Abstract
Dentin sialophosphoprotein (DSPP) is processed into dentin sialoprotein (DSP) and dentin phosphoprotein. A molecular variant of rat DSP, referred to as “HMW-DSP”, has been speculated to be a proteoglycan form of DSP. To determine if HMW-DSP is the proteoglycan form of DSP and to identify the glycosaminoglycan side-chain attachment site(s), we further characterized HMW-DSP. Chondroitinase ABC treatment reduced the migration rate for portions of rat HMW-DSP to the level of DSP. Disaccharide analysis showed that rat HMW-DSP contains glycosaminoglycan chains made of chondroitin-4-sulfate and has an average of 31-32 disaccharides/mol. These observations confirmed that HMW-DSP is the proteoglycan form of DSP (renamed “DSP-PG”). Edman degradation and mass spectrometric analyses of tryptic peptides from rat DSP-PG, along with substitution analyses of candidate Ser residues in mouse DSPP, confirmed that 2 glycosaminoglycan chains are attached to Ser241 and Ser253 in the rat, or Ser242 and Ser254 in the mouse DSPP sequence.
Keywords: dentin extracellular matrix, dentin sialophosphoprotein, dentin sialoprotein, post-translational modification, proteoglycan
Introduction
Human and mouse genetic studies have demonstrated the importance of dentin sialophosphoprotein (DSPP) in biomineralization (Xiao et al., 2001; Zhang et al., 2001; Sreenath et al., 2003; Verdelis et al., 2009). DSPP is a precursor protein that is proteolytically processed to form dentin sialoprotein (DSP) and dentin phosphoprotein (DPP) originating from the 5′ end and 3′ end sequences of the DSPP transcript, respectively (MacDougall et al., 1997). Although derived from the same mRNA, the chemical structures of DSP and DPP are very different. Rat DSP, which contains few phosphates, is highly glycosylated, while rat DPP is highly phosphorylated, but not glycosylated (Qin et al., 2004).
Previously, we identified a high-molecular-weight variant of DSP from the extracellular matrix (ECM) of rat dentin; this molecular species of DSP, which has an anionic strength greater than that of DSP, was referred to as “HMW-DSP” (Qin et al., 2003). Two research groups subsequently isolated the proteoglycan form of DSP from the ECM of porcine and bovine dentin (Yamakoshi et al., 2005; Sugars et al., 2006), which appeared to be similar to the rat dentin HMW-DSP identified earlier by our group. The glycosaminoglycan (GAG) side-chain(s) for the proteoglycan form of porcine DSP is made of chondroitin-6-sulfate, while the GAG of bovine DSP proteoglycan seems to consist of chondroitin-4-sulfate. It was proposed that the GAG chain attachment site(s) for the proteoglycan form of porcine DSP might be Ser253 and/or Ser265 in the porcine DSPP sequence (Yamakoshi et al., 2005), but this hypothesis has not been confirmed. In this study, we determined if rat HMW-DSP is the proteoglycan form of DSP (renamed DSP-PG) and identified the GAG chain attachment site(s) of DSP-PG.
Materials & Methods
Separation of Dentin Non-collagenous Proteins and Purification of DSP-PG
The non-collagenous proteins of rat incisor dentin were extracted and separated, as previously described (Qin et al., 2003). After the ion-exchange chromatography separated the DSP-PG from the DSP, an anti-DSP affinity column was used to purify the DSP-PG from the fractions containing the proteoglycan. The DSP-PG isolated in this manner was highly pure and free of any contaminating proteins.
The experimental procedures involving the use of animal tissues were approved by the Animal Welfare Committee of Baylor College of Dentistry, Texas A&M Health Science Center.
Amino Acid Analysis
Aliquots from a DSP-PG sample in water were analyzed with an ABI 420A Amino Acid Analyzer (Applied Biosystems, Foster City, CA, USA). We performed 3 independent analyses to calculate the average values. Based on these analyses, the molar concentration of DSP-PG in the water solution was determined, and we used this value to calculate the number of sulfates and disaccharides for each mole of DSP-PG. The amino acid composition of DSP-PG was very similar to that calculated from the cDNA-deduced sequence of rat DSP, which further confirmed the purity of the sample.
Treatment with Chondroitinase ABC (ChABC)
An aliquot of DSP-PG was incubated with ChABC (Sigma, St. Louis, MO, USA) in reaction buffer containing 50 mM Tris-HCl and 60 mM sodium acetate (pH, 8.0) at 37ºC for 2 hrs, as reported previously (Qin et al., 2006).
Sulfate and Disaccharide Analyses
The analyses for protein-associated sulfates and GAG disaccharides were performed by the Glycotechnology Core Facility of the University of California at San Diego, as previously described (Qin et al., 2006). Aliquots taken from the DSP-PG sample used for amino acid analysis were used for these analyses. For the sulfate analysis, anion chromatography of sulfate was performed in an Ion Pac AS4A-SC column (Dionex, Sunnyvale, CA, USA). For the disaccharide analysis, the GAG in DSP-PG was first released from the protein by β-elimination. Then, the chondroitin disaccharides liberated by chondroitinase ABC were separated by high-performance liquid chromatography (HPLC). The chondroitin disaccharides derived for fluorescence detection were measured by a Jasco fluorescence detector (Essex, UK) set at excitation λ 346 nm and emission λ 410 nm. We used 3 independent values for each sample to calculate the sulfate and disaccharide contents. We used the molar concentrations of the 2 components to calculate the numbers of sulfates and disaccharides for each mole of DSP-PG, using the molar concentration of the core protein as the denominator.
Separation of DSP-PG Tryptic Peptides; Edman Degradation and Mass Spectrometric Analyses
The tryptic peptides from 900 µg of DSP-PG were separated according to size on a Superdex 75 HR 10/30 column, as previously reported (Qin et al., 2006). The 30-mL elution volume was collected as 60 0.5-mL fractions. The GAG-containing peptides eluted in the void volume fractions of the chromatography were first sequenced by Edman degradation and then analyzed by mass spectrometry (MS).
For MS analyses, first, the GAG-containing tryptic peptides dissolved in 5% formic acid were directly applied to an HPLC system (Dionex, Sunnyvale, CA, USA) connected to a QStar XL mass spectrometer (Applied Biosystems); this set of experiments failed to generate any useful data. Second, the GAG-containing tryptic peptides were digested with endoproteinase Glu-C (Roche Applied Science, Mannheim, Germany) in 25 mM NH4HCO3 solution at 25ºC for 5 hrs. The digestion products were applied to HPLC, and individual peaks containing the Glu-C peptides were sequenced by MS. Third, the GAG-containing tryptic peptides were incubated with endoproteinase Asp-N in 5 mM NH4HCO3 solution at 37ºC for 18 hrs, and the Asp-N peptides separated by HPLC were sequenced by MS. Amino acid sequences obtained by MS from the Glu-C or Asp-N digestion products were compared against the rat DSPP sequence.
Substitutions of Ser Residues that were Potential GAG Chain Attachment Sites
Chondroitin-sulfate GAG chains of proteoglycans are usually attached to Ser residues in the Ser-Gly dipeptide motifs. Based on the results obtained from the Edman degradation and MS analyses (see below), we believe that the attachment sites for the GAG chain(s) of rat DSP-PG must be among the 3 residues Ser200, Ser241, and Ser253 in the rat DSPP sequence, which correspond to Ser201, Ser242, and Ser254 in the mouse DSPP. To identify the specific GAG chain attachment site(s), we performed site-directed mutagenesis to substitute 1 of the residues, 2 of the residues, and all 3 Ser residues in the mouse DSPP. For these substitution experiments, we made 8 pcDNA3-DSPP constructs using full-length mouse cDNA (from Dr. Shuo Chen, the University of Texas-Health Science Center at San Antonio). These 8 pcDNA3-DSPP constructs were: (1) normal DSPP, (2) S201G-DSPP (encoding a protein in which S201 was replaced by G201), (3) S242G-DSPP, (4) S254G-DSPP, (5) S201G+S242G-DSPP (S201 and S242 were replaced by G), (6) S201G+S254G-DSPP, (7) S242G+S254G-DSPP, and (8) S201G+S242G+ S254G-DSPP (all 3 candidate Ser residues were replaced by G). The generation of pcDNA-DSPP constructs and site-directed mutagenesis were performed as previously reported (Peng et al., 2009). These 8 pcDNA-DSPP constructs were used to transfect human embryonic kidney 293-EBNA (HEK293-EBNA) cells, and the culture medium was collected for the evaluation of DSP and DSP-PG by Western immunoblotting with the anti-DSP antibody (Butler et al., 1992).
Results
ChABC Treatment
After ChABC treatment, the migration rate for a portion of the DSP-PG molecules was reduced to the same level as that of DSP (Fig. 1). While it was not possible to estimate the exact proportion of DSP-PG whose GAG chains were broken off, it appeared that the GAG chains for the majority of the DSP-PG molecules were not removed by the enzyme.
Figure 1.
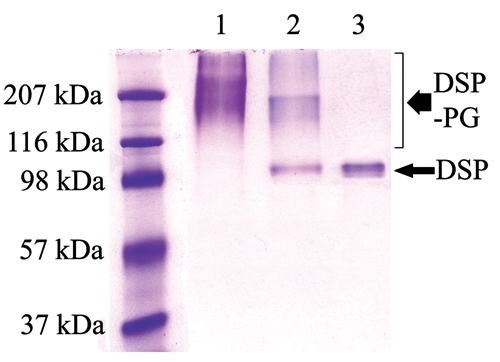
Chondroitinase ABC treatment of DSP-PG (Stains-All staining). DSP-PG was purified by a monoclonal anti-DSP antibody affinity column. Lane 1: the untreated DSP-PG migrating at ~100 - > 207 kDa. Lane 2: DSP-PG after treatment with chondroitinase ABC. Lane 3: DSP isolated from rat dentin was used as a control. Note that portions of the molecules in the DSP-PG sample co-migrated with DSP after treatment with chondroitinase ABC.
Sulfate and Disaccharide Analyses
The sulfate and disaccharide analyses (Table) showed that, on average, each DSP-PG molecule had 37.8 sulfates and 31.6 uronic acid-GalNAc-4-sulfates. The contents of the other types of disaccharides, such as uronic acid-GalNAc-6-sulfate, were negligible, indicating that the GAG chains of DSP-PG are exclusively made of chondroitin-4-sulfate.
Table.
Analyses for Sulfates and Disaccharides in DSP-PG
| Components | Residues / Mol |
|---|---|
| SO4 | 37.80 |
| Uronic acid*-GalNAc-4-sulfate | 31.60 |
| Uronic acid-GalNAc-6-sulfate | 0.00 |
| Uronic acid-GalNAc-4-6-sulfate | 0.08 |
| Uronic acid-GalNAc (without sulfate) | 0.54 |
| Uronic acid-2-sulfate-GalNAc-4-sulfate | 0.11 |
| Uronic acid-2-sulfate-GalNAc-6-sulfate | 0.00 |
| Uronic acid-2-sulfate-GalNAc-4-6-sulfate | 0.00 |
| Uronic acid-2-sulfate-GalNAc | 0.00 |
Glucuronic acid could not be distinguished from iduronic acid by the disaccharide analysis.
Separation of DSP-PG Tryptic Peptides; Edman Degradation and Mass Spectrometry Analyses
Certain tryptic peptides from DSP-PG eluted at the void volume of the Superdex 75 HR 10/30 column (Fig. 2). Our previous experiments with the proteoglycan form of dentin matrix protein 1 (DMP1-PG) had demonstrated that the GAG-containing peptides from the trypsin digestion products of DMP1-PG eluted at the void volume of the Superdex 75 column (Qin et al., 2006). Based on our previous experience with the DMP1-PG tryptic peptides, we believed that the DSP-PG tryptic peptides eluted at the void volume must contain the GAG chains, which gave the peptides a much greater molecular size and prevented them from entering the bead pores of the size-exclusion column. The DSP-PG tryptic peptides from 3 void volume fractions (fractions 12-14) of the Superdex 75 column were sequenced by Edman degradation, which showed that these fractions contained 2 peptides, in major and minor amounts, with the partial N-terminal sequences: major, 212EGEGSENQGA-; minor, 162TGLASD TSQN-. Taking into consideration the trypsin cleavage sites predicted from rat DSPP cDNA, we concluded that the complete structures of these 2 peptides must be: major, 212EGEGSENQGAEVTPSIGEGAGLDNTEGSPSGNGIEEDE DTGSGDGVGADAGDGR265; minor, 162TGLASDTSQN GDATLVQENEPQVAGSKNSTNHEVGTHGSGVAAQETT PQR211 (sequences obtained from Edman degradation are in italics; SG motifs are in bold). Because the attachment sites for chondroitin-sulfate GAG chains usually involve Ser residues in the Ser-Gly motifs, we believe that the GAG chain attachment site(s) of rat DSP-PG must be among these 3 residues: Ser200, Ser241, and Ser253.
Figure 2.
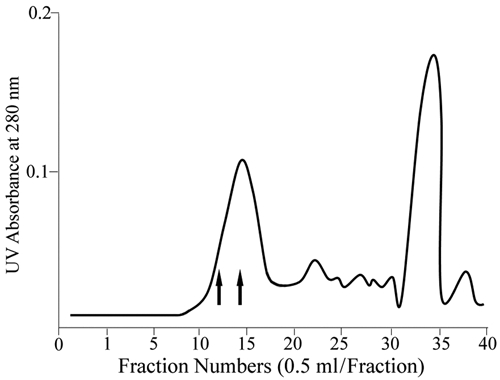
Superdex 75 HR 10/30 column separation of tryptic peptides from rat DSP-PG. Fractions eluted at the void volume (fractions 11-16) contained DSP-PG tryptic peptides to which GAG chains were attached. The fractions between the 2 arrows (12-14) were used for Edman degradation and MS analyses; these fractions contained peptides with amino acid sequences from the Thr162 to Arg265 region of rat DSPP (see Fig. 3A).
MS analyses of the Glu-C products of the GAG-containing tryptic peptides showed that the amino acid sequences were from the region of Thr162 to Arg265 in the rat DSPP sequence (Fig. 3A). In the MS analyses of the Glu-C digestion products, the unmodified, wild type of peptide VGTHGSGVAAQE (residues 195-206) was detected with a strong signal, indicating that S200 was unlikely to be modified. No wild-type forms for the peptides GSPSGNGIE (residues 238-246) and DTGSGDGVGADAG DGR (residues 250-265) were detected from the Glu-C digestion products, suggesting that they might be lost due to modifications of Ser241 and Ser253. In the MS analyses of the Asp-N digestion products, the wild-type forms of the peptides DNTEGSPSGNGIEE (residues 234-247) and DTGSG (residues 250-254) were not detected, further suggesting that Ser241 and Ser253 might be modified. Thus, the MS analyses suggested that the GAG chain attachment sites of rat DSP-PG are very likely to be Ser241 and Ser253.
Figure 3A.
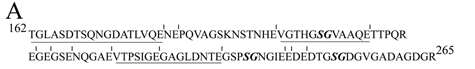
MS sequencing analyses of the Glu-C products showed that the sequences of the GAG-containing peptides were from the Thr162 to Arg265 region in the rat DSPP sequence. Vertical bars indicate cleavage sites of Glu-C. The sequences obtained from MS analyses were underlined. Note that the wild-type forms of Glu-C peptides containing Ser241 and Ser253 were not detected, while the wild-type form of Glu-C peptide containing Ser200 was observed with a strong signal in the MS analyses. These observations suggested that Ser241 and Ser253 might be modified, while Ser200 was unlikely to be modified.
Substitutions of Ser Residues that were Potential GAG Chain Attachment Sites
Next, we used site-directed mutagenesis analyses to examine if there would be a loss and/or quantitative reduction of DSP-PG due to any of the substitutions for Ser201, Ser242, and Ser254 in the mouse DSPP. When both Ser242 and Ser254 were substituted, DSP-PG was completely lost, while if only 1 of these 2 Ser residues was substituted, the quantity of DSP-PG was reduced (Fig. 3B). The substitution of S201 did not reduce the quantity of DSP-PG at all. These substitution studies showed unequivocally that 2 GAG chains are attached to the Ser242 and Ser254 in the mouse DSPP sequence. It is worth noting that, among the 9 Ser-Gly dipeptides in the mouse DSPP sequence, only Ser242-Gly243 and Ser254-Gly255 are completely conserved among mouse, rat, porcine, and human DSPP (Fig. 3C).
Figure 3B.
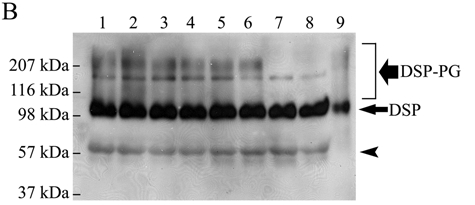
Substitutions of candidate Ser residues confirmed that 2 GAG chains are attached to the Ser242 and Ser254 in the mouse DSPP sequence. Lane 1, culture medium from HEK293-EBNA cells transfected with the pcDNA3-DSPP construct expressing normal mouse DSPP. Lane 2, from cells transfected with the construct expressing S201G-DSPP. Lane 3, from cells transfected with the construct expressing S242G-DSPP. Lane 4, from cells transfected with the construct expressing S254G-DSPP. Lane 5, from cells transfected with the construct expressing S201G+S242G-DSPP. Lane 6, from cells transfected with the construct expressing S201G+S254G-DSPP. Lane 7, from cells transfected with the construct expressing S242G+S254G-DSPP. Lane 8, from cells transfected with the construct expressing S201G+S242G+S254G-DSPP. Lane 9, 0.1 µg of pure DSP isolated from rat dentin (used as a positive control). The samples were treated with β-mercaptoethanol before being loaded onto the gels, and Western immunoblotting was carried out with the anti-DSP polyclonal antibody. The black smears migrating from ~ 100 kDa to > 207 kDa represent DSP-PG. A defined protein band migrating between the 116 and 207 kDa molecular-weight markers represents full-length mouse DSPP. Note that when both Ser242 and Ser254 were substituted (lanes 7 and 8), DSP-PG was completely lost; in contrast, if 1 of these 2 Ser residues was substituted, the amount of DSP-PG was reduced. The substitution of S201 did not reduce the quantity of DSP-PG. These findings confirmed that 2 GAG chains are attached to the Ser242 and Ser254 in the mouse DSPP sequence. Note that the amount of DSP (long/thin arrow) was very similar, but that of DSP-PG was different among lanes 1-8. The protein band (arrowhead) migrating above the 57-kDa molecular marker is likely to be a low-molecular-weight variant of mouse DSP (Goldberg et al., 2000; Qin et al., 2003).
Figure 3C.
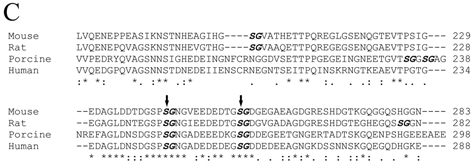
Alignment of amino acid sequences for the GAG chain attachment region of DSPP from the mouse (MacDougall et al., 1997), rat (Ritchie et al., 2001), porcine (Yamakoshi et al., 2003), and human (Gu et al., 2000). We selected a portion corresponding to mouse DSPP residues 176-283 (around the 2 GAG-linking sites) for this alignment. The names of the species are written in the left column. The amino acids are numbered in the right column. The SG dipeptides are in bold and italics. The 2 confirmed GAG attachment sites, separated by 11 amino acid residues, are marked with 2 vertical arrows. “*” indicates that the residues in that column are identical for all sequences in the alignment. “:” indicates conserved substitutions. “.” indicates semi-conserved substitutions. “-” indicates that a deletion is observed compared with the other species. Blank spaces denote the absence of any substitution, indicating that these regions are not conserved. Note that the 2 GAG-linking SG dipeptides are completely conserved among these species. There is also a high level of conservation for the 11 amino acids separating the 2 GAG-linking Ser residues as well as for the region immediately N-terminal to the first GAG-linking site, Ser242.
Discussion
Although derived from the same DSPP mRNA, DPP is remarkably more abundant than DSP in the ECM of rat dentin (Butler, 1998). The recent discovery of DSP-PG revealed that portions of the DSP molecules are proteoglycans, resulting in the quantitative reduction of the core protein form for the N-terminal fragment of DSPP. The fact that DSP-PG elutes in a greater number of chromatographic fractions than does DSP during the ion-exchange separation of rat dentin extracts, and also that the former is shown as broad smears, while the latter occurs as defined bands on SDS-PAGE (Qin et al., 2003), indicates that DSP-PG is more abundant than DSP. While we could not accurately determine whether the combination of DSP and DSP-PG would equal the number of DPP, we believe that the number of molecules with amino acid sequences from the N-terminal region of DSPP is likely to be close, if not equal, to that of DPP in the rat dentin matrix. Our estimated ratio between the N-terminal and C-terminal fragment of DSPP in rat dentin is close to that reported in a previous study showing that, in porcine dentin, these 2 DSPP-derived fragments are present in approximately equal molar amounts (Yamakoshi et al., 2006). Analysis of data obtained through in vitro mineralization studies indicates that DPP is an important initiator and modulator of the formation and growth of hydroxyapatite crystals (Boskey et al., 1990; Saito et al., 1997), while DSP (without GAG) has little or no effect on in vitro mineralization (Boskey et al., 2000). The abundance of DSP-PG suggests that this proteoglycan might be the functional form of the N-terminal fragment of DSPP in the process of biomineralization. Clearly, further studies are warranted to examine the roles of DSP-PG.
Western immunoblotting with an antibody against the cleaved GAG “stubs” revealed that porcine DSP-PG is made of chondroitin-6-sulfate (Yamakoshi et al., 2005). Using the cellulose acetate electrophoresis approach, Sugars et al. (2006) showed that, after proteinase treatment, bovine DSP-PG migrated as a band comparable with the chondroitin-4-sulfate standard. The disaccharide analyses in this investigation demonstrated clearly that the GAG chains of rat DSP-PG are exclusively made of chondroitin-4-sulfates. The variations in the GAG composition of DSP-PG among porcine, bovine, and rat may be due to species differences.
Since the xylosyltransferase that catalyzes the initiation of GAG glycosylation targets Ser residues, followed by Gly (except for keratan sulfate proteoglycans), the GAG chains of proteoglycans are nearly universally attached to Ser residues in the Ser-Gly motifs (Roden et al., 1985; Kjellen and Lindahl, 1991; Winzen et al., 2003). There are 9 Ser-Gly dipeptides in the mouse DSPP, 9 in the rat, 7 in the porcine, and 6 in the human DSPP sequence. Yamakoshi et al. (2005) proposed that the GAG chain(s) might be attached to Ser253 and/or Ser265 in the porcine DSPP, which correspond to Ser242 and Ser254 in the mouse DSPP sequence. The present investigation confirmed that 2 GAG chains, 11 amino acid residues apart, are attached to Ser242 and Ser254 in the mouse DSPP sequence (i.e., Ser241 and Ser253 in the rat DSPP sequence). The fact that Ser242-Gly243 and Ser254-Gly255 in the mouse DSPP are highly conserved among a broad spectrum of species suggests that there may be an important role in the GAG-attachment domain of DSP-PG.
Footnotes
This work was supported by National Institutes of Health, Grant DE 005092 (to CQ).
References
- Boskey A, Spevak L, Tan M, Doty SB, Butler WT. (2000). Dentin sialoprotein (DSP) has limited effects on in vitro apatite formation and growth. Calcif Tissue Int 67:472-478 [DOI] [PubMed] [Google Scholar]
- Boskey AL, Maresca M, Doty S, Sabsay B, Veis A. (1990). Concentration-dependent effects of dentin phosphophoryn in the regulation of in vitro hydroxyapatite formation and growth. Bone Miner 11:55-65 [DOI] [PubMed] [Google Scholar]
- Butler WT. (1998). Dentin matrix proteins. Eur J Oral Sci 106(Suppl 1):204-210 [DOI] [PubMed] [Google Scholar]
- Butler WT, Bhown M, Brunn JC, D’Souza RN, Farach-Carson MC, Happonen RP, et al. (1992). Isolation, characterization and immunolocalization of a 53-kDal dentin sialoprotein (DSP). Matrix 12:343-351 [DOI] [PubMed] [Google Scholar]
- Goldberg M, Boskey A, Robison C, editors (2000). Chemistry and biology of mineralized tissues: Butler W, Brown J. Two molecular variants of mouse dentin sialoprotein (DSP). Proceedings of the Sixth International Conference on the Chemistry and Biology of Mineralized Tissues, Nov 1-6, 1998, Vittel, France Rosemont, IL: American Academy of Orthopaedics Surgeons [Google Scholar]
- Gu K, Chang S, Ritchie HH, Clarkson BH, Rutherford RB. (2000). Molecular cloning of a human dentin sialophosphoprotein gene. Eur J Oral Sci 108:35-42 [DOI] [PubMed] [Google Scholar]
- Kjellen L, Lindahl U. (1991). Proteoglycans: structures and interactions. Annu Rev Biochem 60:443-475 [DOI] [PubMed] [Google Scholar]
- MacDougall M, Simmons D, Luan X, Nydegger J, Feng J, Gu TT. (1997). Dentin phosphoprotein and dentin sialoprotein are cleavage products expressed from a single transcript coded by a gene on human chromosome 4. Dentin phosphoprotein DNA sequence determination. J Biol Chem 272:835-842 [DOI] [PubMed] [Google Scholar]
- Peng T, Huang B, Sun Y, Lu Y, Bonewald L, Chen S, et al. (2009). Blocking of proteolytic processing and deletion of glycosaminoglycan side chain of mouse DMP1 by substituting critical amino acid residues. Cells Tissues Organs 189:192-197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C, Brunn JC, Baba O, Wygant JN, McIntyre BW, Butler WT. (2003). Dentin sialoprotein isoforms: detection and characterization of a high molecular weight dentin sialoprotein. Eur J Oral Sci 111:235-242 [DOI] [PubMed] [Google Scholar]
- Qin C, Baba O, Butler WT. (2004).Posttranslational modifications of SIBLING proteins and their roles in osteogenesis and dentinogenesis. Crit Rev Oral Biol Med 15:126-136 [DOI] [PubMed] [Google Scholar]
- Qin C, Huang B, Wygant JN, McIntyre BW, McDonald CH, Cook RG, et al. (2006). A chondroitin sulfate chain attached to the bone dentin matrix protein 1 NH2-terminal fragment. J Biol Chem 281:8034-8040 [DOI] [PubMed] [Google Scholar]
- Ritchie HH, Wang LH, Knudtson K. (2001). A novel rat 523 amino acid phosphophoryn: nucleotide sequence and genomic organization. Biochim Biophys Acta 1520:212-222 [DOI] [PubMed] [Google Scholar]
- Roden L, Koerner T, Olson C, Schwartz NB. (1985). Mechanisms of chain initiation in the biosynthesis of connective tissue polysaccharides. Fed Proc 44:373-380 [PubMed] [Google Scholar]
- Saito T, Arsenault AL, Yamauchi M, Kuboki Y, Crenshaw MA. (1997). Mineral induction by immobilized phosphoproteins. Bone 21:305-311 [DOI] [PubMed] [Google Scholar]
- Sreenath T, Thyagarajan T, Hall B, Longenecker G, D’Souza R, Hong S, et al. (2003). Dentin sialophosphoprotein knockout mouse teeth display widened predentin zone and develop defective dentin mineralization similar to human dentinogenesis imperfecta-III. J Biol Chem 278:24874-24880 [DOI] [PubMed] [Google Scholar]
- Sugars RV, Olsson ML, Waddington R, Wendel M. (2006). Substitution of bovine dentine sialoprotein with chondroitin sulfate glycosaminoglycan chains. Eur J Oral Sci 114:89-92 [DOI] [PubMed] [Google Scholar]
- Verdelis K, Ling Y, Sreenath T, Haruyama N, MacDougall M, van der Meulen MC, et al. (2009). DSPP effects on in vivo bone mineralization. Bone 43:983-990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzen U, Cole GJ, Halfter W. (2003). Agrin is a chimeric proteoglycan with the attachment sites for heparan sulfate/chondroitin sulfate located in two multiple serine-glycine clusters. J Biol Chem 278:30106-30114 [DOI] [PubMed] [Google Scholar]
- Xiao S, Yu C, Chou X, Yuan W, Wang Y, Bu L, et al. (2001). Dentinogenesis imperfecta 1 with or without progressive hearing loss is associated with distinct mutations in DSPP. Nat Genet 27:201-204 [DOI] [PubMed] [Google Scholar]
- Yamakoshi Y, Hu JC, Liu S, Zhang C, Oida S, Fukae M, et al. (2003). Characterization of porcine dentin sialoprotein (DSP) and dentin sialophosphoprotein (DSPP) cDNA clones. Eur J Oral Sci 111:60-67 [DOI] [PubMed] [Google Scholar]
- Yamakoshi Y, Hu JC, Fukae M, Iwata T, Kim JW, Zhang H, et al. (2005). Porcine dentin sialoprotein is a proteoglycan with glycosaminoglycan chains containing chondroitin 6-sulfate. J Biol Chem 280:1552-1560 [DOI] [PubMed] [Google Scholar]
- Yamakoshi Y, Hu JC, Iwata T, Kobayashi K, Fukae M, Simmer JP. (2006). Dentin sialophosphoprotein is processed by MMP-2 and MMP-20 in vitro and in vivo. J Biol Chem 281:38235-28243 [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhao J, Li C, Gao S, Qiu C, Liu P, et al. (2001). DSPP mutation in dentinogenesis imperfecta Shields type II. Nat Genet 27:151-152 [DOI] [PubMed] [Google Scholar]


