Abstract
Ameloblastin is processed by protease(s) during enamel formation. We tested the hypothesis that MMP-20 (enamelysin) catalyzes the cleavages that generate secretory-stage ameloblastin cleavage products. We isolated a 23-kDa ameloblastin cleavage product from developing enamel and determined its N-terminus sequence. Ameloblastin was stably expressed and secreted from HEK293-H cells, purified, and digested with MMP-20 or Klk4 (kallikrein 4). The digests were analyzed by SDS-PAGE and Western blotting, and cleavage products were characterized by N-terminal sequencing. Six fluorescent peptides were digested with MMP-20 and Klk4 and analyzed by RP-HPLC and by mass spectrometry. MMP-20 cleaved each peptide exactly at the sites corresponding to ameloblastin cleavages catalyzed in vivo. Klk4 cleaved ameloblastin and the fluorescent peptides at sites not observed in vivo, and cleaved at only a single correct site: before Leu171. We conclude that MMP-20 is the enzyme that processes ameloblastin during the secretory stage of amelogenesis, and we present a hypothesis about the sequence of ameloblastin cleavages.
Keywords: proteases, enamel, tooth, fluorescent peptides
Introduction
Dental enamel is the product of ameloblasts and requires the secretion of 3 matrix proteins—amelogenin, ameloblastin, and enamelin—which are hydrolyzed by matrix metalloproteinase 20 (enamelysin, MMP-20) (Bartlett et al., 1996) and kallikrein 4 (Klk4) (Simmer et al., 1998) and removed from the matrix (Smith, 1998). Loss-of-function mouse models have been generated for each matrix constituent, and all exhibit severe enamel defects in the homozygous defective condition (Gibson et al., 2001; Caterina et al., 2002; Fukumoto et al., 2004; Hu et al., 2008; Simmer et al., 2009). These components are individually necessary for proper dental enamel formation, but how they work together to establish the highly organized and mineralized enamel layer is unknown. It is also not known if the intact proteins and/or their cleavage products are important functionally.
Enamel formation progresses through a secretory and a maturation stage. MMP-20 is expressed during the secretory and early-maturation stages, while Klk4 expression starts in the transition between the secretory and maturation stages and continues throughout enamel maturation (Hu et al., 2000; Simmer et al., 2004), although PCR analyses have detected low levels of Klk4 mRNA in enamel organ epithelia dissected from the secretory stage (Nagano et al., 2003). The cleavage of amelogenin during the secretory stage has been well-characterized for the pig (Yamakoshi et al., 1994). Analyses of how MMP-20 and Klk4 cleave amelogenin in vitro indicate that MMP-20 can catalyze all of the main cleavages (Ryu et al., 1999; Nagano et al., 2009), while Klk4 catalyzes other cleavages that are not observed among the accumulated amelogenins in secretory-stage enamel (Ryu et al., 2002).
Intact ameloblastin is a trace component of developing enamel and has never been isolated in vivo. Porcine ameloblastin is modified as it passes through the secretory pathway. The 395-amino-acid secreted protein (Ambn395) is O-glycosylated at Ser86 (Kobayashi et al., 2007) and Thr348 (Yamakoshi et al., 2001), hydroxylated at Pro11 (Hu et al., 1997) and Pro324 (Yamakoshi et al., 2001), and has 4 serines (Ser15, Ser17, Ser209, Ser210) in the required context for phosphorylation by Golgi casein kinase (Brunati et al., 2000). Because of these modifications, recombinant ameloblastin that is similar to the native protein must be expressed and secreted in eukaryotic cells (Zeichner-David et al., 2006; Iwata et al., 2007).
The in vivo pattern of porcine ameloblastin cleavages is not as well-defined as that of amelogenin, but a fair number of cleavage products and cleavage sites have been characterized. The initial cleavages of ameloblastin release 3 products from the ameloblastin N-terminal region. The 17-kDa product extends from Val1 to Arg170, the 15-kDa from Val1 to Gln130, and the 13-kDa from Met32 to Gln130 (Fukae et al., 2006). Cleavage after Leu301 in the C-terminal region generates the 27- and 29-kDa calcium-binding proteins (Fukae and Tanabe, 1987; Murakami et al., 1997) that are subsequently processed after Arg319, Lys379 (Yamakoshi et al., 2001), and Tyr343 (Yamakoshi et al., 2006a), although the final cleavages may occur in later stages. Given the availability of active forms of MMP-20 and Klk4, and knowledge of the sites where ameloblastin is cleaved in vivo, we tested the hypothesis that MMP-20, but not Klk4, can catalyze the cleavages that generate ameloblastin products that accumulate during the secretory stage of amelogenesis, and present a hypothesis about ameloblastin cleavage progression as it may occur in vivo.
Materials & Methods
Protocol Approval
Animal protocols were reviewed and approved by the University of Michigan Institutional Animal Care and Use Committee.
Purification of the 23-kDa Ameloblastin
The 23-kDa ameloblastin cleavage product was isolated from the outer enamel shavings from the unerupted second molars of 6-month-old pigs. The protein was isolated from the neutral extract in the 40 to 65% saturation ammonium sulfate precipitate. The acid-soluble part of the pellet was fractionated by reversed-phase high-performance liquid chromatography (Appendix Fig. 1).
Expression, Purification, and Characterization of Recombinant Ameloblastin (rAmbn)
Recombinant porcine Ambn395 that had previously been transiently expressed as a secreted glycoprotein fused to a C-terminal V5-poly-histidine tag (Iwata et al., 2007) was transfected into HEK293-H cells with Lipofectamine 2000 (Invitrogen Corp., Carlsbad, CA, USA) and selected for stable integration with 0.3 µg/mL blasticidin S HCl (Invitrogen). The highest-expressing HEK293-H line among 54 stable transfectants was identified by Western blot analyses. Secreted rAmbn was isolated from the medium by precipitation with 50% ammonium sulfate, then re-suspended in sodium phosphate buffer and bound to a metal affinity column, eluted with imidazole, desalted, lyophilized, and finally purified by RP-HPLC. To demonstrate that the rpAmbn was O-glycosylated, we incubated it with sialidase and O-glycosidase (QA-Bio, San Mateo, CA, USA) at 37°C for 2 hrs. The sialidase was included because O-glycosidase cleaves only unsubstituted Gal-β(1-3)GalNAc-alpha chains attached to Ser or Thr. The digests were analyzed on SDS-PAGE stained with CBB, Stains-all, Pro-Q Emerald 488 (Molecular Probes, Eugene, OR, USA), and by Western blot analyses with N- and C-terminal-specific ameloblastin antibodies (Appendix Fig. 2A).
Isolation of Klk4 from Mature Porcine Enamel
Klk4 was purified from unerupted 6-month-old pig second molars obtained from the Michigan State University Meat Laboratory (East Lansing, MI, USA) and processed as described previously (Yamakoshi et al., 2006b). Klk4 was isolated from the neutral extract in the 40 to 65% saturation ammonium sulfate precipitate. The serine protease was isolated with benzamidine sepharose 4 Fast Flow beads, followed by RP-HPLC. No protein bands were observed in a Klk4 aliquot on CBB-stained SDS-PAGE, but the Klk4 protease was detected as a doublet at 34 and 36 kDa by Western blotting and gelatin zymography (Appendix Fig. 2B).
Isolation of Recombinant MMP-20 (rMMP-20)
Porcine MMP-20 was expressed from the pProEx1 in bacteria fused to an N-terminal histidine tag (Bartlett et al., 1998). The bacterial lysate was applied to a metal affinity column, washed, eluted, desalted/buffer-exchanged, and activated with 10 mM calcium. The purified rMMP-20 consisted of an active catalytic domain migrating at 22 kDa on a casein zymogram (Appendix Fig. 2C).
Digestion of rAmbn with rMMP-20 and Klk4
Full-length rAmbn was incubated with either rMMP-20 or Klk4 at an approximate enzyme-to-substrate ratio of 1:100 for various time intervals in 10 mM Tris-HCl with 10 mM Ca2+, pH 7.4, at 37°C. Reactions were quenched by the addition of sample buffer (Laemmli Sample Buffer, Bio-Rad, Hercules, CA, USA) and immediate freezing at -80°C.
N-terminal Sequencing
Digests of rAmbn were separated by SDS-PAGE and transblotted to a polyvinylidene fluoride (PVDF) membrane. The membrane was lightly stained with CBB, and selected bands were excised and sent to the W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University for N-terminal sequencing.
Synthetic Ameloblastin Peptides
Fluorescence resonance energy transfer (FRET) peptides containing 11 to 14 amino acids of porcine ameloblastin sequences centering on known ameloblastin cleavage sites were commercially synthesized and purified (JPT Peptide Technologies, Berlin, Germany; Gen Script, Piscataway, NJ, USA). Separation of the fluorescence emitter (Abz) from the quencher (nitroTyr or Lys-Dnp) by enzymatic cleavages induced fluorescence (excitation at 320 nm and emission at 420 nm). Each peptide (100 µg) was suspended in 50 mM Tris, pH 7.4, 10 mM CaCl2, and incubated with rMMP-20 or Klk4 (10-µL aliquot) for 36-48 hrs at 37°C in a total reaction volume of 100 µL. The reactions were stopped by the addition of sample buffer (Laemmli Sample Buffer, Bio-Rad) and freezing at -80°C. Half of each digest was analyzed by RP-HPLC in a C18 column, and the other half was analyzed by LC-MS/MS (NextGen Sciences, Ann Arbor, MI, USA).
Results
A 23-kDa ameloblastin cleavage product was isolated from the outer layer of porcine secretory-stage enamel and shown to have the N-terminal sequence of YGAMFPGFGGMRPNLGGMP PNSAKGGDFxLEFD (Appendix Fig. 1), which demonstrates that ameloblastin is cleaved on the N-terminal side of Tyr223 in vivo. An expression vector engineered to express recombinant porcine ameloblastin was stably integrated into the HEK293-H cell line and used to isolate rAmbn. The recombinant protein stained positive with Emerald 488 and migrated faster on SDS-PAGE following digestion with O-glycosidase (Appendix Fig. 2A). This proves that the recombinant protein is O-glycosylated. Western blot analyses with anti-peptide antibodies against the segments near the N- and C- termini of ameloblastin were positive, indicating that the recombinant protein is full-length and not degraded (Appendix Fig. 2A).
The recombinant Ambn (rAmbn) was incubated with MMP-20 and Klk4 in vitro, and the digestions were analyzed by SDS-PAGE and Western blotting with Ambn63 and Ambn381 antibodies (Fig. 1). An important finding was that rMMP-20 digested rAmbn at multiple sites, but the early cleavages were always on the N-terminal side of the protein (Fig. 1). This can be inferred from the observation that only smaller bands (7- to 25-kDa) were detected by the Ambn63 antibody, while larger products (mainly 45- to 65-kDa) were detected by the Ambn381 antibody. Furthermore, the larger C-terminal products were Stains-all-positive, suggesting that they are calcium-binding.
Figure 1.
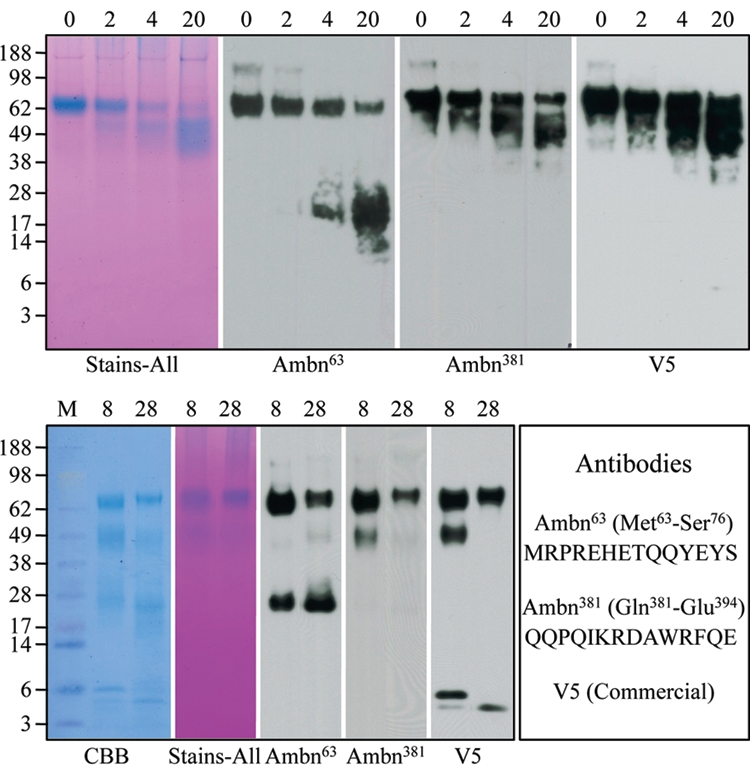
Digestion of rAmbn with rMMP-20 and Klk4. (Top) The rAmbn was digested by MMP-20 for 0, 2, 4, and 20 hrs. From left to right: Samples from these digests were separated by SDS-PAGE and stained with Stains-all, or transferred to a membrane for Western blotting with the primary antibodies Ambn63, Ambn381, or the V5 antibody, which recognizes the extreme C-terminus of the ameloblastin fusion protein. Note that the Ambn63 antibody immunostained the intact protein at ~ 65-kDa and a smear of bands ranging from ~13- to 28-kDa, while the Ambn381 and V5 antibodies recognized the intact protein and a smear of bands ranging from ~45- to 60-kDa. This shows that MMP-20 cleaved rAmbn at multiple sites on the N-terminal side of the protein. (Bottom) The rAmbn was digested by Klk4 for 8 and 28 hrs. From left to right: Samples from these digests were separated by SDS-PAGE and stained with CBB or Stains-all, or transferred to a membrane for Western blotting with the primary antibodies Ambn63, Ambn381, or the V5 antibody. The peptides used to synthesize the Ambn63 and Ambn381 are shown at the right.
The N-terminal sequences of 9 cleavage products from the rMMP-20 digestion and 5 from the Klk4 digestion of rAmbn were determined by N-terminal sequencing (Fig. 2). Five MMP-20 products, ranging in apparent molecular weight from 22- to 37-kDa, gave the ameloblastin N-terminal sequence, 1-VPAFPRQP. The larger MMP-20 cleavage products (48- to 57-kDa) were cleaved on the N-terminal sides of Tyr223, Leu171, and Gln131, respectively. All of these cleavages are known to occur in vivo. Analysis of these data, taken together, suggests that rMMP-20 cleaves rAmbn initially at 3 sites, releasing the following pairs of cleavage products: N-terminus to Ala222/Tyr223 to C-terminus; N-terminus-Arg170/Leu171 to C-terminus; and N-terminus to Gln130/Gln131 to C-terminus. Klk4, in contrast, cleaves rAmbn on the N-terminal sides of Gly29, Ser44, Leu171, Met192, and Phe392. Only the cleavage at Leu171 has been observed in vivo, and MMP-20 also cleaves at this site.
Figure 2.
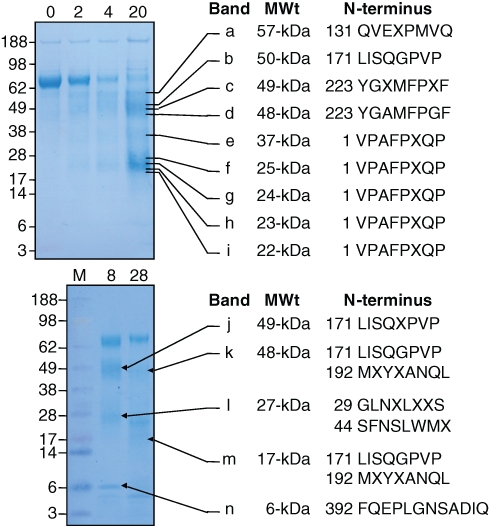
N-terminal sequencing of rAmbn digestion products. (Top) The rAmbn was digested by MMP-20 for 0, 2, 4, and 20 hrs, and samples from these digests were separated by SDS-PAGE and stained with CBB, or transferred to a membrane, which was lightly stained with CBB, and 9 bands (a through i) were excised. The apparent molecular weight and N-terminal sequence of each band are indicated. Bands f, h, and i contained the sequences indicated, starting with Val1, as well as sequences starting with Ala3. It is not known if MMP-20 makes the cleavage before Ala3 or if the signal peptidase alternatively cleaves after Val1 and Ala3. (Bottom) The rAmbn was digested by Klk4 for 8 and 28 hrs, and samples from these digests were separated by SDS-PAGE and stained with CBB, or transferred to a membrane, which was lightly stained with CBB, and 5 bands (j through n) were excised. The N-terminal sequences of the bands are indicated. Bands k, l, and m gave 2 sequences.
Klk4 and rMMP-20 were incubated with 6 FRET-Ambn synthetic peptides that covered the cleavage sites best characterized from in vivo studies. The products were analyzed by HPLC and by mass spectroscopy (Fig. 3). The HPLC profiles show that both enzymes were able to digest all of the peptides, with the lone exception that Klk4 did not digest the 15-kDa peptide centering on Gln131. Mass spectrometry identified the sites cleaved within the peptides. MMP-20 cleaved all 6 peptides at the exact sites that have been observed on ameloblastin cleavage products isolated in vivo. Klk4 cleaved at other sites, with the lone exception that Klk4 cleaved on the N-terminal side of Leu171 on the 17-kDa peptide. A summary of these findings is shown in Fig. 4.
Figure 3.

FRET peptide cleavage sites identified by mass spectrometry. Arrowheads above each peptide sequence indicate sites cleaved by MMP-20. Arrowheads below each peptide sequence indicate sites cleaved by Klk4. MMP-20 cleaved each peptide exactly at the sites corresponding to ameloblastin cleavages catalyzed in vivo: on the N-terminal sides of Met32, Gln131, Leu171, Tyr223, Leu301, and Tyr343.
Figure 4.
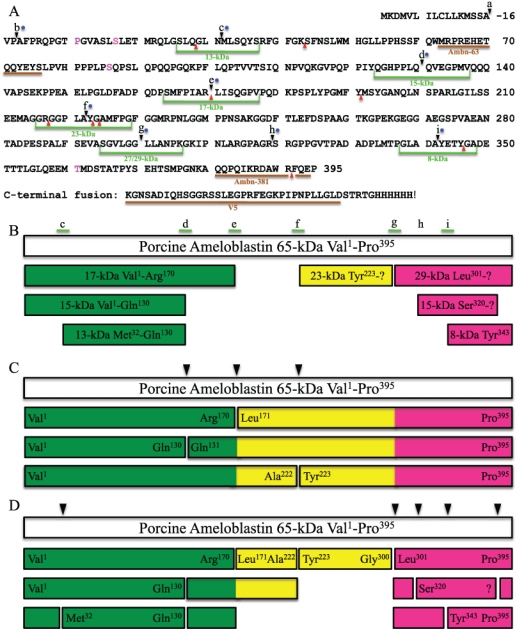
Ambn395 sequence and cleavage sites. (A) Sites where ameloblastin is cleaved in vivo are lettered: (a) Val1, signal peptidase cleavage site (Uchida et al., 1995); (b) Ala3, possible alternative signal peptide cleavage site or MMP-20 site (Iwata et al., 2007); (c) Met31, N-terminus of the 13-kDa (Hu et al., 1997); (d) Gln131, C-terminus of the 13- and 15-kDa (Hu et al., 1997); (e) Leu171, C-terminus of the 17-kDa (Fukae et al., 2006); (f) Tyr223 (this report); (g) Leu301, N-terminus of 27- and 29-kDa calcium-binding proteins (Murakami et al., 1997); (h) N-terminus of the 13- and 15-kDa calcium-binding proteins (Yamakoshi et al., 2001); and (i) N-terminus of an 8-kDa peptide (Yamakoshi et al., 2006a). Green lines indicate synthetic peptides used in this study. Blue dots indicate sites shown to be cleaved by MMP-20 in vitro. Red arrowheads indicate sites shown to be cleaved by Klk4 in vitro. Brown lines indicate antigen-binding sites for antibodies used in this study. (B) Porcine ameloblastin cleavage products that have been isolated and characterized in vivo. White bar shows the porcine ameloblastin 65-kDa protein; colored bars are cleavage products. Colors in bars indicate the N-terminal region (Val1 to Arg170; green), middle (Leu171 to Gly300; yellow), and C-terminal region (Leu301 to Pro395; mauve). (C) Initial cleavage products based upon the digestion of rAmbn by MMP-20 in vitro. Arrowheads indicate initial cleavage sites. (D) Secondary MMP-20 cleavages (arrowheads) continue the processing of ameloblastin to yield additional cleavage products. Labeled ends of bars have been confirmed experimentally; unlabeled bars are generated by multiple MMP-20 cleavages, but have not been confirmed experimentally.
Discussion
We identified the cleavage site that generates the N-terminus of the 23-kDa porcine ameloblastin cleavage product, and have developed a stable cell line that expresses and secretes full-length, glycosylated ameloblastin. N-terminal sequencing of rAmbn cleavage products generated by MMP-20 demonstrated that MMP-20 cleaves rAmbn at the same sites as have been shown to occur in vivo. Mass spectrometric analyses of 6 FRET peptides digested by MMP-20 and Klk4 showed that MMP-20 specifically catalyzed all of the cleavages that have been identified by the characterization of ameloblastin cleavage products isolated from developing teeth, and that Klk4 can catalyze only one of these cleavages. This bolsters the conclusion that MMP-20, but not Klk4, catalyzes the processing of enamel proteins during the secretory stage of amelogenesis.
Recombinant MMP-20 cleaved rAmbn starting on the N-terminal side of the protein, so that the N-terminal cleavage products are smaller than the C-terminal cleavage products, which mimics the in vivo pattern (Murakami et al., 1997; Uchida et al., 1997). Based upon what can be deduced from in vivo and in vitro analyses, we propose that Ambn395 is digested by MMP-20 initially at one of three sites—after Gln130, Arg170, and Ala222—generating 6 cleavage products. These initial products are then cleaved a second or third time at these same sites, as well as at secondary sites that are located mostly near the C-terminus, after Gly300, Arg319, Ala342, and Asn31. The end result is that N-terminal ameloblastin cleavage products (13-, 15-, and 17-kDa) accumulate in the sheath space throughout the enamel layer (Uchida et al., 1991), while calcium-binding ameloblastin C-terminal cleavage products (13- and 15-kDa) (Yamakoshi et al., 2001) are in the rods and are not detectable beyond a depth of 50 µm (Murakami et al., 1997). The cleavage pattern is consistent with an ab initio 3D model of human ameloblastin that portrays ameloblastin as having N- and C-terminal domains that are connected by an unstructured linker that is susceptible to degradation (Vymetal et al., 2008).
The functions of secreted ameloblastin and its MMP-20 cleavage products are not known. Some reports suggest that ameloblastin is important for cell adhesion (Cerny et al., 1996; Fukumoto et al., 2004), possibly by binding heparin (Sonoda et al., 2009) or fibronectin (Beyeler et al., 2010). The evidence, however, is not strong. In the ameloblastin mutant mice, the enamel surface is so pathological that cell attachment to it might fail even if ameloblastin was not part of the attachment apparatus. Heparin and fibronectin have not been localized along the distal membrane of secretory-stage ameloblasts, and fibronectin was not among the secreted and membrane proteins identified in the rat incisor enamel organ by signal-trap screening (Moffatt et al., 2006). The putative ameloblastin cell-binding motifs (VTKG and KRH-rich motifs for heparin and VPIMDFADPQFPT for fibronectin) are not well-conserved among species. Glycosylated rAmbn showed only weak cell adhesion properties equal to those of amelogenin (Zeichner-David et al., 2006). Others have noted that ameloblastin has growth-factor-like properties (Fukae et al., 2006; Zeichner-David et al., 2006). Future studies with glycosylated ameloblastin isolated from stable cell lines may help clarify the function of ameloblastin.
Supplementary Material
Acknowledgments
We thank Mr. Tom Forton, Manager of the Michigan State University Meat Laboratory, for assistance in obtaining fresh developing molars from pigs slaughtered at that facility; Dr. Myron Crawford, Director of the W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University, and Nancy Williams for protein sequencing; and David Allen of NextGen Sciences, Inc., for mass spectrometry.
Footnotes
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
This research was supported by USPHS Research Grants DE015846 and DE0 16276 (NIDCR/NIH).
All authors declare that there are no conflicting interests.
References
- Bartlett JD, Simmer JP, Xue J, Margolis HC, Moreno EC. (1996). Molecular cloning and mRNA tissue distribution of a novel matrix metalloproteinase isolated from porcine enamel organ. Gene 183:123-128 [DOI] [PubMed] [Google Scholar]
- Bartlett JD, Ryu OH, Xue J, Simmer JP, Margolis HC. (1998). Enamelysin mRNA displays a developmentally defined pattern of expression and encodes a protein which degrades amelogenin. Connect Tissue Res 39:101-109 [DOI] [PubMed] [Google Scholar]
- Beyeler M, Schild C, Lutz R, Chiquet M, Trueb B. (2010). Identification of a fibronectin interaction site in the extracellular matrix protein ameloblastin. Exp Cell Res [Epub ahead of print], January 4, 2010 (in press) [DOI] [PubMed] [Google Scholar]
- Brunati AM, Marin O, Bisinella A, Salviati A, Pinna LA. (2000). Novel consensus sequence for the Golgi apparatus casein kinase, revealed using proline-rich protein-1 (PRP1)-derived peptide substrates. Biochem J 351(Pt 3):765-768 [PMC free article] [PubMed] [Google Scholar]
- Caterina JJ, Skobe Z, Shi J, Ding Y, Simmer JP, Birkedal-Hansen H, et al. (2002). Enamelysin (matrix metalloproteinase 20)-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem 277:49598-49604 [DOI] [PubMed] [Google Scholar]
- Cerny R, Slaby I, Hammarström L, Wurtz T. (1996). A novel gene expressed in rat ameloblasts codes for proteins with cell binding domains. J Bone Miner Res 11:883-891 [DOI] [PubMed] [Google Scholar]
- Fukae M, Tanabe T. (1987). 45Ca-labeled proteins found in porcine developing dental enamel at an early stage of development. Adv Dent Res 1:261-266 [DOI] [PubMed] [Google Scholar]
- Fukae M, Kanazashi M, Nagano T, Tanabe T, Oida S, Gomi K. (2006). Porcine sheath proteins show periodontal ligament regeneration activity. Eur J Oral Sci 114(Suppl 1):212-218 [DOI] [PubMed] [Google Scholar]
- Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G, et al. (2004). Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol 167:973-983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, et al. (2001). Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem 276:31871-31875 [DOI] [PubMed] [Google Scholar]
- Hu CC, Fukae M, Uchida T, Qian Q, Zhang CH, Ryu OH, et al. (1997). Sheathlin: cloning, cDNA/polypeptide sequences, and immunolocalization of porcine enamel sheath proteins. J Dent Res 76:648-657 [DOI] [PubMed] [Google Scholar]
- Hu JC, Zhang C, Sun X, Yang Y, Cao X, Ryu O, et al. (2000). Characterization of the mouse and human PRSS17 genes, their relationship to other serine proteases, and the expression of PRSS17 in developing mouse incisors. Gene 251:1-8 [DOI] [PubMed] [Google Scholar]
- Hu JC, Hu Y, Smith CE, McKee MD, Wright JT, Yamakoshi Y, et al. (2008). Enamel defects and ameloblast-specific expression in Enam knock-out/lacz knock-in mice. J Biol Chem 283:10858-10871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata T, Yamakoshi Y, Hu JC, Ishikawa I, Bartlett JD, Krebsbach PH, et al. (2007). Processing of ameloblastin by MMP-20. J Dent Res 86:153-157 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Yamakoshi Y, Hu JC, Gomi K, Arai T, Fukae M, et al. (2007). Splicing determines the glycosylation state of ameloblastin. J Dent Res 86:962-967 [DOI] [PubMed] [Google Scholar]
- Moffatt P, Smith CE, Sooknanan R, St-Arnaud R, Nanci A. (2006). Identification of secreted and membrane proteins in the rat incisor enamel organ using a signal-trap screening approach. Eur J Oral Sci 114(Suppl 1):139-146 [DOI] [PubMed] [Google Scholar]
- Murakami C, Dohi N, Fukae M, Tanabe T, Yamakoshi Y, Wakida K, et al. (1997). Immunochemical and immunohistochemical study of 27 and 29 kDa calcium binding proteins and related proteins in the porcine tooth germ. Histochem Cell Biol 107:485-494 [DOI] [PubMed] [Google Scholar]
- Nagano T, Oida S, Ando H, Gomi K, Arai T, Fukae M. (2003). Relative levels of mRNA encoding enamel proteins in enamel organ epithelia and odontoblasts. J Dent Res 82:982-986 [DOI] [PubMed] [Google Scholar]
- Nagano T, Kakegawa A, Yamakoshi Y, Tsuchiya S, Hu JC, Gomi K, et al. (2009). MMP-20 and Klk4 cleavage site preferences for amelogenin sequences. J Dent Res 88:823-828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu O, Hu JC, Yamakoshi Y, Villemain JL, Cao X, Zhang C, et al. (2002). Porcine kallikrein-4 activation, glycosylation, activity, and expression in prokaryotic and eukaryotic hosts. Eur J Oral Sci 110:358-365 [DOI] [PubMed] [Google Scholar]
- Ryu OH, Fincham AG, Hu CC, Zhang C, Qian Q, Bartlett JD, et al. (1999). Characterization of recombinant pig enamelysin activity and cleavage of recombinant pig and mouse amelogenins. J Dent Res 78:743-750 [DOI] [PubMed] [Google Scholar]
- Simmer JP, Fukae M, Tanabe T, Yamakoshi Y, Uchida T, Xue J, et al. (1998). Purification, characterization, and cloning of enamel matrix serine proteinase 1. J Dent Res 77:377-386 [DOI] [PubMed] [Google Scholar]
- Simmer JP, Sun X, Yamada Y, Zhang CH, Bartlett JD, Hu JC-C. (2004). Enamelysin and kallikrein-4 expression in the mouse incisor. In: Biomineralization: formation, diversity, evolution and application. Proceedings of the 8th International Symposium on Biomineralization, Niigata, Japan, Sept 25-28, 2001 Kobayashi I, Ozawa H, editors. Hadano, Japan: Tokai University Press, pp. 348-352 [Google Scholar]
- Simmer JP, Hu Y, Lertlam R, Yamakoshi Y, Hu JC. (2009). Hypomaturation enamel defects in Klk4 knockout/LacZ knockin mice. J Biol Chem 284:19110-19121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE. (1998). Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med 9:128-161 [DOI] [PubMed] [Google Scholar]
- Sonoda A, Iwamoto T, Nakamura T, Fukumoto E, Yoshizaki K, Yamada A, et al. (2009). Critical role of heparin binding domains of ameloblastin for dental epithelium cell adhesion and ameloblastoma proliferation. J Biol Chem 284:27176-27184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida T, Tanabe T, Fukae M, Shimizu M, Yamada M, Miake K, et al. (1991). Immunochemical and immunohistochemical studies, using antisera against porcine 25 kDa amelogenin, 89 kDa enamelin and the 13-17 kDa nonamelogenins, on immature enamel of the pig and rat. Histochemistry 96:129-138 [DOI] [PubMed] [Google Scholar]
- Uchida T, Fukae M, Tanabe T, Yamakoshi Y, Satoda T, Murakami C, et al. (1995). Immunochemical and immunocytochemical study of a 15 kDa non-amelogenin and related proteins in the porcine immature enamel: proposal of a new group of enamel proteins sheath proteins. Biomed Res 16:131-140 [Google Scholar]
- Uchida T, Murakami C, Dohi N, Wakida K, Satoda T, Takahashi O. (1997). Synthesis, secretion, degradation and fate of ameloblastin during the matrix formation stage of the rat incisor as shown by immunocytochemistry and immunochemistry using region-specific antibodies. J Histochem Cytochem 45:1329-1340 [DOI] [PubMed] [Google Scholar]
- Vymetal J, Slaby I, Spahr A, Vondrasek J, Lyngstadaas SP. (2008). Bioinformatic analysis and molecular modelling of human ameloblastin suggest a two-domain intrinsically unstructured calcium-binding protein. Eur J Oral Sci 116:124-134 [DOI] [PubMed] [Google Scholar]
- Yamakoshi Y, Tanabe T, Fukae M, Shimizu M. (1994). Porcine amelogenins. Calcif Tissue Int 54:69-75 [DOI] [PubMed] [Google Scholar]
- Yamakoshi Y, Tanabe T, Oida S, Hu CC, Simmer JP, Fukae M. (2001). Calcium binding of enamel proteins and their derivatives with emphasis on the calcium-binding domain of porcine sheathlin. Arch Oral Biol 46:1005-1014 [DOI] [PubMed] [Google Scholar]
- Yamakoshi Y, Hu JC-C, Fukae M, Iwata T, Simmer JP. (2006a). How do MMP-20 and KLK4 process the 32 kDa enamelin? Eur J Oral Sci 114(Suppl 1):45-51 [DOI] [PubMed] [Google Scholar]
- Yamakoshi Y, Hu JC, Zhang H, Iwata T, Yamakoshi F, Simmer JP. (2006b). Proteomic analysis of enamel matrix using a two-dimensional protein fractionation system. Eur J Oral Sci 114(Suppl 1):266-271 [DOI] [PubMed] [Google Scholar]
- Zeichner-David M, Chen LS, Hsu Z, Reyna J, Caton J, Bringas P. (2006). Amelogenin and ameloblastin show growth-factor like activity in periodontal ligament cells. Eur J Oral Sci 114(Suppl 1):244-253 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


