Abstract
Research involving bacterial pathogens often requires enumeration of bacteria colonies. Here we present a low-cost, high-throughput colony counting system consisting of colony counting software and a consumer-grade digital camera or document scanner. We demonstrate that this software, called “NICE” (NIST's Integrated Colony Enumerator), can count bacterial colonies as part of a high-throughput multiplexed opsonophagocytic killing assay (MOPA) used to characterize pneumococcal vaccine efficacy. The results obtained with NICE correlate well with the results obtained from manual counting, with a mean difference of less than 3 %. NICE is also rapid; it can count colonies from multiple reaction wells within minutes and export the results to a spreadsheet for data processing. As this program is freely available from NIST, NICE should be helpful in bacteria colony enumeration required in many microbiological studies, and in standardizing colony counting methods.
Keywords: Colony counting, pneumoniae, bacteria, vaccine, software, imaging, MOPA
Introduction
Accurate colony counting of bacteria is crucial for quantitative, precise assessment of pathogens in clinical research and diagnosis. While manual counting remains the gold standard, this tedious process is difficult to implement for high throughput assays due to low speed and inconsistent results. Automation of colony counting has been of increasing interest for many decades (1-3), and these methods have been shown to be more consistent than manual counting (1,4). Herman et al. demonstrated that automated colony counts had significantly less variation when reanalyzing plates than those manually determined by individual or multiple observers (5). Similarly, Lumley et al. attributed the significant disparity in colony forming unit granulocyte-macrophage assay results across laboratories to differences in colony counting (6). Commercial products exist to facilitate accurate colony counting, ranging from manual counting aids (e.g., counting pens) to all-in-one platforms including image acquisition, processing, and analysis. Yet, fully automated counting systems also capable of batch processing multiple images at once can be prohibitively expensive for small labs and large facilities may necessitate multiple counting instruments. For these reasons, alternatives to commercial products using consumer-grade equipment, such as document scanners (7-13) or digital cameras (11,14-17), have been explored.
Colony counting can be affected by numerous parameters related to the physical properties of the colony: size, shape, contrast, and overlapping colonies. Common pitfalls in colony identification, such as separation of clustered colonies, minimum colony sizing and contrast, and analysis near the plate periphery, have been widely discussed (2,4,8). Determinations of overlapping colonies have been deduced by shape analysis (10), object-recognition using the Fuzzy formalism (8), and principal components analysis (12). Alternatively, instrumental techniques can be used to improve the contrast and separation of colonies, such as using the colony to act as a lens (2) or reflective surface (18). Rigorous standardization of the colony counting procedure based on imaging techniques requires full assessment of the optical response performance (e.g., background noise, contrast, resolution, etc.) of the image acquisition tools. Furthermore, any quantitative image analysis software must be based on a flexible algorithm with tunable parameters to optimize the results. When considering automated colony counting, or manual counting from a digital image, the process in which the image is acquired introduces other important factors: resolution, file size/data management, sample lighting, and instrument uniformity. Counting of bacterial colonies has been of increasing importance in recent efforts in the determination of the vaccine efficacy against Streptococcus pneumoniae, a leading cause of infectious disease in children and the elderly (19,20). There is a continued need for the expansion and modification of the vaccines to account for the changing prevalence of pneumococcal serotypes (21). Changing the constituents of the vaccines requires that the new formulation be proven as effective as a previous formulation by characterization of the antibody response.
The opsonophagocytic killing assay (OPA) is a widely accepted method for the quantitation of functional antibodies against S. pneumonia (22). Unlike the enzyme-linked immunosorbent assay (ELISA), which measures both functional and nonfunctional antibodies (23), the OPA directly measures antibodies that both coat the surface of the bacteria (opsonization) and induce the subsequent uptake and killing (phagocytosis) of pneumococcal strains. A high-throughput multiplexed opsonophagocytic killing assay (MOPA) was developed to reduce serum requirements by allowing multiple serotypes to be measured in parallel (24,25). Analysis by MOPA requires the counting of bacterial colonies within multiple spots on a Petri dish. For the analysis of multiple dishes, manual counting of each plate is not feasible due to the sheer number of plates involved. Therefore, a significant need exists for colony counting techniques that are amenable to the MOPA format.
When implementing a colony counting protocol for MOPA using optical imaging techniques, the complete process of sample handling, image acquisition and analysis must be carefully considered. Since the ideal image acquisition and analysis parameters may differ across various species of bacteria, we will tailor our initial analytical methods for the pneumococcal colonies observed in MOPA. Intensive algorithms involving distance transforms (4,13), model-fitting (26), or user-guided watershed algorithms (27) are currently non-ideal for measuring the several thousand colonies per assay plate that may exist in MOPA. Here, we focus on developing simple image acquisition methods that can be readily assembled and provide a user-friendly image processing program designed for the MOPA platform. Precise colony enumeration within 10 % is vital, but does not encompass the entirety of the requirements. High throughput analysis is critical; users must be able to image plates quickly, and image analysis should be performed with minimal intervention. We detail the validation of document scanner- and digital camera-based colony counting protocols. Additionally, we describe and make a publicly available colony counting software package, NIST's Integrated Colony Enumerator (NICE).
Materials and Methods
Preparation of agar plates containing bacterial colonies
As an initial model system, colonies of non-pathogenic Escherichia coli (strain DH5α) were used. Various dilutions of E. coli were prepared in Hanks' Balanced Salt Solution, and 10 μL of the diluted bacteria were spotted onto multiple areas of an agar plate (Todd-Hewitt broth containing 0.5 % yeast extract and 1.5 % agar). After absorption, an agar overlay (Todd-Hewitt broth containing 0.5 % yeast extract and 0.75 % agar, plate size is 10 cm2) containing 2,3,5,-triphenyltetrazolium chloride (TTC), a dye indicative of cellular metabolism that stains the colonies deep red with no coloration of the agar (24,28), was added. These E. coli colony samples were used for the initial development and adjustment of the image acquisition protocol and for initial testing of the image analysis software.
Later development and characterization of NICE utilized plates containing pneumococcal colonies from the MOPA. The MOPA protocol has been described previously (24,25,29,30) Briefly, HL-60 cells, test serum, baby rabbit complement and pneumococcal target strains are mixed. After incubation, 10 μL of the final reaction mixture from multiple wells are spotted onto agar plates (same as above). Plates are then overlaid with agar (same as above) containing TTC, incubated overnight, imaged, and the colonies enumerated.
Image acquisition
Two methods were used to acquire images of agar plates. In the first, plates were placed on a scanner (HP Scanjet G4010, Hewlett-Packard, Palo Alto, CA), covered by soft gloss paper (HP LaserJet soft gloss paper), and 8-bit grayscale images of the plates were obtained at 300 dpi (dots per inch; ≈118 dots per cm), 600 dpi (≈236 dots per cm) and 1200 dpi (≈472 dots per cm). In the second approach, images were acquired with two digital cameras: a Sony Cyber-shot DSC-H2 (6 megapixel) or a Canon PowerShot SX110 IS (9 megapixel). A camera was mounted on an imaging stand (Mini Repo, Industria Fototechnica Firenze, Calenzano, Italy) with the sample placed on top of an illuminator (Mighty Bright Visible, Hoefer Scientific Instruments, San Francisco, CA) as shown in Figure 1.a. The distance from the camera to the sample was approximately 15 cm for 24-spot plates. The shutter speed, aperture and focus were operated manually. Optimal settings were shutter speed of 1/100 s, f4.0, and ISO setting of 80. Images were acquired at the maximum resolution of the camera with no zoom.
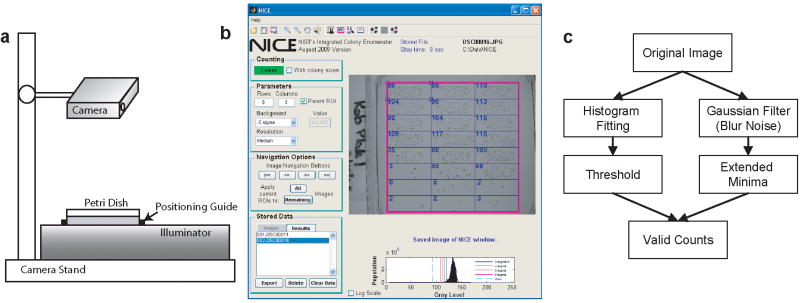
Figure 1.
a) A schematic of an imaging station using a digital camera. The camera was positioned so that the Petri dish barely filled the view field. A positioning guide enabled fast placement of plates. b) Example of the NICE after an image. Displayed is an image with 24 ROIs with minor position adjustments. Counts for each ROI are displayed in blue. c) General procedure used to count colonies in NICE. The threshold is calculated dynamically for each image. Minima are compared with this threshold to determine their validity.
Software
NICE was programmed in MATLAB (Mathworks, Natick, MA), and has been compiled to run using the distributable MATLAB compiler engine. Details regarding software operation are extensively described in the NICE manual (ftp://ftp.nist.gov/pub/physics/mlclarke/NICE). Here, we provide a brief synopsis. The main window of NICE is presented in Figure 1.b. Users can load a single image or a batch of images. Images imported into the software will be converted to grayscale (using the image luminance). Modifications to the counting algorithm can be performed by changing the threshold or changing the resolution function (degree of smoothing and expected minima size). The latter is designed to optimize the counting algorithm for scanned images of high (1200 dpi) to low resolution (300 dpi). A grid of analysis regions of interest (ROIs) is created by entering the desired row and column dimension (e.g., 8 × 3 for a plate containing 24 spots). The user can define a parent ROI, which will confine image analysis to a subregion of the image. ROIs can be moved and resized with ease, and it is possible to scroll between all of the loaded images. All images can then be counted without further user intervention. Once counting has completed, the user can review each counted image if desired, and export the results to a text or Microsoft Excel file.
NICE operates by combining extended minima and thresholding algorithms (Figure 1.c). A Gaussian smoothing function is applied to the image to remove pixel noise prior to minima analysis. The extended minima function is used to find the center of the colonies which can be viewed as regional dark (low) points. Choosing this regional method allows for colonies to be identified based on the local contrast, and therefore is not significantly influenced by contrast changes due to artifacts such as shadows. Additionally, the minima function can also distinguish touching colonies where the darker colony centers are still separable.
Thresholding is performed by automatic analysis of the image. Signal from the background (agar) dominates the image histogram. This signal is fitted to a Gaussian distribution function and guides the threshold choice. The Gaussian center minus 3 to 6 standard deviations (σ) are selectable thresholds. Alternatively the Otsu threshold function (31) or a manually entered value can be used as the threshold. Automated thresholding allows for plates with varied contrast due to changes in lighting or agar thickness to be measured with minimal user intervention. The identified minima are slightly dilated and the mean intensity within the minima of each colony is compared against the threshold. If the mean is below the threshold it is considered to be a valid colony. Colonies vary dramatically in size depending on the colony density and other growth conditions. For this reason, size was not considered as a selecting factor for colony analysis. This allows for a greater variety of colonies to be analyzed within single plates and among plates in batch processing.
Results
TTC and Contrast
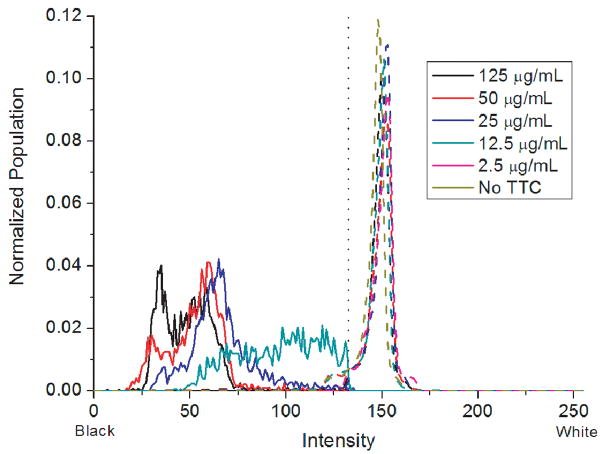
Any protocol for colony counting by staining must consider the impact of dye concentration on the final image contrast. Plates with and without E. coli were stained using TTC concentrations ranging from 0 to 125 μg/mL. These plates were scanned at 1200 dpi and further analyzed. Histograms of the colony intensities (determined from the mean of the extended minima for each colony) were calculated and compared to the histogram of the entire plate (Figure 2). Corroborating the visual inspection, the histogram confirms that TTC concentration has no effect on the background intensity. TTC concentrations below 12.5 μg/mL produced insufficient contrast for automated counting and were not studied further. Contrast further improves until 50 μg/mL TTC at which point more dye does not improve the image. By this analysis we conclude that a TTC concentration of 25 μg/mL is sufficient to produce an image with high contrast between the colonies and the agar.
Figure 2.
Histograms of the mean intensities of colony minima (solid lines) and histogram of the entire plate (dashed lines) as a function of TTC concentration. The threshold line of the Gaussian center minus 5σ is displayed as a dotted line. Colonies stained with 2.5 μg/mL TTC or no TTC did not possess a significant contrast and are not presented here for clarity. A minimum TTC concentration of 25 μg/mL is required for high contrast between the background and colonies. TTC concentration has no significant effect on the histogram of the background intensities.
Scanner validation
Scanner image uniformity was evaluated by scanning a standard reflectance plaque (Avian Technologies, Sunapee NH, data not shown). The image area displayed exceptional uniformity (0.12 % relative standard deviation). There is higher deviation (15 %) at the vertical edges due to diffraction, rendering about 2 mm of either side non-ideal for colony measurements. These deviations will have negligible effect on colony counting since this deviation region will be occupied by the plate walls. To ensure that local measured intensities (e.g., colonies) are not affected by the nearby background intensity, we scanned a reference density step chart (Edmund Optics, Barrington, NJ) surrounded by either white photo paper or black plastic, corresponding to the scanner's maximum and minimum gray levels, respectively. While the effective luminance is higher when white paper is used, the measured gray level step differences are unaffected. As a final contrast test, we cut out and placed pieces of these grayscale charts on agar plates as mock colonies. These “colonies” retain the same grayscale intensity regardless of their location on the agar or the agar TTC concentration.
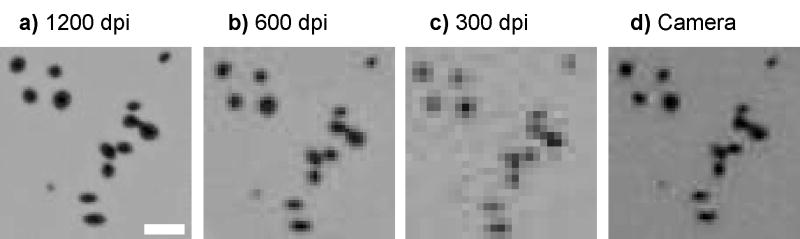
Scanner image dimensions were checked to confirm that the dpi rating matched the physical size. For all resolutions, back-calculated image sizes were within ≈1 % deviation from the actual image sizes, and this deviation is considered to be a negligible factor for colony counting and sizing. Colony sizes vary greatly, and in the MOPA numerous small colonies are observed. Typical colony sizes observed in Figure 3 range from 0.2 mm to 0.4 mm in diameter, but larger and smaller colonies, 0.1 mm to 0.7 mm in diameter, were also observed. Colony sizes determined by the scanner were compared to those found using a 40× magnification confocal laser scanner. The results were similar with the flatbed scanner reporting slightly smaller colony sizes (≈8 % smaller).
Figure 3.
Colony images obtained with a scanner at different resolutions (1200 dpi to 300 dpi). Differences in the signal-to-noise level are readily apparent. Small colonies in the 300 dpi image are difficult to discern. Both the 600 dpi and 1200 dpi images are able to distinguish all the colonies, though these colonies are sharper in the 1200 dpi image. Two digital cameras (9 megapixel shown here) result in similar quality images to 600 dpi resoltuion. Scale bar is 0.5 mm.
Digital Camera Validation
A typical digital camera setup (Figure 1.a) produces higher spatial variance than the document scanner. This is caused by any number of factors such as illuminated field non-uniformity, lens distortion, difference in planar angles between the illuminated plate and the camera, and sources of stray light from ambient lighting. Imaging plates on a backlit illuminator box with dim or no overhead lighting eliminates shadows and glares. A typical field-illuminator results in approximately 5 % standard deviation in intentsity over the imaging area. Illuminating the sample from the same side as the camera, i.e., in reflection mode, is inadvisable due to severe degradation of the quality of the imaging field. Glare or shadow significantly changes both the dynamic range of the image contrast and the background intensity profile, causing difficulties in precisely determining the threshold intensity. In this case, the background intensity histogram exhibits multiple peaks due to the heterogeneous background with regions of shadow or glare. The Gaussian fitting of the background intensity histogram performs poorly and the resultant fit broadens from a full-width at half max of 12 to 28 in the 256-level grayscale. Samples imaged using overhead lighting resulted in ≈11 % fewer counted colonies compared to the sample image using a field-illuminator (n=24).
Image Acquisition Factors
We scanned the plates at different resolutions (75 dpi to 1200 dpi), and took images using the digital cameras (Figure 3). Examining the quality of the images at these different resolutions illustrates the necessity for high resolution imaging of the small colonies. The digital camera images are similar in quality to the 600 dpi resolution scan. The time to scan an image of a 24-spot plate is shown in Table 1. Compared to the 600 dpi image, the contrast in the 300 dpi image is severely reduced. While it may be possible to count the majority of colonies present, the result will be highly dependent on setting the parameters for the counting algorithm, more so than for the higher resolution (600 dpi or 1200 dpi) samples. The contrast of the 600 dpi image should be sufficient for accurate counting of colonies in this study. Despite the clearer image at 1200 dpi, the longer acquisition time, larger file size and increased processing time become significant barriers to high throughput analyses.
Table 1.
Acquisition and analysis parameters of the document scanner and camera (9 megapixel model) for images of a 24-spot plate. Time does not include other acquisition steps (e.g., sample placement, initial scan to find plate location, focusing).
| Resolution (DPI) | Acquisition Time (s) | Analysis Time (s) | File Size (Mb) |
|---|---|---|---|
| 300 | 17 | 3 | 0.5 |
| 600 | 40 | 8 | 1.7 |
| 1200 | 126 | 24 | 6.4 |
| Camera | < 1 | 12 | 2.9 |
We then compared the counting results under different resolutions and analyzed the sources of discrepancy. To this end, the same plates were scanned at 300 dpi, 600 dpi, and 1200 dpi and the results were analyzed. The 1200 dpi scan was considered to represent the true count and sizing, and the counts from this high resolution image were compared to the counts of the 600 dpi and 300 dpi scans. For one 24-spot plate with a high CFU/spot (137 CFU/spot on average) a total of 3311 colonies were found with a mean colony area of 0.054 ± 0.024 mm2 at the 1200 dpi resolution. When the resolution was lowered the number of colonies decreased. We consider there to be two sources of undercounting: exclusion and combination. Exclusion refers to a colony that simply is not counted. Its size and contrast have become insufficient for detection. Combination is the merging of two distinct colonies (as determined by the high resolution image) into one colony in the lower resolution image causing the reduction in total counted colonies. Comparing the 1200 dpi and 300 dpi resolution images, we determine the number of colonies lost from 300 dpi images to exclusion and combination are 393 colonies (11.9 %) and 63 colonies (1.9 %), respectively. The sizes of colonies missed due to exclusion are typically smaller (mean colony area of 0.033 mm2 ± 0.013 mm2) than those of combined colonies (mean colony area of 0.045 mm2 ± 0.011 mm2). In the analysis of 600 dpi images, far fewer colonies are excluded (38 colonies, 1.1 %) or combined (8 colonies, 0.2 %). The colony density has a significant impact on this resolution-dependent counting. When scanning low density colony spots (≈6 CFU/spot), there were no counting differences among the results from the three different resolutions (131 colonies total). Colonies tend to grow to a larger size (0.12 mm2 ± 0.03 mm2) and are more dispersed in such low density spots.
NICE performance
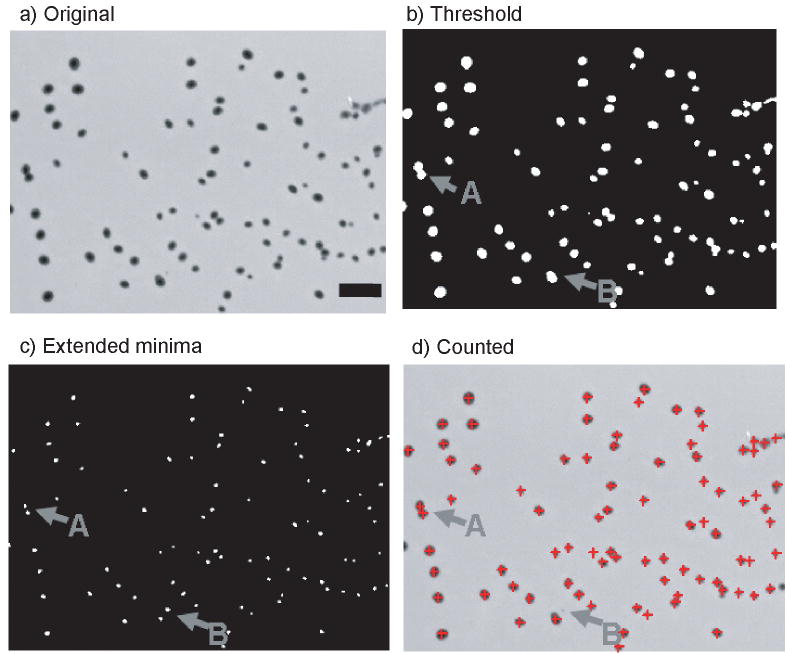
Combining the threshold and extended minima algorithms produced an effective counting process. An example image of the counting results is presented in Figure 4. Bacterial colonies are observed to grow in circular or oval shapes. This image presents challenging features for counting: neighboring colonies have merged (gray arrow A), and colony sizes and contrast vary greatly. The extended minima function allows for clear separation of nearest neighboring colonies (gray arrow A) which is difficult for images analyzed only by thresholding. The mean intensities of these minima are compared against the thresholded image. Comparing the mean intensity of colony minima against the threshold eliminates low contrast non-colony objects such as dust or scratches. A small spot in the image (gray arrow B) was detected by the minima, but rejected by the threshold. The final result is a counted image with high precision.
Figure 4.
Illustration of the colony counting procedure. a) The initial image has many colonies present. b) A thresholded image is shown as an example. The threshold was determined by using the Gaussian center minus 5σ. During analysis the threshold image is never explicitly calculated, but is shown here to foster understanding. c) The extended minima of the original image are shown. The white spots indicate the centers of the colonies. d) The counted colonies are presented with a red crosshair indicating a valid colony. Scale bar is 1.0 mm.
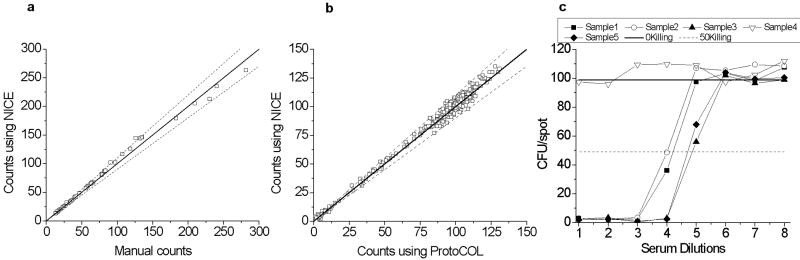
Any colony counting platform must match well with the current gold standard of manually determined counts. As a first step, we compared the counts determined by NICE with the manually counted results from a reference plate. The counts from NICE are in strong agreement with the manual counts (Figure 5.a), with all values within 10 % of the manually determined number. The mean absolute error was 2.7 % (n = 48). Slight over-counting can occur for colony densities of 50 to 150 CFU/spot, while undercounting can occur above 200 CFU/spot. Despite the minima function accounting for many overlapping colonies, if sufficient colony density exists, some colony doublets may be counted as a single colony. While it would be possible to include shape analysis of the colonies to reduce the error caused by such cases, we decided that increased analysis speed was crucial, and that a colony count error below 5 % error with less than 300 CFU/spot was acceptable. The precision of NICE was further evaluated by comparing NICE to the commercial counter, ProtoCOL (Symbiosis, Cambridge, UK). The counts generated by NICE and ProtoCOL are in excellent agreement (n = 480). Some scatter exists between the methods; however, they remain within 10 % deviation (Figure 5.b). In the MOPA, the opsonization index for each test sample is calculated by plotting the CFU/spot as a function of serum concentration (Figure 5.c), and the serum concentration resulting in 50 % killing is interpolated. The opsonization index was determined using the counts from ProtoCOL and NICE for 29 test samples. The mean difference in the ProtoCOL and NICE opsonization index was less than 1 %, with the maximum difference being less than 5 %.
Figure 5.
a) Comparison of colony counts determined by NICE and by manual counting (n = 48). b) The number of colonies determined using a digital camera and NICE were compared to those determined by the commercial ProtoCOL system (n = 480). The solid line represents a slope of 1. Dashed lines are 10 % deviations. c) Example MOPA data for five samples. Samples 1, 2, 3, and 5 have a detectable opsonization index (OI). Sample 4 has no detectable OI since it did not kill at least 50 % of the bacteria.
In addition to counting accuracy, techniques such as MOPA demand the ability to analyze large numbers of plates. We used NICE to count 80 agar plates (the number of agar plates in a standard MOPA experiment), and the program was able to handle this number of images without difficulty. The time to count plates was fairly linear: the image processing times for 5 plates, 20 plates and 80 plates were 58 s, 233 s, and 962 s, respectively. While this time only includes the processing time, it should be noted that no user intervention is required.
Discussion
Document scanners have an exceptionally simple setup enabling automated colony counting with minimal user intervention and no additional equipment. In general, there is no need to control data acquisition parameters except for the image resolution. The uniform illumination through the sample plate eliminates most artifacts, such as shadows and glares which are often seen in the non-optimized camera-based platform. While digital cameras capture images of the exposed agar plate from the top, document scanners image the plate from the bottom and therefore are more susceptible to errors caused by scratches in the plate bottom. Typically, scratches appear lighter in the image than the agar and are unlikely to be mistaken as colonies. Rarely, a line scratch crossing over a single colony resulted in double-counting. Illumination shadows caused by the plate edge can obstruct colony counting in either technique; however provided that the spots are modestly removed from the plate edges, this was typically insignificant. Artifacts in the agar matrix itself, such as bubbles in the media or contaminants, affect both techniques equally. The main disadvantage of the document scanner is the long imaging time. While this time can be decreased by scanning multiple plates simultaneously (10), it is still a comparatively slow method at higher resolutions (600 dpi or 1200 dpi). Digital cameras require a more advanced setup and additional equipment, but the imaging time can be much less. Finding the optimal positioning and manual focus settings can be challenging due to the small colony size. However, after this initial setup, a large number of images can be acquired without further adjustment.
While no single imaging and analysis platform is ideal for all colony measurements, freely distributable programs allow for ready comparison for the development and refinement of other imaging methodologies. NICE has the advantage that it can be freely distributed and can serve as a standard reference counting algorithm among laboratories. NICE reads standard image formats, and therefore may be used in conjunction with many imaging systems. NICE is not without limitation: imaging and processing are separate components of the process, and it relies on the initial careful setup of an imaging platform. However, with the simple cost-effective imaging platform design and software, NICE can serve as a preliminary counting tool, a supplement to commercial counters, or as a routinely used program for clinical research.
Although designed for MOPA, NICE may be used or adapted for other colony counting systems. NICE may be readily applied to the counting of any bacterial colony possessing optical characteristics similar to E. coli and S. pneumoniae. Adaptations to the image processing algorithms would allow NICE to count colonies of substantially different appearance, such as those with poorly-defined boundaries. Colonies imaged via fluorescent markers may be analyzed by inverting the image contrast. NICE could be applied to general bactericidal assays (32), and vaccination efficacy studies for other bacteria, such as cholera (33), pertussis (34) or salmonella (35).
Acknowledgments
This project was supported in part by funding from PATH and NIH (N01 AI-30021 to MHN). The views expressed by the authors do not necessarily reflect the views of PATH. The authors thank Kyung Hyo Kim (Ewha Womans University, Seoul, South Korea) for beta testing NICE.
Footnotes
Publisher's Disclaimer: Disclaimer: Certain commercial equipment, instruments, or materials are identified in this paper to foster understanding and does not imply recommendation or endorsement by NIST, nor does it imply that the materials or equipment identified are necessarily the best available for the purpose.
References
- 1.Mansberg HP. Automatic particle and bacterial colony counter. Science. 1957;126(3278):823–827. doi: 10.1126/science.126.3278.823. [DOI] [PubMed] [Google Scholar]
- 2.Ingels NB, Daughters GT, Burzio A. New design for an automated bacterial colony counter. Rev Sci Instrum. 1968;39(1):115–120. [Google Scholar]
- 3.Thielmann HW, Hagedorn R, Freber W. Evaluation of colony-forming ability experiments using normal and DNA repair-deficient human fibroblast strains and an automatic colony counter. Cytometry. 1985;6(2):130–136. doi: 10.1002/cyto.990060208. [DOI] [PubMed] [Google Scholar]
- 4.Barber PR, Vojnovic B, Kelly J, Mayes CR, Boulton P, Woodcock M, Joiner MC. Automated counting of mammalian cell colonies. Phys Med Biol. 2001;46(1):63–76. doi: 10.1088/0031-9155/46/1/305. [DOI] [PubMed] [Google Scholar]
- 5.Herman CJ, Pelgrim OE, Kirkels WJ, Verheijen R, Debruyne FMJ, Kenemans P, Vooijs GP. In-use evaluation of the omnicon automated tumor colony counter. Cytometry. 1983;3(6):439–442. doi: 10.1002/cyto.990030609. [DOI] [PubMed] [Google Scholar]
- 6.Lumley MA, Burgess R, Billingham LJ, McDonald DF, Milligan DW. Colony counting is a major source of variation in GFU-GM results between centres. Brit J Haematol. 1997;97(2):481–484. doi: 10.1046/j.1365-2141.1997.492695.x. [DOI] [PubMed] [Google Scholar]
- 7.Parry RL, Chin TW, Donahoe PK. Computer-aided cell colony counting. Biotechniques. 1991;10(6):772–774. [PubMed] [Google Scholar]
- 8.Marotz J, Lubbert C, Eisenbeiss W. Effective object recognition for automated counting of colonies in Petri dishes (automated colony counting) Comput Meth Prog Bio. 2001;66(2-3):183–198. doi: 10.1016/s0169-2607(00)00128-0. [DOI] [PubMed] [Google Scholar]
- 9.Biston MC, Corde S, Camus E, Marti-Battle R, Esteve F, Balosso J. An objective method to measure cell survival by computer-assisted image processing of numeric images of Petri dishes. Phys Med Biol. 2003;48(11):1551–1563. doi: 10.1088/0031-9155/48/11/305. [DOI] [PubMed] [Google Scholar]
- 10.Dahle J, Kakar M, Steen HB, Kaalhus O. Automated counting of mammalian cell colonies by means of a flat bed scanner and image processing. Cytom Part A. 2004;60A(2):182–188. doi: 10.1002/cyto.a.20038. [DOI] [PubMed] [Google Scholar]
- 11.Putman M, Burton R, Nahm MH. Simplified method to automatically count bacterial colony forming unit. J Immunol Methods. 2005;302(1-2):99–102. doi: 10.1016/j.jim.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Masala GL, Bottigli U, Brunetti A, Carpinelli M, Diaz N, Fiori PL, Golosio B, Oliva P, Stegel G. Automatic cell colony counting by region-growing approach. Nuovo Cimento C. 2007;30(6):633–644. [Google Scholar]
- 13.Bewes JM, Suchowerska N, McKenzie DR. Automated cell colony counting and analysis using the circular hough image transform algorithm (CHiTA) Phys Med Biol. 2008;53(21):5991–6008. doi: 10.1088/0031-9155/53/21/007. [DOI] [PubMed] [Google Scholar]
- 14.Dobson K, Reading L, Scutt A. A cost-effective method for the automatic quantitative analysis of fibroblastic colony-forming units. Calcified Tissue Int. 1999;65(2):166–172. doi: 10.1007/s002239900677. [DOI] [PubMed] [Google Scholar]
- 15.Wang XD, Yamaguchi N, Someya T, Nasu M. Rapid and automated enumeration of viable bacteria in compost using a micro-colony auto counting system. J Microbiol Meth. 2007;71(1):1–6. doi: 10.1016/j.mimet.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 16.Sieuwerts S, de Bok FAM, Mols E, de Vos WM, Vlieg JETV. A simple and fast method for determining colony forming units. Lett Appl Microbiol. 2008;47(4):275–278. doi: 10.1111/j.1472-765X.2008.02417.x. [DOI] [PubMed] [Google Scholar]
- 17.Chen WB, Zhang CC. An automated bacterial colony counting and classification system. Inform Syst Front. 2009;11(4):349–368. [Google Scholar]
- 18.Corkidi G, Diaz-Uribe R, Folch-Mallol JL, Nieto-Sotelo J. Covasiam: An image analysis method that allows detection of confluent microbial colonies and colonies of various sizes for automated counting. Appl Environ Microb. 1998;64(4):1400–1404. doi: 10.1128/aem.64.4.1400-1404.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Obaro SK, Monteil MA, Henderson DC. The pneumococcal problem. Brit Med J. 1996;312(7045):1521–1525. doi: 10.1136/bmj.312.7045.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bryce J, Boschi-Pinto C, Shibuya K, Black RE. WHO estimates of the causes of death in children. Lancet. 2005;365(9465):1147–1152. doi: 10.1016/S0140-6736(05)71877-8. [DOI] [PubMed] [Google Scholar]
- 21.Singleton RJ, Hennessy TW, Bulkow LR, Hammitt LL, Zulz T, Hurlburt DA, Butler JC, Rudolph K, Parkinson A. Invasive pneumococcal disease caused by nonvaccine serotypes among Alaska native children with high levels of 7-valent pneumococcal conjugate vaccine coverage. J Am Med Assoc. 2007;297(16):1784–1792. doi: 10.1001/jama.297.16.1784. [DOI] [PubMed] [Google Scholar]
- 22.Romero-Steiner S, Libutti D, Pais LB, Dykes J, Anderson P, Whitin JC, Keyserling HL, Carlone GM. Standardization of an opsonophagocytic assay for the measurement of functional antibody activity against Streptococcus pneumoniae using differentiated HL-60 cells. Clin Diagn Lab Immun. 1997;4(4):415–422. doi: 10.1128/cdli.4.4.415-422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nahm MH, Olander JV, Magyarlaki M. Identification of cross-reactive antibodies with low opsonophagocytic activity for Streptococcus pneumoniae. J Infect Dis. 1997;176(3):698–703. doi: 10.1086/514093. [DOI] [PubMed] [Google Scholar]
- 24.Kim KH, Yu JU, Nahm MH. Efficiency of a pneumococcal opsonophagocytic killing assay improved by multiplexing and by coloring colonies. Clin Diagn Lab Immun. 2003;10(4):616–621. doi: 10.1128/CDLI.10.4.616-621.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burton RL, Nahm MH. Development and validation of a fourfold multiplexed opsonization assay (mopa4) for pneumococcal antibodies. Clin Vaccine Immunol. 2006;13(9):1004–1009. doi: 10.1128/CVI.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernard R, Kanduser M, Pernus F. Model-based automated detection of mammalian cell colonies. Phys Med Biol. 2001;46(11):3061–3072. doi: 10.1088/0031-9155/46/11/320. [DOI] [PubMed] [Google Scholar]
- 27.Malpica N, deSolorzano CO, Vaquero JJ, Santos A, Vallcorba I, Garcia-Sagredo JM, delPozo F. Applying watershed algorithms to the segmentation of clustered nuclei. Cytometry. 1997;28(4):289–297. doi: 10.1002/(sici)1097-0320(19970801)28:4<289::aid-cyto3>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 28.Line JE. Development of a selective differential agar for isolation and enumeration of campylobacter spp. J Food Protect. 2001;64(11):1711–1715. doi: 10.4315/0362-028x-64.11.1711. [DOI] [PubMed] [Google Scholar]
- 29.Romero-Steiner S, Frasch C, Concepcion N, Goldblatt D, Kayhty H, Cakevainen M, Laferriere C, Wauters D, Nahm MH, Schinsky MF, et al. Multilaboratory evaluation of a viability assay for measurement of opsonophagocytic antibodies specific to the capsular polysaccharides of Streptococcus pneumoniae. Clin Diagn Lab Immun. 2003;10(6):1019–1024. doi: 10.1128/CDLI.10.6.1019-1024.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu BT, Yu XH, Jones TR, Kirch C, Harris S, Hildreth SW, Madore DV, Quataert SA. Approach to validating an opsonophagocytic assay for Streptococcus pneumoniae. Clin Diagn Lab Immun. 2005;12(2):287–295. doi: 10.1128/CDLI.12.2.287-295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otsu N. Threshold selection method from gray-level histograms. IEEE T Syst Man Cyb. 1979;9(1):62–66. [Google Scholar]
- 32.Liu X, Wang S, Sendi L, Caulfield MJ. High-throughput imaging of bacterial colonies grown on filter plates with application to serum bactericidal assays. J Immunol Methods. 2004;292(1-2):187–193. doi: 10.1016/j.jim.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 33.Yang JS, Kim HJ, Yun CH, Kang SS, Im J, Kim HS, Han SH. A semi-automated vibriocidal assay for improved measurement of cholera vaccine-induced immune responses. J Microbiol Meth. 2007;71(2):141–146. doi: 10.1016/j.mimet.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 34.Prior S, Fleck RA, Gillett ML, Rigsby PR, Corbel MJ, Stacey GN, Xing DKL. Evaluation of adenyl cyclase toxin constructs from Bordetella pertussis as candidate vaccine components in an in vitro model of complement-dependent intraphagocytic killing. Vaccine. 2006;24(22):4794–4803. doi: 10.1016/j.vaccine.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 35.Shaio MF, Rowland H. Bactericidal and opsonizing effects of normal serum on mutant strains of Salmonella typhimurium. Infect Immun. 1985;49(3):647–653. doi: 10.1128/iai.49.3.647-653.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]