Summary
The outer membrane channel TolC is a key component of multidrug efflux and type I secretion transporters in Escherichia coli. Mutational inactivation of TolC renders cells highly susceptible to antibiotics and leads to defects in secretion of protein toxins. Despite impairment of various transport functions, no growth defects were reported in cells lacking TolC. Unexpectedly, we found that the loss of TolC notably impairs cell division and growth in minimal glucose medium. The TolC-dependent phenotype was further exacerbated by the loss of ygiB and ygiC genes expressed in the same operon as tolC and their homologs yjfM and yjfC located elsewhere on the chromosome. Our results show that this growth deficiency is caused by depletion of the critical metabolite NAD+ and high NADH/NAD+ ratios. The increased amounts of PspA and decreased rates of NADH oxidation in ΔtolC membranes indicated stress on the membrane and dissipation of a proton motive force. We conclude that inactivation of TolC triggers metabolic shut-down in E. coli cells grown in minimal glucose medium. The ΔtolC phenotype is partially rescued by YgiBC and YjfMC, which have parallel functions independent from TolC.
Introduction
TolC is the universal outer membrane portal for export of toxins and drug efflux in Enterobacteriacea species (Andersen et al., 2001). Previous studies showed that E. coli cells lacking tolC are highly susceptible to multiple antibiotics and fail to secrete microcin J25, colicin V, hemolysin and heat-stable enterotoxin II (Gilson et al., 1990, Holland et al., 1990, Yamanaka et al., 2008). TolC was also found to be involved in cysteine tolerance, as well as in secretion of porphyrins and enterobactin (Cosme et al., 2008, Tatsumi & Wachi, 2008, Wiriyathanawudhiwong et al., 2009, Bleuel et al., 2005). More recently, TolC-deficient cells were shown to overproduce MarA, SoxR and Rob transcriptional regulators (Rosner & Martin, 2009). This finding suggested that TolC might be involved in extrusion of metabolites. In many human pathogens, TolC is required for host invasion but specific substrates secreted within hosts remain unknown (Ferhat et al., 2009, Platz et al., 2009, Webber et al., 2009).
The diverse functionality of TolC is based on its ability to interact with multiple transporters and their accessory proteins located in the inner membrane and the periplasm (Tikhonova et al., 2009, Zgurskaya, 2009). When assembled, the TolC-dependent transport complexes are believed to span the entire envelope of E. coli and expel their substrates from the cytoplasm or the periplasm into the external medium. Some of these complexes, such as type I secretion systems, are assembled only transiently when the substrate is ready to be translocated across the outer membrane (Thanabalu et al., 1998). In contrast, the constitutively produced multidrug transporters exemplified by AcrAB form more stable associations with TolC that are independent from the presence of substrates (Tikhonova & Zgurskaya, 2004, Touze et al., 2004).
In the genome of E. coli and other enterobacteria, the chromosomal tolC locus contains three genes tolC, ygiB and ygiC, which are presumably expressed in a single operon (Fig. 1A). However, functions of YgiB and YgiC as well as any functional link between these proteins and TolC remain unknown. Sequence analyses identified YgiB as an outer membrane lipoprotein containing a putative type II signal peptide. YgiB is highly conserved in cyanobacteria and proteobacteria but its homologs are also present in some Gram-positive species where they precede the ygiC-like genes.
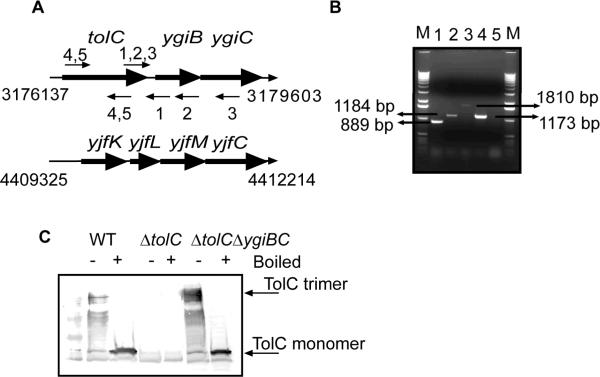
Figure 1.
TolC is expressed in a single operon with ygiBC. A. Structures of tolC-ygiBC and yjfKLMC operons. Annealing regions of primers used in Reverse Transcriptase (RT)-PCR reactions are indicated as small arrows. B. RT-PCR analysis of RNA isolated from E. coli BW25113 (WT) strain. The same RNA sample was used in all reactions. Lanes 1–3: RT-PCR reactions with forward and reverse primers that anneal to tolC-ygiB transcript (primers 1, 2 and 3 in panel A). Lane 4: RT-PCR reaction using forward and reverse primers that anneal to tolC transcript (primers 4, 5 in panel A). Lane 5 is the same reaction as in lane 4 but RT was omitted (no DNA control). C. Immunoblotting analysis of TolC in WT (BW25113), ΔtolC (GD100) and ΔygiBC (GD101) cells. 20 μg of total membrane proteins were separated on 12% SDS-PA gel. TolC was visualized using polyclonal anti-TolC antibody.
YgiC shares significant homology with the C-terminal domain of E. coli GspS, a bifunctional enzyme possessing both glutathionylspermidine (GSP) synthetase and GSP amidase activities (Bollinger et al., 1995). The C-terminal domain of GspS is a GSP synthetase belonging to the ATP-Grasp ATPases family of proteins, which also include the human glutathione (GSH) synthetase (Pai et al., 2006). The high conservation of catalytically important residues in YgiC suggested that this protein could be a GSP synthetase.
The GSP conjugate was initially detected in E. coli under stationary or anaerobic growth conditions (Tabor & Tabor, 1975). Subsequently GSP was identified in parasites as a precursor of trypanothione, a critical redox-active, thiol-containing biomolecule (Fairlamb et al., 1986). The physiological function of bacterial GSP however is unclear and E. coli cells lacking GspS synthetase do not have any phenotype under laboratory conditions (Bollinger et al., 1995). In agreement with this observation, mutants that do not produce either GSH or spermidine (SPE), the two constituents of GSP, also lack growth phenotype, presumably because thioredoxins and other polyamines can replace GSH and SPE in their numerous functions (Fuchs, 1995, Tabor et al., 1978, Xie et al., 1993).
In this study, we investigated how the lack of TolC impacts cell physiology and discovered a functional link between YgiBC/YjfMC and TolC. We report that inactivation of TolC causes stress in the inner membrane. The membrane stress leads to metabolic shut-down, inhibition of NADH dehydrogenases and growth arrest. The function of YgiBC/YjfMC partially protects membranes from the damage and enables ΔTolC cells to grow in the minimal medium with glucose.
Results
tolC mRNA contains ygiB and ygiC sequences
To confirm that tolC is transcribed as a part of an operon we performed RT-PCR analysis of RNA isolated from E. coli BW25113 cells (WT). Fig. 1B shows that primers complementary to tolC and either to ygiB or ygiC regions amplify the expected products only when reverse transcriptase is present in the reaction mixture. Thus, all three tolC, ygiB and ygiC genes are transcribed together.
We next tested whether ygiBC genes are involved in regulation of TolC expression or assembly. Immunoblotting analysis with anti-TolC antibody detected no changes in TolC levels or trimerization in the absence of ygiBC (Fig. 1C). This result implied that ygiBC products are not required for the post-transcriptional expression of TolC.
TolC-dependent growth defect of ΔygiBCΔyjfMC mutant
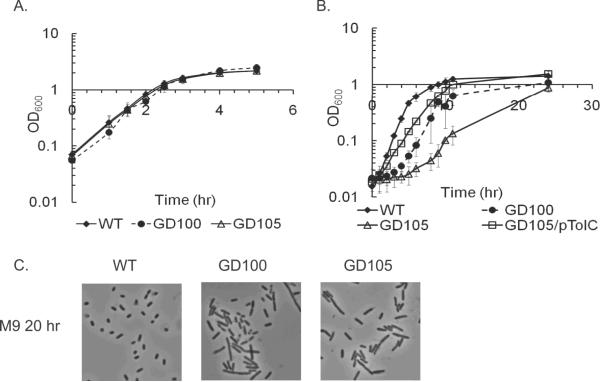
The arrangement in a single operon suggested that functions of YgiB and YgiC could be linked to that of TolC. Therefore, we investigated growth phenotypes of cells lacking tolC alone, ygiBC alone or the entire operon. Consistent with previous results, no defects were found when tolC mutant (GD100) was grown in Luria-Bertani (LB) broth (Fig. 2A). In the minimal M9 medium however GD100 cells delayed exiting from the stationary phase for at least four hours and in the exponential phase grew 30% slower than the WT cells. The E. coli mutant with the deletion of the entire tolC-ygiBC operon (GD102) was phenotypically similar to ΔtolC (Table 1).
Figure 2.
Growth and cell division defects of ΔtolC cells. Overnight cultures of WT, GD100 (ΔtolC) and GD105 (ΔtolC–ygiBC ΔyjfMC) grown either in LB (A) or M9 (B) were re-inoculated (1:100) into the fresh matching medium. The growth was monitored by measuring absorbance at 600 nm. The initial cell densities were adjusted to approximately the same value. For each growth curve the average of three independent experiments is shown. Error bars are standard deviations (SD, n=3). C. Phase contrast microscopy. Cells were grown overnight in LB, diluted 1:100 into fresh M9 medium and incubated at 37°C for twenty hours.
Table 1.
Growth rates of E. coli mutants in LB and minimal M9 media (hr−1)
| Name | Genotypea | LB | M9 (Lag time, hr) |
|---|---|---|---|
| BW25113 | WT | 1.22±0.13 | 0.62±0.05 |
| GD100 | Δ tolC | 1.12±0.08 | 0.44±0.05b (4±1.8) |
| GD101 | Δ ygiBC | 1.14±0.18 | 0.63±0.01 |
| GD102 | Δ tolC-ygiBC | 1.04±0.003 | 0.55±0.05 (5.3±0.6) |
| GD103 | Δ yjfMC | 1.17±0.04 | 0.63±0.02 |
| GD104 | ΔygiBC ΔyjfMC∷kan | 1.03±0.01 | 0.65±0.03 |
| GD105 | ΔtolC-ygiBC ΔyjfMC∷kan | 1.11±0.03 | 0.34±0.10 (7.8±1.7) |
| GD111 | ΔtolC∷kan ΔyjfMC | 1.01±0.007 | 0.43±0.08 (1.3±0.5) |
| JW0117-1 | Δ speE ∷ kan | NDc | 0.52±0.04 |
| JW2914-1 | Δ gshB ∷ kan | ND | 0.57±0.04 |
| JW2956-1 | Δ gspS ∷ kan | ND | 0.55±0.04 |
| JW2663-1 | Δ gshA ∷ kan | ND | 0.58 |
| GD106 | ΔspeE ΔtolC∷kan | ND | 0.49±0.06 (1.8±0.6) |
| GD107 | ΔgshB ΔtolC∷kan | ND | 0.52±0.06 (3±1.4) |
| GD108 | ΔygiBC ΔyjfMC ΔgspS∷kan | ND | 0.58±0.03 |
| GD109 | ΔspeE ΔygiBC∷kan | ND | 0.61±0.02 |
Positions of kanamycin cassette when applicable are indicated
Here and in all other Tables, values in bold are significantly different from the BW25113 strain
ND, no data. Averages of three independent experiments and standard deviations are shown.
The BLAST analysis showed that E. coli chromosome contains two genes yjfM and yjfC homologous to ygiB and ygiC, respectively. The yjfM gene shares 23% identity and 40% similarity with ygiB, whereas yjfC and ygiC are 50% identical. The yjfMC genes are preceded by two additional genes of unknown functions, which together with yjfMC form an yjfKLMC operon (Fig. 1A). Since yjfMC are highly homologous to ygiBC, functions of these two sets of genes could be complementary to each other. Therefore, we constructed ΔtolC mutants containing deletions in yjfMC alone (GD111) and in both ygiBC and yjfMC (GD105). Similar to ΔtolC and ΔtolC-ygiBC strains, the LB grown GD111 had the WT phenotype, but in M9 these cells experienced a two-hour lag when exiting the stationary phase and grew slower than the WT (Table 1). This growth deficiency phenotype was significantly exacerbated in GD105 cells. Upon re-inoculation into a fresh M9 medium (M9-to-M9), the overnight grown GD105 cells experienced approximately eight hours of lag time and once resumed, the growth rate was only half of the WT (0.34 ±0.1 h−1, Table 1). Therefore, the deletion of either YgiBC or YjfMC alone does not further compromise the ΔtolC phenotype. However, when both gene sets are deleted, ΔtolC cells have significant growth defects in M9. This result suggested that YgiBC and YjfMC have complementary functions that enable growth of ΔtolC cells in M9.
Reciprocally, the growth phenotype of ΔygiBCΔyjfMC cells depended on TolC. The TolC+ cells lacking ygiBC and yjfMC (GD104) had the WT phenotype in both LB and M9 media (Table 1). Furthermore, the growth phenotype of GD105 cells was rescued by the plasmid carrying tolC alone (Fig. 2B). Taken together these results suggest that TolC and YgiBC/YjfMC have parallel, independent functions. When both functions are compromised E. coli is unable to grow efficiently in the minimal medium.
Determination of colony forming units and the LIVE/DEAD BacLight staining of cells showed that in stasis and during extended lags ΔtolC cells are viable. For all strains, we found that the number of dead cells is less than 5% and remains the same in the exponential and stationary phases. However, GD100 and GD105 cells when grown in M9 were notably longer than the WT indicating cell division defects (Fig. 2C).
GD105 cells are more susceptible to antibiotics
To investigate whether inactivation of ygiBC and/or yjfMC affects antibiotic susceptibility of E. coli cells we measured minimal inhibitory concentrations (MICs) of antibiotics erythromycin, novobiocin and norfloxacin and detergent SDS (Table 2). Consistent with previous studies, ΔtolC cells were highly susceptible to all tested compounds. However, deletions of either ygiBC or yjfMC, alone and in combination, in the WT and ΔtolC backgrounds did not increased susceptibility to antibiotics. The only notable change was the 2–4 fold decrease of MICs of novobiocin and norfloxacin in GD105 (ΔtolC–ygiBC ΔyjfMC). We conclude that ygiBC and/or yjfMC do not contribute to intrinsic levels of antibiotic resistance but their absence aggravates the ΔtolC phenotype.
Table 2.
Susceptibility of E.coli strains to antimicrobial agents
| Strain | Genotype | ERYa | NOVa | NORa | SDSa |
|---|---|---|---|---|---|
| BW25113 | WT | 32 | 32 | 0.04 | 10240 |
| ETBW | Δ tolC | 1 | 1 | 0.01 | 5 |
| GD101 | Δ ygiBC | 64 | 128 | 0.04 | 10240 |
| GD102 | Δ tolC-ygiBC | 1 | 1 | 0.01 | 5 |
| GD103 | Δ yjfMC | 64 | 64 | 0.04 | 10240 |
| GD104 | ΔygiBC ΔyjfMC∷kan | 64 | 128 | 0.04 | 10240 |
| GD105 | ΔtolC-ygiBC ΔyjfMC∷kan | 1 | 0.25 | 0.005 | 5 |
ERY-Erythromycin, NOV- Novobiocin, NOR- Norfloxacin, SDS- sodium dodecyl sulfate; numbers are minimal inhibitory concentrations, MICs, in μg/ml. All experiments were repeated at least three times.
ΔtolC cells are depleted of essential metabolites
To investigate why ΔtolC cells have problems growing in M9, we measured concentrations of NAD+ and NADH during the LB-to-M9 transition and in the stationary phase. NAD+ and NADH are the major players in microbial metabolism and their absolute and relative amounts report on the metabolic and redox states of cells (Gennis & Stewart, 2006). Consistent with their growth phenotypes, the NAD+ concentrations were at the same ≈1.5 mM levels in WT and mutants growing in LB (Table 3). After transition into stationary phase, the concentration of NAD+ decreased in all strains but GD105 cells contained the lowest 0.3 mM NAD+.
Table 3.
NAD+ and NADH concentrations in exponential and stationary phase cells
| Strains | LB | M9 | ||||
|---|---|---|---|---|---|---|
| 1 h | 12 h | 1 h | 12 h | 20h | ||
| BW25113 | NAD+, mM | 1.4 ± 0.28 | 0.77 ±0.29 | 1.5±0.06 | 1.1± 0.15 | 0.78 ± 0.29 |
| NADH, mM | 0.48±0.2 | 0.21 ±0.06 | 0.38±0.13 | 0.63±0.08 | 0.61±0.27 | |
| NADH/NAD+ | 0.3 | 0.3 | 0.2 | 0.5 | 0.8 | |
|
| ||||||
| GD104 | NAD+, mM | 1.7±0.03 | 0.43±0.01 | 1.5±0.1 | 1.1±0.14 | 0.52±0.007 |
| NADH, mM | 0.97±0.04 | 0.21±0.07 | 0.25±0.07 | 0.5±0.007 | 0.36±0.01 | |
| NADH/NAD+ | 0.6 | 0.5 | 0.2 | 0.4 | 0.7 | |
|
| ||||||
| GD100 | NAD+, mM | 1.2±0.3 | 0.79±0.2 | 1.4±0.08 | 1.1±0.01 | 0.34±0.01 |
| NADH, mM | 0.80±0.4 | 0.19±0.06 | 0.5±0.09 | 0.4±0.06 | 0.35±0.06 | |
| NADH/NAD+ | 0.7 | 0.2 | 0.4 | 0.4 | 1.0 | |
|
| ||||||
| GD105 | NAD+, mM | 1.35±0.18 | 0.29±0.2 | 1.57±0.3 | 0.47±0.04 | 0.08±0.02 |
| NADH, mM | 0.94±0.47 | 0.34±0.18 | 0.36±0.06 | 0.29±0.01 | 0.23±0.01 | |
| NADH/NAD+ | 0.7 | 1.2 | 0.2 | 0.6 | 2.9 | |
|
| ||||||
| GD105/pTolC | NAD+, mM | 1.51±0.01 | 0.52±0.05 | 1.2±0.1 | 0.99±0.01 | 0.6±0.05 |
| NADH, mM | 1.1±0.01 | 0.26 ±0.05 | 0.3±0.003 | 0.51±0.06 | 0.51±0.01 | |
| NADH/NAD+ | 0.7 | 0.5 | 0.3 | 0.5 | 0.8 | |
All strains contained similar concentrations of NAD+ and NADH one hour after the shift into M9 medium (Table 3). However, transition into the stationary phase caused a significant drop of NAD+ concentration in GD105. Extension of stasis for 20 hours led to further decrease of NAD+ to 0.08 mM in GD105 cells and the threefold drop to 0.3 mM of NAD+ in GD100. Thus, when growing in M9, albeit to a different degree, GD100 and GD105 cannot efficiently synthesize NAD+. This leads to dilution and eventual depletion of this critical cofactor in the stationary phase.
Interestingly, WT and ΔtolC mutants also responded differently to stasis in terms of concentration of NADH. Although amounts of NADH increased in the M9 12 and 20 hr samples of the WT cells reflecting the reduced respiration and starvation, the amounts of this cofactor did not change significantly in GD100 and GD105 cells. As a result, despite the large difference in absolute concentrations, the NADH/NAD+ ratio was close to one in the M9 20 hr samples of WT and GD100 but increased to almost three in GD105 cells due to dramatic depletion of NAD+. This result strongly suggests that in M9 medium, GD105 is not only deficient in biosynthetic activities but cannot oxidize NADH as well.
The NAD+ depletion in stationary GD105 cells was rescued by a plasmid carrying tolC alone suggesting that expression of TolC is required and sufficient to maintain normal metabolism in M9 medium (Table 3). In agreement, GD104 cells, which are TolC+ but lack ygiBC and yjfMC, contained normal NADH/NAD+ ratios.
Taken together these results show that inactivation of TolC causes depletion of essential metabolites when cells enter stasis. The depletion of NAD+ is notably accelerated in GD105 cells and in minimal medium. The unusually high NADH/NAD+ ratio and low concentrations of NAD+ imply that stationary GD105 cells are in the state of the metabolic shutdown.
NADH dehydrogenases are inhibited in ΔtolC cells
During aerobic metabolism, the major pool of NADH is re-oxidized on the membrane by two membrane-bound NADH dehydrogenases NDH-I and NDH-II (Yun et al., 2005). Thus, either one or both of these proteins could be inhibited in GD105 cells. Therefore, we next measured the NADH oxidase activity in membrane fractions isolated from WT and ΔtolC mutants grown in LB and M9 media. We found that exponential WT and mutant cells contained similar levels of the membrane-bound NADH oxidase activity (Table 4). Consistent with previous reports this activity did not depend on the growth medium and was the same for the LB and M9 grown cells (Dancey et al., 1976). In the stationary WT and GD100 cells, the NADH oxidase activity decreased by 25–35%. In contrast, the NADH oxidase activity of GD105 membranes dropped 2–3 folds when cells entered stasis in either medium and was 40–60% lower than in the stationary WT cells. Thus, in accord with NADH/NAD+ measurements (Table 3), GD105 membranes cannot efficiently oxidize NADH. We conclude that inhibition of membrane-bound NADH dehydrogenases contributes to metabolic shutdown in ΔtolC cells.
Table 4.
Specific NADH oxidase activity of WT and ΔtolC membranes
| Medium, time of incubation | BW25113 | GD100 | GD105 |
|---|---|---|---|
| NADH oxidase activity (U/mg) | |||
| LB, 1 hr | 0.39 ± 0.06 | 0.46 ± 0.09 | 0.39 ± 0.03 |
| LB, 12 hr | 0.28 ± 0.02 | 0.28 ± 0.04 | 0.13 ± 0.01 |
| M9, 1 hr | 0.49 ± 0.09 | 0.33 ± 0.04 | 0.52 ± 0.1 |
| M9, 12 hr | 0.31 ± 0.1 | 0.36 ± 0.05 | 0.21 ± 0.01 |
| M9, 20 hr | 0.35 ±0.007 | 0.24 ± 0.07 | 0.20 ± 0.05 |
Deletion of TolC creates stress in the inner membrane
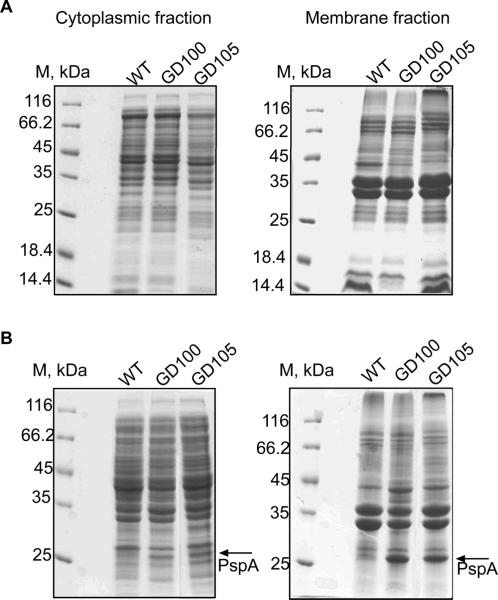
To investigate how ΔtolC cells respond to metabolic shutdown, we compared protein profiles of cytoplasmic and membrane fractions isolated from exponential and stationary phase cells. Protein compositions of the exponential WT and ΔtolC mutant cells were very similar. However, upon transition into the stationary phase ΔtolC cells significantly overproduced a 26 kDa membrane protein (Fig. 3). No major changes in protein composition were found in GD104 cells lacking only ygiBC/yjfMC (data not shown). The 26 kDa protein from ΔtolC membranes was readily solubilized with 1% sarcosyl indicating that the protein is associated with the inner membrane. The N-terminal protein sequencing identified this protein as a phage shock protein A, PspA. PspA is a peripheral inner membrane protein, which is induced specifically by membrane stress and dissipation of a proton motive force (PMF) (Kobayashi et al., 2007). Thus, inactivation of TolC creates stress on the cytoplasmic membrane of E. coli.
Figure 3.
Stationary phase ΔtolC cells overproduce the membrane stress protein PspA. SDS-PAGE analysis of the cytoplasmic and membrane fractions isolated from the exponential (A) and stationary (B) cultures of WT and ΔtolC mutants.
Growth defects of GD105 cells are suppressed by L-serine
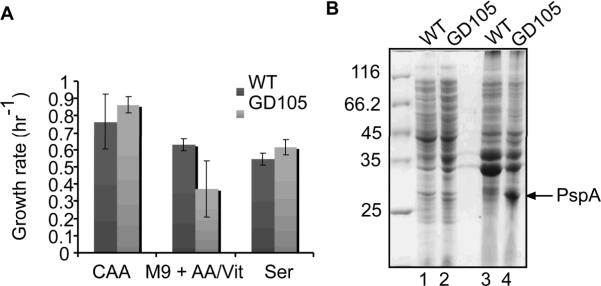
The experiments described above established that the membrane stress and metabolic shutdown in GD100 and GD105 cells are aggravated in the M9 medium, whereas LB medium suppresses these defects. This result suggested that certain nutrients or vitamins might alleviate the problems these cells experience in M9. Indeed, the growth rate and rapid exit from stasis of GD105 strain was restored by supplementing M9 medium with 0.4% casamino acids (Fig. 4A).
Figure 4.
Growth defects, but not the membrane stress of GD105 cells, are rescued by L-serine. A. Growth rates of WT and GD105 cells in M9 supplemented with casamino acids, individual amino acids or vitamins. B. SDS-PAGE analysis of the cytoplasmic (lanes 1 and 2) and membrane (lanes 3 and 4) fractions isolated from the stationary (48 hrs) cultures of WT and GD105.
We next tested various vitamins and amino acids for their ability to restore normal growth of GD105 in M9. Among six tested amino acids only L-serine restored completely the growth phenotypes of these cells to that of the WT (Fig. 4A). Although glycine and cysteine are synthesized from L-serine, neither of these two amino acids could restore the GD105 growth in M9. Furthermore, consistent with previous reports (Wiriyathanawudhiwong et al., 2009), we found that cysteine inhibits the growth of all ΔtolC mutants (data not shown). Thus, in M9 medium ΔtolC and GD105 cells became auxotrophic for L-serine.
Interestingly, despite the normal growth in the M9 medium supplemented with L-serine, the stationary phase ΔtolC cells still contained the elevated amounts of PspA protein (Fig. 4B). We conclude that L-serine supplementation does not prevent membrane stress caused by the inactivation of TolC but likely enables cells to repair membranes and resume growth in the presence of glucose.
Cells lacking glutathione, spermidine and glutathionylspermidine do not have the TolC-dependent growth defect
In all assays described above, deletions of ygiBC and yjfMC significantly aggravated the metabolic problems of the stationary phase ΔtolC cells. Since YgiC/YjfC and GspS share significant homology, all three proteins might be involved in synthesis of GSP. Furthermore, previous studies suggested that GSP accumulates in the stationary phase E. coli cells (Smith et al., 1995). To investigate whether the lack of GSP contributes to growth defects of GD105, we constructed E. coli cells lacking all three GSP synthetases: gspS, ygiC and yjfC (GD108). However, GD108 cells had the WT phenotype when growing in M9 (Table 1). Thus, the lack of GSP alone does not cause growth defects in M9.
To further investigate the putative functional link between GSP synthetases and TolC we constructed ΔtolC mutants that are also deficient in the synthesis of either GSH (ΔgshB and ΔgshA mutants lacking GSH synthetase and γ-glutamate-cysteine ligase, respectively) or SPE (ΔspeE lacking spermidine synthase). Previous studies confirmed that these mutants do not produce GSH and SPE, respectively (Xie et al., 1993, Fuchs, 1995). We found however that these cells grow in M9 medium with the same rate as ΔtolC cells (Table 1). Furthermore, double ΔgshBΔspeE mutant producing neither GSH nor SPE does not have any significant growth defect in M9 medium. These results strongly suggested that metabolic defects of GD100 and GD105 are not linked to the biosynthetic pathways of GSH, SPE or GSP.
ΔtolC cells deplete glutathione
Earlier studies suggested that both GSH and GSP are expelled from the E. coli and Salmonella stationary cells (Smith et al., 1995). To investigate whether inactivation of TolC in GD100 and GD105 strains leads to accumulation of these metabolites, we compared the amounts and composition of non-protein thiols in cells grown either in LB or M9 medium. As a negative control, we used ΔgshA cells, which do not produce GSH and its precursor γ-Glu-Cys (Helbig et al., 2008).
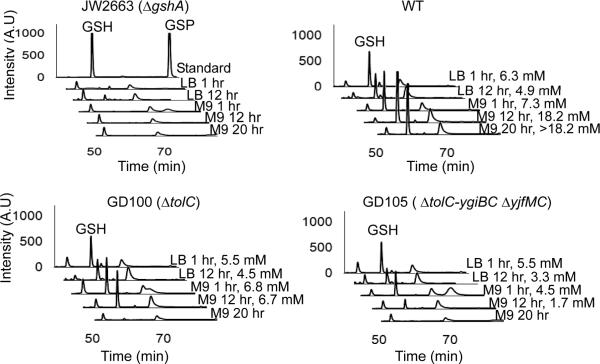
We first determined whether there is any change in the composition of the reduced thiols. For this purpose, intracellular thiols were derivatized with monobromobimane (MBB) and analyzed by HPLC (Fairlamb et al., 1986). Fig. 5 shows that the composition of non-protein thiols is identical in the WT and ΔtolC mutants growing in LB, with the major peak identified as GSH. Although previous studies suggested that GSH is converted into GSP upon transition into the stationary phase (Smith et al., 1995, Tabor & Tabor, 1975), we did not find larger amounts of GSP in the stationary phase cells (Fig. 5). When incubated in M9, the major difference between the WT and ΔtolC cells was a significant decrease in the amounts of GSH in GD105 cells even one hour after the shift into M9 (M9 1 hr). Furthermore, twenty hours of incubation in M9 (M9 20 hr) led to the drop of GSH amounts below detection limits in both GD100 and GD105 cells (Figs. 5C, D).
Figure 5.
The M9 grown ΔtolC cells deplete endogenous GSH. Exponentially growing WT and mutant cells were collected by centrifugation, washed and resuspended either in LB or in M9 medium. At indicated time points, intracellular non-protein thiols were derivatized with MBB and separated by HPLC. A. Top lane, GSH and GSP standards derivatized with MBB; bottom lanes, MBB-labeled thiols from ΔgshA cells. B, C, D. MBB-labeled non-protein thiols from the WT, GD100 and GD105, respectively. The intracellular concentrations of GSH when detectable were calculated from peak area using the calibration curve generated with GSH. These are indicated for each profile where GSH is detectable.
To quantify GSH and its oxidized form glutathione disulfide (GSSG) we used the glutathione reductase assay (Anderson, 1985). In agreement with HPLC data, one hour after LB-to-M9 shift, GD100 maintained [GSH+GSSG] at levels similar to those in the WT but the amounts of [GSH+GSSG] decreased more than eight fold to ≈1.8 mM in the stationary M9 20 hr cells (Table 5). The depletion of GSH was accelerated in GD105 cells. These cells lost half of their [GSH+GSSG] one hour after transfer into M9 and were down to 0.3 mM of [GSH+GSSG] in the stationary phase (M9 20 hr). Thus, similar to NAD+/NADH, the amounts of GSH are rapidly depleted when ΔtolC mutants enter the stationary phase in M9. The concentrations of GSSG were extremely low in all cells (less than 1 μM, data not shown) indicating that the dramatic drop of GSH concentration in stationary GD100 and GD105 cells was not caused by its oxidation.
Table 5.
Intracellular (in) and excreted (out) concentrations of glutathione in the WT and ΔtolC mutants
| Medium, time of incubation | BW25113 | GD100 | GD105 | |||
|---|---|---|---|---|---|---|
| GSH+GSSG, mM | GSH+GSSG, mM | GSH+GSSG, mM | ||||
| in | out | in | out | in | out | |
| LB, 1 hr | 10.4±0.1 | NDa | 9.7±0.8 | ND | 7.6±1.9 | ND |
| LB, 12 hrs | 8.1±0.4 | ND | 8.3±0.01 | ND | 7.8±1.2 | ND |
| M9, 1 hr | 8.8±0.7 | -b | 7.1±0.3 | - | 4.2±0.3 | - |
| M9, 12 hr | 17.7±1.9 | 0.001±0.002 | 8.5±0.1 | 0.008±0.0003 | 2.4±0.2 | 0.009±0.001 |
| M9, 20 hr | 15.1±0.6 | 0.019±0.001 | 1.8±0.5 | 0.014±0.001 | 0.3±0.1 | 0.005±0.0004 |
ND – no data
- below detection levels
Micromolar concentrations of GSH were detected in supernatants of all stationary phase cultures except ΔgshA cells that do not produce GSH (Table 5). Interestingly, despite lower intracellular GSH levels and lack of TolC, GD100 and GD105 cells excreted up to ten times more GSH than the WT cells, with GD105 releasing the largest amounts of GSH. Thus, TolC is not required for GSH efflux. Furthermore, the higher concentrations of GSH in supernatants suggest that ΔtolC cells leak GSH into medium.
Discussion
In this study, we found that TolC-deficient E. coli cells are defective when grown in minimal medium with glucose. The growth defect manifests itself in the decreased growth rate, the prolonged lag when exiting the stationary phase and altered morphology. This phenotype is further exacerbated by the lack of ygiBC and yjfMC gene products, which share homology with GSP synthetases. Based on the results presented here and extant literature we propose that stress on the cytoplasmic membrane and inhibition of NADH dehydrogenases are the major causes of the TolC-dependent growth defects.
We found that membrane fractions of the stationary ΔtolC cells contain large amounts of PspA (Fig. 3). PspA is known to be overproduced in response to various extracytoplasmic challenges such as osmotic shock, misfolded membrane proteins or presence of ionophore CCCP (Kleerebezem et al., 1996). A well-established commonality among various Psp-activating signals is reduction in the membrane electrochemical potential, which drives the PMF. Therefore, membranes of the ΔtolC cells are under the stress that dissipates PMF. This stress is most profound when ΔtolC cells enter the stationary phase (Table 3, Fig. 3). The stationary phase in general is stressful to E. coli as evidenced by activation of various stress response systems in starving cells (Nystrom, 2004). Furthermore, one of the most damaging starvation-induced stresses is oxidative stress (Dukan & Nystrom, 1999). Interestingly, recent study reported that cells lacking TolC overproduce stress response proteins SoxS, MarA and Rob (Rosner & Martin, 2009). Among these factors, SoxRS regulon is known to specifically respond to the intracellular accumulation of superoxide. Thus, ΔtolC cells cannot efficiently detoxify this oxygen derivative, which could further aggravate the oxidative stress in starving cells.
In addition to membrane stress, ΔtolC cells are severely depleted in essential metabolites. The decrease of GSH concentration in GD105 is notable already one hour after transfer into M9 medium (Fig. 5 and Table 5). However, it is in the stationary phase the concentrations of both GSH and NAD+ become very low in both GD100 and GD105 cells (Tables 3 and 5). The degree of metabolite depletion correlates with the time required for these two mutants to exit the stasis and resume growth in the fresh medium. GD105 cells cannot resume the growth for up to eight hours after transfer into a fresh medium, whereas GD100 start growing after two-four hours of delay (Fig. 2). Since the number of live cells is the same in WT and ΔtolC cells, we suggest that ΔtolC cells cannot exit the stasis because of the stress on the membrane and low concentration of NAD+ but resume growth when they replenish depleted metabolites and repair the membranes.
Growth of ΔtolC cells is also significantly hindered by abnormal NADH/NAD+ ratios. In the stationary GD105 cells the concentration of NADH exceeds that of NAD+ almost three times (Table 3) and this problem is serious in both LB and M9 media. For comparison, shift under anaerobic conditions leads to change in the NADH/NAD+ ratios in E. coli cells from 0.2 to only 0.67 (Leonardo et al., 1996). High levels of NADH impact E. coli physiology on several levels. First, large NADH/NAD+ ratios inhibit key enzymes, e.g. citrate synthase and malate dehydrogenase, shared by the tricarboxylic acid cycle (TCA) and the glyoxylate shunt (Pruss et al., 1994). Therefore, in GD105 these metabolic pathways are disabled and these cells cannot efficiently catabolize or synthesize metabolites that require the TCA cycle.
Second, high concentrations of NADH despite the drop in amounts of NAD+ indicate that activity of NADH dehydrogenases is inhibited as well. Indeed, we found that rates of NADH oxidation in GD105 membranes decrease significantly upon entry into stasis and remain only at 50% level of the WT (Table 4). NADH oxidation in E. coli is carried out by multiple enzymes (Gennis & Stewart, 2006). However, previous studies showed that mutational inactivation of the membrane-bound NADH dehydrogenase NDH-I leads to growth defects during transition into the stationary phase (Pruss et al., 1994). These defects of NDH-I mutants can be suppressed by serine. Similarly, we found that serine suppresses the growth defects of ΔtolC cells (Fig. 4). In addition, NDH-I couples NADH oxidation to production of a PMF. The overproduction of PspA in ΔtolC membranes indicates a decrease in a PMF. Taken together these findings suggest that TolC inactivation leads to inhibition of NDH-I, which in turn results in metabolic shutdown and dissipation of a PMF.
Finally, NADH acts as an iron reductant in a Fenton reaction and drives DNA damage by iron and peroxide in vivo (Brumaghim et al., 2003). Previous studies showed that raising the NADH level in vivo by eliminating or negatively regulating NADH dehydrogenase activity dramatically sensitizes E. coli to killing by peroxide (Imlay & Linn, 1988). Thus, high levels of NADH in GD105 cells could contribute significantly to oxidative stress in these cells. We suggest that rising levels of NADH are the major reasons for the induction of SoxS, MarA and Rob stress responses in the cells lacking TolC.
It remains unclear whether inactivation of one specific TolC-dependent function or a cumulative effect of the loss of multiple transport activities is responsible for the membrane stress and growth phenotypes of ΔtolC cells. It is often assumed that diverse phenotypes of ΔtolC cells are caused by accumulation of toxic metabolites or by the lack of secretion (Rosner & Martin, 2009, Wiriyathanawudhiwong et al., 2009). We found however, that cells lacking the major drug efflux pump AcrAB and its homolog AcrEF have the WT phenotype (data not shown). Our results suggest that stress on the cytoplasmic membrane and metabolic defects in ΔtolC cells, rather than loss of efflux, contribute significantly to most of the previously reported phenotypes.
All ΔtolC phenotypes are significantly aggravated by the lack of YgiBC and YjfMC proteins. YgiC and YjfM share highly conserved catalytic residues with GspS and are likely to conjugate GSH and spermidine in the cytoplasm. However, their counterparts YgiB and YjfM are peripheral membrane proteins located in the periplasm. The function of GSP and YgiBC and YjfMC proteins are unknown. Our studies suggest that activities of these proteins are not dependent on the GSH and spermidine biosynthetic pathways (Table 1) but are needed for protection of the inner membrane. We noticed that the PspA-like transcriptional regulator is located immediately upstream of the yjfKLMC operon, further strengthening the notion that the function of these proteins might be related to the maintenance or protection of the cytoplasmic membrane.
Experimental procedures
Strains and plasmids
All E. coli strains constructed in this study are derivatives of BW25113 (lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78) (Datsenko & Wanner, 2000). The BW25113 derivatives JW0117-1 (ΔspeE), JW2914-1 (ΔgshB), JW2956-1 (ΔgspS), JW2663 (ΔgshA) were obtained from the Keio collection (distributed by Coli Genetic Stock Center at Yale University) and confirmed with PCR. All mutants were constructed using λ red recombinase system and antibiotic resistance genes were removed as described in (Datsenko & Wanner, 2000). The genotypes of strains and positions of antibiotic resistance cassettes, when appropriate, are indicated in Table 1. All gene disruptions including purchased strains were confirmed with PCR. The plasmid pTolC carrying tolC under IPTG-inducible Ptac promoter was a gift from Dr. Rajeev Misra.
Growth media and conditions
Cells were grown in a rotary shaker set at 220 rpm and 37°C either in LB broth (10 g of Bacto tryptone, 10 g of yeast extract, and 5 g of NaCl per liter) or in M9 minimal medium (5×M9 salts (Difco), 0.4% glucose, 2 mM MgSO4 and 0.1 mM CaCl2). Growth rates were measured after 1:100 dilution of the overnight grown cultures into fresh LB or M9 media. The sequential second and third re-inoculation into M9 medium showed that the GD105 phenotype is stable and that resumption of the growth is not caused by suppressor mutants. Changes in cell density were monitored by measuring absorbance at 600 nm. Growth rates were calculated from the linear slopes of the logarithmic plots of growth curves. P-values were calculated using Microsoft Office Excel and were found to be 0.011 and 0.010 for GD100 and GD105, respectively. To determine the amino acid requirements, strains were grown in M9 medium supplemented with either 0.4% casamino acids or one of the following amino acids: 0.2 g/L L-aspartic acid, 0.6 g/L L-glutamic acid, 0.1 g/L L-serine, 0.2 g/L L-methionine, 0.2 g/L L-glycine, 0.1 g/L L-cysteine.
Minimal Inhibitory Concentrations
MICs of various antimicrobial agents were measured in 96-well plates (Tikhonova et al., 2002). Approximately 104 exponentially growing cells were inoculated in LB medium supplemented with two-fold increasing concentrations of antibiotic. Plates were incubated overnight at 37°C without shaking.
Reverse Transcriptase PCR
RT-PCR was performed by using QIAGEN OneStep RT-PCR kit. Total RNA was isolated from exponentially growing cells (LB broth) using QIAGEN RNeasy kit. 2 μg of purified total RNA were used for RT-PCR. To detect the tolC transcript (reaction 4 in Fig. 1), the forward primer 5'-AATGCAAATGAAGAAATTGC and the reverse primer 5'-CGCGTACCGACGCAGTAGCCCGCTTC were used to yield a product of 1173 bp. To detect the ygiB and ygiC mRNA (reactions 1, 2, and 3 on Fig. 1) the forward primer 5'-CAAAGCTCATTATGCGCGATGGAAGCG and the reverse primers (1) 5'-CATCATGTAACCGGCCATCAG 3', (2) 5'-GATGTAGAGGAACTCAGTAGC 3' and (3) 5'-GTATCAGCCACCCATTGAACG 3' were used to yield products of 889 bp, 1184 bp and 1810 bp, respectively.
Protein Expression Profiles
WT and mutants grown overnight in M9 were diluted 1:100 into fresh M9 medium and incubated at 37°C with shaking at 220 rpm. Cells were collected by centrifugation in the exponential phase (OD600= 0.6) and 48 hrs after inoculation into the fresh medium. Cells were then washed and re-suspended in the buffer containing 10 mM Tris-HCl (pH 8.0) and 5 mM EDTA and then treated with 100 μg/ml lysozyme for 40 min. Sonication was used to lyse cells. Unbroken cells were removed by low speed centrifugation and membranes were collected by ultracentrifugation for 1 hr at 100,000×g. Membrane pellets were resuspended in buffer containing 50 mM Tris-HCl (pH 8.0), 100 mM NaCl and 1 mM PMSF. The cytoplasmic and membrane fractions (20 μg of total protein as estimated using Bradford assay with bovine serum albumin (BSA) as a standard) in the SDS-sample buffer were boiled for 5 min and then analyzed using standard 12% SDS PAGE and Coomassie brilliant blue staining.
NAD+ and NADH measurements
NADH recycling assay was used to determine intracellular concentrations of NAD+ and NADH (Matsumura & Miyachi, 1980). Cells growing in LB (OD600 ~ 1.0) were collected by centrifugation and either re-suspended in the same volume of fresh LB or washed and then re-suspended in fresh M9. Cells were incubated at 37°C with shaking (220 rpm) and were collected by centrifugation at one, twelve and twenty hours post re-inoculation. For the assay, ~5×109 cells were collected by centrifugation at 15,700×g for 2 min, the cell pellets were frozen in dry ice ethanol bath and stored at −80°C until use. 250 μl of 0.2 M NaOH (for NADH extraction) or 250 μl of 0.2 M HCl (for NAD+ extraction) were added to the frozen pellets, samples were boiled for 10 min and then centrifuged at 9,300×g for 5 min. The NAD+ and NADH supernatants were immediately used for assay.
The reaction mixture contained 30 μl of 1.0 M BICINE-NaOH (pH 8.0), 75 μl of cell extract, 75 μl of 0.1 M NaOH for NAD+ samples or 0.1 M HCl for NADH samples, 30 μl of 16.6 mM phenazine ethosulfate, 30 μl of 4.2 mM 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT), 30 μl of absolute ethanol and 30 μl of 40 mM EDTA (pH 8.0). The reaction mix was pre-incubated for 3 min at room temperature and 6 μl of 500 U/ml yeast alcohol dehydrogenase enzyme was added to start the reaction. The rate of reduction of MTT, which is directly proportional to amounts of NAD+ and NADH in samples, was monitored at 570 nm for 5 min. Standard curves were generated by using 0.125 to 1.25 nmoles of NAD+ and NADH (Sigma), respectively.
NADH oxidase assay
Cell membrane preparations and the NADH oxidase assay were performed as described before (Osborn et al., 1972, Tikhonova & Zgurskaya, 2004). Briefly, cells were incubated in LB and M9 media for 1, 12 and 20 hrs as described above. Spheroplasts were prepared by EDTA-lysozyme treatment. After sonication, unbroken cells were removed by low speed centrifugation and membranes were collected by ultracentrifugation for 1 hr at 100,000×g. Membrane pellet was resuspended in 10 mM Tris-HCl (pH 8.0), 0.25 M sucrose. Protein concentration of membranes was measured using Bradford assay with BSA as a standard. 50 μg of total protein was used for the assay. The assay mixture contained 50 mM Tris-HCl (pH 7.5), 0.3 mM DTT and 0.60 μM NADH (Sigma). Rate of decrease in absorbance at 340 nm was measured for one minute.
Determination of GSP, GSH and GSSG content
Two methods were used to determine the composition and concentration of non-protein thiols. HPLC was used to analyze intracellular thiols derivatized with monobromobimane (MBB) as described in (Fairlamb et al., 1986, Helbig et al., 2008). Cells were incubated in LB and M9 media for 1, 12 and 20 hrs as described above. 1×109 cells (1.0 OD600) were washed with fresh M9 medium and re-suspended in buffer containing 40 mM HEPES-KOH (pH 7.0) and 5 mM EDTA. Derivatization with MBB (73.7 μM final concentration) was carried out at 70°C for 3 min. HPLC analyses were done on a Zorbax C-18 column (Hewlett Packard) and Shimadzu LC-10Ai HPLC system with RF-10AXL fluorescence detector. The fluorescence of MBB derivatized thiols was detected at 380 nm excitation and 490 nm emission wavelengths. Mobile phase A contained 0.25% (w/v) d-camphorsulfonate in water, pH 2.7. Mobile phase B contained 0.25% (w/v) d-camphorsulfonate and 25% isopropanol.
Three peaks were detected in all samples (Fig. 5). By comparison to standards and ΔgshA cells, the major peak of the WT cells is GSH. The early 52 min peak was present in the control sample containing dye alone and likely to be a contaminant (data not shown). The third peak (70 min) contained an unidentified thiol compound, which is also present in ΔgshA cells and thus, does not originate from GSH. In addition, all cells contained small variable amounts of the fourth minor peak, which by its mobility could correspond to GSP (85 min).
The GSSG reductase recycling assay was used to determine the total GSH+GSSG and GSSG only concentrations in vivo and in culture supernatants (Anderson, 1985). Approximately 1×109 cells growing in LB or M9 medium were re-suspended in 20 μl of buffer containing 143 mM sodium phosphate (pH 7.4) and 6.3 mM EDTA and 10 μl of 10% 5-sulfosalicylic acid. Samples were centrifuged at 15,700×g for 5 min to remove protein precipitates. For each reaction, 210 μl of 0.25 mg/ml NADPH (Sigma) and 30 μl of 6 mM 5,5'-dithiobis(2-nitrobenzoic acid) (DTNB, Sigma) were mixed together and incubated at 30°C for 15 minutes. The reaction was started by addition of 60 μl of sample and 3 μl of 266 U/ml yeast glutathione reductase enzyme (Sigma). For GSSG measurements, 20 μl of each sample were treated with 0.4 μl of 2-vinylpyridine. The pH of sample was adjusted to 6–7 by triethanolamine. The rate of absorbance increase at 412 nm was monitored for 5 min. The assay was calibrated with GSH and GSSG standards (Sigma), in the range of 0.15 to 1.2 nmoles. The same reactions were set to measure GSH and GSSG in culture supernatants, except 25 μl of each supernatant were used in the reaction.
Phase contrast and fluorescence microscopy
For phase contrast microscopy, E. coli strains were grown overnight in LB medium and diluted 1:100 into fresh M9 medium and grown at 37°C for twenty hours. 1 μl of culture was spread onto the slide and a coverslip was placed on the top. The slides were viewed and photographed with Spot (Insight) camera mounted onto an Olympus BX50 microscope.
The LIVE/DEAD BacLight Bacterial Viability kit (Invitrogen) was used to score live and dead cells. Samples were processed and stained as recommended by the manufacturer.
Acknowledgement
This work was supported by the National Institutes of Health Grant 2RO1-AI052293 to H.I.Z.
References
- Andersen C, Hughes C, Koronakis V. Protein export and drug efflux through bacterial channel-tunnels. Curr Opin Cell Biol. 2001;13:412–416. doi: 10.1016/s0955-0674(00)00229-5. [DOI] [PubMed] [Google Scholar]
- Anderson ME. Determination of glutathione and glutathione disulfide in biological samples. Methods Enzymol. 1985;113:548–555. doi: 10.1016/s0076-6879(85)13073-9. [DOI] [PubMed] [Google Scholar]
- Bleuel C, Grosse C, Taudte N, Scherer J, Wesenberg D, Krauss GJ, Nies DH, Grass G. TolC Is Involved in Enterobactin Efflux across the Outer Membrane of Escherichia coli. J. Bacteriol. 2005;187:6701–6707. doi: 10.1128/JB.187.19.6701-6707.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger JM, Jr., Kwon DS, Huisman GW, Kolter R, Walsh CT. Glutathionylspermidine metabolism in Escherichia coli. Purification, cloning, overproduction, and characterization of a bifunctional glutathionylspermidine synthetase/amidase. J Biol Chem. 1995;270:14031–14041. doi: 10.1074/jbc.270.23.14031. [DOI] [PubMed] [Google Scholar]
- Brumaghim JL, Li Y, Henle E, Linn S. Effects of hydrogen peroxide upon nicotinamide nucleotide metabolism in Escherichia coli: changes in enzyme levels and nicotinamide nucleotide pools and studies of the oxidation of NAD(P)H by Fe(III) J Biol Chem. 2003;278:42495–42504. doi: 10.1074/jbc.M306251200. [DOI] [PubMed] [Google Scholar]
- Cosme AM, Becker A, Santos MR, Sharypova LA, Santos PM, Moreira LM. The outer membrane protein TolC from Sinorhizobium meliloti affects protein secretion, polysaccharide biosynthesis, antimicrobial resistance, and symbiosis. Mol Plant Microbe Interact. 2008;21:947–957. doi: 10.1094/MPMI-21-7-0947. [DOI] [PubMed] [Google Scholar]
- Dancey GF, Levine AE, Shapiro BM. The NADH dehydrogenase of the respiratory chain of Escherichia coli. I. Properties of the membrane-bound enzyme, its solubilization, and purification to near homogeneity. J Biol Chem. 1976;251:5911–5920. [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukan S, Nystrom T. Oxidative stress defense and deterioration of growth-arrested Escherichia coli cells. J Biol Chem. 1999;274:26027–26032. doi: 10.1074/jbc.274.37.26027. [DOI] [PubMed] [Google Scholar]
- Fairlamb AH, Henderson GB, Cerami A. The biosynthesis of trypanothione and N1-glutathionylspermidine in Crithidia fasciculata. Mol Biochem Parasitol. 1986;21:247–257. doi: 10.1016/0166-6851(86)90130-1. [DOI] [PubMed] [Google Scholar]
- Ferhat M, Atlan D, Vianney A, Lazzaroni JC, Doublet P, Gilbert C. The TolC protein of Legionella pneumophila plays a major role in multi-drug resistance and the early steps of host invasion. PLoS One. 2009;4:e7732. doi: 10.1371/journal.pone.0007732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs JA. Glutathione mutants. Methods Enzymol. 1995;252:83–92. doi: 10.1016/0076-6879(95)52011-2. [DOI] [PubMed] [Google Scholar]
- Gennis RB, Stewart V. Respiration. In: Bock A, Curtis R III, Kaper JB, Neidhardt FC, Nystrom T, Rudd KE, Squires CL, editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press; Washington, DC: 2006. [Google Scholar]
- Gilson L, Mahanty HK, Kolter R. Genetic analysis of an MDR-like export system: the secretion of colicin V. Embo J. 1990;9:3875–3894. doi: 10.1002/j.1460-2075.1990.tb07606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig K, Bleuel C, Krauss GJ, Nies DH. Glutathione and transition-metal homeostasis in Escherichia coli. J Bacteriol. 2008;190:5431–5438. doi: 10.1128/JB.00271-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland IB, Blight MA, Kenny B. The mechanism of secretion of hemolysin and other polypeptides from gram-negative bacteria. J Bioenerg Biomembr. 1990;22:473–491. doi: 10.1007/BF00763178. [DOI] [PubMed] [Google Scholar]
- Imlay JA, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- Kleerebezem M, Crielaard W, Tommassen J. Involvement of stress protein PspA (phage shock protein A) of Escherichia coli in maintenance of the protonmotive force under stress conditions. Embo J. 1996;15:162–171. [PMC free article] [PubMed] [Google Scholar]
- Kobayashi R, Suzuki T, Yoshida M. Escherichia coli phage-shock protein A (PspA) binds to membrane phospholipids and repairs proton leakage of the damaged membranes. Mol Microbiol. 2007;66:100–109. doi: 10.1111/j.1365-2958.2007.05893.x. [DOI] [PubMed] [Google Scholar]
- Leonardo MR, Dailly Y, Clark DP. Role of NAD in regulating the adhE gene of Escherichia coli. J Bacteriol. 1996;178:6013–6018. doi: 10.1128/jb.178.20.6013-6018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura H, Miyachi S. Cycling assay for nicotinamide adenine dinucleotides. Meth. Enzymol. 1980;69:465–470. [Google Scholar]
- Nystrom T. Stationary-phase physiology. Annu Rev Microbiol. 2004;58:161–181. doi: 10.1146/annurev.micro.58.030603.123818. [DOI] [PubMed] [Google Scholar]
- Osborn MJ, Gander JE, Parisi E, Carson J. Mechanism of Assembly of the outer membrane of Salmonella typhimurium. J. Biol. Chem. 1972;247:3962–3972. [PubMed] [Google Scholar]
- Pai CH, Chiang BY, Ko TP, Chou CC, Chong CM, Yen FJ, Chen S, Coward JK, Wang AH, Lin CH. Dual binding sites for translocation catalysis by Escherichia coli glutathionylspermidine synthetase. Embo J. 2006;25:5970–5982. doi: 10.1038/sj.emboj.7601440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platz GJ, Bublitz DC, Mena P, Benach JL, Furie MB, Thanassi DG. A tolC mutant of Francisella tularensis is hypercytotoxic and elicits increased proinflammatory responses from host cells. Infect Immun. 2009 doi: 10.1128/IAI.00992-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss BM, Nelms JM, Park C, Wolfe AJ. Mutations in NADH:ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. J. Bacteriol. 1994;176:2143–2150. doi: 10.1128/jb.176.8.2143-2150.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner JL, Martin RG. An excretory function for the Escherichia coli outer membrane pore TolC: upregulation of marA and soxS transcription and Rob activity due to metabolites accumulated in tolC mutants. J Bacteriol. 2009;191:5283–5292. doi: 10.1128/JB.00507-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, Borges A, Ariyanayagam MR, Fairlamb AH. Glutathionylspermidine metabolism in Escherichia coli. Biochem J. 1995;312(Pt 2):465–469. doi: 10.1042/bj3120465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor CW, Tabor H, Hafner EW. Escherichia coli mutants completely deficient in adenosylmethionine decarboxylase and in spermidine biosynthesis. J Biol Chem. 1978;253:3671–3676. [PubMed] [Google Scholar]
- Tabor H, Tabor CW. Isolation, characterization, and turnover of glutathionylspermidine from Escherichia coli. J Biol Chem. 1975;250:2648–2654. [PubMed] [Google Scholar]
- Tatsumi R, Wachi M. TolC-dependent exclusion of porphyrins in Escherichia coli. J Bacteriol. 2008 doi: 10.1128/JB.00595-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanabalu T, Koronakis E, Hughes C, Koronakis V. Substrate-induced assembly of a contiguous channel for protein export from E.coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. Embo J. 1998;17:6487–6496. doi: 10.1093/emboj/17.22.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonova EB, Dastidar V, Rybenkov VV, Zgurskaya HI. Kinetic control of TolC recruitment by multidrug efflux complexes. Proc Natl Acad Sci U S A. 2009;106:16416–16421. doi: 10.1073/pnas.0906601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonova EB, Wang Q, Zgurskaya HI. Chimeric Analysis of the Multicomponent Multidrug Efflux Transporters from Gram-Negative Bacteria. J Bacteriol. 2002;184:6499–6507. doi: 10.1128/JB.184.23.6499-6507.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonova EB, Zgurskaya HI. AcrA, AcrB, and TolC of Escherichia coli Form a Stable Intermembrane Multidrug Efflux Complex. J Biol Chem. 2004;279:32116–32124. doi: 10.1074/jbc.M402230200. [DOI] [PubMed] [Google Scholar]
- Touze T, Eswaran J, Bokma E, Koronakis E, Hughes C, Koronakis V. Interactions underlying assembly of the Escherichia coli AcrAB-TolC multidrug efflux system. Mol Microbiol. 2004;53:697–706. doi: 10.1111/j.1365-2958.2004.04158.x. [DOI] [PubMed] [Google Scholar]
- Webber MA, Bailey AM, Blair JM, Morgan E, Stevens MP, Hinton JC, Ivens A, Wain J, Piddock LJ. The global consequence of disruption of the AcrAB-TolC efflux pump in Salmonella enterica includes reduced expression of SPI-1 and other attributes required to infect the host. J Bacteriol. 2009;191:4276–4285. doi: 10.1128/JB.00363-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiriyathanawudhiwong N, Ohtsu I, Li ZD, Mori H, Takagi H. The outer membrane TolC is involved in cysteine tolerance and overproduction in Escherichia coli. Appl Microbiol Biotechnol. 2009;81:903–913. doi: 10.1007/s00253-008-1686-9. [DOI] [PubMed] [Google Scholar]
- Xie QW, Tabor CW, Tabor H. Deletion mutations in the speED operon: spermidine is not essential for the growth of Escherichia coli. Gene. 1993;126:115–117. doi: 10.1016/0378-1119(93)90598-w. [DOI] [PubMed] [Google Scholar]
- Yamanaka H, Kobayashi H, Takahashi E, Okamoto K. MacAB is involved in the secretion of Escherichia coli heat-stable enterotoxin II. J Bacteriol. 2008;190:7693–7698. doi: 10.1128/JB.00853-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun N-R, San K-Y, Bennett GN. Enhancement of lactate and succinate formation in adhE or pta-ackA mutants of NADH dehydrogenase-deficient Escherichia coli. Journal of Applied Microbiology. 2005;99:1404–1412. doi: 10.1111/j.1365-2672.2005.02724.x. [DOI] [PubMed] [Google Scholar]
- Zgurskaya HI. Multicomponent drug efflux complexes: architecture and mechanism of assembly. Future Microbiol. 2009;4:919–932. doi: 10.2217/fmb.09.62. [DOI] [PMC free article] [PubMed] [Google Scholar]