- Minelli A. Limbs and tail as evolutionarily diverging duplicates of the main body axis. Evol Dev. 2000;2:157–65. doi: 10.1046/j.1525-142x.2000.00054.x. [DOI] [PubMed] [Google Scholar]
- Moczek AP, Rose D, Sewell W, Kesselring BR. Conservation, innovation, and the evolution of horned beetle diversity. Dev Genes Evol. 2006;216:655–65. doi: 10.1007/s00427-006-0087-2. [DOI] [PubMed] [Google Scholar]
- Moczek AP, Rose DJ. Differential recruitment of limb patterning genes during development and diversification of beetle horns. Proc Natl Acad Sci U S A. 2009;106:8992–7. doi: 10.1073/pnas.0809668106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–7. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban G, Irvine SM, Lowe C, Roehl H, Corley LS, Sherbon B, Grenier JK, Fallon JF, Kimble J, Walker M, Wray GA, Swalla BJ, Martindale MQ, Carroll SB. The origin and evolution of animal appendages. Proc Natl Acad Sci U S A. 1997;94:5162–6. doi: 10.1073/pnas.94.10.5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban G, Rubenstein JL. Developmental functions of the Distal-less/Dlx homeobox genes. Development. 2002;129:4371–86. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- Pueyo JI, Couso JP. Parallels between the proximal-distal development of vertebrate and arthropod appendages: homology without an ancestor? Curr Opin Genet Dev. 2005;15:439–46. doi: 10.1016/j.gde.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Schaeper ND, Prpic NM, Wimmer EA. A conserved function of the zinc finger transcription factor Sp8/9 in allometric appendage growth in the milkweed bug Oncopeltus fasciatus. Dev Genes Evol. 2009 doi: 10.1007/s00427-009-0301-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shubin N, Tabin C, Carroll S. Fossils, genes and the evolution of animal limbs. Nature. 1997;388:639–48. doi: 10.1038/41710. [DOI] [PubMed] [Google Scholar]
- Silver SJ, Rebay I. Signaling circuitries in development: insights from the retinal determination gene network. Development. 2005;132:3–13. doi: 10.1242/dev.01539. [DOI] [PubMed] [Google Scholar]
- Tabin C, Carroll S, Panganiban G. Out on a Limb: Parallels in Vertebrate and Invertebrate Limb Patterning and the Origin of Appendages. American Zoologist. 1999;39:650–663. [Google Scholar]
- Valentine JW. On the origin of phyla. University of Chicago Press; Chicago: 2004. [Google Scholar]
- Wu J, Cohen SM. Proximodistal axis formation in the Drosophila leg: subdivision into proximal and distal domains by Homothorax and Distal-less. Development. 1999;126:109–17. doi: 10.1242/dev.126.1.109. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Nassif C, Green P, Hartenstein V. Early neurogenesis of the Drosophila brain. J Comp Neurol. 1996;370:313–29. doi: 10.1002/(SICI)1096-9861(19960701)370:3<313::AID-CNE3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Zakany J, Duboule D. The role of Hox genes during vertebrate limb development. Curr Opin Genet Dev. 2007;17:359–66. doi: 10.1016/j.gde.2007.05.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.