Abstract
Bursicon is an insect neuropeptide hormone that is secreted from the central nervous system into the hemolymph and initiates cuticle tanning. The receptor for bursicon is encoded by the rickets (rk) gene and belongs to the G protein-coupled receptor (GPCR) superfamily. The bursicon and its receptor regulate cuticle tanning as well as wing expansion after adult eclosion. However, the molecular action of bursicon signaling remains unclear. We utilized RNA interference (RNAi) and microarray to study the function of the bursicon receptor (Tcrk) in the model insect, Tribolium castaneum. The data included here showed that in addition to cuticle tanning and wing expansion reported previously, Tcrk is also required for development and expansion of integumentary structures and adult eclosion. Using custom microarrays, we identified 24 genes that are differentially expressed between Tcrk RNAi and control insects. Knockdown in the expression of one of these genes, TC004091, resulted in the arrest of adult eclosion. Identification of genes that are involved in bursicon receptor mediated biological processes will provide tools for future studies on mechanisms of bursicon action.
Keywords: Tanning, wing expansion, ecdysis, RNAi, microarray, Tribolium castaneum
Introduction
During the postembryonic development, insects shed their old exoskeletons through stereotyped ecdysis behavior to accommodate growth or change in shape. Immediately after each ecdysis, the new and soft cuticles undergo tanning (sclerotization and melanization) processes to resume their protection functions. More than four decades ago, a blood-borne tanning hormone was discovered in blowflies and was named bursicon (Cottrell, 1962; Fraenkel and Hsiao, 1962; Honegger et al., 2008). However, many attempts to purify and identify this hormone failed until the first bursicon gene was cloned from the fruit fly, Drosophila melanogaster in 2004 (Dewey et al., 2004). In a molecular genetics study, Baker and Truman (2002) discovered that bursicon regulated cuticle tanning and wing expansion through a leucine-rich repeats containing G protein-coupled receptor 2 (LGR2) encoded by the gene rickets (rk) in D. melanogaster. Recently, two parallel studies discovered that the active form of bursicon consists of a heterodimer of two cystine knot peptides encoded by CG13419 (named burs) and CG15284 (named pburs) (Luo et al., 2005; Mendive et al., 2005). Recombinant bursicon binds to the bursicon receptor (rk) with high affinity (Kd = 2.5 × 10−9 M) (Luo et al., 2005). Heterodimeric burs and pburs stimulated intracellular cAMP levels in rk-expressing cells, while burs or pburs alone have no effect on cAMP levels (Luo et al., 2005; Mendive et al., 2005). The discovery of bursicon (hereafter bursicon will be used to refer to the heterodimer of burs and pburs) and its receptor has been reviewed recently (Honegger et al., 2008).
In several insects, bursicon and the bursicon receptor were found to be involved in cuticle tanning, wing expansion and maturation after adult eclosion (Baker and Truman, 2002; Dai et al., 2008; Dewey et al., 2004; Huang et al., 2007). The rk and burs mutant flies are not able to expand their wings after adult eclosion suggesting that bursicon may regulate the wing expansion motor program (Baker and Truman, 2002; Dewey et al., 2004). RNAi-aided knockdown in the expression of burs in Bombyx mori pupae also led to the defects in wing expansion (Huang et al., 2007). It has been suggested that bursicon initiates wing epithelial cell death leading to the fusion of the ventral and dorsal layers of cuticle (Kimura et al., 2004). Bursicon signaling also triggers epithelial–mesenchymal transition (EMT) leading to the removal of the epithelial cells from the wing (Natzle et al., 2008).
Studies on the expression of bursicon showed that the burs gene is expressed in a subset of crustacean cardioactive peptide (CCAP) neurons in D. melanogaster. Targeted ablation of the CCAP neurons eliminated bursicon bioactivity (Dewey et al., 2004). Crustacean cardioactive peptide is one of the major neuropeptide hormones involved in ecdysis behavior and CCAP-cell ablation resulted in deficiencies in both pupal and adult ecdysis (Park et al., 2003). Recently, Arakane et al. (2008) reported that the knockdown in the expression of genes coding for bursicon and its receptor (Tcrk) in Tribolium castaneum by injecting dsRNA into the pharate pupae caused the wrinkled elytra phenotype, but showed no effect on cuticle tanning. Interestingly, when we injected Tcrk dsRNA into the early stage last instar larvae, cuticle tanning, development and expansion of integumentary structures and the adult eclosion were affected in RNAi insects. Furthermore, we performed microarray analysis to study the molecular mechanism of bursicon action and identified 24 genes that are differentially expressed between Tcrk RNAi and control insects. Knockdown in expression of one of these genes (TC004091) resulted in the arrest of adult eclosion, suggesting TC004091 may play an important role in bursicon receptor mediated biological processes such as adult eclosion in T. castaneum.
Materials and Methods
Beetle rearing and staging
Strain GA-1 of T. castaneum was reared on organic wheat flour containing 10 % yeast at 30°C. The final instar larvae were staged based on untanned white head phenotypes observed immediately after molting.
Double-stranded RNA (dsRNA) synthesis
For preparing dsRNA, primers containing gene-specific sequences and T7 polymerase promoter (TAATACGACTCACTATAGGG) at the 5′-end of both the forward primer and reverse primer were used to amplify a 200–600 bp region of genes (Table S3). The PCR products were used as templates for dsRNA synthesis using the Ambion MEGAscript transcription kit (Ambion, Austin, TX). DsRNAs were treated with DNase I (Ambion, Austin, TX) and purified using a phenol/chloroform extraction followed by ethanol precipitation. DsRNAs were then dissolved in nuclease-free water to a concentration of 3–5 μg/μl. The quality of dsRNAs was checked by running on an agarose gel and the concentration of dsRNAs was measured using a NanoDrop1000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA).
DsRNA Microinjection
One-day-old final instar larvae were anaesthetized using ether vapors for 5 min, and then placed on double-sided sticky tape over a glass slide. 200–500 ng of dsRNA was injected into each larva on the lateral side of the second or third abdominal segments using a aspirator tube assembly (Sigma) fitted with 3.5″ glass capillary tube (Drummond) pulled by a needle puller (Model P-2000, Sutter Instruments Co.). Injected larvae were allowed to recover for one hour at room temperature, and then transferred to 30°C incubator. Control larvae were injected with dsRNA for the malE gene from Escherichia coli.
Imaging and documentation
RNAi phenotypes were photographed using the modular zoom system (Leica Z16 APO, Germany) fitted with JVC 3CCD Digital Camera KY-F75U. The images were documented using Cartograph version 6.1.0 (GT Vision Demonstration) and processed using Archimed version 5.2.2 (Microvision Instruments).
Wing length and pronotum width measurement
Elytras or pronotums from ten two-day-old pupae were dissected in 1 × PBS. Elytras and pronotums were then photographed as described above and the length and width of these structures were measured using NIH ImageJ program (http://rsbweb.nih.gov/ij/). The elytra length refers to the distance from the distal end of the elytra to the base of the elytra along the proximal-distal axis. The pronotum width refers to the maximal distance cross the pupal pronotum.
Behavioral studies
Eclosion behavior was video recorded at 25 °C using a Panasonic digital camera GP-KR222 and JVC SR-DVM70 video recorder. Images were exported from video files at various time points during the adult eclosion. Six to seven individuals from Tcrk RNAi and control groups were analyzed.
Microarray analysis
Total RNA was isolated from pooled wings, legs and abdominal epidermis tissues dissected from 6–7 Tcrk RNAi or control pupae per replicate using RNeasy Kit (Qiagen). Abdominal epidermis was prepared by removing the alimentary canal followed by cleaning of fat body and muscles from the abdomen. The final preparation contains epidermis and some contaminating muscle, trachea and fat body tissues. Two hundred nanograms of total RNA samples from three replicates for each treatment were labeled using Agilent Low RNA input fluorescent linear amplification kit. Labeled cDNAs were purified using RNeasy mini purification columns (Qiagen). Fifteen picomoles of labeled cDNAs were used for hybridization. The 60-mer-oligonucleotide chips were designed based on 15,008 predicted genes and 736 control probe sets (Agilent Technologies). The hybridizations were performed according to the manufacturer’s instructions (Agilent Technologies). The microarray slides were scanned using Typhoon 9410 scanner (GE HealthCare) and the images were analyzed using ArrayVision v.8.0 software (Imaging Research Inc.). The normalization and statistical analysis were done using GeneSpring GX v.9.0.1 software. Normalization was performed using a per-chip 50th percentile method that normalizes each chip on its median, allowing comparison among chips. The expression of each gene was normalized on its median among samples. The differentially expressed genes of significance were evaluated with the aid of volcano plots (p-value vs. fold change). Pair-wise comparison between experimental and control groups used the data derived from the volcano plots. The false discovery rate calculations were performed using Bonferroni or Benjamini-Hochberg multiple testing correction to adjust p-value. The cut-off of differential gene expression between treatments was set as p-value ≤ 0.05 and fold change ≥ 1.2.
Bioinformatics
Hierarchical cluster analysis of gene expression was used to group genes with similar expression pattern. Genes with differential expression pattern between malE control and Tcrk RNAi beetles were selected from the microarray data. The expression data were logarithm transformed and grouped using hierarchical clustering algorithm in Gene Cluster 3.0 program (de Hoon et al., 2004). Heat-map was generated using Java Treeview program (Saldanha, 2004).
Gene Ontology (GO) information of each gene was retrieved using Blast2go program (Gotz et al., 2008). To identify T. castaneum ortholog for each gene identified in D. melanogaster microarray analysis (An et al. 2008), we retrieved the amino acid sequences encoded by bursicon regulated genes identified in D. melanogaster from Flybase (http://flybase.org/). Retrieved D. melanogaster sequences were then used as queries in a BLASTP search program against T. castaneum peptide database (Glean prediction, 05-19-2006 version). The T. castaneum hits with an E value less than 1 × 10−25 and amino acid sequence identity more than 30 % were considered as D. melanogaster ortholog.
cDNA synthesis and quantitative real-time reverse-transcriptase polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the whole bodies, or tissues dissected from the staged larvae and pupae using TRI reagent (Molecular Research Center Inc., Cincinnati, OH). Total RNA was then treated with DNase I (Ambion, Austin, TX) in a 50 μl total reaction volume following the manufacturer’s protocol. cDNA synthesis by reverse transcription was performed using 2 μg of DNase I treated RNA and iScript cDNA synthesis kit (Biorad Laboratories, Hercules, CA) in a 20 μl reaction volume. QRT-PCR was performed using MyiQ single color real-time PCR detection system (Biorad Laboratories, Hercules, CA). qRT-PCR was performed in a 20 μl total reaction volume containing 1 μl of cDNA, 1 μl 10 mM each of forward and reverse gene specific primers (Table S4), 7 μl of H2O and 10 μl of supermix (Biorad Laboratories, Hercules, CA). Standard curve was obtained using a ten-fold serial dilution of pooled cDNAs. Ribosomal protein, Tcrp49 was used as an internal control. The mRNA abundance of each gene was obtained relative to Tcrp49 mRNA by a standard-curve based method (Larionov et al., 2005; Rutledge and Cote, 2003). Mean and standard errors for each time point were obtained from the averages of three independent biological replicates.
In vitro tissue culture assay
The wings were dissected from staged pupae under aseptic conditions and cultured in Grace’s medium (Invitrogen, Grand Island, NY) for 3 hours. The entire medium was replaced with fresh medium containing 1 μM 20-hydroxyecdysone (20E, Sigma chemical company) and incubated for an additional 6 hours. Then, the total RNA was extracted from the wing tissues and used to quantify Tcrk mRNA levels by qRT-PCR.
Statistical analysis
Student’s t-Test and analysis of variance were performed using JMP 8.0 (SAS Institute Inc., Cary, NC) to test for statistical differences among treatments (α = 0.05). Pair-wise comparisons were performed using the Tukey-Kramer HSD method.
Results
Bursicon receptor is required for pupal cuticle tanning
The gene coding for bursicon receptor (Tcrk) of T. castaneum (GenBank accession no. XP_975514) was recently identified (Arakane et al., 2008). However, RNAi-mediated Tcrk gene silencing showed no effect on cuticle tanning. Since the primary function of bursicon and its receptor is the regulation of cuticle tanning, we reexamined whether the Tcrk is involved in the tanning process. The difference between our experiments and the previous study is the time of dsRNA delivery. We injected the Tcrk dsRNA into one-day-old final instar larvae. Tcrk dsRNA was injected into the pharate pupae in the previous study. As shown in Figure 1, the cuticle tanning was delayed in the pupae developed from Tcrk dsRNA injected larvae. The pupae developed from the control larvae injected with malE dsRNA initiated tanning by 8 h after ecdysis into the pupal stage (AEP). The cuticle sclerotization of the posterior edge of each abdominal segment was evident in the control pupae (Fig. 1E). In contrast, very little tanning was observed by 8 h AEP in the pupae developed from the larvae injected with Tcrk dsRNA (Fig. 1F). Although the cuticle tanning gradually increased by 24 h AEP, the overall tanning in the Tcrk RNAi pupae was less than that observed in the control pupae (Fig. 1D, 1D′). These results suggest that Tcrk is required for complete cuticle tanning during the pupal stage.
Fig. 1. Inhibition of cuticle tanning by Tcrk RNAi in T. castaneum.
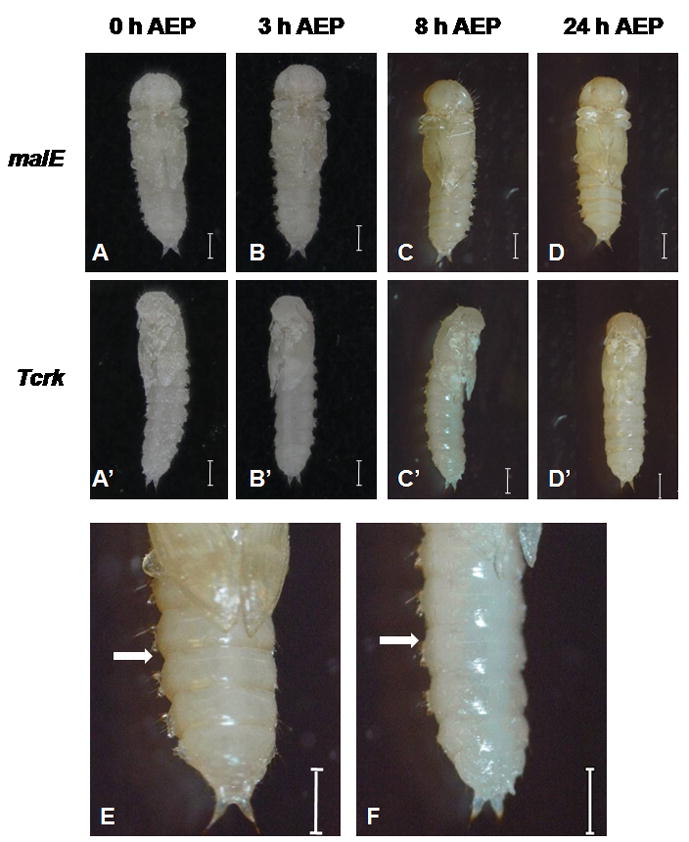
DsRNA for malE (A–D) or Tcrk (A′–D′) were injected into one-day-old final instar larvae. The insects were photographed at various time points after pupation. The cuticle tanning of pupae (at 8 h AEP) developed from larvae injected with malE (E) or Tcrk (F) dsRNA are shown at higher magnification. White color arrow indicates the intersegmental region between two abdominal segments. AEP: hours after ecdysis to the pupal stage. Scale bar: 1.0 mm.
Knockdown in the expression of bursicon receptor affects the development and expansion of integumentary structures and adult eclosion
Pupae developed from the larvae injected with the Tcrk dsRNA showed shortened wings and elongated abdomens (Fig. 2A′). The hind wings (Fig. 2B′) as well as the elytra (data not shown) dissected from the Tcrk RNAi pupae were not fully expanded. Interestingly, the expansion of other integumentary structures such as gin traps, legs and pronotum was also affected in the Tcrk RNAi insects (Fig. 2C′, 2D′, 2E′). As shown in Figure 2F, elytra length and pronotum width were significantly reduced in Tcrk RNAi beetles when compared to the elytra length and pronotum width in the control beetles. The gin trap is a pupal-specific integumentary structure that is formed during the quiescent stage (a stage when final instar larvae stop feeding and change to a “C” like shape prior to pupal ecdysis). The gin traps formed in the Tcrk RNAi pupae were shorter than those in the control pupae. These data suggest that the Tcrk is required for development and expansion of integumentary structures during the pupal stage in T. castaneum.
Fig. 2. Knockdown in the expression of Tcrk blocks the development and expansion of integumentary structures in T. castaneum.
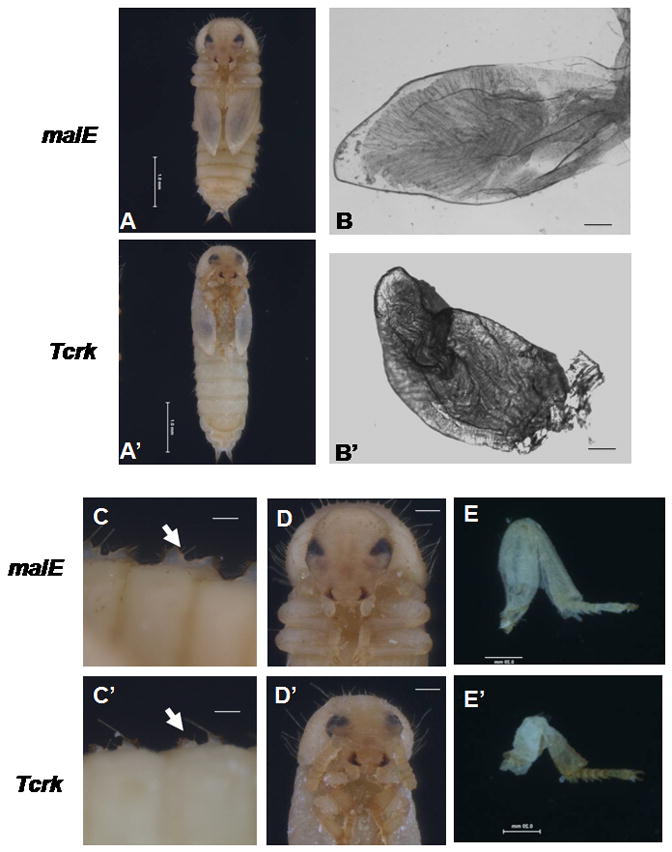
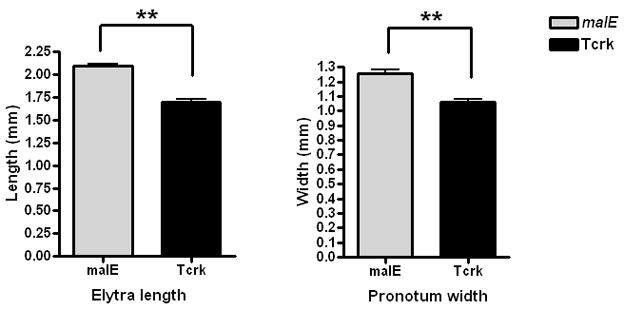
Four-day-old pupae formed after injection of malE (A) or Tcrk (A′) dsRNA into one-day-old final instar larvae were photographed. Hind wings were dissected from the beetles injected with malE (B) or Tcrk (B′) dsRNA at 48 h AEP. Wings were not fully expanded in Tcrk RNAi beetles. In beetles inject with Tcrk dsRNA, gin traps (C′, arrow) and pronotum (D′) are smaller, when comparing to gin traps (C, arrow) and pronotum (D) in malE control insects. Similarly, the first pair of legs are shorter in Tcrk RNAi beetles (E′) than that in malE control beetles (E). AEP: hours after ecdysis to the pupal stage. Scale bar: (A, A′) 1.0 mm; (B, B′, C, C′) 0.1 mm; (D, D′, E, E′) 0.5 mm. (F). Comparison of wing length and pronotum width between malE and Tcrk RNAi beetles. Elytra and pronotum from 10 pupae (1–2 day-old) were photographed and measured using NIH ImageJ program. Mean + SE are shown. Asterisk indicates significant difference between treatments (p ≤ 0.01; Student t-test).
Interestingly, Tcrk RNAi in T. castaneum also caused a lethal arrest during adult eclosion. The adults eclosed from the pupae developed from the larvae injected with the Tcrk dsRNA, showed the exuviae that remained attached to the end of the abdomen (Fig. 3G), while the pupal cuticle covering the head and the thoracic regions was partially shed (Fig. 3H). To further investigate the role of the Tcrk in the ecdysis behavior, we placed 6–7 four-day-old pupae developed from the larvae injected with the Tcrk or malE dsRNA under the microscope with a video camera attached. Live video was recorded throughout the ecdysis process. In both Tcrk and malE RNAi pupae, the normal ecdysis behavior was initiated (Fig. 3A, 3A′) and the wings were inverted to the dorsal side of the body (Fig. 3B, 3B′). However, the adult head and thoracic region were not able to escape from the old cuticle in the Tcrk RNAi insects (Fig. 3D′), while in the control insects shedding of the exuviae was completed by 15 min after the initiation of the ecdysis (Fig. 3D). The expansion of the adult wing was observed at 45 min after the beginning of the ecdysis in control insects (Fig. 3E). In contrast, the insects injected with Tcrk dsRNA were still struggling to shed the old cuticle through the anterior-posterior contractions at 45 min after initiation of ecdysis behavior (Fig. 3E′). Eventually, Tcrk RNAi insects died with an incompletely shed cuticle attached to the body (Fig. 3G, 3H). When Tcrk dsRNA was injected into the newly molted pupae, no shortened wing and eclosion arrest phenotypes were observed and only the wrinkled adult elytra was detected in the RNAi adults (Fig. S1). The wrinkled elytra phenotype observed in the adults is similar to the wrinkled elytra phenotype reported by Arakane et al., 2008. These data suggest that the effects of Tcrk RNAi are stage-dependent and the defects in adult eclosion observed in the Tcrk RNAi insects may be a consequence of the defects in development and expansion of integumentary structure caused by reduced levels of Tcrk.
Fig. 3. Adult eclosion arrest by Tcrk RNAi in T. castaneum.
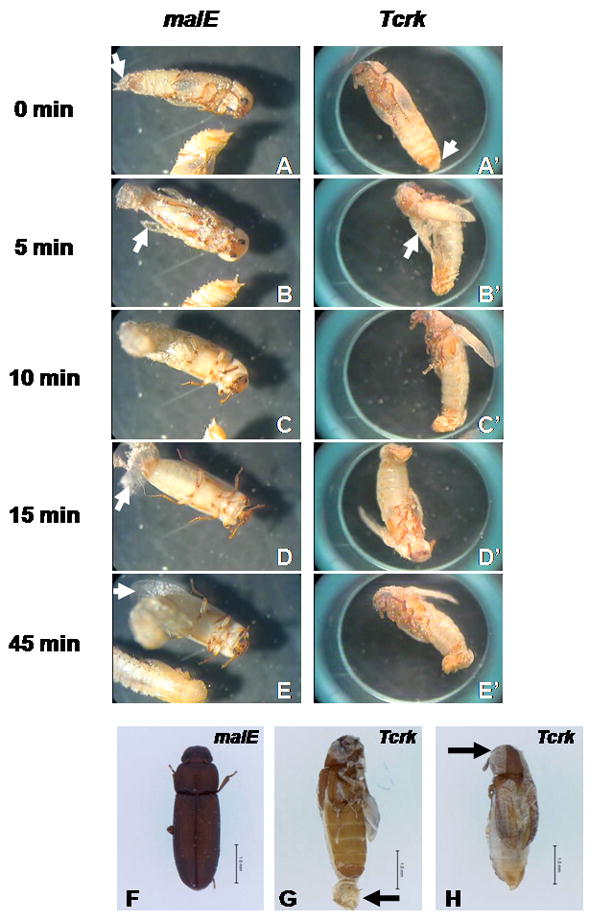
Onset of the ecdysis behavior in beetles injected with dsRNA for malE (A) or Tcrk (A′). Exuviae at the tip of abdomen is indicated with white arrow. Five minutes after onset of the ecdysis behavior, wing inversion (white arrow) was observed in both malE (B) and Tcrk (B′) RNAi beetles. Ten min after onset of the ecdysis behavior, the adult head and thoracic region were free from the old cuticle in malE (C), but not in Tcrk RNAi beetles (C′). Fifteen min after onset of the ecdysis behavior, exuviae (white arrow) was completely shed in malE (D), while Tcrk RNAi beetles (D′) were still struggling to shed the exuviae by continuous anterior-posterior contraction. Forty-five minutes after onset of the ecdysis behavior, fully expanded wings (white arrow) were observed in malE (D), while Tcrk RNAi beetles (D′) continue to undergo anterior-posterior contractions. (F). Dorsal view of control beetles injected with malE dsRNA after adult eclosion. (G). Ventral view of Tcrk RNAi beetles that show the unshed exuviae (arrow) during the adult eclosion. (H). Dorsal view of Tcrk RNAi beetles that show the attached old cuticle in the head region (arrow) during the adult eclosion. Scale bar: 1.0 mm.
To confirm whether the phenotypes shown above are caused by knockdown in the mRNA levels of the Tcrk gene, we performed qRT-PCR to check the knockdown efficiency of Tcrk RNAi (see supporting material for detail). Injection of Tcrk dsRNA caused about an 81% reduction in the Tcrk mRNA levels (Fig. S2) and the Tcrk RNAi showed dose-dependent effects on wing expansion and adult eclosion defects (Tables S1 and S2). To test the specificity of Tcrk RNAi, a new dsRNA targeting a different region of Tcrk gene was produced (named Tcrk-2 dsRNA) and tested. Injection of Tcrk-2 dsRNA caused similar phenotypes observed in Tcrk-1 dsRNA injected insects (see supporting materials section for detail).
Stage- and tissue-specific expression of bursicon receptor
The Tcrk mRNA levels were low at the beginning of the final instar larval stage (0–2 days), then the Tcrk mRNA levels increased by three days after ecdysis into the final instar larval stage (AEFL) (Fig. 4A). These higher levels of Tcrk mRNA were maintained until one day after pupation. Then the Tcrk mRNA levels decreased and the lower levels were detected on the 2nd day after pupation. The Tcrk mRNA levels increased again and reached the maximum levels by the end of the pupal stage. Interestingly, the levels of Tcrk mRNA were higher in the female tissues when compared to their levels in the male tissues. Also, the abdominal epidermis and wing tissues showed the higher levels of Tcrk mRNA in comparison to those in the midgut dissected from the pupae (Fig. 4B). The stage- and tissue-specific expression patterns of Tcrk gene support its roles in the regulation of the development and expansion of integumentary structures.
Fig. 4. Stage and tissue-specific expression pattern of Tcrk in T. castaneum determined by qRT-PCR.
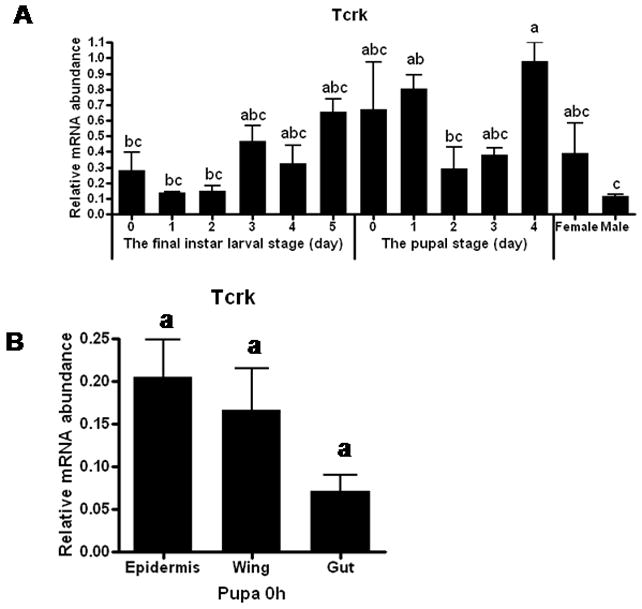
(A). Stage-specific expression pattern of Tcrk. The whole body samples were collected at one-day intervals during the final instar larval and pupal stages, and 3-day-old male and female (B). Tissue-specific expression pattern of Tcrk. The tissue samples were collected at 0 h after the pupation. Total RNA was extracted from pools of three larvae or pupae whole bodies, or tissues dissected from 6–7 beetles per treatment. The expression levels of Tcrk were normalized using Tcrp49 mRNA levels as an internal control. Mean + SE of three independent experiments are shown. Means with the same letter are not significantly different (α = 0.05; ANOVA).
Bursicon receptor is an ecdysone-inducible gene
Ecdysteroids (20-hydroxyecdysone, 20E is the most active form) are one of the important hormones involved in insect molting and metamorphosis. In D. melanogaster, 20E regulates the formation of pupal and adult integumentary structures by up-regulating the expression of the pupal cuticle genes such as Edg78E and Pcp (Zhou and Riddiford, 2002). In vitro experiments showed that the Tcrk mRNA levels in the wing tissues dissected from the newly molted pupae were induced by 1 μM 20E treatment (Fig. 5A). Furthermore, the Tcrk mRNA levels were lower in insects injected with ecdysone receptor (EcR) dsRNA when compared to the Tcrk mRNA levels in control insects injected with malE dsRNA (Fig. 5B). These data suggest that the expression of the Tcrk gene is regulated by ecdysteroids.
Fig. 5. Tcrk is a 20E-inducible gene.
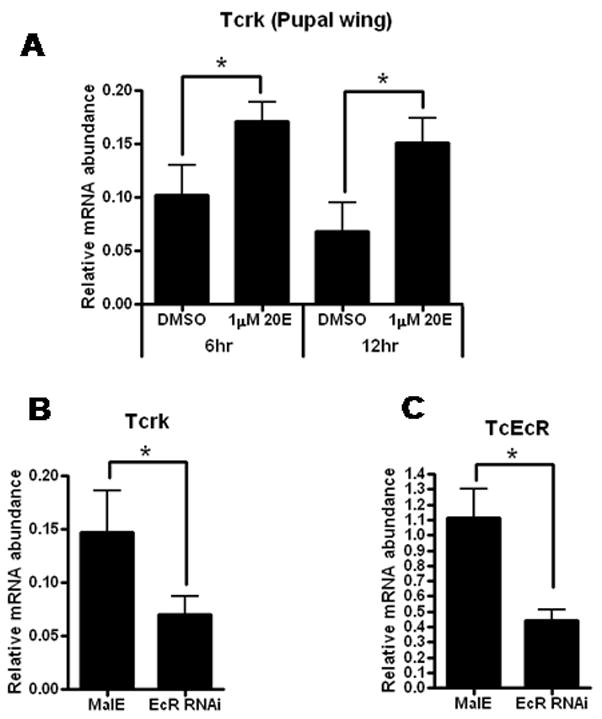
(A). The expression of Tcrk in wing tissues dissected from 0 h pupae was determined using qRT-PCR after 6 and 12 hours incubation with 1 μM 20E in vitro. The expression of Tcrk (B) and TcEcR (C) in whole body samples isolated from beetles injected with malE and TcEcR dsRNA (The sequences of primers used for TcEcR dsRNA synthesis and qRT-PCR were reported by Tan and Palli, 2008). The expression levels of Tcrk and TcEcR were normalized using Tcrp49 mRNA levels as an internal control. Mean + SE of three independent experiments are shown. Asterisk indicates significant difference between treatments (p ≤ 0.05; Student t-test).
Microarray analysis to identify genes whose mRNA levels change in the bursicon receptor RNAi beetles
Microarray analyses were performed to compare the expression of genes in the integumentary tissues between the Tcrk RNAi beetles and the control beetles injected with malE dsRNA. Total RNA samples were isolated from the pool of pupal tissues including wings, legs and abdominal epidermis dissected from the pupae at 0 h AEP and 24 h AEP. T. castaneum custom microarrays containing 15,208 probe sets were hybridized with the probes prepared from these RNA samples. 11,928 probe sets were detected in at least one of the RNA samples tested. Pair-wise comparison analysis applying highly stringent Bonferroni correction to the microarray data identified 43 genes that were differentially expressed between 0 h and 24 h AEP in insects injected with malE (control) dsRNA (Table 1). In contrast, in Tcrk RNAi pupae only five genes were expressed differentially between 0 h and 24 h AEP stages. The microarray data analysis after applying a less stringent Benjamini-Hochberg correction identified 5410 genes that were differentially expressed between 0 h and 24 h AEP in insects injected with malE dsRNA (Table 1). In contrast, this correction identified only 418 genes that were differentially expressed between 0 h and 24 h AEP stages in Tcrk RNAi pupae. These data suggest that global changes in gene expression occur in integumentary tissues between 0 h and 24 h AEP and the presence of Tcrk is required for changes in the expression of at least some of the genes.
Table 1.
Number of genes differentially expressed among treatments with a p-value ≤ 0.05 and a fold change ≥ 1.2
| Correction test | 0 h AEP | 24 h AEP | malE | Tcrk |
|---|---|---|---|---|
| malE vs. Tcrk | malE vs. Tcrk | 0 h vs. 24 h AEP | 0 h vs. 24 h AEP | |
| Bonferroni | 0 | 0 | 43 | 5 |
| Benjamini- Hochberg | 0 | 0 | 5410 | 418 |
| T-test only | 496 | 625 | 6389 | 2377 |
Pair-wise comparison of microarray data identified 496 genes whose mRNA levels changed by more than 1.2-fold with a p-value of less than 0.05 between control and Tcrk RNAi beetles at 0 h AEP. Similar analysis identified 625 genes whose mRNA levels changed by more that 1.2-fold with a p-value of less than 0.05 between control and Tcrk RNAi beetles at 24 h AEP. Among these genes, 18 genes were down-regulated and 6 genes were up-regulated by Tcrk RNAi at both 0 h and 24 h AEP stages (Table S5). Using a hierarchical cluster analysis, the 18 genes that were down-regulated by Tcrk RNAi were grouped into two clusters (clusters 1 and 2), while 6 genes up-regulated by Tcrk RNAi were grouped into one cluster (cluster 3, Fig. 6). The expression of cluster 1 genes is similar between 0 h and 24 h AEP, which is close to the expression pattern of Tcrk. In contrast, the expression of cluster 2 genes is much higher at 0 h AEP when compared to their expression at 24 h AEP. The expression patterns of three genes (TC004091, TC016332 and TC013400) showed the closest relationship to the expression pattern of Tcrk. TC016332 encodes for a gag-pol protein, while no known function or protein motifs were identified in TC004091 and TC013400. The differential expression patterns of these genes in integumentary tissues suggest that these genes may be associated with integumentary defects seen in Tcrk RNAi beetles.
Fig. 6. Hierarchical cluster analysis of gene expression.
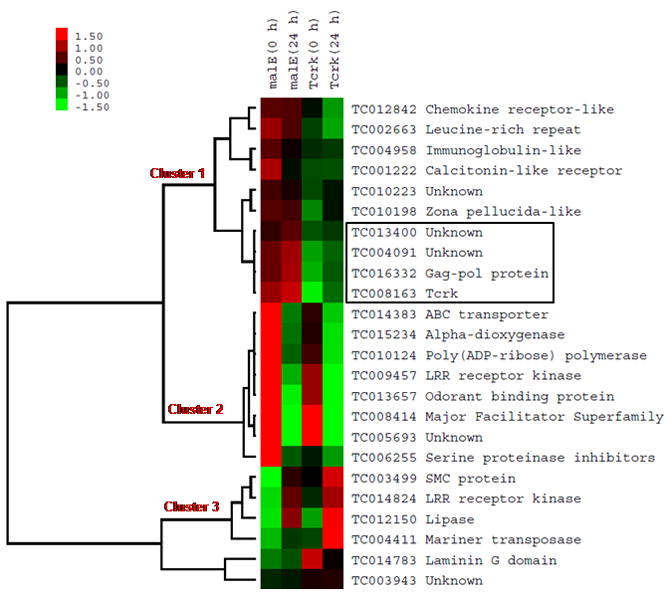
Eighteen down-regulated genes and six up-regulated genes were selected from microarray data for the hierarchical cluster analysis. The expression data were logarithm transformed and grouped using hierarchical clustering algorithm in Gene Cluster 3.0 program. Heat-map was generated using Java Treeview program. Genes down-regulated by Tcrk RNAi were grouped into cluster 1 and 2, while genes up-regulated by Tcrk RNAi were grouped into cluster 3. The expression of cluster 1 genes is similar between 0 h and 24 h AEP. In contrast, the expression of cluster 2 genes is much higher at 0 h AEP stage when compared to the expression at 24 h AEP stage. Three genes were clustered with Tcrk, which indicates that they have similar expression pattern (enclosed in a square).
The expression of most of the sclerotization related genes (e.g. laccase 1 and tyrosine hydroxylase) except for laccase 2 (TcLac2, TC010489) are similar in Tcrk RNAi and control beetles. The TcLac2 mRNA levels were lower in Tcrk RNAi beetles than in the control insects at 0 h AEP (Fig. S3). The reduction in the expression of TcLac2 in Tcrk RNAi beetles suggests that Tcrk may influence pupal cuticle tanning by regulating the expression of TcLac2 in T. castaneum.
We also compared the list of genes that are identified in our microarray analysis as those that require the presence of Tcrk for their expression with the list of bursicon regulated genes identified in D. melanogaster (An et al. 2008). We retrieved the amino acid sequences encoded by bursicon regulated genes identified in D. melanogaster from Flybase (http://flybase.org/). We used these sequences to perform a BLASTP search against T. castaneum peptide database (Glean prediction, 05-19-2006 version). The hits with E value less than 1 × 10−25 and amino acid sequence identity more than 30 % were considered as D. melanogaster orthologs. Then, we searched our microarray data for T. castaneum orthologs of bursicon regulated genes identified in D. melanogaster. These analyses identified five T. castaneum genes (TC008773, TC015214, TC011227, TC016226 and TC006706, Table 2). The results from these analyses suggest that these five genes that required bursicon or its receptor for their expression in two different insects may play important roles in bursicon regulated physiological processes.
Table 2.
Common genes identified in T. castaneum and D. melanogaster bursicon/bursicon receptor microarray studies
| Tribolium Fold change1 | Drosophila Fold change2 | |||||
|---|---|---|---|---|---|---|
| Tribolium ID | Drosophila ortholog | Gene name or domain | 0 h AEP | 24 h AEP | 1 h | 3 h |
| TC008773 | CG32438 | Smc5 | NC | −1.2 | NA | −2.2 |
| TC015214 | CG8920 | Tudor domain | NC | −1.3 | 2.1 | NA |
| TC011227 | CG1522 | Cacophony | NC | 1.3 | 2.0 | NA |
| TC016226 | CG32491 | Modifier of mdg4 | 1.5 | NC | 2.4 | 2.4 |
| TC006706 | CG103693 | Inwardly rectifying potassium channel | −2.5 | NC | NA | 2.3 |
Tribolium fold change refers to fold changes between Tcrk RNAi vs. malE control.
Drosophila fold change refers to fold changes between r-bursicon injection vs. control injection control, based on An et al., 2008.
TC006706 is the closest homolog to DmIrk3 in T. castaneum.
NC=No change.
NA=No data available.
Twenty-four genes were randomly selected for the validation of microarray data using the qRT-PCR method (Fig. S4, S5). The fold change in expression between Tcrk RNAi and control insects for 21 out of 24 genes tested showed high correlation between results obtained by microarray and qRT-PCR (Fig. S6). However, the fold changes of three genes (TC010625, TC007060 and TC011804) observed in microarray analysis were not validated by qRT-PCR.
TC004091 is required for adult eclosion
Five genes (TC002663, TC004091, TC016332, TC009457 and TC13657) that showed similar expression patterns as the Tcrk expression pattern and their mRNA levels were lower (fold change > 2, p < 0.05) in Tcrk RNAi pupae at both 0 h and 24 h AEP when compared to their mRNA levels in the control pupae, were chosen for further functional analysis. Injection of TC002663 or TC004091 dsRNA into the larvae caused high mortality, 70.00% and 90.91% respectively, during the pupal stage (Table 3). Nearly half of the larvae injected with TC002663 dsRNA died during the pupal stage, but no defects in integumentary structures were detected. Interestingly, 12 out of 22 larvae injected with TC004091 dsRNA died during the adult eclosion (Fig. 7B). Unlike, Tcrk RNAi insects, the integumentary structures of TC004091 RNAi insects developed well and are similar to those in control insects. Only a few pupae developed from TC004091 dsRNA injected larvae showed malformed wings (Fig. 7A). These results suggest that TC004091 expression is required for adult eclosion. However, it is not clear whether TC004091 regulates the expansion of integumentary structures.
Table 3.
Mortality caused by RNAi targeting five genes identified in microarray analysis.
| Gene symbol | Protein domain | # injected larvae | # dead larvae | # dead pupae | # dead pharate adults | # alive adults | % Mortality |
|---|---|---|---|---|---|---|---|
| malE | / | 17 | 3 | 0 | 0 | 14 | 17.65 |
| TC002663 | Leucine-rich repeat | 20 | 5 | 8 | 0 | 6 | 70.00 |
| TC009457 | Leucine-rich repeat receptor kinase | 23 | 8 | 0 | 0 | 15 | 34.78 |
| TC016332 | Gag-pol protein | 21 | 5 | 2 | 0 | 14 | 33.33 |
| TC004091 | Unknown | 22 | 4 | 4 | 12 | 2 | 90.91 |
| TC013657 | Odorant binding protein | 15 | 6 | 3 | 0 | 6 | 60.00 |
Fig. 7. Knockdown in the expression of TC004091 results in pupal wing defects (A) and arrest of adult eclosion (B) in T. castaneum.

DsRNA for TC004091 was injected into one-day old final instar larvae and images of two-day old pupae and pharate adults were photographed. Scale bar: 1.0 mm.
Discussion
The data reported here clearly showed that Tcrk is required for pupal cuticle tanning, development and expansion of integumentary structures and adult eclosion. Previous studies showed that the bursicon receptor is involved in cuticle tanning and wing expansion after adult eclosion in D. melanogaster (Baker and Truman, 2002). Bursicon also regulates adult wing expansion in B. mori (Huang et al., 2007) and Manduca sexta (Dai et al., 2008). In a recent study in T. castaneum, injection of dsRNA for bursicon and its receptor into pharate pupae caused a wrinkled elytra phenotype, but showed no effect on cuticle tanning. In the current study, knockdown in the Tcrk mRNA levels during final instar larval and pupal stages by injecting dsRNA into the final instar larvae delayed and reduced cuticle tanning. Tcrk RNAi also affected development and expansion of integumentary structures and blocked adult eclosion. In addition, our qRT-PCR analysis showed higher Tcrk mRNA levels in the epidermis and wing when compared to their levels in the midgut during the pupal stage. Higher levels of Tcrk mRNA were also detected during the prepupal and pupal stage. Taken together, these data suggest the Tcrk functions during the prepupal and pupal stages to facilitate development and expansion of integumentary structures in T. castaneum. Furthermore, complete development and expansion of integumentary structures during the pupal stage is probably required for successful adult eclosion. Therefore, the block in adult eclosion observed in the Tcrk RNAi insects could be caused by defects in the development and expansion of integumentary structures in these insects. These results indicate that bursicon functions not only during the adult stage but also during the late larval and pupal stages. The expression of Dmrk during the larval and pupal stages was reported previously (Luo et al., 2005), these data suggest that Dmrk may function prior to adult eclosion in D. melanogaster as well (Luo et al., 2005). It is intriguing why the bursicon receptor mutants in D. melanogaster do not exhibit defects in development and expansion of integumentary structures and adult eclosion. Alternative loss-of-function approaches such as transgenic RNAi should be used to address this question.
To understand the molecular basis of bursicon action through its receptor in regulation of tanning and wing expansion, several studies have been conducted using fly genetics as a tool. Kimura et al. (2004) reported that the wing epithelial cell death during the wing maturation was inhibited in Dmrk mutant flies. In contrast, a recent study suggests that Dmrk regulates the epithelial-mesenchymal transition (EMT) mediated by armadillo/β-catenin signal pathway (Natzle et al., 2008). Epithelial-mesenchymal transition helps the wing cells change shape and migrate to the wing hinge. In D. melanogaster rk mutant, the wing epidermis was retained and the GFP-tagged armadillo/β-catenin remained longer in these mutant flies when compared to the wild-type flies. A recent study suggests that the bursicon-immunoreactive neurons located in the subesophageal ganglion are essential for the wing expansion behavior in D. melanogaster (Peabody et al., 2008). Furthermore, a genome-wide screen using microarray analysis identified 87 genes whose mRNA levels were affected by the injection of a recombinant bursicon protein into neck-ligated flies (An et al., 2008). In the current study, we used microarray analysis to identify genes that require Tcrk for their expression. A number of genes that showed differences in their mRNA levels between Tcrk RNAi and control insects at 0 h AEP and 24 h AEP were identified by microarray analysis. Interestingly, six genes coding for leucine-rich repeat proteins showed reduced levels of mRNA in Tcrk RNAi pupae when compared to their levels in control pupae at 0 h AEP (data not shown). Two of them, TC002663 and TC009457, showed lower mRNA levels in Tcrk RNAi pupae when compared to their levels in control pupae at both 0 h AEP and 24 h AEP. Leucine-rich repeat domains are known to be involved in protein-protein interactions that are important for ligand binding, cell adhesion and extracellular matrix-binding (Eriksen et al., 2000). The bursicon receptor itself contains a leucine-rich repeat domain at the N-terminal region. These data suggest that the leucine-rich repeat protein family may play important roles in bursicon function. It is possible that the knockdown in the expression of Tcrk disrupts the communication between wing epithelial cells and causes the persistence of cell adhesion and extracellular matrix-binding resulting in blocking of the wing expansion. Nearly half of the larvae injected with TC002663 dsRNA died during the pupal stage. However, we did not observe any defects in integumentary structures. It is possible that TC002663 may regulate other functions critical for survival of pupae. Out of five genes selected for functional analysis, knockdown in the expression of TC004091 resulted in the arrest of adult eclosion but caused no effect on the expansion of integumentary structures. These data suggest that TC004091 may be involved Tcrk-mediated biological processes other than the development and expansion of integumentary structures. Further experiments are required to elucidate the underlying mechanism by which Tcrk regulates the development and expansion of integumentary structures and adult eclosion.
Insect cuticle tanning and sclerotization requires enzymatic reactions such as hydroxylation of tyrosine to 3,4-dihydroxyphenylalanine (DOPA) (Andersen, 2005). Laccase 2 (one of the tyrosinases in T. castaneum) and tyrosine hydroxylase (TH) are two key enzymes that control cuticle sclerotization processes in T. castaneum (Arakane et al., 2005; Gorman and Arakane, 2010). In our microarray analysis, we only detected a decrease in TcLac2 mRNA levels in Tcrk RNAi insects, whereas the mRNA levels of TcLac1 and TcTH did not change between Tcrk RNAi and control insects. Since it has been shown that TcLac2 is required for the cuticle tanning during the pupal and adult stages (Arakane et al., 2005), it is possible that Tcrk effects cuticle sclerotization by regulating the expression of the TcLac2 gene. However, these data cannot exclude the possibility that Tcrk does not regulate transcription of enzymes involved in tanning since most of the enzymes are laid down in the new cuticle prior to ecdysis. In D. melanogaster, it has been shown that the mRNA levels of epidermal TH did not change at eclosion, but the levels of TH enzyme increased rapidly following eclosion (Davis et al., 2007). Although Dmrk mutants showed relatively normal mRNA expression of epidermal TH after eclosion, they failed to phosphorylate TH. It has been suggested that post-translational modifications such as phosphorylation may play an important role in regulation of cuticle tanning by the bursicon receptor (Honegger et al., 2008).
Comparison of microarray data from D. melanogaster (An et al., 2008) and T. castaneum (this study) identified five common genes that require bursicon or Tcrk for their expression. One of them, TC006706, encodes an inwardly-rectifying potassium channel (Irk or Kir). Sequence similarity and phylogenetic analysis showed that TC006706 belongs to human G protein-coupled Kir3 and ATP-sensitive Kir6 subfamily. Both Kir3 and Kir6 channels are activated by G protein subunits (Dascal, 1997; Kubo et al., 2005). In D. melanogaster, the suppression of membrane excitability of CCAP-expressing neurons by overexpressing human Kir2.1 channel in CCAP neurons resulted in head eversion defects, shortened forewings and legs and high pupal lethality. Whereas, overexpression of a modified shaker K+ channel in CCAP neurons blocked the release of bursicon into the hemolymph (Luan et al., 2006). The wide distribution of G protein-coupled Kir in the central nervous system and its regulation of neuronal excitability indicate its important role in neuron physiology (Dascal, 1997). The functions of other genes identified in both D. melanogaster and T. castaneum are not well understood. Further studies on these five common genes will help in understanding the molecular mechanisms of bursicon signaling.
In summary, our studies clearly showed that Tcrk presence during the pupal stage is required for cuticle tanning, development and expansion of not only wing but also other integumentary structures. The proper development and expansion of these integumentary structures might be critical for successful adult eclosion. Some of the genes identified by microarray analysis that showed changes in mRNA levels in Tcrk RNAi pupae may be directly or indirectly involved in development and expansion of integumentary structures. Identification of these genes should help future studies aimed at understanding the molecular mechanisms of bursicon signaling.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (GM070559-06). We thank Dr. Nigel Cooper and Ms. Xiahong Li of University of Louisville for help with microarray experiment. The University of Louisville microarray facility is supported by NCRR IDeA Awards INBRE-P20 RR016481 and COBRE-P20RR018733 We thank Dr. Michael Sharkey and Dicky Yu for help with microscopic facilities, Dr. Kenneth F. Haynes for help with behavioral studies, Dr. Lynn M. Riddiford, Dr. Yoonseong Park and Dr. Qisheng Song for their valuable comments on the manuscript. This is contribution number 10-08-23 from the Kentucky Agricultural Experimental Station.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An S, Wang S, Gilbert LI, Beerntsen B, Ellersieck M, Song Q. Global identification of bursicon-regulated genes in Drosophila melanogaster. BMC Genomics. 2008;9:424. doi: 10.1186/1471-2164-9-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SO. Cuticular Sclerotization and Tanning. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science. Elsevier; Oxford: 2005. pp. 145–170. [Google Scholar]
- Arakane Y, Li B, Muthukrishnan S, Beeman RW, Kramer KJ, Park Y. Functional analysis of four neuropeptides, EH, ETH, CCAP and bursicon, and their receptors in adult ecdysis behavior of the red flour beetle, Tribolium castaneum. Mech Dev. 2008;125:984–995. doi: 10.1016/j.mod.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Arakane Y, Muthukrishnan S, Beeman RW, Kanost MR, Kramer KJ. Laccase 2 is the phenoloxidase gene required for beetle cuticle tanning. Proc Natl Acad Sci U S A. 2005;102:11337–11342. doi: 10.1073/pnas.0504982102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JD, Truman JW. Mutations in the Drosophila glycoprotein hormone receptor, rickets, eliminate neuropeptide-induced tanning and selectively block a stereotyped behavioral program. J Exp Biol. 2002;205:2555–2565. doi: 10.1242/jeb.205.17.2555. [DOI] [PubMed] [Google Scholar]
- Cottrell CB. The imaginal ecdysis of blowflies. The control of cuticular hardening and darkening. J Exp Biol. 1962;39:395–411. [Google Scholar]
- Dai L, Dewey EM, Zitnan D, Luo CW, Honegger HW, Adams ME. Identification, developmental expression, and functions of bursicon in the tobacco hawkmoth, Manduca sexta. J Comp Neurol. 2008;506:759–774. doi: 10.1002/cne.21575. [DOI] [PubMed] [Google Scholar]
- Dascal N. Signalling via the G protein-activated K+ channels. Cell Signal. 1997;9:551–573. doi: 10.1016/s0898-6568(97)00095-8. [DOI] [PubMed] [Google Scholar]
- Davis MM, O’Keefe SL, Primrose DA, Hodgetts RB. A neuropeptide hormone cascade controls the precise onset of post-eclosion cuticular tanning in Drosophila melanogaster. Development. 2007;134:4395–4404. doi: 10.1242/dev.009902. [DOI] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Dewey EM, McNabb SL, Ewer J, Kuo GR, Takanishi CL, Truman JW, Honegger HW. Identification of the gene encoding bursicon, an insect neuropeptide responsible for cuticle sclerotization and wing spreading. Curr Biol. 2004;14:1208–1213. doi: 10.1016/j.cub.2004.06.051. [DOI] [PubMed] [Google Scholar]
- Eriksen KK, Hauser F, Schiott M, Pedersen KM, Sondergaard L, Grimmelikhuijzen CJ. Molecular cloning, genomic organization, developmental regulation, and a knockout mutant of a novel leu-rich repeats-containing G protein-coupled receptor (DLGR-2) from Drosophila melanogaster. Genome Res. 2000;10:924–938. doi: 10.1101/gr.10.7.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraenkel G, Hsiao C. Hormonal and nervous control of tanning in the fly. Science. 1962;138:27–29. doi: 10.1126/science.138.3536.27. [DOI] [PubMed] [Google Scholar]
- Gorman MJ, Arakane Y. Tyrosine hydroxylase is required for cuticle sclerotization and pigmentation in Tribolium castaneum. Insect Biochem Mol Biol. 2010;40:267–273. doi: 10.1016/j.ibmb.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honegger HW, Dewey EM, Ewer J. Bursicon, the tanning hormone of insects: recent advances following the discovery of its molecular identity. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2008;194:989–1005. doi: 10.1007/s00359-008-0386-3. [DOI] [PubMed] [Google Scholar]
- Huang J, Zhang Y, Li M, Wang S, Liu W, Couble P, Zhao G, Huang Y. RNA interference-mediated silencing of the bursicon gene induces defects in wing expansion of silkworm. FEBS Lett. 2007;581:697–701. doi: 10.1016/j.febslet.2007.01.034. [DOI] [PubMed] [Google Scholar]
- Kimura K, Kodama A, Hayasaka Y, Ohta T. Activation of the cAMP/PKA signaling pathway is required for post-ecdysial cell death in wing epidermal cells of Drosophila melanogaster. Development. 2004;131:1597–1606. doi: 10.1242/dev.01049. [DOI] [PubMed] [Google Scholar]
- Kubo Y, Adelman JP, Clapham DE, Jan LY, Karschin A, Kurachi Y, Lazdunski M, Nichols CG, Seino S, Vandenberg CA. International Union of Pharmacology. LIV. Nomenclature and molecular relationships of inwardly rectifying potassium channels. Pharmacol Rev. 2005;57:509–526. doi: 10.1124/pr.57.4.11. [DOI] [PubMed] [Google Scholar]
- Larionov A, Krause A, Miller W. A standard curve based method for relative real time PCR data processing. BMC Bioinformatics. 2005;6:62. doi: 10.1186/1471-2105-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H, Lemon WC, Peabody NC, Pohl JB, Zelensky PK, Wang D, Nitabach MN, Holmes TC, White BH. Functional dissection of a neuronal network required for cuticle tanning and wing expansion in Drosophila. J Neurosci. 2006;26:573–584. doi: 10.1523/JNEUROSCI.3916-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo CW, Dewey EM, Sudo S, Ewer J, Hsu SY, Honegger HW, Hsueh AJ. Bursicon, the insect cuticle-hardening hormone, is a heterodimeric cystine knot protein that activates G protein-coupled receptor LGR2. Proc Natl Acad Sci U S A. 2005;102:2820–2825. doi: 10.1073/pnas.0409916102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendive FM, Van Loy T, Claeysen S, Poels J, Williamson M, Hauser F, Grimmelikhuijzen CJ, Vassart G, Vanden Broeck J. Drosophila molting neurohormone bursicon is a heterodimer and the natural agonist of the orphan receptor DLGR2. FEBS Lett. 2005;579:2171–2176. doi: 10.1016/j.febslet.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Natzle JE, Kiger JA, Jr, Green MM. Bursicon signaling mutations separate the epithelial-mesenchymal transition from programmed cell death during Drosophila melanogaster wing maturation. Genetics. 2008;180:885–893. doi: 10.1534/genetics.108.092908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Schroeder AJ, Helfrich-Forster C, Jackson FR, Ewer J. Targeted ablation of CCAP neuropeptide-containing neurons of Drosophila causes specific defects in execution and circadian timing of ecdysis behavior. Development. 2003;130:2645–2656. doi: 10.1242/dev.00503. [DOI] [PubMed] [Google Scholar]
- Peabody NC, Diao F, Luan H, Wang H, Dewey EM, Honegger HW, White BH. Bursicon functions within the Drosophila CNS to modulate wing expansion behavior, hormone secretion, and cell death. J Neurosci. 2008;28:14379–14391. doi: 10.1523/JNEUROSCI.2842-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge RG, Cote C. Mathematics of quantitative kinetic PCR and the application of standard curves. Nucleic Acids Res. 2003;31:e93. doi: 10.1093/nar/gng093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Zhou X, Riddiford LM. Broad specifies pupal development and mediates the ‘status quo’ action of juvenile hormone on the pupal-adult transformation in Drosophila and Manduca. Development. 2002;129:2259–2269. doi: 10.1242/dev.129.9.2259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


