Abstract
Pigment cells of the zebrafish, Danio rerio, offer an exceptionally tractable system for studying the genetic and cellular bases of cell fate decisions. In the zebrafish, neural crest cells generate three types of pigment cells during embryogenesis: yellow xanthophores, iridescent iridophores and black melanophores. In this study, we present evidence for a model whereby melanophores and iridophores descend from a common precursor whose fate is regulated by an interplay between the transcription factors Mitf and Foxd3. Loss of mitfa, a key regulator of melanophore development, resulted in supernumerary ectopic iridophores while loss of foxd3, a mitfa repressor, resulted in fewer iridophores. Double mutants showed a restoration of iridophores, suggesting that one of Foxd3’s roles is to suppress mitfa to promote iridophore development. Foxd3 co-localized with pnp4a, a novel marker of early iridophore development, and was necessary for its expression. Considerable overlap was found between iridoblast and melanoblast markers but not xanthoblast markers, which resolved as cells began to differentiate. Cell lineage analyses using the photoconvertible marker, EosFP, revealed that both melanophores and iridophores develop from a mitfa+ precursor. Taken together, our data reveal a Foxd3/mitfa transcriptional switch that governs whether a bi-potent pigment precursor will attain either an iridophore or a melanophore fate.
Keywords: zebrafish, neural crest, pigment cell, cell fate regulation
INTRODUCTION
Mechanisms underlying cell fate decisions and the acquisition of specific characteristics required for cells to perform their differentiated functions are of central importance in developmental biology. The neural crest, a multipotent cell population that migrates from the dorsal neural tube and develops into multiple cell types including neurons, glia, craniofacial cartilage and pigment cells, has been a popular model system for cell fate acquisition. Current models suggest a sequential process of fate restriction, with a combination of intrinsic regulators, such as transcription factors, and extrinsic cell signals influencing differentiation decisions. One outstanding question is the relative plasticity of these cell fate decisions. While there is considerable evidence that post-migratory, neural crest-derived stem cells retain multipotency when challenged in vitro (Kim et al., 2003; Morrison et al., 2000; Morrison et al., 1999), back transplantation studies suggest that the cell fate choices of these stem cells have become restricted within the embryo (White et al., 2001). However, the relative plasticity of neural crest cells in vivo, which have begun to express lineage-restricted markers, is not well understood.
The development of the black-pigmented melanocytes, referred to as melanophores in fishes and other aquatic vertebrates, is among the best understood cell fate decisions in neural crest development (Cooper and Raible, 2009). Mitf (microphthalmia-associated transcription factor) is a central player in the specification of melanocytes and is amongst the earliest genes expressed in this lineage (Hodgkinson et al., 1993; Opdecamp et al., 1997). Genetic studies demonstrate that this bHLH-leucine zipper transcription factor is necessary for the differentiation of melanocytes and melanophores in all vertebrate taxa (Hodgkinson et al., 1998; Lister et al., 2001; Mochii et al., 1998; Tassabehji et al., 1994). Mitf directly regulates the expression of multiple genes necessary for melanophore development, including dopachrome tautomerase (dct), tyrosinase, tyrosinase-related protein-1, c-kit and bcl2 (Steingrimsson et al., 2004); furthermore, ectopic misexpression of Mitf is sufficient to confer a melanoblast phenotype (Planque et al., 2004; Tachibana et al., 1996). The Mitf gene is directly regulated by the neural crest transcription factors Sox10 and Pax3 (Bondurand et al., 2000; Elworthy et al., 2003; Lacosta et al., 2005; Lee et al., 2000; Potterf et al., 2000; Watanabe et al., 1998), and is extrinsically activated by the Wnt and cyclic AMP signaling pathways (Busca and Ballotti, 2000; Dorsky et al., 2000; Takeda et al., 2000; Widlund et al., 2002) suggesting a model in which melanocyte cell fate specification is promoted by these factors through activation of Mitf. Expression of Mitf is repressed by the forkhead transcription factor, Foxd3, suggesting that its negative regulation is also important for cell fate specification (Curran et al., 2009; Ignatius et al., 2008; Thomas and Erickson, 2009). Indeed, a recent report provides evidence that Foxd3 expression in the avian neural/glial lineage prevents glial precursors from differentiating as melanocytes (Thomas and Erickson, 2009).
Aquatic vertebrates have additional neural crest-derived pigment cells besides melanophores, including yellow xanthophores and iridescent iridophores. Iridophores are present in amphibians, fish, reptiles and certain invertebrate taxa such as cephalopods (Bagnara et al., 1968; Braasch et al., 2006; Demski, 1992; Kelsh, 2004; Mills and Patterson, 2009; Morrison, 1995). Select wavelengths of light are reflected from stacks of organelle-bound crystallized guanine platelets and are perceived by the viewer as bursts of iridescence (Bagnara et al., 2007; Ziegler, 2003). Electron microscopy studies have identified single cells that contain pigment organelles from each of the three pigment cell types, suggesting the different pigment cells may be derived from a common precursor (Bagnara et al., 1979). However, single cell lineage analyses in zebrafish have not supported a common precursor, or clonal relationship, amongst the three pigment cell types (Dutton et al., 2001; Raible and Eisen, 1994).
In this study, we present a model of pigment cell fate whereby melanophores and iridophores descend from a common precursor cell. Our genetic analysis of iridophore development suggests that mitfa, the zebrafish Mitf orthologue, and Foxd3 both regulate iridophore development. Iridophores are strongly reduced with loss of foxd3 activity, whereas excess iridophores are found with loss of mitfa activity. These phenotypes suggest a model in which melanophores and iridophores derive from a common precursor whose fate is regulated by a Foxd3/mitfa transcriptional switch. Epistasis analyses presented here supports the hypothesis that Foxd3 both promotes iridophore development and blocks melanophore development by repressing mitfa. We then test if well-characterized markers for other pigment lineages overlap with a new marker of early iridoblast development, the purine nucleoside phosphorylase gene pnp4a. We find significant overlap between markers of melanoblasts and iridoblasts, but not xanthoblasts. Finally, we test the lineage relationships of these cells directly using transgenic lines and the photoactivatable protein EosFP. These analyses show that a substantial fraction of mitfa expressing cells will subsequently differentiate as iridophores without cell division. These results indicate that cell fate choices remain plastic even after mitfa expression in zebrafish and support a model in which melanophores and iridophores develop from a common precursor cell.
MATERIALS AND METHODS
Animal husbandry and establishment of transgenic lines
A sox10:nls-eos plasmid was generated by PCR amplification from pN1-eos using a primer set containing the SV40 nuclear localization sequence and attB1/B2 recombination sites. The resulting nls-eos cassette was recombined into pDONR221 using BP cloning (Invitrogen) to yield pME-nls-eos. A three-fragment Gateway LR reaction (Invitrogen) then combined pME-nls-eos, p5E-sox10 (Carney et al., 2006) and the Tol2 kit components: p3E-polyA and pDestTol2pA2 (Kwan et al., 2007) to yield sox10:nls-eos. The foxd3zdf10/mitfaw2 double mutant was generated by crossing homozygous mutant animals for the mitfaw2 allele with heterozygous carriers for the foxd3zdf10 allele. Mutant foxd3 carriers were identified by PCR with previously described primers (Stewart et al., 2006) and carriers intercrossed. mitfa−/− offspring were raised to identify zdf10 carriers. Since both genes are located on chromosome 6 (30 cM apart), only a small fraction (5 recombinants out of 37 mitfa−/− fish screened) carried the mutant foxd3 allele. Adult fish of the *AB strain, carrying alleles of foxd3zdf1 (sym1; Stewart et al., 2006), ltkty82 (shd; Kelsh et al., 1996), the transgenic reporter line Tg(mitfa:gfp)w47 (Curran et al., 2009), or mitfaw2 (nacre; Lister et al., 1999) were maintained on a 14 h/10 h light/dark cycle at 28.5 °C. Embryos for all experiments were obtained through natural crosses and staged according to (Kimmel et al., 1995). In some experiments phenylthiocarbamide (PTU; Sigma) was added to embryo medium at a final concentration of 0.2 mM to inhibit melanin synthesis.
pnp4a phylogenetic tree
Alignment of PNP amino acid sequences was performed with ClustalX 2.0.10 (www.clustal.org). Phylogenetic trees were drawn with FigTree v1.2.3 (http://tree.bio.ed.ac.uk/software/figtree/). The following sequences were used for alignment: Zebrafish Pnp4a (NP_001002102.1; ZDB-GENE-040625-83), Zebrafish Pnp4b (NP_991206; ZDB-GENE-040426-1887), Zebrafish Pnp5a (NP_998476;ZDB-GENE-040426-2553), Zebrafish Pnp5b (NP_001004628;ZDB-GENE-040912-54), Zebrafish Pnp6 (NP_991218;ZDB-GENE-040426-1800), Human PNP (NP_000261.2), Mouse Pnp1 (AAC37635), Mouse Pnp2 (NP_001116843), predicted Mouse Pnp3 (XP_001474586), Drosophila Pnp (NP_647727), Yeast Pnp (NP_013310), E. coli PNP(NP_416902), Yeast Mtap (NP_013117), Drosophila Mtap (NP611208), Zebrafish Mtap (NP_956848), Mouse Mtap (NP_077753), Human Mtap (CAG46471).
In situ hybridizations and immunohistochemistry
Digoxigenin-labeled riboprobes for the genes pnp4a (ZDB-GENE-040625-83; Thisse et al., 2004), dct (ZDB-GENE-000508-1; Kelsh et al., 2000b), csf1r (fms, ZDB-GENE-001205-1; Parichy et al., 2000), aox3 (ZDB-GENE-001205-2; Parichy et al., 2000) have been characterized previously. In situ hybridization was performed as described previously (Lister et al., 1999), using NBT/BCIP as a chromogenic substrate. Fluorescent in situ hybridization was performed as described previously (Julich et al., 2005) using anti-Dig POD for pnp4a and dct, anti-Fluor POD for pnp4a, dct, csf1r and aox3, Alexa-Fluor tyramide substrate 568 and 488 (Invitrogen) and Roche blocking reagent and buffer. The following antibodies were used for immunohistochemistry at the indicated dilutions: rabbit polyclonal anti-Foxd3 (Lister et al., 2006), 1:500; mouse monoclonal anti-Pax3/7 (DP312; Davis et al., 2005), 1:500; mouse monoclonal anti-Green Fluorescent Protein (Invitrogen), 1:1000; anti-mouse (Alexa 488) and anti-rabbit (Alexa 568) secondary antibodies (Molecular Probes) were used at 1:750. Brightfield images were obtained on a Nikon dissecting microscope with a Spot RT Slider digital camera (Diagnostic Instruments). Fluorescent confocal images were obtained on a LSM 5 Pascal confocal microscope (Zeiss). Images were processed for color balancing and brightness/contrast using Photoshop CS4 (Adobe) and formatted with Illustrator CS4 (Adobe).
EosFP photoconversion and cell lineage tracing
Photoconversion experiments were performed on individual cells expressing the sox10:nls-eos and mitfa:gfp transgenes. sox10:nls-eos plasmid was injected into one-cell Tg(mitfa:gfp)w47 embryos. The resulting embryos express GFP signal throughout the cytoplasm of mitfa positive cells and transiently express photoconvertible green Eos in the nuclei of a sub-set of sox10 cells. At 24 hpf, embryos were de-chorionated, anesthetized with MESAB and individually mounted for examination on a Zeiss Axioplan2 compound scope. Using a 20X objective with a constricted diaphragm, a single double-positive sox10:nls-eos/mitfa:gfp cell per zebrafish was briefly exposed (3–5 seconds) to ultraviolet light (405nm). The resulting cell displayed a photoconverted red sox10:nls-eos nucleus and maintained the green mitfa:gfp cytoplasm. Photoconverted zebrafish were returned to embryo media in a 28.5 °C incubator. At 48 and 72 hpf, photoconverted zebrafish were analyzed with brightfield light and incident light to identify cell fate.
Iridophore cell counts and morpholino oligonucleotide injection
Tail iridophores (those appearing caudal to the cloaca) were counted at approximately 51–54 hpf on a dissecting microscope with epi-illumination from a fiber optic light source. foxd3 and mitfa antisense morpholino oligonucleotides have been previously described: foxd3, (Lister et al., 2006); mitfa (Nasevicius and Ekker, 2000).
RESULTS
pnp4a is a novel iridoblast marker
The first iridophores terminally differentiate by 42 hpf, as revealed by iridescence of their organelle-bound reflecting platelets. These initial iridophores sparsely populate the dorsal stripe and the surface of the eye (Fig. 1A). By 72 hpf, differentiated iridophores more densely populate the dorsal stripe, ventral stripe, ventral yolk stripe and the eye (Fig. 1B). To characterize earlier events during iridophore development we needed to identify an early marker of iridophore precursors. By microarray screening, we found that transcript for the purine nucleoside phosphorylase gene, pnp4a, is enriched in skin with attached pigment cells in wild-type zebrafish as compared to the closely related iridophore-deficient species, D. albolineatus (D.M. Parichy, unpublished). pnp4a is one of five highly conserved pnp genes found in zebrafish, compared to three genes expressed in mouse and one in human (see Fig. S1 in the supplementary material).
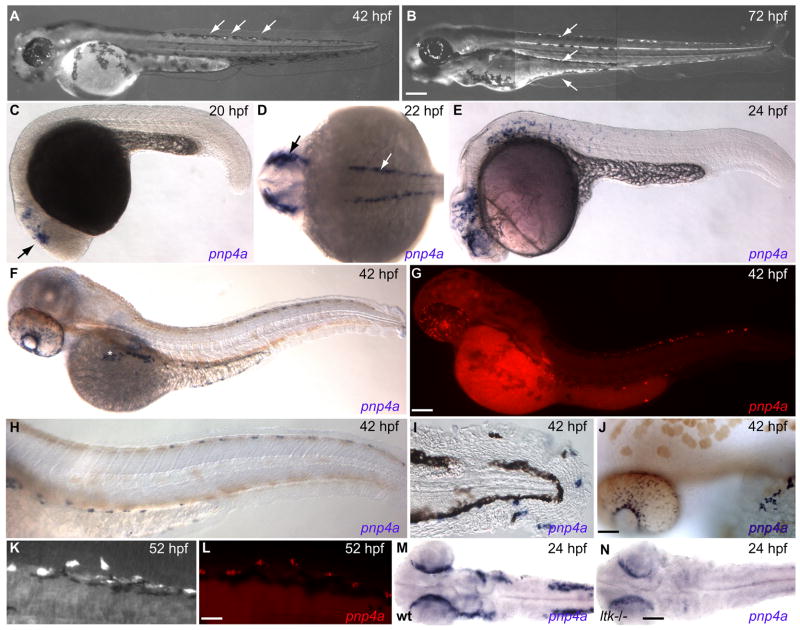
Fig. 1. Expression pattern of iridophores and pnp4a throughout embryonic development.
(A,B) Terminally differentiated iridophores illuminate under incident light. (A) At 42 hpf, three iridophores first reach terminal differentiation along dorsal stripe (arrows), iridophores scatter across the surface of retina (*). (B) By 72 hpf, iridophores more densely populate dorsal, ventral and ventral yolk stripes (arrows). Eye iridophores coalesce into a ring surrounding the lens (*). (C–J,L,M) In situ hybridization reveals pnp4a expression at different embryonic stages. (C) pnp4a first appears in anterior head region at 20hpf, behind primordial eye (arrow). (D) By 22 hpf, pnp4a expresses exclusively in neural crest domains: lateral dorsal stripes along anterior trunk region (white arrow) and cranial region (black arrow). (E) At 24 hpf, pnp4a positive cells migrate posteriorly and ventrally. (F,H,J) Embryos treated with 1× PTU to inhibit melanin synthesis. (F) pnp4a positive cells have organized along the dorsal, ventral and ventral yolk stripes. A patch of pnp4a positive cells scatter across eye and congregate along presumptive swim bladder iridophore patch on dorsal side of yolk ball (*). (H) Close-up of trunk and tail reveal pnp4a positive cells migrate along similar pathway as melanophores in dorsal and ventral stripes, (20X). (I) Close-up of pnp4a positive cells in tail peripheral to v-stripe of melanophores, (20X). (J) pnp4a positive cells coalesce around lens in eye and along yolk ball, (20X). (G,L) pnp4a in situ fluorescence, Red: pnp4a. (K) wild-type embryo illuminated with incident light to reveal iridophore pattern then (L) fixed and processed for pnp4a fluorescent in situ hybridization. (M,N) Dorsal view of head and anterior trunk region of 24 hpf zebrafish. (M) pnp4a expression in wild-type embryo (heterozygous sibling). (N) pnp4a expression in ltk−/− (shd) mutant. Scale bars: (A,B) 300 μm; (C-G) 150 μm; (H–J) 75 μm; (K,L) 25 μm; (M,N) 80 μm.
We determined the spatiotemporal pattern of pnp4a gene expression by whole-mount mRNA in situ hybridization (Figs. 1C–J,L,M). We found that expression of pnp4a begins at 22 hpf, approximately 20 hours prior to iridophore terminal differentiation. pnp4a is expressed solely in a subset of neural crest cells (Figs. 1C–E) and does not appear in other tissue types in the developing larval zebrafish (Figs. 1F,G). pnp4a first appears at 20 hpf in the head (Fig. 1C). A dorsal view (22hpf) reveals staining restricted to the neural crest dorsolateral stripes (Fig. 1D). At 24 hpf, pnp4a+ cells are observed along characteristic neural crest migratory pathways (Figs. 1E,M). At 42 hpf, pnp4a positive cells are restricted to a spatial pattern that is characteristic of differentiated iridophores: the dorsal stripe, ventral stripe, ventral yolk stripe and eye, as seen with NBT/BCIP in situ hybridization (Fig. 1F) and fluorescent in situ hybridization (Fig. 1G). High magnification images reveal pnp4a positive cells in the trunk (Fig. 1H), tail (Fig. 1I), and the surface of the eye and dorsal yolk ball (Fig. 1J). To confirm that terminally differentiated iridophores express pnp4a, we imaged the dorsal stripe iridophore pattern of individual WT embryos at 52 hpf (Fig. 1K), processed embryos individually for pnp4a mRNA expression and imaged the resulting pattern of pnp4a staining (Fig. 1L; note that native iridescence of iridophores is lost following histological processing). The positions of pnp4a+ cells and terminally differentiated iridophores correspond precisely, thereby illustrating the specificity of pnp4a to iridophores.
If pnp4a is an enzyme present in iridoblasts, one would expect a pnp4a reduction in an iridophore mutant. Leukocyte tyrosine kinase (ltk), is critical for early iridophore development (Lopes et al., 2008). We assayed pnp4a expression in homozygous ltk mutant embryos by in situ hybridization. Heterozygous ltk mutant parents were crossed and 26% (6 of 23) of offspring exhibited a dramatic reduction of pnp4a signal (Figs. 1M,N). pnp4a signal in the eye was moderately reduced while trunk and anterior trunk staining were strongly reduced. Similar reductions in pnp4a signal were observed at 30 hpf in offspring from heterozygous ltk mutant parents: 25% (7 of 28; data not shown). Taken together, these results confirm that pnp4a is a specific cell marker for iridoblasts.
Foxd3 is necessary for pnp4a expression
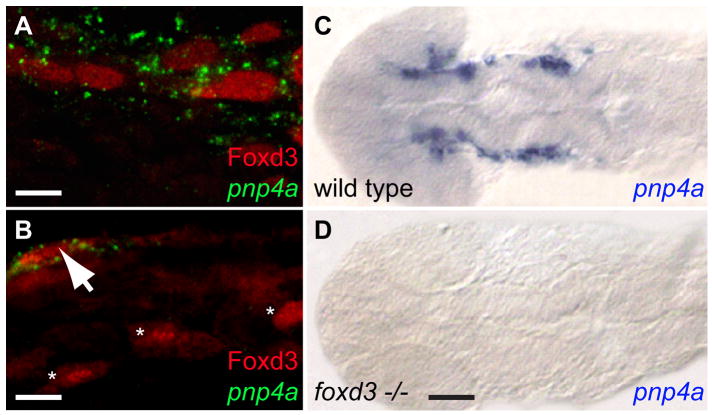
The transcriptional repressor Foxd3 is necessary for iridophore differentiation (Lister et al., 2006). While Foxd3 expression was not previously found in overtly differentiated iridophores, we showed that GFP expression persists in iridophores in a Tg(foxd3:gfp) transgenic line (Curran et al., 2009), suggesting this protein may be expressed at earlier stages in the iridophore lineage. To further explore the Foxd3/iridophore relationship, we co-stained migratory neural crest at 24 hpf with Foxd3 antibody and pnp4a probe. We found that 63% of pnp4a+ cells (n=78) co-expressed Foxd3 protein (Fig. 2A). Reciprocally, 24% of Foxd3+ cells (n=123) co-expressed pnp4a transcript (Fig. 2B). This would be expected as Foxd3 is expressed in multiple neural crest derivatives, including glia, enteric neurons and dorsal root ganglion neurons (Gilmour et al., 2002; Lister et al., 2006). To test if Foxd3 is necessary for pnp4a expression, we next assayed pnp4a staining in homozygous mutant foxd3 embryos at 22–24 hpf by in situ hybridization (Figs. 2C,D). Heterozygous parents were crossed and 23% (6 of 26) of progeny exhibited dramatically reduced pnp4a signal. Flat mounted wild-type larvae reveal pnp4a signal in the lateral stripes flanking the neural tube in the head and anterior trunk region (Fig. 2C). pnp4a signal is strikingly absent in the foxd3 mutants (Fig. 2D). These results demonstrate that Foxd3 is necessary for initial pnp4a expression.
Fig. 2. Foxd3 is necessary for pnp4a expression.
(A,B) Wild-type fish co-stained with pnp4a and Foxd3, 24 hpf, anterior trunk. Green: pnp4a mRNA, Red: Foxd3 antibody (A) Punctate, cytoplasmic pnp4a mRNA signal surrounds Foxd3 positive nuclei, 63X. (B) Field reveals a pnp4a+/Foxd3+ cell (arrow) adjacent to three pnp4a−/Foxd3+ cells (*), 40X. (C,D) Flat mounted head and trunk stained with pnp4a riboprobe, 22 hpf, dorsal view, anterior left, 10X. (C) wild-type (D) foxd3 −/− mutant (sym1). Scale bars: (A) 10 μm; (B) 20 μm; (C,D) 70 μm.
Rescue of iridophores in foxd3/mitfa double mutants
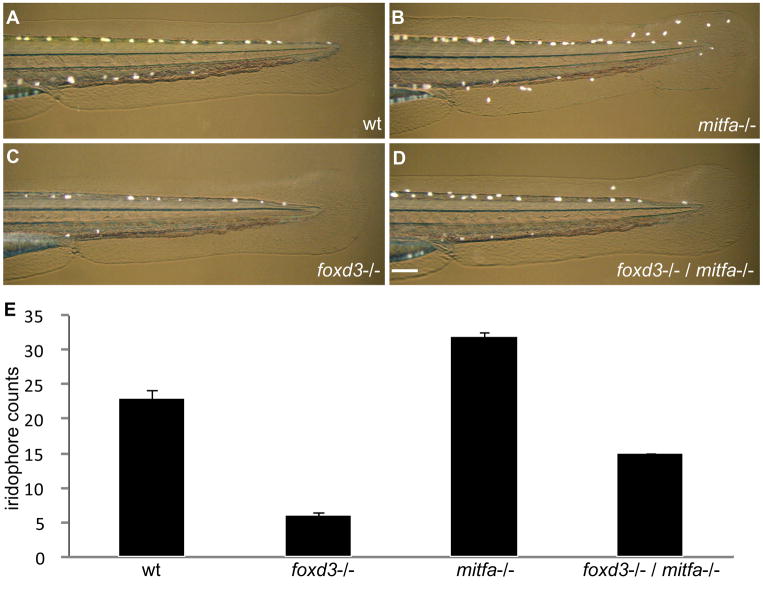
To better understand the role of Foxd3 and mitfa in iridophore development, we performed an epistasis analysis on the iridophore phenotype. We and others previously showed that Foxd3 negatively regulates Mitf expression (Curran et al., 2009; Ignatius et al., 2008; Thomas and Erickson, 2009), while loss of foxd3 function results in a decrease in iridophores and loss of mitfa results in ectopic iridophores (Lister et al., 2006; Lister et al., 1999). Therefore, we predicted that if Foxd3 promotes iridophore development by repressing mitfa, then embryos doubly mutant for foxd3 and mitfa should have more iridophores than foxd3 single mutants. Moreover, if Foxd3 also promotes iridophore development independently of its repressive effects on mitfa, then foxd3/mitfa double mutants should have fewer iridophores than mitfa single mutants. Compared to wild-type animals (Figs. 3A,E), loss of mitfa results in supernumerary iridophores (Figs. 3B,E) while loss of foxd3 strongly reduces iridophore counts (Figs. 3C,E), as previously reported. Notably, significantly more iridophores are observed in double mutant embryos (Figs. 3D,E). As expected, the loss of melanophore phenotype observed in mitfa−/− was not rescued in double mutants, as mitfa is necessary for melanophore specification and differentiation (Lister et al., 1999). Additionally, other neural crest derivatives, including enteric neurons, dorsal root ganglia, jaw cartilage and glial cells remained reduced in the double mutant (data not shown). Therefore, mitfa is epistatic to foxd3 with respect to the iridophore phenotype. These results demonstrate that Foxd3 repression of mitfa is a necessary step in iridophore development and suggest the hypothesis that melanophores and iridophores derive from a common precursor.
Fig. 3. foxd3/mitfa double mutant exhibits partial rescue of iridophores.
(A–D) Incident light reveals iridophores on trunk and tail of 51–54 hpf zebrafish, lateral view, anterior left, 5X. (A) Wild-type zebrafish displays normal numbers of iridophores. (B) mitfa −/− (nacre) displays supernumerary iridophores. (C) foxd3 −/− (sym1) displays iridophore reduction. (D) foxd3/mitfa double mutant exhibits partial rescue of iridophore phenotype as compared to foxd3−/− reduction alone. (E) Iridophores were counted along the dorsal and ventral stripes from the posterior tail region, between the cloacae and tail tip. Cell counts taken from 51 zebrafish for each genetic background: Total cell counts (wild-type: 1144), (foxd3−/−: 305), (mitfa−/−: 1615), (foxd3/mitfa double mutant: 748). Mean iridophore cell counts: (wild-type: 22.9), (foxd3−/−: 6.1), (mitfa−/−: 32.3), (foxd3/mitfa double mutant: 15.0). Bars = s.d. Scale bar: (A–D) 100 μm.
To address the question of whether eye iridophores are regulated by Foxd3 in a similar manner to trunk iridophores, we performed iridophore cell counts on the eyes of 48 hpf zebrafish under various conditions (see Fig. S2 in the supplementary material). In agreement with trunk iridophores, we observed ectopic eye iridophores in mitfa −/− mutants. However, in contrast to trunk iridophores, we did not observe a reduction in eye iridophores in either the foxd3 MO or after loss of function of mitfa and foxd3. These results contrast with initial expression of pnp4a in the eye at early stages (Fig. 2). However we see later expression of pnp4a in foxd3 mutants wherever iridophores are found (data not shown). We conclude that Foxd3 does exert transcriptional control on the timing and intensity of pnp4a eye expression; however Foxd3 is dispensable for eye iridophore differentiation.
mitfa-expressing neural crest cells reacquire Foxd3 expression
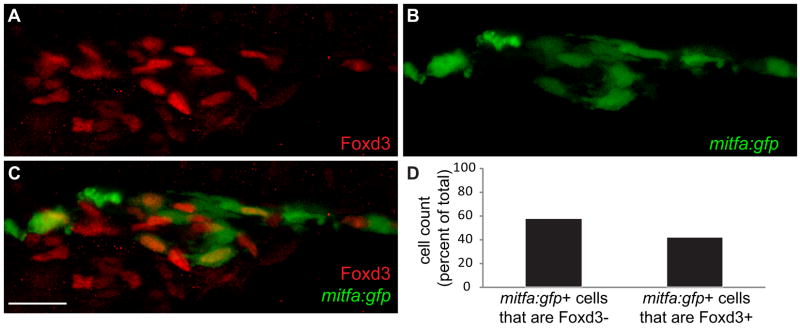
Foxd3 expression serves as a robust marker for pre-migratory neural crest (Hromas et al., 1999; Labosky and Kaestner, 1998; Odenthal and Nusslein-Volhard, 1998; Pohl and Knochel, 2001; Sasai et al., 2001), however, as development proceeds Foxd3 is down-regulated in the head and anterior trunk then later reappears in specific neural crest derivatives, such as glia associated with the lateral line (Kelsh et al., 2000a; Gilmour et al., 2002; Lister et al., 2006). We find that Foxd3 exhibits similar biphasic expression in mitfa+ chromatoblasts. Previously, we demonstrated that the vast majority (greater than 90%) of 18 hpf neural crest cells, which begin to express mitfa, are Foxd3 negative (Curran, 2009). To test if Foxd3 is reactivated in a subset of mitfa+ cells, we used an identical assay to examine Foxd3 expression in mitfa:gfp transgenic animals at 24 hpf. We have previously shown that all mitfa:gfp+ cells express endogenous mitfa transcript (Curran et al., 2009). Foxd3 displayed nuclear expression in a substantial proportion of mitfa+ neural crest cells (Figs. 4A–C). Cell counts reveal 48% of mitfa+ cells (n=748) are Foxd3 positive at 24 hpf (Fig. 4D). Thus, despite the downregulation of Foxd3 in the vast majority of mitfa+ cells at 18 hpf, nearly half of mitfa+ cells have begun to re-express Foxd3 six hours later. These results suggest the hypothesis that re-expression of Foxd3 in a subset of mitfa+ cells promotes their differentiation as iridophores.
Fig. 4. mitfa positive neural crest cells re-acquire Foxd3 expression.
(A–C) Confocal images taken from lateral aspect of anterior trunk, 40X. (A) Foxd3, (B) mitfa:gfp, (C) Merge: Red channel: Foxd3. Green channel: mitfa:gfp. (D) Cell counts of mitfa:gfp positive cells that are either Foxd3 positive or negative, counts derived from 40X confocal images at 24 hpf, numbers given as percent of total. 52% of mitfa:gfp+ cells are Foxd3− (432/748). 48% of mitfa:gfp+ cells are Foxd3+ (316/748). Scale bar=30 μm.
Iridoblast marker co-localizes with melanoblast markers
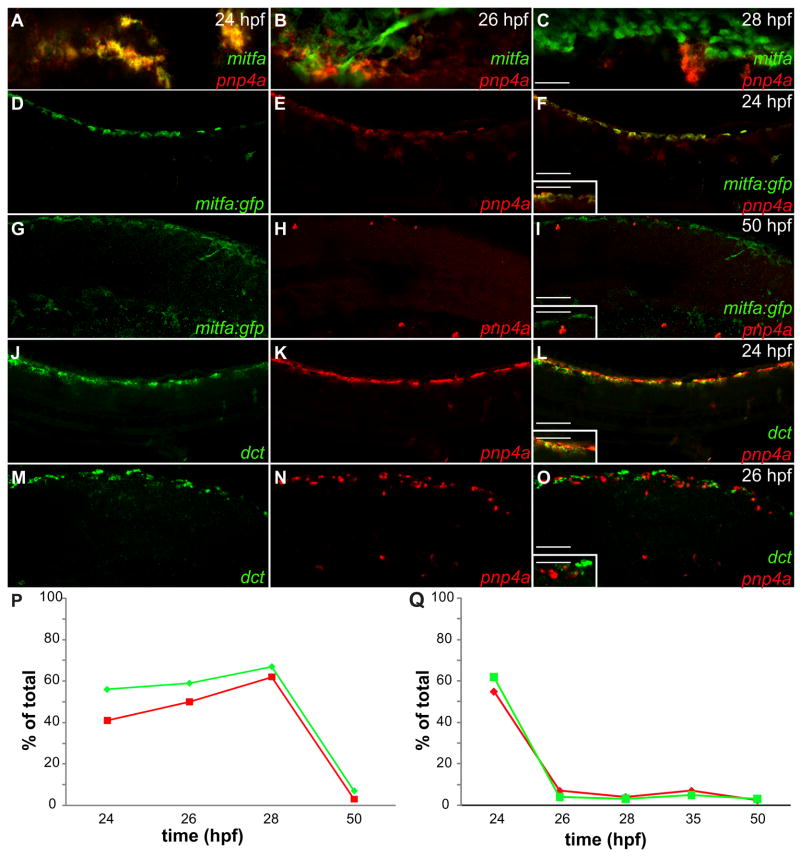
To assess the cell lineage relationship between melanoblasts and iridoblasts we quantified the degree of overlap between the iridoblast marker, pnp4a, and melanoblast genes, mitfa and dct. All cell counts were collected from confocal images from the lateral aspect of the anterior tail region of fixed zebrafish. Cells co-stained to reveal both mitfa and pnp4a mRNA reveal considerable overlap at 24 hpf (Fig. 5A), this co-localization progressively diminishes as development proceeds (Figs. 5B,C). To further quantify mitfa expression, we took advantage of mitfa:gfp transgenic animals. At 24 hpf, 42% of pnp4a+ cells stain for mitfa:gfp and 57% of mitfa+ cells stain for pnp4a mRNA (Figs. 5D–F,P; Table 1). A similar degree of overlap between mitfa:gfp and pnp4a mRNA persists until 28 hpf (Fig. 5P; Table 1). At 50 hpf the population of co-localized cells drops considerably; only 3% of pnp4a+ cells stain for mitfa and 8% of mitfa+ cells stain for pnp4a (Figs. 5G–I,P; Table 1). To further test if melanoblasts and iridoblasts share developmental genes, we examined overlap with a second melanoblast marker, dct (Kelsh et al., 2000b). A similar pattern of overlap was observed with pnp4a and dct, however, dct and pnp4a resolve somewhat earlier than mitfa and pnp4a. At 24 hpf, 55% of pnp4a+ cells stain positive for dct mRNA and 62% of dct+ cells stain positive for pnp4a (Figs. 5J–L,Q; Table 1). By 26 hpf, two hours later, these cell markers are largely resolved; only 7% of pnp4a+ cells stain for dct and 4% of dct+ cells are pnp4a+ (Figs. 5M–O,Q; Table 1). These temporal differences in dct and mitfa:gfp expression may reflect increased stability of GFP protein relative to transcript. In conclusion, we observed that approximately 50% of melanoblasts and iridoblasts share developmental genes at 24 hpf, while the markers resolve into their respective cell types at 50 hpf.
Fig. 5. Iridoblast marker co-localizes with melanoblast markers.
(A–O) Confocal images collected from lateral aspect of anterior tail region of fixed zebrafish, 20X. (A–C) Cells co-stained with mitfa riboprobe (red) and pnp4a riboprobe (green) reveal considerable overlap at 24 hpf (A) and diminishing overlap as development proceeds (B,C). mitfa:gfp transgenic reveals mitfa+ cells overlap with pnp4a expression at 24 hpf (D–F) and resolve at 50 hpf (G–I). (D,G) mitfa:gfp (E,H) pnp4a (F,I) Color merge: Green: GFP expression, Red: pnp4a mRNA (inset 40x). Wild-type embryos reveal dct+ cells overlap with pnp4a expression at 24 hpf (J–L) then resolve at 26 hpf (M–O). (J,M) dct (K,N) pnp4a (L,O) Color merge: Green: dct mRNA, Red: pnp4a mRNA (inset-40x). (P,Q) Percent of overlap between chromatoblast markers (see Table 1). (P) Green line= % of mitfa:gfp+ cells that are mitfa:gfp+/pnp4a+. Red line= % of pnp4a+ cells that are mitfa:gfp+/pnp4a+. (Q) Green line= % of dct+ cells that are dct+/pnp4a+. Red line= % of pnp4a+ cells that are dct+/pnp4a+. Scale bars: (A–C) 40 μm; (D–O) 60 μm; (F,I,L,O inset) 30 μm.
Table 1.
Iridoblasts and melanoblast co-localization analysis
| time-points(hpf) | double positive cells(pnp4a+/mitfa+) | pnp4a+ only | mitfa+ only | % of pnp4a+ that are pnp4a+/mitfa+ | % of mitfa+ that are pnp4a+/mitfa+ |
|---|---|---|---|---|---|
| 24 | 135 | 188 | 124 | 42 | 57 |
| 26 | 128 | 128 | 89 | 50 | 59 |
| 28 | 142 | 87 | 70 | 62 | 67 |
| 50 | 6 | 194 | 259 | 3 | 8 |
| time- points(hpf) | double positive cells (pnp4a+/dct+) | pnp4a+ only | dct+ only | % of pnp4a+ that are pnp4a+/dct+ | % of dct+ that are pnp4a+/dct+ |
| 24 | 215 | 176 | 132 | 55 | 62 |
| 26 | 9 | 120 | 216 | 7 | 4 |
| 28 | 4 | 96 | 129 | 4 | 3 |
| 35 | 7 | 93 | 133 | 7 | 5 |
| 50 | 11 | 264 | 356 | 4 | 3 |
Neither iridoblast nor melanoblast markers co-localize with xanthoblast markers
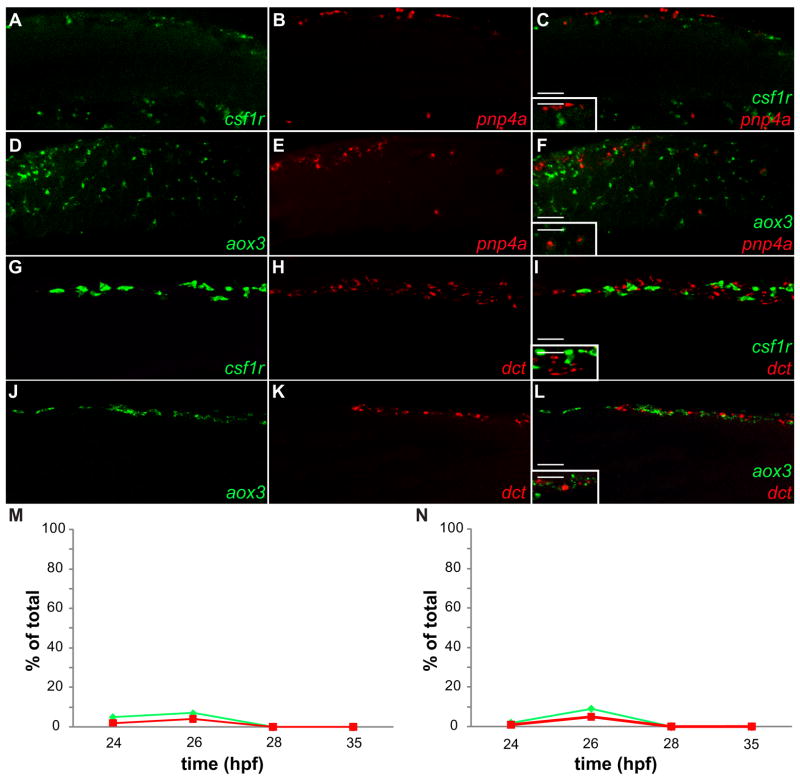
To assess the cell lineage relationship between xanthoblasts and the other chromatoblasts, iridoblasts and melanoblasts, we quantified the degree of overlap between the xanthoblast markers colony stimulating factor-1 receptor (csf1r, formerly fms) and aldehyde oxidase 3 (aox3, formerly xanthine dehydrogenase) with both the iridoblast marker, pnp4a, and the melanoblast markers, dct and mitfa. All cell counts were collected from confocal images from the lateral aspect of the anterior tail region of fixed zebrafish. csf1r encodes a receptor tyrosine kinase and is essential for early larval xanthophore development, whereas aox3 catalyses the synthesis of the xanthophore pteridine pigment, xanthopterin (Parichy et al., 2000). At 24 hpf, 2% of pnp4a+ cells stain positive for csf1r and 5% of csf1r+ cells stain positive for pnp4a (Figs. 6A–C,M; Table 2). By 28 hpf, there is no overlap between csf1r and pnp4a (Fig. 6M; Table 2). aox3 and pnp4a display a similar pattern of overlap. At 24 hpf, only 1% of pnp4a+ cells stain for aox3 and 2% of aox3+ cells stain for pnp4a (Figs. 6D–F,N; Table 2), while at 28 hpf there is no overlap between markers (Fig. 6N; Table 2). In conclusion, we observed no significant overlap of the xanthoblast development genes, csf1r and aox3, with the iridoblast gene, pnp4a. The Pax3/7 sub-family of genes encodes paired box transcription factors. They are critical for xanthophore development but express broadly early in the neural crest (Minchin and Hughes, 2008). In contrast to csf1r and aox3, Pax3/7 antibody staining shows significant overlap with pnp4a until 50 hpf (see Figs. S2 [A–F,J] and Table S1 in the supplementary material). Pax3/7 also co-localizes strongly with mitfa:gfp positive cells between 24 and 32 hpf (see Figs. S3 [G–I,K] and Table S1 in the supplementary material).
Fig. 6. Neither iridoblast nor melanoblast markers co-localize with xanthoblast markers.
(A–L) Confocal images collected from lateral aspect of anterior tail region of fixed zebrafish, 20X. (A–C,M) Wild-type embryo reveals csf1r signal not localized with pnp4a expression at 24 hpf (A) csf1r (B) pnp4a (C) Color merge: Green: csf1r mRNA, Red: pnp4a mRNA (inset 40x). (D–F,N) Wild-type embryo reveals aox3 signal is not localized with pnp4a expression at 24 hpf (D) aox3 (E) pnp4a (F) Color merge: Green: aox3 mRNA, Red: pnp4a mRNA (inset-40x). (G–I) Wild-type embryo reveals csf1r signal is not localized with dct expression at 24 hpf (G) csf1r (H) dct (I) Color merge: Green: csf1r mRNA, Red: dct mRNA (inset 40x). (J–L) Wild-type embryo reveals aox3 signal is not localized with dct expression at 24 hpf (J) aox3 (K) dct (L) Color merge: Green: aox3 mRNA, Red: dct mRNA (inset 40x). (M,N) Percent of overlap between chromatoblast markers (see Table 2,3). (M) Green line= % of csf1r+ cells that are pnp4a+/csf1r+. Red line= % of pnp4a+ cells that are pnp4a+/csf1r+. (N) Green line= % of aox3+ cells that are pnp4a+/aox3+. Red line= % of pnp4a+ cells that are pnp4a+/aox3+. Scale bars: (AL) 60 μm; (C,F,I,L inset) 30 μm.
Table 2.
Iridoblast and xanthoblast co-localization analysis
| time-points (hpf) | double positive cells (pnp4a+/csf1r+) | pnp4a+ only | csf1r + only | % of pnp4a+ that are pnp4a+/csf1r + | % of csf1r + that are pnp4a+/csf1r + |
|---|---|---|---|---|---|
| 24 | 3 | 147 | 57 | 2 | 5 |
| 26 | 5 | 120 | 66 | 4 | 7 |
| 28 | 0 | 234 | 210 | 0 | 0 |
| 35 | 0 | 145 | 153 | 0 | 0 |
| time-points (hpf) | double positive cells (pnp4a+/aox3+) | pnp4a+ only | aox3+ only | % of pnp4a+ that are pnp4a+/aox3+ | % of aox3+ that are pnp4a+/aox3+ |
| 24 | 4 | 396 | 196 | 1 | 2 |
| 26 | 6 | 114 | 61 | 5 | 9 |
| 28 | 0 | 189 | 215 | 0 | 0 |
| 35 | 0 | 109 | 165 | 0 | 0 |
The percentage of overlap between melanoblast markers and xanthoblast markers remains below 1% at each time point. At 24 hpf, only 0.6% of dct+ cells stain for csf1r and 0.4% of csf1r+ cells stain for dct (Figs. 6G–I; Table 3). The percentage of overlap between dct and csf1r falls to 0% at 28 hpf (Table 3). At 24 hpf, 0.7% of dct positive cells stain positive for aox3 and 0.5% of aox3+ cells stain for dct (Figs. 6J–L; Table 3). The percentage of overlap between dct and aox3 also arrives at 0% at 28 hpf (Table 3). In conclusion, we observed no significant overlap of the xanthoblast development genes, csf1r and aox3, with the melanoblast development gene, dct, between 24 and 35 hpf.
Table 3.
Melanoblast and xanthoblast co-localization analysis
| time-points (hpf) | double positive cells (dct+/csf1r +) | dct+ only | csf1r + only | % of dct+ that are dct+/csf1r + | % of csf1r + that are dct+/csf1r + |
|---|---|---|---|---|---|
| 24 | 1 | 166 | 249 | 0.6 | 0.4 |
| 26 | 0 | 132 | 123 | 0 | 0 |
| 28 | 0 | 89 | 78 | 0 | 0 |
| 35 | 0 | 65 | 68 | 0 | 0 |
| time points (hpf) | double positive cells (dct+/aox3+) | dct+ only | aox3+ only | % of dct+ that are dct+/aox3+ | % of aox3+ that are dct+/aox3+ |
| 24 | 1 | 142 | 199 | 0.7 | 0.5 |
| 26 | 0 | 132 | 99 | 0 | 0 |
| 28 | 0 | 56 | 78 | 0 | 0 |
| 35 | 0 | 83 | 56 | 0 | 0 |
Melanophores and iridophores arise from a common mitfa+ precursor
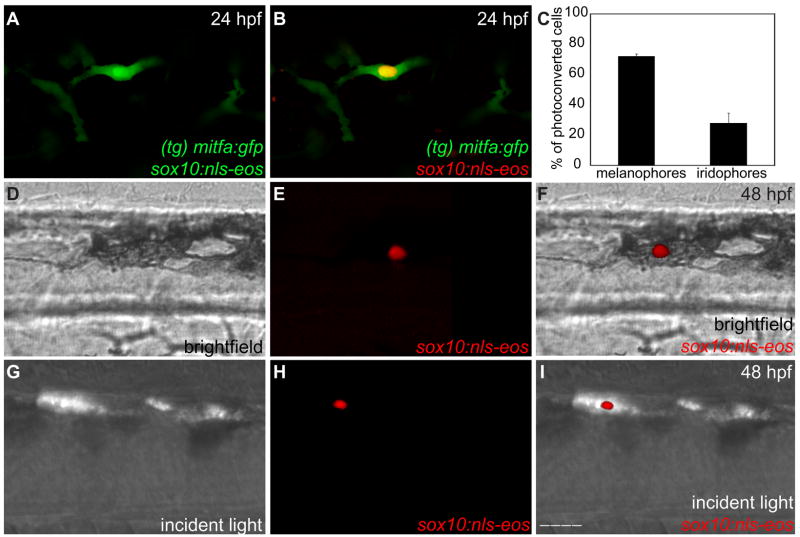
If a subset of pigment cell precursors express both iridoblast and melanoblast genes at 24 hpf, it is possible that these precursors maintain the potential to acquire either an iridophore or a melanophore fate. To explore this cell lineage relationship, we employed the photoconvertible gene reporter, EosFP. EosFP emits green fluorescence (516 nm) that changes to red (581 nm) upon near-UV irradiation (≈405 nm) due to a photo-induced modification involving a break in the peptide backbone adjacent to the chromophore (Wiedenmann et al., 2004). Sox10, a high mobility group transcription factor, directly activates the zebrafish mitfa proximal promoter and is necessary to specify all zebrafish non-ectomesenchymal neural crest derivatives (Elworthy et al., 2003; Lee et al., 2000; Verastegui et al., 2000). Injecting sox10:nls-eos into mitfa:gfp transgenic fish allows tracking of mitfa+ neural crest cells. We photoconverted individual cells that expressed both nuclear sox10:nls-eos and cytoplasmic mitfa:gfp at 24 hpf (Fig. 7A). The resulting photoconverted cell displayed a red sox10:nls-eos nuclear signal and a green cytoplasmic mitfa:gfp signal (Fig. 7B). This distinct color marking allowed individual cells from 24 hpf mitfa:gfp zebrafish to be tracked over a 48 hour time period, thereby permitting us to assess their fates. Brightfield light overlaid with red fluorescence allowed photoconverted cells that accumulated melanin to be scored as melanophores (Figs. 7D–F). In a similar fashion, incident light overlaid with red fluorescence allowed photoconverted cells that iridesced to be scored as iridophores (Figs. 7G–I). Photoconverted cells were successfully tracked (n=144) and identified as attaining either a melanophore or iridophore fate (see Table 4 for all values). 104 photoconverted cells acquired a melanophore fate (72% of scored cells); 40 photoconverted cells acquired an iridophore fate (28% of scored cells) (Fig. 7C; Table 4). We conclude that both melanophores and iridophores are derived from a mitfa+ neural crest precursor. Notably, none of the labeled cells divided, suggesting mitfa:gfp/sox10:nls-eos cells were post-mitotic at 24 hpf. To confirm that mitfa:gfp+ cells are not mitotically active, we co-stained with anti-phosphohistone H3, which detects mitotic cells in M-phase. 579 (99%) of mitfa:gfp cells were phosphohistone H3 negative; 6 (1%) of mitfa:gfp cells were phosphohistone H3 positive (n=585) (see Fig. S4 in the supplementary material). These results demonstrate that post-mitotic, chromatoblast cell fate decisions remain plastic.
Fig. 7. Melanoblasts and iridoblasts share a mitfa+ bipotent precursor.
(A,B) Confocal image of a double positive sox10:nls-eos/mitfa:gfp cell surrounded by mitfa:gfp cells, lateral view, anterior trunk, 24 hpf. 40X. (A) Unconverted, pre-UV exposure. (B) Photoconverted, post-UV exposure. (C) Bar graph: 72% of identified photoconverted cells acquire a melanophore fate; 28% of identified photoconverted cells acquire an iridophore fate (n= 144) Bars=s.d. (for all values see Table 4). (D–F) Photoconverted sox10:nls-eos/mitfa:gfp cell acquires melanophore fate, lateral view, anterior trunk, 48 hpf, 40X. (D) Brightfield. (E) Red channel. (F) Merge Brightfield/Red channel. (G–I) Photoconverted sox10:nls-eos/mitfa:gfp cell acquires iridophore fate, lateral view, anterior trunk, 48 hpf, 40X. (G) Incident light. (H) Red channel. (I) Merge Incident/Red channel. Scale bars: (A,B, D–I) 30 μm.
Table 4.
Photoconversion lineage tracing
| EosFP photoconversion data | Exp. 1 | Exp. 2 | Exp. 3 | Totals |
|---|---|---|---|---|
| photoconverted cells | 88 | 73 | 70 | 231 |
| photoconverted cells that could be tracked | 40 | 51 | 53 | 144 |
| Melanophore descendants | 34 | 36 | 33 | 103 |
| Iridophore descendants | 6 | 15 | 20 | 41 |
| % of photoconverted cells that become melanophores | 85 | 71 | 62 | 72 |
| % of photoconverted cells that become iridophores | 15 | 29 | 38 | 28 |
DISCUSSION
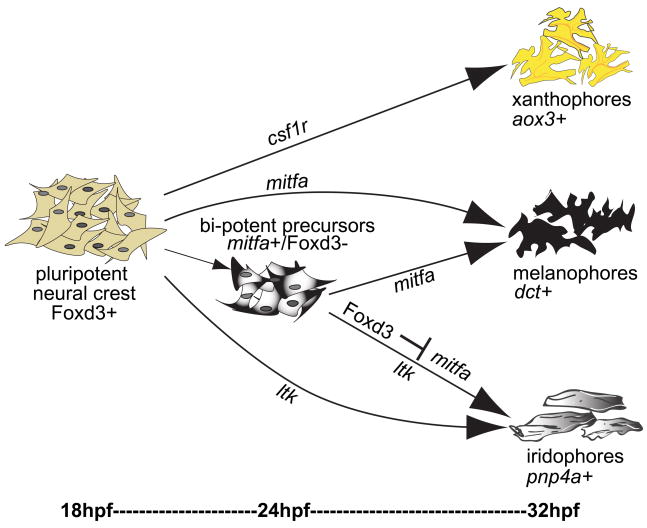
The results of this study identify a partially fate-restricted neural crest precursor, marked by mitfa expression, with the capacity to produce both melanophores and iridophores in zebrafish embryos. We propose there are two avenues for neural crest cells to acquire the melanophore or iridophore fate (Figure 8). A cell can either develop directly into one of the two chromatophores or it may pass through a bipotent stage before acquiring its ultimate fate. Our results suggest that cells expressing mitfa are still plastic: some cells will continue to express this gene and become melanophores while other cells will eventually repress mitfa and form iridophores. We refer to these mitfa+ precursors as bi-potent, since they retain the potential to produce either melanophores or iridophores. As indicated by both photolabeling and phosphohistone H3 staining, each cell is post-mitotic, and therefore gives rise to either an iridophore or a melanophore, but not both. We hypothesize that the cell fate decision is decoupled from cell division and that cells remain plastic after their final mitosis. Foxd3 acts as a molecular switch upon this precursor population by repressing melanophore fate and promoting iridophore fate, in part by repressing mitfa expression. We note that there is no evidence for any biochemical difference between a chromatophore that passes though a double-positive phase and one that develops directly; both sets of precursors express pnp4a.
Fig. 8. Model for chromatophore development from the neural crest.
We propose a model for pigment cell lineages that includes two pathways for melanophores and iridophores to differentiate. Pigment cells may develop directly from neural crest cells or transit through a bi-potent pigment precursor stage. Between 18–24 hpf, neural crest cells begin to express lineage-specific markers. Xanthoblasts require csf1r and commit to the pteridine pigment synthesis pathway, as indicated by aox3 expression. Iridoblasts require ltk and continue along the purine synthesis pathway as indicated by pnp4a expression. Melanoblasts require mitfa and continue along the melanin synthesis pathway as indicated by dct expression. In addition, some mitfa+ cells will retain the capacity to produce either melanophores or iridophores, a process regulated by expression of Foxd3. Foxd3 is initially expressed in all neural crest cells at 18 hpf, then downregulated. Foxd3 reappears in approximately half of mitfa+ bi-potent precursors at 24 hpf, resulting in repression of mitfa, activation of pnp4a and promotion to an iridophore fate. A reciprocal population will remain Foxd3 negative, continue mitfa expression and follow the melanophore path.
pnp4a as a novel iridophore marker
Our study of early iridophore development was facilitated by the identification of pnp4a as a specific marker, originally identified as enriched in the iridophore inter-stripe region of adult zebrafish dermal tissue. pnp4a is a highly conserved member of the purine nucleoside phosphorylase family with nearly 50% similarity to the E. coli protein (see Fig. S1 in the supplementary material). The biochemical function of pnp4a has not been tested in zebrafish, however, functional experiments with PNPase orthologs have been performed in both mouse and E. coli (Della Ragione et al., 1996; Seeger et al., 1995). PNPase is an enzyme involved in purine synthesis, which metabolizes guanosine into guanine, a nitrogenous base, and ribose phosphate, a sugar (Bzowska et al., 2000). Therefore, it would follow that pnp4a would be of paramount importance for zebrafish iridescent pigmentation, which results from light reflecting off accumulated guanine crystals. Taken together, pnp4a seems to act as an early expressing enzyme in iridophore differentiation, analogous to the manner with which dct is used in melanin biosynthesis in early melanoblasts and melanocyte stem cells. ltk, a leukocyte tyrosine kinase known to be critical for early iridophore development (Lopes et al., 2008) is necessary for pnp4a expression. The very low level of ltk expression precluded further co-localization analysis with pnp4a and other pigment cell markers.
A bipotent melanophore/iridophore precursor in the chromatoblast lineage
EosFP photoactivation experiments demonstrate that mitfa:gfp+ cells are capable of forming either melanophores or iridophores, allowing us to infer that expression of mitfa is not sufficient to drive cells towards the melanophore fate. Our results also suggest, however, that not all mitfa:gfp+/pnp4a+ cells become iridophores. Comparing cell ratios between marker co-localization assays and cell-labeling experiments reveals significantly more double positive mitfa:gfp+/pnp4a+ cells than the number of mitfa+ cells that become iridophores (p<0.001 by Chi-square analysis; see Table S2 in the supplementary material). These findings suggest a model in which mitfa+/pnp4a+ cells are precursors for both melanophores and iridophores. However, we do not believe that all melanophores and iridophores are derived from this cell type. There are many more melanophores than double-positive pnp4a+/mitfa+ cells, suggesting an additional melanophore source. Moreover, while there is an iridophore increase with loss of mitfa, the number of ectopic iridophores is much lower than the number of melanophores lost (data not shown). Similarly, there is not a complete loss of iridophores in foxd3 mutants and the number of iridophores is not completely restored in foxd3/mitfa double mutants, suggesting some iridophores are regulated by independent mechanisms. As our data shows that all differentiated iridophores express pnp4a, these iridoblasts may be represented by the mitfa:gfp−/pnp4a+ population.
A close relationship between iridophores and melanophores has been proposed previously. Bagnara et al. (1979) first suggested the idea that pigment cells share a common origin after observing single chromatophores housing multiple pigment organelles. Iridophores from the iris of the dove contained partially melanized reflecting platelets, likewise, single chromatophores from the tapetum lucidum of the teleost fish, Dasyatis sabina, revealed melanosomes and reflecting platelets bound within the same intracellular membrane. This chromatophore mosaicism suggested pigment cells, specifically iridophores and melanophores, share a common precursor capable of activating multiple pigment synthesis pathways. Ide and Bagnara found proliferating bullfrog melanophores in clonal culture lose melanosomes and form reflecting platelets (iridosomes) when cultured in a medium containing high guanosine content, suggesting developing melanoblasts maintain the capacity to trans-differentiate to an iridophore fate (Ide and Bagnara, 1980). Previous work in zebrafish has also demonstrated a close developmental relationship between iridophores and melanophores. parade (pde), a pigment phenotype mutant, accumulates ectopic, mosaic chromatophores that contain organelles characteristic of both iridophores (iridosomes) and melanophores (melanosomes) (Kelsh et al., 1996). More recently, Lopes et al. (2008) presented a progressive fate restriction model, which describes a sox10+/ltk+ pigment precursor prepared to adopt either an iridophore or melanophore fate. Since ltk positive cells accumulate in the posterior trunk and tail region of sox10 mutants, the authors hypothesize the iridoblasts are trapped in a multipotent pigment precursor state. Our data builds on these reports to support a chromatophore lineage model based on an iridoblast/melanoblast precursor that is separate from xanthophore precursors.
Our temporal analysis of genetic markers revealed substantial overlap of iridoblast and melanoblast markers. In contrast, there is less than 1% overlap between either of the xanthoblast markers, csf1r and aox3, and markers of either iridoblasts or melanoblasts. This result suggests the xanthoblast lineage is distinct and, in agreement with previous clonal analysis (Dutton et al., 2001; Raible and Eisen, 1994), does not provide convincing evidence for a tri-potent chromatophore precursor capable of producing all three chromatoblast cells. However, it remains possible that the observed 0.4% overlap between csf1r+ and dct+ cells and the 0.5% overlap between aox3+ and dct+ cells represents a small population of tri-potent chromatoblast precursors. Expression of Pax3 has previously been shown to be required for xanthophore development but not for melanophores or iridophores. Pax3/7 antibody labels xanthoblasts (Minchin and Hughes, 2008), however, in our hands this antibody additionally labels the majority of mitfa+ and pnp4a+ cells, demonstrating it is not specific to xanthoblasts.
It remains possible that other cell types, including glia and xanthophores, could descend from mitfa+ precursors. Thomas and Erickson (2009) reported MITF repression causes cells to acquire glial characteristics. Furthermore, a recent report using chick embryos shows myelinating krox20+ Schwann cells retain the competence to differentiate into melanocytes (Adameyko et al., 2009). These results illustrate a close lineage relationship between glial cells and melanoblasts. Minchin and Hughes (2008) found that Pax3 knockdown led to a loss of xanthophores accompanied by a small increase in melanophores, supporting the possibility of a common chromatoblast precursor between melanophores and xanthophores. We neither observed photoconverted cells adopt xanthophore or glial morphology nor migrate in a manner characteristic of either cell type. To remain spatially unbiased, we photoconverted cells from all expression domains (dorsal trunk, brain, yolk ball, lateral trunk). However, by only photoconverting cells at a single time point (24 hpf) we introduced a temporal bias that may have limited the diversity of descendents; partially restricted glial/melanophore or xanthophore/melanophore precursors could theoretically exist before the onset of mitfa expression.
Foxd3 as a molecular switch between melanophores and iridophores
We propose that Foxd3 promotes pnp4a expression in cells that are initially positive for mitfa. We demonstrate Foxd3 is necessary for pnp4a expression at 24 hpf and, using in situ fluorescence imaging, we reveal Foxd3 positive cells express pnp4a transcript. Foxd3 often serves as a transcriptional repressor (Curran et al., 2009; Pohl and Knochel, 2001; Stewart et al., 2006; Yaklichkin et al., 2007), however, Foxd3 transcriptional activation is not unprecedented. Lee et al. observed Foxd3 directly bind the myf5 promoter so as to maintain myf5 expression in somites and adaxial cells, thus driving somitogenesis (Lee et al., 2006). It currently remains unclear whether Foxd3 directly binds the pnp4a promoter to activate its expression.
Foxd3 has previously demonstrated the capacity to bind and repress the zebrafish mitfa promoter, thereby preventing melanophore fate (Curran et al., 2009; Ignatius et al., 2008) or to indirectly inhibit avian MITF transcription by binding activators (Thomas and Erickson, 2009). Accordingly, we have previously shown mitfa expression is initially exclusive with Foxd3. At 18hpf, more than 90% of mitfa expressing neural crest cells are Foxd3 negative (Curran et al., 2009). In contrast, by 24 hpf, approximately 50% of mitfa expressing cells stain positive for Foxd3, suggesting a subset of mitfa+ cells have reacquired Foxd3 activity. This transcriptional control of chromatophore fate is further illustrated with our iridophore count experiments. Loss of mitfa resulted in ectopic iridophore development, loss of foxd3 caused a loss of iridophores, while the foxd3/mitfa double mutants allowed a partial rescue, suggesting Foxd3 promotes iridophore development, in part, by repressing mitfa. It should be noted that neither the foxd3zdf10 mutant (Stewart et al., 2006) nor the Foxd3 morphant (Lister et al., 2006) resulted in a complete loss of iridophores, and we observed some corresponding pnp4a+ cells in foxd3zdf10 mutants at 28 and 32 hpf (data not shown). Residual Foxd3 function is likely to remain after MO injection and while the foxd3zdf10mutation was originally reported as a null, recent work from the Kessler lab has demonstrated that mutant Foxd3 protein retains some function (Chang and Kessler, 2010). Low levels of Foxd3 activity may be sufficient for the few iridophores that specify in Foxd3 mutant and morpholino-injected embryos.
Our study demonstrates a remarkable flexibility in development after the onset of expression of a key differentiation regulator. The Mitf transcription factor has previously been thought of as a “master regulatory” gene for melanocyte differentiation, with its expression both necessary and sufficient for acquisition of melanoblast characteristics. Our results contrast with recent reports that suggest mitfa expression is sufficient to commit a neural crest cell towards the melanophore fate (Ignatius et al., 2008; Thomas and Erickson, 2009). However direct lineage tracing has not previously been described. It has been shown previously that mammalian melanoblasts can be maintained in an undifferentiated state after the onset of Mitf expression through the continued action of Pax3 (Lang et al., 2005). We speculate that neural crest cells in this state might retain the flexibility to differentiate along multiple lineage pathways.
Supplementary Material
Phylogenetic relationship between purine nucleoside phosphorylases. Members of the Pnp family were aligned along with closely related methylthioadenosine phosphorylase (Mtap) polypeptides. Zebrafish (Dr) sequences clustered together separately from other vertebrate (human, Hs; mouse, Mm) genes, and separate from Drosophila (Dm), yeast (Sc) and bacterial (Ec) polypeptides.
(A–D) Incident light reveals iridophores on eyes of 48 hpf zebrafish, lateral view, 10X. (A) Wild-type zebrafish eye displays normal numbers of eye iridophores. (B) Foxd3 morphant displays normal number of eye iridophores. (C) mitfa−/− eye displays supernumerary iridophores. (D) foxd3MO/mitfa mutant eye exhibits supernumerary iridophores. (E) Eye iridophore cell counts collected from 20 zebrafish for each genetic background. Mean iridophore cell counts: (wild-type: 39), (foxd3 MO: 42), (mitfa/−: 78), (foxd3 MO/mitfa−/−: 74). Bars = s.d. For an internal control, trunk iridophores were counted along the dorsal and ventral stripes from the posterior tail region, between the cloacae and tail tip at 72 hpf. Mean trunk iridophore cell counts: (wild-type: 28), (foxd3 MO: 16), (mitfa−/−: 48), (foxd3 MO/mitfa−/−: 38). Trunk iridophore standard deviation: (wild-type: +/−5), (foxd3 MO: +/−5), (mitfa−/−: +/−4), (foxd3 MO/mitfa−/− double mutant: +/−4).
(A–I) Confocal images collected from lateral aspect of anterior tail region of fixed zebrafish, 20X. (A–C,J) Wild-type embryo reveals Pax3/7+ cells overlap with pnp1 expression at 24 hpf then resolve at 50 hpf (D–F). (A,D) Pax3/7 (B,E) pnp1 (C,F) Color merge: Red: Pax3/7 antibody, Green: pnp1 mRNA (inset 40x). (G–I) Wild-type embryo reveals Pax3/7 signal strongly localized with mitfa:gfp expression at 24 hpf (G) Pax3/7 (H) mitfa:gfp (I) Color merge: Green: mitfa:gfp, Red: Pax3/7 (inset 40x). (J,K) Line graphs depicting percent of overlap between chromatoblast markers (see Table S1 in supplementary material). (J) Red line= % of Pax3/7+ cells that are pnp4a+/Pax3/7+. Green line= % of pnp4a+ cells that are pnp4a+/Pax3/7+. (K) Red line= % of Pax3/7+ cells that are mitfa:gfp+/Pax3/7+. Green line= % of mitfa:gfp+ cells that are mitfa:gfp+/Pax3/7+. Scale bars: (A–I) 20 μm; (C,F,I inset) 10 μm.
(A–C) Confocal images collected from lateral aspect of anterior tail region of 24 hpf wild-type zebrafish, anterior left, 20X. (A) mitfa:gfp (B) phosphohistone H3 (C) Color merge: Green: mitfa:gfp, Red: phosphohistone H3. Scale bars: (A–C) 25 μm.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adameyko I, Lallemend F, Aquino JB, Pereira JA, Topilko P, Muller T, Fritz N, Beljajeva A, Mochii M, Liste I, Usoskin D, Suter U, Birchmeier C, Ernfors P. Schwann cell precursors from nerve innervation are a cellular origin of melanocytes in skin. Cell. 2009;139:366–79. doi: 10.1016/j.cell.2009.07.049. [DOI] [PubMed] [Google Scholar]
- Bagnara JT, Fernandez PJ, Fujii R. On the blue coloration of vertebrates. Pigment Cell Res. 2007;20:14–26. doi: 10.1111/j.1600-0749.2006.00360.x. [DOI] [PubMed] [Google Scholar]
- Bagnara JT, Matsumoto J, Ferris W, Frost SK, Turner WA, Jr, Tchen TT, Taylor JD. Common origin of pigment cells. Science. 1979;203:410–5. doi: 10.1126/science.760198. [DOI] [PubMed] [Google Scholar]
- Bagnara JT, Taylor JD, Hadley ME. The dermal chromatophore unit. J Cell Biol. 1968;38:67–79. doi: 10.1083/jcb.38.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondurand N, Pingault V, Goerich DE, Lemort N, Sock E, Le Caignec C, Wegner M, Goossens M. Interaction among SOX10, PAX3 and MITF, three genes altered in Waardenburg syndrome. Hum Mol Genet. 2000;9:1907–17. doi: 10.1093/hmg/9.13.1907. [DOI] [PubMed] [Google Scholar]
- Braasch I, Salzburger W, Meyer A. Asymmetric evolution in two fish-specifically duplicated receptor tyrosine kinase paralogons involved in teleost coloration. Mol Biol Evol. 2006;23:1192–202. doi: 10.1093/molbev/msk003. [DOI] [PubMed] [Google Scholar]
- Busca R, Ballotti R. Cyclic AMP a key messenger in the regulation of skin pigmentation. Pigment Cell Res. 2000;13:60–9. doi: 10.1034/j.1600-0749.2000.130203.x. [DOI] [PubMed] [Google Scholar]
- Bzowska A, Kulikowska E, Shugar D. Purine nucleoside phosphorylases: properties, functions, and clinical aspects. Pharmacol Ther. 2000;88:349–425. doi: 10.1016/s0163-7258(00)00097-8. [DOI] [PubMed] [Google Scholar]
- Carney TJ, Dutton KA, Greenhill E, Delfino-Machin M, Dufourcq P, Blader P, Kelsh RN. A direct role for Sox10 in specification of neural crest-derived sensory neurons. Development. 2006;133:4619–30. doi: 10.1242/dev.02668. [DOI] [PubMed] [Google Scholar]
- Chang LL, Kessler DS. Foxd3 is an Essential Nodal-Dependent Regulator of Zebrafish Dorsal Mesoderm Development. Dev Biol. 2010 doi: 10.1016/j.ydbio.2010.03.017. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper CD, Raible DW. Mechanisms for reaching the differentiated state: Insights from neural crest-derived melanocytes. Semin Cell Dev Biol. 2009;20:105–10. doi: 10.1016/j.semcdb.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran K, Raible DW, Lister JA. Foxd3 controls melanophore specification in the zebrafish neural crest by regulation of Mitf. Dev Biol. 2009;332:408–17. doi: 10.1016/j.ydbio.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis GK, D’Alessio JA, Patel NH. Pax3/7 genes reveal conservation and divergence in the arthropod segmentation hierarchy. Dev Biol. 2005;285:169–84. doi: 10.1016/j.ydbio.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Della Ragione F, Takabayashi K, Mastropietro S, Mercurio C, Oliva A, Russo GL, Della Pietra V, Borriello A, Nobori T, Carson DA, Zappia V. Purification and characterization of recombinant human 5′-methylthioadenosine phosphorylase: definite identification of coding cDNA. Biochem Biophys Res Commun. 1996;223:514–9. doi: 10.1006/bbrc.1996.0926. [DOI] [PubMed] [Google Scholar]
- Demski LS. Chromatophore systems in teleosts and cephalopods: a levels oriented analysis of convergent systems. Brain Behav Evol. 1992;40:141–56. doi: 10.1159/000113909. [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Raible DW, Moon RT. Direct regulation of nacre, a zebrafish MITF homolog required for pigment cell formation, by the Wnt pathway. Genes Dev. 2000;14:158–62. [PMC free article] [PubMed] [Google Scholar]
- Dutton KA, Pauliny A, Lopes SS, Elworthy S, Carney TJ, Rauch J, Geisler R, Haffter P, Kelsh RN. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001;128:4113–25. doi: 10.1242/dev.128.21.4113. [DOI] [PubMed] [Google Scholar]
- Elworthy S, Lister JA, Carney TJ, Raible DW, Kelsh RN. Transcriptional regulation of mitfa accounts for the sox10 requirement in zebrafish melanophore development. Development. 2003;130:2809–18. doi: 10.1242/dev.00461. [DOI] [PubMed] [Google Scholar]
- Gilmour DT, Maischein HM, Nusslein-Volhard C. Migration and function of a glial subtype in the vertebrate peripheral nervous system. Neuron. 2002;34:577–88. doi: 10.1016/s0896-6273(02)00683-9. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson E, Copeland NG, Jenkins NA, Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Nakayama A, Li H, Swenson LB, Opdecamp K, Asher JH, Jr, Arnheiter H, Glaser T. Mutation at the anophthalmic white locus in Syrian hamsters: haploinsufficiency in the Mitf gene mimics human Waardenburg syndrome type 2. Hum Mol Genet. 1998;7:703–8. doi: 10.1093/hmg/7.4.703. [DOI] [PubMed] [Google Scholar]
- Hromas R, Ye H, Spinella M, Dmitrovsky E, Xu D, Costa RH. Genesis, a Winged Helix transcriptional repressor, has embryonic expression limited to the neural crest, and stimulates proliferation in vitro in a neural development model. Cell Tissue Res. 1999;297:371–82. doi: 10.1007/s004410051365. [DOI] [PubMed] [Google Scholar]
- Ide H, Bagnara JT. The differentiation of leaf frog melanophores in culture. Cell Differ. 1980;9:51–61. doi: 10.1016/0045-6039(80)90007-x. [DOI] [PubMed] [Google Scholar]
- Ignatius MS, Moose HE, El-Hodiri HM, Henion PD. colgate/hdac1 Repression of foxd3 expression is required to permit mitfa-dependent melanogenesis. Dev Biol. 2008;313:568–83. doi: 10.1016/j.ydbio.2007.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julich D, Hwee Lim C, Round J, Nicolaije C, Schroeder J, Davies A, Geisler R, Lewis J, Jiang YJ, Holley SA. beamter/deltaC and the role of Notch ligands in the zebrafish somite segmentation, hindbrain neurogenesis and hypochord differentiation. Dev Biol. 2005;286:391–404. doi: 10.1016/j.ydbio.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Kelsh RN. Genetics and evolution of pigment patterns in fish. Pigment Cell Res. 2004;17:326–36. doi: 10.1111/j.1600-0749.2004.00174.x. [DOI] [PubMed] [Google Scholar]
- Kelsh RN, Brand M, Jiang YJ, Heisenberg CP, Lin S, Haffter P, Odenthal J, Mullins MC, van Eeden FJ, Furutani-Seiki M, Granato M, Hammerschmidt M, Kane DA, Warga RM, Beuchle D, Vogelsang L, Nusslein-Volhard C. Zebrafish pigmentation mutations and the processes of neural crest development. Development. 1996;123:369–89. doi: 10.1242/dev.123.1.369. [DOI] [PubMed] [Google Scholar]
- Kelsh RN, Dutton K, Medlin J, Eisen JS. Expression of zebrafish fkd6 in neural crest-derived glia. Mech Dev. 2000a;93:161–4. doi: 10.1016/s0925-4773(00)00250-1. [DOI] [PubMed] [Google Scholar]
- Kelsh RN, Schmid B, Eisen JS. Genetic analysis of melanophore development in zebrafish embryos. Dev Biol. 2000b;225:277–93. doi: 10.1006/dbio.2000.9840. [DOI] [PubMed] [Google Scholar]
- Kim J, Lo L, Dormand E, Anderson DJ. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron. 2003;38:17–31. doi: 10.1016/s0896-6273(03)00163-6. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–99. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Labosky PA, Kaestner KH. The winged helix transcription factor Hfh2 is expressed in neural crest and spinal cord during mouse development. Mech Dev. 1998;76:185–90. doi: 10.1016/s0925-4773(98)00105-1. [DOI] [PubMed] [Google Scholar]
- Lacosta AM, Muniesa P, Ruberte J, Sarasa M, Dominguez L. Novel expression patterns of Pax3/Pax7 in early trunk neural crest and its melanocyte and non-melanocyte lineages in amniote embryos. Pigment Cell Res. 2005;18:243–51. doi: 10.1111/j.1600-0749.2005.00238.x. [DOI] [PubMed] [Google Scholar]
- Lang D, Lu MM, Huang L, Engleka KA, Zhang M, Chu EY, Lipner S, Skoultchi A, Millar SE, Epstein JA. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature. 2005;433:884–7. doi: 10.1038/nature03292. [DOI] [PubMed] [Google Scholar]
- Lee HC, Huang HY, Lin CY, Chen YH, Tsai HJ. Foxd3 mediates zebrafish myf5 expression during early somitogenesis. Dev Biol. 2006;290:359–72. doi: 10.1016/j.ydbio.2005.11.032. [DOI] [PubMed] [Google Scholar]
- Lee M, Goodall J, Verastegui C, Ballotti R, Goding CR. Direct regulation of the Microphthalmia promoter by Sox10 links Waardenburg-Shah syndrome (WS4)-associated hypopigmentation and deafness to WS2. J Biol Chem. 2000;275:37978–83. doi: 10.1074/jbc.M003816200. [DOI] [PubMed] [Google Scholar]
- Lister JA, Close J, Raible DW. Duplicate mitf genes in zebrafish: complementary expression and conservation of melanogenic potential. Dev Biol. 2001;237:333–44. doi: 10.1006/dbio.2001.0379. [DOI] [PubMed] [Google Scholar]
- Lister JA, Cooper C, Nguyen K, Modrell M, Grant K, Raible DW. Zebrafish Foxd3 is required for development of a subset of neural crest derivatives. Dev Biol. 2006;290:92–104. doi: 10.1016/j.ydbio.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Lister JA, Robertson CP, Lepage T, Johnson SL, Raible DW. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development. 1999;126:3757–67. doi: 10.1242/dev.126.17.3757. [DOI] [PubMed] [Google Scholar]
- Lopes SS, Yang X, Muller J, Carney TJ, McAdow AR, Rauch GJ, Jacoby AS, Hurst LD, Delfino-Machin M, Haffter P, Geisler R, Johnson SL, Ward A, Kelsh RN. Leukocyte tyrosine kinase functions in pigment cell development. PLoS Genet. 2008;4:e1000026. doi: 10.1371/journal.pgen.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills MG, Patterson LB. Not just black and white: pigment pattern development and evolution in vertebrates. Semin Cell Dev Biol. 2009;20:72–81. doi: 10.1016/j.semcdb.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchin JE, Hughes SM. Sequential actions of Pax3 and Pax7 drive xanthophore development in zebrafish neural crest. Dev Biol. 2008;317:508–22. doi: 10.1016/j.ydbio.2008.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochii M, Ono T, Matsubara Y, Eguchi G. Spontaneous transdifferentiation of quail pigmented epithelial cell is accompanied by a mutation in the Mitf gene. Dev Biol. 1998;196:145–59. doi: 10.1006/dbio.1998.8864. [DOI] [PubMed] [Google Scholar]
- Morrison RL. A transmission electron microscopic (TEM) method for determining structural colors reflected by lizard iridophores. Pigment Cell Res. 1995;8:28–36. doi: 10.1111/j.1600-0749.1995.tb00771.x. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Csete M, Groves AK, Melega W, Wold B, Anderson DJ. Culture in reduced levels of oxygen promotes clonogenic sympathoadrenal differentiation by isolated neural crest stem cells. J Neurosci. 2000;20:7370–6. doi: 10.1523/JNEUROSCI.20-19-07370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, White PM, Zock C, Anderson DJ. Prospective identification, isolation by flow cytometry, and in vivo self-renewal of multipotent mammalian neural crest stem cells. Cell. 1999;96:737–49. doi: 10.1016/s0092-8674(00)80583-8. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–20. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Odenthal J, Nusslein-Volhard C. fork head domain genes in zebrafish. Dev Genes Evol. 1998;208:245–58. doi: 10.1007/s004270050179. [DOI] [PubMed] [Google Scholar]
- Opdecamp K, Nakayama A, Nguyen MT, Hodgkinson CA, Pavan WJ, Arnheiter H. Melanocyte development in vivo and in neural crest cell cultures: crucial dependence on the Mitf basic-helix-loop-helix-zipper transcription factor. Development. 1997;124:2377–86. doi: 10.1242/dev.124.12.2377. [DOI] [PubMed] [Google Scholar]
- Parichy DM, Ransom DG, Paw B, Zon LI, Johnson SL. An orthologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a subpopulation of adult melanocytes in the zebrafish, Danio rerio. Development. 2000;127:3031–44. doi: 10.1242/dev.127.14.3031. [DOI] [PubMed] [Google Scholar]
- Planque N, Raposo G, Leconte L, Anezo O, Martin P, Saule S. Microphthalmia transcription factor induces both retinal pigmented epithelium and neural crest melanocytes from neuroretina cells. J Biol Chem. 2004;279:41911–7. doi: 10.1074/jbc.M404964200. [DOI] [PubMed] [Google Scholar]
- Pohl BS, Knochel W. Overexpression of the transcriptional repressor FoxD3 prevents neural crest formation in Xenopus embryos. Mech Dev. 2001;103:93–106. doi: 10.1016/s0925-4773(01)00334-3. [DOI] [PubMed] [Google Scholar]
- Potterf SB, Furumura M, Dunn KJ, Arnheiter H, Pavan WJ. Transcription factor hierarchy in Waardenburg syndrome: regulation of MITF expression by SOX10 and PAX3. Hum Genet. 2000;107:1–6. doi: 10.1007/s004390000328. [DOI] [PubMed] [Google Scholar]
- Raible DW, Eisen JS. Restriction of neural crest cell fate in the trunk of the embryonic zebrafish. Development. 1994;120:495–503. doi: 10.1242/dev.120.3.495. [DOI] [PubMed] [Google Scholar]
- Sasai N, Mizuseki K, Sasai Y. Requirement of FoxD3-class signaling for neural crest determination in Xenopus. Development. 2001;128:2525–36. doi: 10.1242/dev.128.13.2525. [DOI] [PubMed] [Google Scholar]
- Seeger C, Poulsen C, Dandanell G. Identification and characterization of genes (xapA, xapB, and xapR) involved in xanthosine catabolism in Escherichia coli. J Bacteriol. 1995;177:5506–16. doi: 10.1128/jb.177.19.5506-5516.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- Stewart RA, Arduini BL, Berghmans S, George RE, Kanki JP, Henion PD, Look AT. Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Dev Biol. 2006;292:174–88. doi: 10.1016/j.ydbio.2005.12.035. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Takeda K, Nobukuni Y, Urabe K, Long JE, Meyers KA, Aaronson SA, Miki T. Ectopic expression of MITF, a gene for Waardenburg syndrome type 2, converts fibroblasts to cells with melanocyte characteristics. Nat Genet. 1996;14:50–4. doi: 10.1038/ng0996-50. [DOI] [PubMed] [Google Scholar]
- Takeda K, Yasumoto K, Takada R, Takada S, Watanabe K, Udono T, Saito H, Takahashi K, Shibahara S. Induction of melanocyte-specific microphthalmia-associated transcription factor by Wnt-3a. J Biol Chem. 2000;275:14013–6. doi: 10.1074/jbc.c000113200. [DOI] [PubMed] [Google Scholar]
- Tassabehji M, Newton VE, Read AP. Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat Genet. 1994;8:251–5. doi: 10.1038/ng1194-251. [DOI] [PubMed] [Google Scholar]
- Thisse B, Heyer V, Lux A, Alunni V, Degrave A, Seiliez I, Kirchner J, Parkhill JP, Thisse C. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 2004;77:505–19. doi: 10.1016/s0091-679x(04)77027-2. [DOI] [PubMed] [Google Scholar]
- Thomas AJ, Erickson CA. FOXD3 regulates the lineage switch between neural crest-derived glial cells and pigment cells by repressing MITF through a non-canonical mechanism. Development. 2009;136:1849–58. doi: 10.1242/dev.031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verastegui C, Bille K, Ortonne JP, Ballotti R. Regulation of the microphthalmia-associated transcription factor gene by the Waardenburg syndrome type 4 gene, SOX10. J Biol Chem. 2000;275:30757–60. doi: 10.1074/jbc.C000445200. [DOI] [PubMed] [Google Scholar]
- Watanabe A, Takeda K, Ploplis B, Tachibana M. Epistatic relationship between Waardenburg syndrome genes MITF and PAX3. Nature Genet. 1998;18:283–6. doi: 10.1038/ng0398-283. [DOI] [PubMed] [Google Scholar]
- White PM, Morrison SJ, Orimoto K, Kubu CJ, Verdi JM, Anderson DJ. Neural crest stem cells undergo cell-intrinsic developmental changes in sensitivity to instructive differentiation signals. Neuron. 2001;29:57–71. doi: 10.1016/s0896-6273(01)00180-5. [DOI] [PubMed] [Google Scholar]
- Widlund HR, Horstmann MA, Price ER, Cui J, Lessnick SL, Wu M, He X, Fisher DE. Beta-catenin-induced melanoma growth requires the downstream target Microphthalmia-associated transcription factor. J Cell Biol. 2002;158:1079–87. doi: 10.1083/jcb.200202049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenmann J, Ivanchenko S, Oswald F, Schmitt F, Rocker C, Salih A, Spindler KD, Nienhaus GU. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc Natl Acad Sci U S A. 2004;101:15905–10. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaklichkin S, Steiner AB, Lu Q, Kessler DS. FoxD3 and Grg4 physically interact to repress transcription and induce mesoderm in Xenopus. J Biol Chem. 2007;282:2548–57. doi: 10.1074/jbc.M607412200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler I. The pteridine pathway in zebrafish: regulation and specification during the determination of neural crest cell-fate. Pigment Cell Res. 2003;16:172–82. doi: 10.1034/j.1600-0749.2003.00044.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic relationship between purine nucleoside phosphorylases. Members of the Pnp family were aligned along with closely related methylthioadenosine phosphorylase (Mtap) polypeptides. Zebrafish (Dr) sequences clustered together separately from other vertebrate (human, Hs; mouse, Mm) genes, and separate from Drosophila (Dm), yeast (Sc) and bacterial (Ec) polypeptides.
(A–D) Incident light reveals iridophores on eyes of 48 hpf zebrafish, lateral view, 10X. (A) Wild-type zebrafish eye displays normal numbers of eye iridophores. (B) Foxd3 morphant displays normal number of eye iridophores. (C) mitfa−/− eye displays supernumerary iridophores. (D) foxd3MO/mitfa mutant eye exhibits supernumerary iridophores. (E) Eye iridophore cell counts collected from 20 zebrafish for each genetic background. Mean iridophore cell counts: (wild-type: 39), (foxd3 MO: 42), (mitfa/−: 78), (foxd3 MO/mitfa−/−: 74). Bars = s.d. For an internal control, trunk iridophores were counted along the dorsal and ventral stripes from the posterior tail region, between the cloacae and tail tip at 72 hpf. Mean trunk iridophore cell counts: (wild-type: 28), (foxd3 MO: 16), (mitfa−/−: 48), (foxd3 MO/mitfa−/−: 38). Trunk iridophore standard deviation: (wild-type: +/−5), (foxd3 MO: +/−5), (mitfa−/−: +/−4), (foxd3 MO/mitfa−/− double mutant: +/−4).
(A–I) Confocal images collected from lateral aspect of anterior tail region of fixed zebrafish, 20X. (A–C,J) Wild-type embryo reveals Pax3/7+ cells overlap with pnp1 expression at 24 hpf then resolve at 50 hpf (D–F). (A,D) Pax3/7 (B,E) pnp1 (C,F) Color merge: Red: Pax3/7 antibody, Green: pnp1 mRNA (inset 40x). (G–I) Wild-type embryo reveals Pax3/7 signal strongly localized with mitfa:gfp expression at 24 hpf (G) Pax3/7 (H) mitfa:gfp (I) Color merge: Green: mitfa:gfp, Red: Pax3/7 (inset 40x). (J,K) Line graphs depicting percent of overlap between chromatoblast markers (see Table S1 in supplementary material). (J) Red line= % of Pax3/7+ cells that are pnp4a+/Pax3/7+. Green line= % of pnp4a+ cells that are pnp4a+/Pax3/7+. (K) Red line= % of Pax3/7+ cells that are mitfa:gfp+/Pax3/7+. Green line= % of mitfa:gfp+ cells that are mitfa:gfp+/Pax3/7+. Scale bars: (A–I) 20 μm; (C,F,I inset) 10 μm.
(A–C) Confocal images collected from lateral aspect of anterior tail region of 24 hpf wild-type zebrafish, anterior left, 20X. (A) mitfa:gfp (B) phosphohistone H3 (C) Color merge: Green: mitfa:gfp, Red: phosphohistone H3. Scale bars: (A–C) 25 μm.