Abstract
Background
EGFR intron1 has a polymorphic region of CA-repeats which is believed to be associated with increased EGFR expression, tumor aggressiveness, and worse survival in cancer patients.
Methods
We investigated a large population of pancreatic adenocarcinoma patients to evaluate this polymorphism as a potential prognostic marker of clinical outcome. We included DNA obtained from 50 resected pancreatic adenocarcinomas and from 85 diagnostic EUS-FNA corresponding to patients with unresectable tumors. The correlation between CA-repeats length and EGFR mRNA levels was also examined.
Results
Analysis of the 135 patients revealed no correlation between EGFR intron 1 CA repeats length and tumor stage. There was no difference in overall patient survival when stratified by allele length. A correlation between EGFR intron 1 length and EGFR transcript and protein levels could not be established.
Conclusions
The length of the EGFR intron1 CA repeats does not correlate with levels of EGFR expression and can not be employed as marker of clinical prognosis in pancreatic cancer patients.
Keywords: EGFR, pancreatic cancer, intron 1 polymorphism
Introduction
Pancreatic adenocarcinoma remains the fourth deadliest cancer in the United States with 42,470 new cases and 35,240 deaths estimated in 2009(1). The five year survival rate remains dismal at roughly 5% despite aggressive therapy(1). There is a critical need to identify molecular markers of prognosis that could improve patient selection for surgical treatment and which may also be employed in the identification of candidates for targeted systemic therapeutic strategies. The epidermal growth factor receptor (EGFR) is one of four members of the ErbB receptor family and is believed to play an integral part in tumorigenesis of multiple epithelial cancers including pancreatic cancer (2). In addition, EGFR is overexpressed in pancreatic cancer (3) and overexpression has been shown to correlate with aggressive tumor behavior (4) and decreased overall survival (5). The EGFR gene intron 1 has a polymorphic region of CA dinucleotide repeats, ranging from 9 to 26 repeats, which is believed by some researchers to affect EGFR transcription efficiency, influence clinical prognosis and modulate anti-EGFR drug sensitivity in colorectal (6), head and neck (7), and breast cancers (8).
We have previously demonstrated that short length of the EGFR intron 1 CA repeats is associated with decreased overall survival among a small number of patients undergoing pancreatic cancer resection (9). Recent reports, however, have challenged the role of EGFR CA repeat length in the regulation of EGFR transcription and its potential role as a predictive indicator of cancer patient survival, tumor aggressiveness, and response to anti-EGFR therapy in colorectal cancer and osteosarcoma (10, 11).
In the present study, we sought to expand the analysis of EGFR intron 1 length in pancreatic cancer by significantly increasing our patient population size and the duration of its clinical follow-up. In this analysis, we have included tissue from patients with locally advanced and/or metastatic pancreatic cancer collected at the time of diagnostic endoscopic ultrasoundguided fine needle apsiration (EUS-FNA). We thus performed an analysis of the relationship between EGFR intron 1 length and clinical outcome in the entire spectrum of pancreatic adenocarcinoma clinical presentations including pancreatic cancer patients with unresectable tumors which constitute the majority. The objectives of our study were to correlate the length of the EGFR intron 1 CA repeats with EGFR mRNA and protein expression levels, tumor characteristics, patient demographics and overall survival in a large cohort of pancreatic cancer patients while attempting to validate the role of EGFR intron 1 length as predictor of clinical outcome.
Materials and Methods
Study subjects
After IRB-approval and informed consent were obtained, tumor specimens were collected from 135 pancreatic cancer patients evaluated at the University of Alabama at Birmingham between 4/1999 and 5/2007. Patients were staged using helical computed tomography with triple phase intravenous contrast pancreatic protocol as well as endoscopic ultrasound. There were 50 patients who underwent laparotomy with curative intent. Tumor specimens were collected at the time of operation, snap-frozen, and stored in liquid nitrogen for later analysis. This group of 50 patients includes a subset (n=30) which has already been described in our previous report (9). In addition, 85 patients who underwent diagnostic EUS-FNA were determined to have unresectable disease by imaging studies. FNA-acquired tumor specimens were collected, snap-frozen, and stored in liquid nitrogen for later analysis. Clinical follow-up was obtained from hospital records.
Human pancreatic cancer cell lines
S2-013 and S2-VP10 cell lines, cloned sublines of SUIT-2 (12) (a gift from Dr. Michael Hollingsworth, University of Nebraska Medical Center), were cultured in DMEM supplemented with L-glutamine and 10% FBS, in a 37 °C incubator with 5% CO2. ASPC-1, BxPC-3, CAPAN-1, HPAC, HPAF-II, MIA PaCa-2, and PANC-1 were obtained from the American Type Culture Collection (Rockville, MD) and propagated according to provider’s recommendations.
Tumor and cell lines DNA isolation
We have previously demonstrated that EGFR intron 1 polymorphism can be reliably measured in any source of patient genomic DNA (9). Tumor DNA from resected pancreatic cancer was isolated using the AquaPure Genomic DNA Isolation Kit (Bio-Rad, Hercules, CA). Tumor genomic DNA from EUS-FNA material was isolated by incubating the entire FNA specimen with 50 μL of a DNA extraction solution at 56 C overnight. The DNA extraction solution consisted of 100 mM Tris-HCl, 2 mM EDTA, 1% Tween-20, and 0.42 mg/mL Proteinase K. The DNA was subsequently purified using Wizard Plus DNA purification system (Promega, Madison, WI). Genomic DNA from nine pancreatic cancer cell lines was isolated using the AquaPure Genomic DNA Isolation Kit (Bio-Rad, Hercules, CA).
Laser capture microdissection of resected tumor samples
Fresh-frozen samples were embedded in Optimal Cutting Temperature (OCT) compound and 8-μm sections were stained with hematoxyllin and eosin. Sections were reviewed by a pancreatic pathologist (N.C.J.) to localize and verify the presence of cancer cells on each slide. Tumor cells were collected using a PixCell II Laser Capture Microdissection (LCM) System (Arcturus Molecular Devices, Sunnydale, CA). Approximately 3,000 tumor cells were captured on each LCM cap for RNA extraction.
RNA isolation and real time RT-PCR analysis
Total RNA from the microdissected sections and cell lines was isolated with the RNAqueous Micro Kit (Ambion, Austin, TX) using the manufacturer protocol. A gross yield of 18 μL of RNA solution per sample was subsequently stored at −80 C. cDNA was synthesized using a High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA). Using patient cDNA, real-time quantitative PCR was performed using TaqMan Gene Expression EGFR Assay-on-Demand (Applied Biosystems, Foster, CA) in an ABI Prism 7700 Detection System. RPLPO was used as an endogenous reference gene.
Immunoblotting
Cell line protein lysates were prepared, and standard SDS-PAGE was performed as previously described (13). Anti-EGFR (Sigma-Aldrich) was diluted 1:1000.
Immunohistochemical (IHC) analysis of EGFR expression
Serial 5 μm sections were cut one day prior to immunostaining from the representative formalin-fixed and paraffin-embedded blocks and mounted on Superfrost/Plus slides (Fisher Scientific, Pittsburgh, PA). Sections were then incubated overnight with anti-EGFR monoclonal antibodies (Santa Cruz biotechnology, Santa Cruz, CA). Secondary detection was performed using a multi-species detection system (Signet Lab Inc., Dedham, MA). Sections were incubated in biotinylated anti-mouse antibodies for 20 min, then incubated with peroxidase-labeled streptavidin for 20 min (Signet Lab Inc.). Antigen–antibody complexes were visualized by incubation with 3,3′-diaminobenzidine substrate (BioGenex, San Ramon, CA) and counterstained with diluted Harris hematoxylin. The stained slides were systematically evaluated by a pathologist for the staining intensity and sub-cellular localization. The intensity was scored from 0 (equals no staining) to 3 (strongest staining, equivalent to A431 cell line control). The H-score was calculated by multiplying staining intensity by percent of cells positive (0 to 100). All negative control slides (omitted primary antibodies) were negative for staining.
EGFR intron 1 polymorphism (CA repeat)
After extraction, 25 ng of DNA was amplified by PCR reaction using an unlabeled forward EGFR primer (5′-GGGCTCACAGCAAACTTCTC-3′) and a fluorescent HEX-labeled reverse EGFR primer (5′-AAGCCAGACTCGCTCATGTT-3′). Conditions were as follows: initial denaturation at 95°C for 5 min; 30 cycles of denaturation at 95°C for 45 seconds, annealing at 60°C for 45 seconds, and extension at 72°C for 45 seconds, and final extension of 72°C for 10 min. Final concentrations in a 50 μL PCR reaction were 5 μL of 10x PCR Buffer II (Applied Biosystems, ABI), 2.5 μM MgCl2, 200 μM each dNTP, 0.25 μM forward primer, 0.25 μM labeled reverse primer, 1.25 units AmpliTaq Gold Polymerase (ABI), and 1 μL DMSO. One to two μL of the amplified PCR products were diluted in 20 μL of water (high-performance liquid chromatography grade) containing 0.5 μL of 400 HD fluorescent size standard. Genotypes were resolved on an ABI Prism 3130XL Genetic Analyzer (Applied Biosystems) to determine allele lengths and number of CA repeats. All analyses were performed at least in duplicates of independent PCRs.
Statistical analysis
Statistical analysis was performed with SPSS (SPSS, Chicago, IL). Patient overall survival was measured from the day of diagnosis by either EUS-FNA cytology or computed tomography. Kaplan-Meier analysis with log-rank test was employed to analyze patient survival and to test differences between patients who underwent pancreatic cancer resection and patients with unresectable tumors. Multivariate Cox regression analysis was applied to evaluate effect on survival of multiple factors simultaneously. Fisher exact test was used to estimate significance of associations between EGFR intron 1 length and various factors (age, race, stage, resection with curative intent, microscopic margins for surgical patients, perineural invasion for surgical patients, adjuvant chemotherapy for surgical patients, and radiation therapy for surgical patients). Spearman correlation coefficient was employed to estimate the association between EGFR intron 1 length and expression levels of EGFR transcript and EGFR protein. Statistical significance was defined at p<0.05.
Results
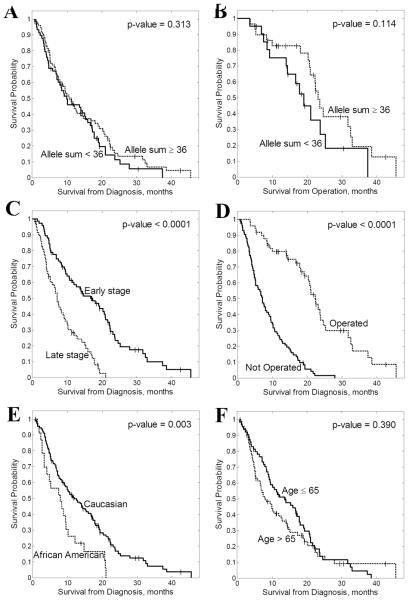
The median allele-specific length of the EGFR intron 1 CA repeats for all the analyzed patients was 18 (range 14-24), with 16 repeats being the most frequent allele-specific length (63% patients) followed by 20 repeats (41% patients). This corroborated the interethnic distribution seen by Liu et al. (14), who found that CA repeat length 16 was the most common allele-specific CA repeat length among Americans (42-43% of all alleles). When allele-specific CA repeats were added, the median sum of repeats (Asum) was 36 (range 29-44), with 43% patients having less than 36 repeats. As described in our previous report, CA repeat length shorter than 36 was classified as short EGFR intron 1 length and CA repeat length ≥ 36 was considered as long EGFR intron 1 length. There was no difference in the proportion of patients with short EGFR intron 1 length between the resectable and unresectable pancreatic cancer subgroups.
We then analyzed the correlation between EGFR intron 1 short (<36) and long (≥36) Asum and patient demographics as well as clinical outcome (Table 1). There was no association between the length of the EGFR intron 1 CA repeats and overall survival in the combined group of pancreatic cancer patients or in the subset of unresectable patients (Figure 1). Interestingly, the association of short length of the EGFR intron 1 CA repeats (Asum<36) with worse overall survival (19.7 vs. 23.1 months, p=0.114) did not reach statistical significance in the subset of resectable patients, and this expanded analysis failed to corroborate our previous findings (9).
Table I.
| Factor | Split | N | Non- censored N |
Median Survival (months) |
Log-rank p-value |
Hazard Ratio |
95% CI | N Allele Sum <36 |
N Allele Sum ≥36 |
Fisher p-value |
|---|---|---|---|---|---|---|---|---|---|---|
|
Allele Sum
(Surgical Patients) |
≥36 | 29 | 19 | 23.1 | 0.114 | 1.86 | 0.86-3.99 | |||
| <36 | 21 | 14 | 19.1 | |||||||
|
Allele Sum
(Combined Patients) |
≥36 | 77 | 65 | 10.6 | 0.313 | 1.22 | 0.83-1.78 | |||
| <36 | 58 | 50 | 9.9 | |||||||
| Min Allele | ≥18 | 35 | 29 | 12.2 | 0.253 | 1.27 | 0.84-1.90 | |||
| <18 | 100 | 86 | 10.1 | |||||||
| Max Allele | ≥20 | 71 | 61 | 10.6 | 0.525 | 1.13 | 0.78-1.64 | |||
| <20 | 64 | 54 | 9.9 | |||||||
| Age | ≤65 | 64 | 57 | 13.6 | 0.390 | 1.18 | 0.81-1.70 | 24 | 40 | 0.296 |
| >65 | 71 | 58 | 7.9 | 34 | 37 | |||||
| Ract | Caucasian | 111 | 93 | 11.8 | 0.003 | 2.46 | 1.35-4.47 | 49 | 62 | 0.818 |
| African Am | 23 | 22 | 7.9 | 9 | 14 | |||||
| Sex | Female | 71 | 61 | 12.2 | 0.456 | 1.12 | 0.78-1.62 | 35 | 36 | 0.163 |
| Male | 64 | 54 | 9.1 | 23 | 41 | |||||
| Stage | I - II | 76 | 61 | 16.7 | < 0.0001 | 3.47 | 2.230-5.39 | 33 | 43 | 1.000 |
| III - IV | 59 | 54 | 7.1 | 25 | 34 | |||||
|
Resection with
Curative Intent |
Yes | 50 | 33 | 22.3 | < 0.0001 | 4.76 | 3.18-7.14 | 21 | 29 | 1.000 |
| No | 85 | 82 | 7.1 | 37 | 48 |
Figure 1.
Kaplan-Meier analysis of overall survival for (A) all pancreatic cancer patients stratified by EGFR intron 1 total CA repeat length; (B) patients who underwent pancreatic cancer resection stratified by EGFR intron 1 total CA repeat length.
Median patient follow-up ranged from 0.7 to 42.4 months. Median overall survival in the pancreatic cancer patient group who underwent surgical resection was 21 months (range 0.8 – 42 months) which is consistent with described pancreatic cancer surgical outcomes (15). Median overall survival for patients with unresectable pancreatic cancer was 7.1 months (range 0.7-27.9 months) (p<0.0001).
To validate the consistency of our pancreatic cancer population as a representative sample, we analyzed the correlation between overall survival and pancreatic cancer patient demographics (Table 1). As expected, earlier tumor stage (I-II), Caucasian race, and ability to undergo resection with curative intent were factors associated with significantly improved overall survival in our cohort. In surgical patients, microscopic margins, peri-neural invasion, adjuvant chemotherapy and radiation therapy were not associated with either improved overall survival or length of EGFR intron 1 CA repeats.
We also performed multivariate Cox regression analysis to test the simultaneous influence of Asum, age, race, sex, tumor stage and surgical treatment on overall patient survival. Our analysis confirmed that stage, race and ability to undergo surgical treatment were the only significant factors correlated with clinical outcome (p<0.0003, p<0.002 and p<10−7, respectively). We did find that age becomes a significant prognostic factor (p<0.0003) when other factors are taken into account.
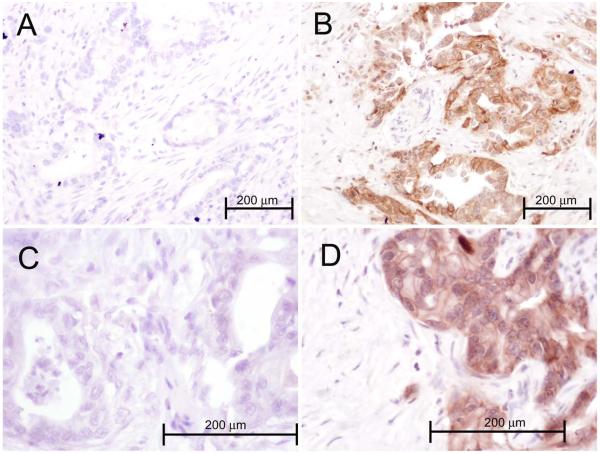
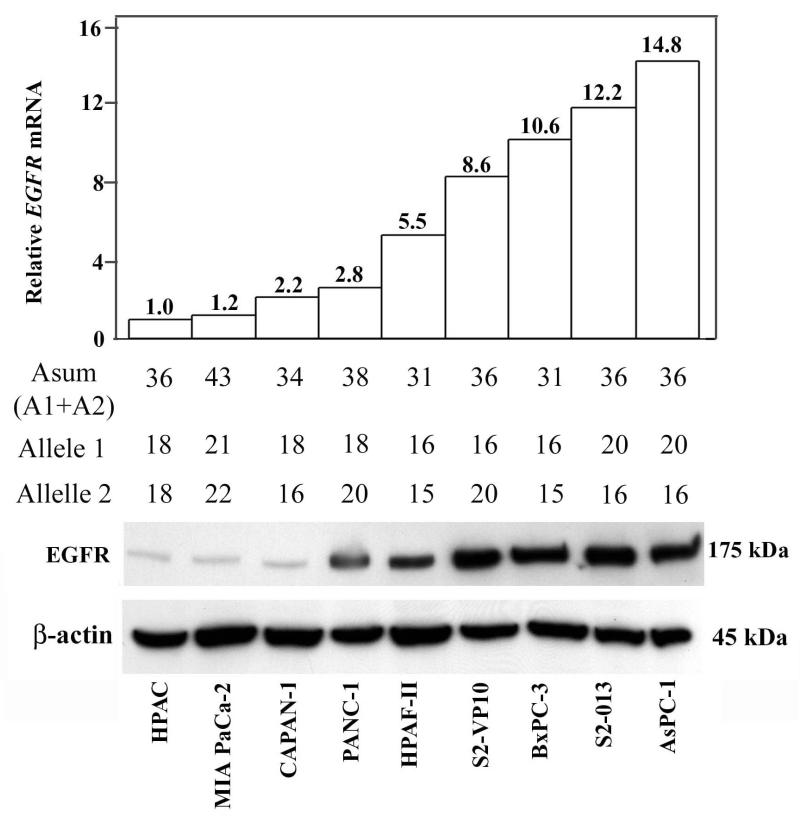
Lastly, we wanted to test if shorter EGFR intron 1 length is indeed associated with increased EGFR transcript and protein expression levels. To test this hypothesis, we microdissected cancer cells in tumor specimens from a subset of 23 patients who had undergone pancreatic cancer resection. This procedure enabled us to separate carcinoma cells from surrounding stromal elements and to exclusively isolate tumor-specific RNA. Real time RT-PCR analysis of EGFR expression levels was then performed and correlated to the EGFR intron 1 length in the same group of patients. The calculated Spearman coefficient found no significant correlation (rho=0.099, p=0.57) between EGFR intron 1 length and EGFR mRNA levels in pancreatic adenocarcinoma patients. We then obtained paraffin blocks for the same 23 patients and performed IHC staining for EGFR protein (Figure 2). As predicted, there was a correlation between transcript and protein expression (rho=0.55, p=0.03), however, there was still no significant correlation between EGFR intron 1 length and EGFR protein expression (rho=0.15, p=0.56). To further prove the absence of correlation between EGFR intron 1 length and EGFR protein expression, we tested the intron 1 length and EGFR expression (both transcript and protein) in a panel of nine representative pancreatic cancer cell lines (Figure 3). The nine pancreatic cancer cell lines displayed the expected spectrum of CA repeat lengths (allele-specific range 15-22). There was a strong correlation between EGFR protein level and EGFR transcript (rho=0.821, p= 0.0067). However, average expression of EGFR protein did not vary significantly according to EGFR intron 1 length.
Figure 2.
The photographs show representative samples of pancreatic adenocarcinoma from 4 patients, stained using immunohistochemistry for EGFR. Each sections shows malignant glands composed of plump epithelial cells with nuclear enlargement and pleomorphism, and the intervening desmoplastic reaction of small fibroblasts. Panels A and C (magnification 200 and 400X, respectively) were scored as 0 (cytoplasmic), 0 (membranous). Panels B and D represent EGFR expressing cancers and were scored as 140 (cytoplasmic) and 60 (membranous). The stromal elements were negative in all samples.
Figure 3.
Expression of EGFR protein, EGFR transcript and EGFR intron 1 total CA repeat length in a panel of nine pancreatic cancer cell lines. Allele-specific and total CA repeats are displayed for each cell line.
Discussion
Increased EGFR protein expression is associated with decreased survival in several epithelial cancers but the mechanisms regulating levels of EGFR expression in cancer are not fully understood. EGFR gene intron 1 length has been proposed as a determinant of EGFR overexpression by improving EGFR transcription efficiency. The molecular implications of this EGFR intron 1 polymorphism have not been completely characterized, but it has been suggested that this area of the intron acts like a joint, bringing the promoter in proximity to a putative repressor protein bound downstream of the CA repeat sequence (16). (6-8). In that regard, EGFR intron 1 length has been shown to affect gene transcription in skin, mammary, head and neck, and gastric cell lines, and this evidence resulted in enough momentum to justify testing its significance in vivo (6, 17).
Analysis of EGFR intron 1 length is a test that can be easily performed in peripheral blood DNA, making it inexpensive and practical as a predictive assay. Because of the described ease of measurement, EGFR intron 1 length has been postulated as an extremely attractive marker not only of EGFR expression but also as a potential predictor of response to anti-EGFR therapy (6). Via its role as a putative regulator of EGFR transcription and EGFR protein expression levels, EGFR intron 1 length has been described as a predictive marker of clinical outcome for a number of carcinomas. Buerger et al. described EGFR intron 1 length in breast cancer (8). Their report showed decreased EGFR transcription activity with increasing number of CA repeats. Since then, a growing number of publications have critically examined the functional meaning of EGFR intron 1 CA repeat length with conflicting results. Etienne-Grimaldi et al noticed that normal tissue EGFR concentrations were not significantly influenced by the length of the CA repeats while such correlation did exist in tumor samples (7). McKay et al showed no association between the distribution of EGFR intron 1 length in colorectal tumors and EGFR protein levels. More intriguingly, intron 1 genotype identified in normal colon samples was not predictive of EGFR mRNA expression levels (18). An additional observation by Buisine et al later confirmed these findings (10). Recent studies in osteosarcoma patients have failed to verify the functional importance of EGFR intron 1 length in EGFR expression (11). Possible reasons for this discordance include lack of tumor-specific EGFR expression regulation and variable significance of EGFR signaling in carcinogenesis and tumor progression depending on cancer type. While the scope of this manuscript was not to demonstrate the functional correlation between EGFR intron 1 length and EGFR expression, our results provide further evidence that CA repeats in intron 1 are unlikely regulators of EGFR gene expression. While this fact was originally postulated based on in vitro experiments, a growing body of literature indicates that there is no functional evidence supporting an association between intron 1 length and EGFR gene expression levels. The lack of relationship with clinical outcome demonstrated in our results further supports this claim, s suggesting the existence of alternative transcription control mechanisms which might account for the purported role of EGFR expression as clinical prognostic marker (11, 19).
We have previously reported that EGFR intron 1 length could be potentially employed to predict pancreatic cancer aggressiveness in a group of 30 surgically resected patients (9). In the present study, we expanded our population size and increased the statistical power of our analysis by by providing extended clinical patient follow-up, and by including patients with unresectable pancreatic cancer. The inclusion of the latter patient subpopulation is of particular significance since over 70% of pancreatic cancer patients present with advanced disease and therefore are not candidates for surgical resection. This majority of patients are often excluded from pancreatic cancer studies due to the lack of available tumor tissue for analysis.
To our knowledge, this is the first comprehensive evaluation of EGFR intron 1 length as a potential surrogate marker of pancreatic cancer clinical outcome in a large population of patients. It is also the first attempt to correlate EGFR intron 1 length with EGFR mRNA and protein levels among patients with both resectable and unresectable pancreatic adenocarcinoma.
Our findings further challenge the putative role of EGFR intron 1 length as a modulator of EGFR transcription efficiency that can influence clinical outcome. The described analysis indicates that EGFR intron 1 length does not reliably discriminate patients with resectable pancreatic cancer from those with unresectable disease as there was no significant difference between the respective groups’ CA repeat lengths. Longer EGFR intron 1 length could not be associated with prolonged patient survival. Furthermore, and at a functional level, EGFR intron 1 length does not seem to influence EGFR transcription efficiency. In the subset of pancreatic cancer patients in whom we analyzed EGFR transcript levels, we found no association between EGFR intron 1 length and either EGFR mRNA or protein expression.
The discrepancy between our current findings and what we have previously reported can be explained by the increase in the sample size and the extension of clinical follow-up. The previously reported improved patient survival with longer EGFR intron 1 length did not hold statistical significance in our updated analysis when 20 patients were added to the resected group and also when a large number of patients with unresectable pancreatic cancer were included. These findings suggest that the length of the CA repeat has no influence on EGFR transcription and pancreatic cancer tumor progression. There are several possible explanations for the discrepancies among studies of EGFR intron 1 length, including the complexity of pancreatic cancer tumorigenesis with multiple signaling pathways that play a critical role in its inception and that may be relatively independent from the EGFR cascade. There is also a wide array of highly variable tumor-host interactions that probably influence tumor progression and that cannot be fully characterized or predicted by analysis of a single gene polymorphism.
We conclude that pancreatic adenocarcinoma EGFR intron 1 does not contribute to the regulation of EGFR transcription. We also conclude that the length of the EGFR intron 1 CA repeats cannot be employed as marker for clinical prognosis in patients with pancreatic cancer.
Summary.
Analysis of the 135 pancreatic cancer patieints revealed no creelation between the length of polymorphic region of CA-repeats in EGFR intron 1 tumor stage. The length of the EGFR intron1 CA repeats also does not correlate with levels of EGFR expression and can not be employed as marker of clinical prognosis in pancreatic cancer patients.
Acknowledgments
This study was supported by Pancreatic SPORE grant P20 CA10195-01 and by the Robert E. Reed Gastrointestinal Oncology Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009 Jul-Aug;59(4):225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Normanno N, De Luca A, Bianco C, Strizzi L, Mancino M, Maiello MR, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006 Jan 17;366(1):2–16. doi: 10.1016/j.gene.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 3.Lemoine NR, Hughes CM, Barton CM, Poulsom R, Jeffery RE, Kloppel G, et al. The epidermal growth factor receptor in human pancreatic cancer. The Journal of pathology. 1992 Jan;166(1):7–12. doi: 10.1002/path.1711660103. [DOI] [PubMed] [Google Scholar]
- 4.Yamanaka Y, Friess H, Kobrin MS, Buchler M, Beger HG, Korc M. Coexpression of epidermal growth factor receptor and ligands in human pancreatic cancer is associated with enhanced tumor aggressiveness. Anticancer research. 1993 May-Jun;13(3):565–9. [PubMed] [Google Scholar]
- 5.Ueda S, Ogata S, Tsuda H, Kawarabayashi N, Kimura M, Sugiura Y, et al. The correlation between cytoplasmic overexpression of epidermal growth factor receptor and tumor aggressiveness: poor prognosis in patients with pancreatic ductal adenocarcinoma. Pancreas. 2004 Jul;29(1):e1–8. doi: 10.1097/00006676-200407000-00061. [DOI] [PubMed] [Google Scholar]
- 6.Amador ML, Oppenheimer D, Perea S, Maitra A, Cusati G, Iacobuzio-Donahue C, et al. An epidermal growth factor receptor intron 1 polymorphism mediates response to epidermal growth factor receptor inhibitors. Cancer Res. 2004 Dec 15;64(24):9139–43. doi: 10.1158/0008-5472.CAN-04-1036. [DOI] [PubMed] [Google Scholar]
- 7.Etienne-Grimaldi MC, Pereira S, Magne N, Formento JL, Francoual M, Fontana X, et al. Analysis of the dinucleotide repeat polymorphism in the epidermal growth factor receptor (EGFR) gene in head and neck cancer patients. Ann Oncol. 2005 Jun;16(6):934–41. doi: 10.1093/annonc/mdi189. [DOI] [PubMed] [Google Scholar]
- 8.Buerger H, Gebhardt F, Schmidt H, Beckmann A, Hutmacher K, Simon R, et al. Length and loss of heterozygosity of an intron 1 polymorphic sequence of egfr is related to cytogenetic alterations and epithelial growth factor receptor expression. Cancer Res. 2000 Feb 15;60(4):854–7. [PubMed] [Google Scholar]
- 9.Tzeng CW, Frolov A, Frolova N, Jhala NC, Howard JH, Vickers SM, et al. Pancreatic cancer epidermal growth factor receptor (EGFR) intron 1 polymorphism influences postoperative patient survival and in vitro erlotinib response. Ann Surg Oncol. 2007 Jul;14(7):2150–8. doi: 10.1245/s10434-007-9409-5. [DOI] [PubMed] [Google Scholar]
- 10.Buisine MP, Wacrenier A, Mariette C, Leteurtre E, Escande F, Aissi S, et al. Frequent mutations of the CA simple sequence repeat in intron 1 of EGFR in mismatch repair-deficient colorectal cancers. World J Gastroenterol. 2008 Feb 21;14(7):1053–9. doi: 10.3748/wjg.14.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kersting C, Agelopoulos K, Schmidt H, Korsching E, August C, Gosheger G, et al. Biological importance of a polymorphic CA sequence within intron 1 of the epidermal growth factor receptor gene (EGFR) in high grade central osteosarcomas. Genes, chromosomes & cancer. 2008 Aug;47(8):657–64. doi: 10.1002/gcc.20571. [DOI] [PubMed] [Google Scholar]
- 12.Iwamura T, Katsuki T, Ide K. Establishment and characterization of a human pancreatic cancer cell line (SUIT-2) producing carcinoembryonic antigen and carbohydrate antigen 19-9. Jpn J Cancer Res. 1987 Jan;78(1):54–62. [PubMed] [Google Scholar]
- 13.Frolov A, Chahwan S, Ochs M, Arnoletti JP, Pan ZZ, Favorova O, et al. Response markers and the molecular mechanisms of action of Gleevec in gastrointestinal stromal tumors. Mol Cancer Ther. 2003 Aug;2(8):699–709. [PubMed] [Google Scholar]
- 14.Liu W, Innocenti F, Chen P, Das S, Cook EH, Jr., Ratain MJ. Interethnic difference in the allelic distribution of human epidermal growth factor receptor intron 1 polymorphism. Clin Cancer Res. 2003 Mar;9(3):1009–12. [PubMed] [Google Scholar]
- 15.Cameron JL, Riall TS, Coleman J, Belcher KA. One thousand consecutive pancreaticoduodenectomies. Ann Surg. 2006 Jul;244(1):10–5. doi: 10.1097/01.sla.0000217673.04165.ea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gebhardt F, Burger H, Brandt B. Modulation of EGFR gene transcription by a polymorphic repetitive sequence--a link between genetics and epigenetics. Int J Biol Markers. 2000 Jan-Mar;15(1):105–10. doi: 10.1177/172460080001500120. [DOI] [PubMed] [Google Scholar]
- 17.Gebhardt F, Zanker KS, Brandt B. Modulation of epidermal growth factor receptor gene transcription by a polymorphic dinucleotide repeat in intron 1. J Biol Chem. 1999 May 7;274(19):13176–80. doi: 10.1074/jbc.274.19.13176. [DOI] [PubMed] [Google Scholar]
- 18.McKay JA, Murray LJ, Curran S, Ross VG, Clark C, Murray GI, et al. Evaluation of the epidermal growth factor receptor (EGFR) in colorectal tumours and lymph node metastases. Eur J Cancer. 2002 Nov;38(17):2258–64. doi: 10.1016/s0959-8049(02)00234-4. [DOI] [PubMed] [Google Scholar]
- 19.Zhang W, He L, Liu W, Sun C, Ratain MJ. Exploring the relationship between polymorphic (TG/CA)n repeats in intron 1 regions and gene expression. Human genomics. 2009 Apr;3(3):236–45. doi: 10.1186/1479-7364-3-3-236. [DOI] [PMC free article] [PubMed] [Google Scholar]