Abstract
This tutorial review describes recent progress in modeling the active sites of carboxylate-rich non-heme diiron enzymes that activate dioxygen to carry out several key reactions in nature. The chemistry of soluble methane monooxygenase, which catalyzes the selective oxidation of methane to methanol, is of particular interest for (bio)technological applications. Novel synthetic diiron complexes that mimic structural, and, to a lesser extent, functional features of these diiron enzymes are discussed. The chemistry of the enzymes is also briefly summarized. A particular focus of this review is on models that mimic characteristics of the diiron systems that were previously not emphasized, including systems that contain (i) aqua ligands, (ii) different substrates tethered to the ligand framework, (iii) dendrimers attached to carboxylates to mimic the protein environment, (iv) two N-donors in a syn-orientation with respect to the iron-iron vector, and (v) a N-rich ligand environment capable of accessing oxygenated high-valent diiron intermediates.
1 Introduction
Biology has adopted geologically abundant iron with its inherent electronic properties, including Lewis acidity and accessible redox states for selective O2 binding and/or activation, in heme and non-heme enzymes.1 A subfamily of non-heme enzymes contains a carboxylate-bridged non-heme diiron active site, which is responsible for many different biochemical O2 utilization pathways including (i) biomineralization of iron as an oxide in ferritin (Ft),2 (ii) DNA biosynthesis via the generation of an essential tyrosyl radical in the ribonucleotide reductase subunit R2 (RNR-R2),3 (iii) fatty acid desaturation in Δ9 stearoyl-acyl carrier protein desaturase (Δ9D),4 (iv) regulation of cell proliferation via the biosynthesis of hypusine in human deoxyhypusine hydroxylase (hDOHH),5 and (v) hydrocarbon oxidation in the hydroxylase components of bacterial multicomponent monooxygenases (BMMs).6 Enzymes belonging to the BMM family include soluble methane monooxygenase (sMMO),7,8 toluene/o-xylene monooxygenase (ToMO),9 and phenol hydroxylase (PH).10 The chemistry in these non-heme diiron enzymes occurs at a common structural motif, a diiron active site that is embedded in a four-helix bundle of an α-helical protein. Each diiron center is coordinated by four carboxylates from glutamate or aspartate residues and two imidazoles from histidine side chains that are bound in a syn-disposition to the diiron vector.
Hemerythrin, responsible for reversible O2 binding in marine invertebrates,1 myo-inositol oxygenase, which catalyzes the ring-opening glycol cleavage of myo-inositol by a radical O2 activation pathway,11 and flavo-diiron proteins, which function as O2- and/or NO-scavenging reductases,12 are some additional members of the extended family of carboxylate-bridged diiron enzymes, but they contain more than two histidine residues per diiron center.
Of special interest is to create synthetic analogs of the BMMs because several biological reactions catalyzed by these enzymes are of potential technological relevance. An example is the activation/utilization of petrochemical hydrocarbons. The carboxylate-rich diiron motif of the enzyme active site has often been considered as a template for achieving comparable chemistry and efficiency in biomimetic chemistry.13 An excellent example is the conversion of the simplest and least reactive of the saturated hydrocarbons, methane, into methanol, a liquid fuel. Such a chemical transformation would allow for the exploitation of vast natural gas resources in remote areas, often found alongside crude oil or as methane clathrates in deep-sea water and permafrost regions, and its use as a safe and transportable fuel, methanol. Natural gas is an energy-rich and low-cost chemical feedstock and it has a lower carbon footprint (CO2 emission) than coal and oil when burnt as a fuel. Today, methanol is produced from methane in an energy-intensive and costly process by the intermediate production of synthesis gas, which involves complete dehydrogenation of methane, on a large scale by the petroleum industry (eqs. 1 and 2).14-16
-
industrial CH3OH production
(1) (2) -
biological CH3OH production (MMOH)
(3)
Methanotrophic micro-organisms rely on methane as their only carbon and energy source. These aerobic proteobacteria contain the sophisticated enzyme machinery of sMMO to hydroxylate the nonpolar and strong C–H bond (ΔH°C–H = 414 kJ mol-1) in CH4 using O2, protons, and electrons in the first step of methane metabolism under mild conditions (eq. 3). 17,18 The diiron active site of the hydroxylase component of the enzyme, MMOH, is an attractive target for biomimetic synthetic chemists, having the potential for achieving hydrocarbon oxidation catalysis such as methane hydroxylation. In this review, we present enzymatic features that facilitate methane hydroxylation as a guide to, and for comparison with current, strategies for the preparation of MMOH model complexes.
2 Bacterial Multicomponent Monooxygenases
Much effort has been spent to understand the mechanistic details by which BMMs achieve their biological transformations, including detailed structural, spectroscopic, and kinetic studies.6,7,19,20 We focus in this section on the sMMO system, because it is well-studied and can convert methane into methanol.7 Methane hydroxylation also occurs in particulate MMO (pMMO), a membrane-bound copper-containing MMO from methanotropic bacteria that is beyond the scope of this review.21,22
Dioxygen activation and CH4 oxidation in sMMO involves a complex multi-component protein system. The three components (Figure 1) are (i) a hydroxylase (MMOH), (ii) an NADH oxidoreductase (MMOR), and (iii) a regulatory protein (MMOB). The hydroxylase is a 251 kDa heart-shaped heterodimer consisting of two αβγ protomers with an almost entirely α-helical secondary structure. The hydroxylating diiron active site is embedded in a four-helix bundle in each of two identical α-subunits. MMOH is only active in the presence of a protein cofactor, MMOB, which forms a specific complex with MMOH that indirectly affects the structure and reactivity of the diiron site. The required electron equivalents are transferred to MMOH from MMOR, which contains a bound flavin adenine dinucleotide (FAD) and a [2Fe-2S]-ferredoxin (Fd) cofactor.7 In this multicomponent system, four substrates, namely the hydrocarbon, dioxygen, electrons, and protons, are transported selectively and separately (to avoid quenching of the high-valent state by electrons) to the diiron active site by biologically well-engineered substrate tunnels or pockets.20
Figure 1.
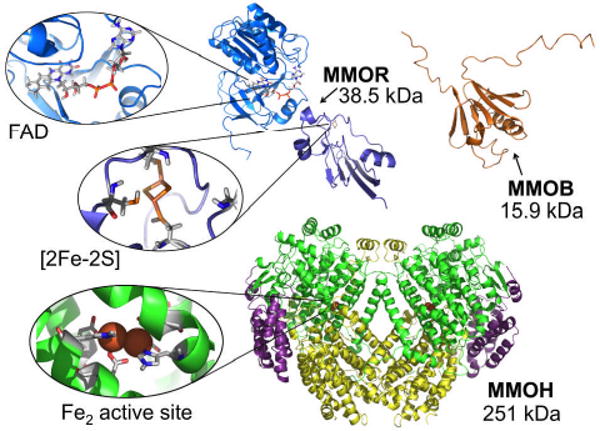
The multicomponent enzyme system of sMMO from Methylococcus capsulatus (Bath) consists of a hydroxylase (MMOH, pdb reference 1MTY), an oxidoreductase (MMOR; consisting of FAD domain, 1TVC, and [2Fe2S]-Fd domain, 1JQ4), and a regulatory (binding) protein (MMOB, 1CKV). The ribbon diagram representation of MMOH is based on X-ray coordinates and those of MMOB and the two truncated MMOR fragments, on NMR structures.
Although all components of sMMO are required for CH4 oxidation, O2 activation and C–H bond functionalization occur at the diiron site of MMOH, which is the central catalyst of the enzyme system. The diiron site can be accessed in the following three O2-free high-spin oxidation states: FeIIIFeIII (MMOHox), FeIIIFeII (MMOHmv), and FeIIFeII (MMOHred), but only the last, diiron(II), state can react directly with O2. Thus, before MMOH reacts with O2, the resting, oxidized di(μ-hydroxo)(μ-carboxylato)diiron(III) species, MMOHox, must be activated by a two-electron reduction with NADH via MMOR to form MMOHred (Figure 2, Scheme 1). Reduction occurs simultaneously with a carboxylate shift of a terminally coordinated semi-bridging glutamate (E243) residue in MMOHox, resulting in protonation and displacement of both bridging hydroxide ions and formation of a μ-η1:η1-bridging glutamate with a concomitant decrease in Fe–Fe distance from ca. 3.3 Å (in FeIII2) to ca. 3.0–3.1 Å (in FeII2).6
Figure 2.
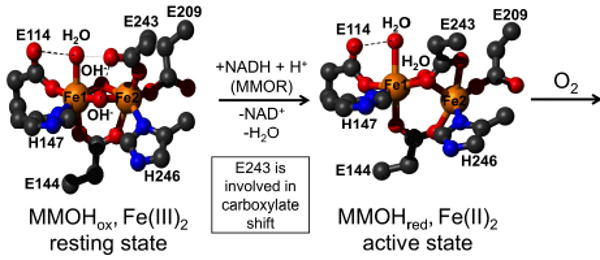
The inactive (resting state) diiron(III) site of MMOHox (left) is activated by two-electron reduction and a carboxylate shift of E243 to the diiron(II) state (MMOHred), which can then react with O2 in the presence of MMOB to form high-valent diiron-oxo species. Ball and stick structures of MMOHox and MMOHred adopted from D. A. Kopp, S. J. Lippard Curr. Opin. Chem. Biol. 2002, 6, 568-576.
Scheme 1.
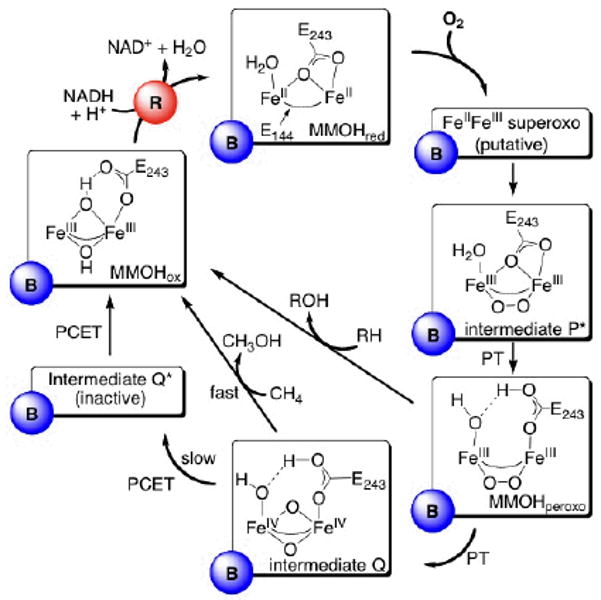
Catalytic cycle of O2 activation and CH4 hydroxylation in sMMO. The oxidized diiron(III) state (MMOHox) is activated via two-electron reduction by MMOR (R, red circle) to a diiron(II) state (MMOHred), which reacts in the presence of MMOB (B, blue circle) with dioxygen to form intermediate P*, presumably via a superoxo species. Intermediate P* then transforms via proton transfer (PT) into MMOHperoxo, which can either decay to MMOHox via oxidation of electrophilic substrates RH (e.g. ethers), or form the diiron(IV) intermediate Q, which is responsible for CH4 hydroxylation. In the absence of CH4, intermediate Q decays slowly to intermediate Q*, which is not on the methane activation pathway, and then to MMOHox. The bridging glutamates (E144 and E243) are also shown. Characteristic physical parameters of the intermediates can be found in the text.
The dioxygen activation pathway has been studied extensively in MMOH from M. capsulatus Bath (Mc) and M. trichosporium OB3b (Mt). Kinetic analyses in both systems have revealed a minimum of four oxygenated intermediates in a multi-step reaction pathway. The detailed proposed mechanism for O2 activation and substrate oxidation in Mc is illustrated in Scheme 1. Dioxygen reacts with the reduced, diiron active site of MMOHred to form intermediate P*, presumably by intermediate formation of a spectroscopically silent and/or very short-lived superoxodiiron(III,II) species analogous to superoxoiron(III) units in the oxidation of reduced heme proteins23 and synthetic iron porphyrin complexes.24 Experimental evidence for a superoxo species in MMOH has not been obtained, however. Intermediate P* is the precursor to the peroxodiiron(III) species, MMOHperoxo, having optical spectroscopic features (720 nm, ε ≈ 1250 M−1 cm−1 and 420 nm, ε ≈ 3500 M−1 cm−1) nearly identical to those of this intermediate, suggesting that they have very similar oxygenated diiron core structures. MMOHperoxo is tentatively assigned as a cis-μ-1,2-peroxodiiron(III) species and it features peroxo ligand-to-iron(III) charge transfer (LMCT) bands centered at 720 nm (ε ≈ 1350 M−1 cm−1) and 420 nm (ε ≈ 3880 M−1 cm−1) and Mössbauer parameters of δ = 0.66 mm s−1 and ΔEQ = 1.51 mm s−1, characteristic of two antiferromagnetically coupled iron atoms.7,25
MMOHperoxo is competent for oxidation of electron-rich hydrocarbons, such as diethyl ether and propylene, but not CH4. 26,27 Following O–O bond cleavage, MMOHperoxo converts to a high-valent iron species, the methane-oxidizing intermediate Q.7,19,28,29 The mechanism of this step is not well understood, however, and it is still under debate whether the O–O bond is cleaved homolytically or heterolytically.25 A di(μ-oxo)diiron(IV) “diamond core” structure with a short Fe–Fe distance of 2.46 Å was derived by EXAFS spectroscopy for intermediate Q, which has intense optical absorption bands at 420 nm (ε ≈ 7200 M−1 cm-1) and 350 nm (ε ≈ 3600 M−1 cm−1).30 pH dependence studies demonstrated that proton transfer is necessary for conversion of Hperoxo to Q.25,31 The origin of the protons is most likely an active site water molecule, because there are no suitable amino acids within reasonable proton-transfer distance to the diiron site.
Hydroxylation presumably occurs by a mechanism whereby intermediate Q abstracts a hydrogen atom from CH4 with concomitant electron transfer to an iron atom followed by recombination of the bound methyl radical with a bridging oxygen atom as a second electron transfers to the other iron atom.28 The cycle completes upon release of methanol from the hydrophobic substrate-binding pocket and formation of the resting-state MMOHox, thereby completing the catalytic cycle. Rate-limiting in this reaction is presumably product release, as demonstrated for the hydroxylation of nitrobenzene.31 In the absence of hydrocarbon substrate, intermediate Q gradually decays to intermediate Q*, which does not react with methane, to form the resting state of the enzyme, MMOHox. The structure of Q* has not yet been elucidated. Its optical spectrum contains an absorption band centered around 420 nm and a broad shoulder at 455 nm.
Studies of the hydroxylase component of ToMO (ToMOH) revealed that reaction of the reduced form of the enzyme with O2 results in the formation of a putative peroxodiiron(III) species that is responsible for hydrocarbon oxidation. There is no evidence for a high-valent, Q-type intermediate in this system.32 In contrast to the MMOHperoxo species, the oxygenated intermediate in ToMOH has distinctive Mössbauer parameters of δ = 0.54 mm s−1 and ΔEQ = 0.67 mm s-1, and it lacks an observable UV-vis absorption band.20 The intermediate has an isomer shift that lies within the range of peroxodiiron(III) species. The quadrupole splitting parameter, however, is more than 0.3 mm s−1 smaller. This EPR-silent intermediate was tentatively assigned as a peroxodiiron(III) species having a different coordination mode and/or protonation state than peroxodiiron(III) species typically observed in non-heme diiron enzymes. An oxygenated intermediate formed by the hydroxylase of PH (PHH) has nearly identical spectroscopic features, implying a structure related to that of ToMOH.33
Dioxygen activation in other non-heme diiron enzymes, such as ferritin, RNR-R2, Δ9D, and human deoxyhypusine hydroxylase,5 occurs by formation of a peroxodiiron(III) species. Spectroscopic studies of these peroxo intermediates revealed Mössbauer parameters δ = 0.55−0.68 mm s−1 and ΔEQ > 0.9 mm s−1 and optical bands similar to those of MMOHperoxo. For these enzymes, a cis-μ-1,2-peroxo binding mode is suggested.
RNR-R2 is the only non-heme diiron enzyme besides sMMO where a high-valent iron center has been observed on the pathway of O2 activation, as shown in Scheme 2. The reduced diiron(II) species, R2red, reacts with O2 to form a peroxodiiron(III) species. R2peroxo then transforms to the mixed-valent diiron(III,IV) intermediate X, which abstracts an electron from a neighboring tyrosine residue involved in the reduction of ribonucleotides to desoxyribonucleotides.3
Scheme 2.
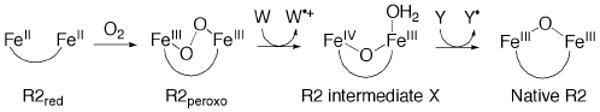
Catalytic cycle of O2 activation and tyrosyl radical formation in RNR-R2. The diiron(II) species reacts with O2 to form the peroxo intermediate R2peroxo, which oxidizes a tryptophan residue (Trp48) to form intermediate X. This FeIIIFeIV species then generates a tyrosine (Tyr122) radical and restores the resting diiron(III) state, which can be activated again by a two-electron reduction to form R2red.
Dioxygen binding in MMOH most likely occurs by substitution of a weakly coordinating, bridging water molecule distal to the syn-histidines. This site directly faces a hydrophobic substrate-binding cavity with a volume of approximately 185 Å3 adjacent to the diiron center, which favors the binding of hydrophobic guests, such as methane and O2.34 Product molecules like MeOH are released from the active site upon reduction of MMOHox. Furthermore, in order for chemistry to occur at the diiron center, electrons, protons, dioxygen, and hydrocarbon substrates all need to be provided through processes that are tightly regulated. Such regulation is finely tuned by the tertiary structure of the MMOH/MMOB complex.7,10
Although many structural and mechanistic details about sMMO have been clarified, providing information that forms the basis for the assembly of corroborative and functional biomimetic compounds, there is another important consideration to be taken into account in the design of future biomimetic catalysts. X-ray crystal structures of MMOHred have been solved and are used to help design biomimetic target constructs, but it must be remembered that the enzyme has diminished activity in the absence of MMOB.35 Thus, MMOB is presumably not only responsible for modulating the MMOH tertiary structure to control access of substrates to the active site, but may also affect the first coordination sphere of the diiron center. The geometry of MMOHred and, in particular, its diiron active site that reacts with O2 may differ from that seen in any of the known crystal structures.
3 Biomimetic Carboxylate-rich Diiron Complexes
3.1 Classical biomimetic MMOH systems containing bulky monocarboxylate building blocks
Iron carboxylate complexes are kinetically labile and have a strong tendency to form polymeric species in the absence of steric shielding. In order to isolate discrete diiron complexes, carboxylates with a finely tuned degree of steric bulk must be used. Too bulky ligands result in mononuclear complexes and too little steric constraint results in formation of oligo- and polynuclear iron species (Figure 3). Sterically open carboxylates (e.g., −O2Cph = benzoate) can only be incorporated into discrete structures as bridging ligands when they co-coordinate with other, sterically demanding ligands (e.g., in [Fe2(Ph-bimp)(O2Cph)](BF4)2; Ph-bimp = 2,6-bis[bis{2-(1-methyl-4,5-diphenylimidazolyl)-methyl}-aminomethyl]-4-methylpheno-late),36 preventing their use in carboxylate-rich diiron systems. One example of a polyiron(II) species is [FeII(μ-O2CCF3)2(HO2CCF3)2]n (−O2CCF3 = trifluoroacetate), which has bridging trifluoroacetate ligands arranged in a windmill configuration.37 The asymmetric 2-biphenylcarboxylate ligand (−O2Cbiph) provides sufficient steric bulk to avoid polymerization and maintains the ability to facilitate the assembly of discrete planar tetra-, linear tri-, and paddlewheel diiron species [Fen(μ-O2Cbiph)2nL2-4] (n = 2–4), depending on experimental conditions and the donor L.38
Figure 3.
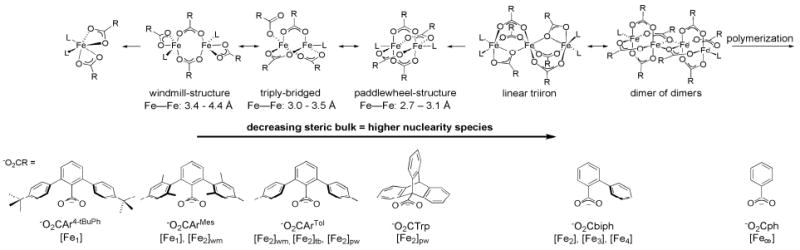
The nuclearity of carboxylate-rich iron complexes is sterically controlled. Sterically highly demanding ligands like −O2CAr4-tBuPh form monoiron complexes, [Fe1], whereas sterically open ligands like −O2Cph form polymeric species, [Fe∞]. Reversible cluster interconversions occur between windmill, [Fe2]wm, and paddlewheel, [Fe2]pw, complexes (presumably via a triply bridged species, [Fe2]tb) with −O2CArTol and between triiron, [Fe3], and tetrairon complexes, [Fe4], with −O2Cbiph. Interconversions must occur via carboxylate shifts (see text).
The preparation of diiron complexes containing the m-terphenylcarboxylate ligands 2,6-di(p-mesityl)benzoate (−O2CArMes) and 2,6-di(p-tolyl)benzoate (−O2CArTol) having the general formula [Fe2(O2CR)4(L)2] marked a considerable breakthrough in MMOH diiron core modeling.39,40 These compounds not only resemble the first coordination sphere of the MMOH active site stoichiometrically, but also recapitulate important aspects of MMOH chemistry, like the carboxylate shift, formation of high valent diiron species upon reaction with O2, and encapsulation of the diiron core with a hydrophobic shell allowing for mimicking of the protein interior.41,42
Diiron complexes with four −O2CArTol carboxylates and different ligands L were isolated either as doubly (windmill), triply, or quadruply (paddlewheel) bridged iron complexes in the solid state.43 Moreover, an equilibrium between doubly- and quadruply-bridged species in solution was found by variable-temperature solution 19F NMR spectroscopic studies of [Fe2(O2CAr4–FPh)4(THF)2], −O2CAr4-FPh = 2,6-di(p-fluoro-phenyl)benzoate, thereby simulating an important feature of the MMOH active site – the carboxylate shift.43 This ability can be attributed to the rotational flexibility of the carboxylate ligand. Triply bridged diiron species are possible intermediates in this equilibrium. Triptycene carboxylates (−O2CTrp) only support paddlewheel complexes due to the interlocking geometry of the triptycene units, and no windmill structures have yet been isolated. The bulky −O2CArMes carboxylate, on the other hand, exclusively facilitates the formation of a doubly bridged diiron compound, e.g. [Fe2(μ-O2CArMes)2(O2CArMes)2(MeCN)2], which disso-ciates into mononuclear species upon addition of various pyridine donors. A further increase in steric bulk, as in 2,6-di(4-tert-butylphenyl)benzoate (−O2CAr4-tBuPh), affords only mononuclear iron complexes.44
A short Fe–Fe distance may be crucial for O2 activation in MMOHred (see above), and metal-metal distances in diiron(II) models usually vary between 2.7 and 4.4 Å. The actual value depends on the number of bridging ligands (Figure 3), but exceptions have recently been reported. The windmill complexes [Fe2(μ-O2CArXyl)2(O2CArXyl)2(NH2(CH2)3SCH3)2] (−O2CArXyl = 2,6-di(3,5-dimethylphenyl)-benzoate),45 [Fe2(μ-O2CArXyl)2(O2CArXyl)2(NH2(CH2)3CCH)2],45 and [Fe2(μ-O2CArPh,Xyl)2(O2CArPh,Xyl)2(Py)2] (−O2CArPh,Xyl = asymmetric 3,5-dimethyl-1,1′:3′1″-terphenyl-2′-carboxylate)46 feature very short windmill Fe–Fe distances, between 3.25 and 3.46 Å, comparable to those found in MMOHred. In diiron complexes with mixed carboxylates, [Fe2(μ-O2CArTol)2(O2CArPh,Xyl)2-(Py)2] (Fe–Fe = 4.0 Å), the bridging 2,6-di-(p-tolyl)benzoate prevents shortening of the Fe–Fe distances.46 Thus, the shortening of the metal-metal distance compared to analogous −O2CArTol complexes is presumably due to diminished steric repulsion of the flanking methyl groups in the bridging −O2CArXyl and −O2CArPh,Xyl ligands. The introduction of bulky N-donors in [Fe2(μ-O2CTrp)4(L)2] complexes, which cannot convert into the more open windmill configuration (see above), results in elongated Fe–Fe distances for paddlewheel diiron complexes, e.g. [Fe2(μ-O2CTrp)4(2-PhIm)2] with Fe–Fe = 3.0 Å.47
Diiron(II) complexes of the type [Fe2(μ-O2CArTol)4(L)2] (L = 4-tBuPy and Py) form a deep green solution upon reaction with O2 at −78 °C in CH2Cl2 or toluene.40,48 The closed paddlewheel complex is in equilibrium with the corresponding open windmill complex, which can react quickly with O2. Detailed analyses of the oxygenated product confirmed the presence of an equal mixture of the quadruply bridged diiron(II,III), and a high-valent diiron(III,IV) species.49,50 The proposed reaction pathway is depicted in Scheme 3. A peroxo species forms by exposing the diiron(II) complex to O2, which may convert to a high-valent diiron(IV) species. The latter acts as one-electron oxidant toward the diiron(II) starting material, which leads to the simultaneous formation of a 1:1 mixture of two mixed-valent species with S = 1/2 (FeIIIFeIV) and S = 9/2 (FeIIIFeII), as demonstrated by EPR spectroscopy. The diiron(III,IV) species effects the one-electron oxidation of substituted phenol substrates. This process closely resembles the mechanism in RNR-R2, in which the diiron(III,IV) intermediate X oxidizes a neighboring tyrosine residue.51
Scheme 3.
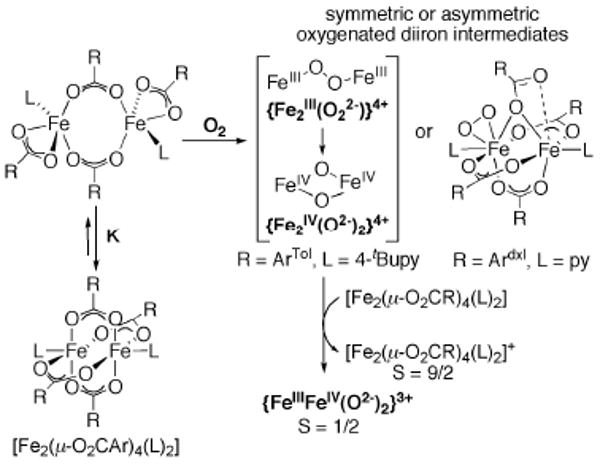
Formation of a high-valent FeIIIFeIV-oxo intermediate upon oxygenation of carboxylate-rich diiron(II) of [Fe2(O2CR)4(L)2] type complexes at −78 °C in CH2Cl2 or toluene.
Oxygenation reactions with [Fe2(μ-O2CArMes)2(O2C-ArMes)2(MeCN)2] at low temperatures yielded a purple-colored intermediate, which was spectroscopically assigned as a symmetrically bridged peroxo species.39 The quadruply bridged diiron(II) complex with benzyl-substituted benzoate ligands dxlCO2−, [Fe2(μ-O2Cdxl)4(py)2], reacts with O2 to generate an asymmetrically bound peroxo species. One possible structure based on spectroscopic analysis is depicted in Scheme 3.52
3.2 Considerations and strategies for modeling advanced features of the MMOH active site
An intrinsic difficulty in the understanding and, even more, modeling carboxylate-bridged diiron protein systems is that, although the active sites are largely conserved, they promote a variety of different reactions with O2. Thus, subtle changes in the coordination number, carboxylate binding mode, ligation by water or hydroxide, active site hydrophobicity, ligand protonation states including those of intermediates, e.g. peroxo vs. hydroperoxo, as well as electronic and structural contributions from the surrounding protein environment, play various roles in tuning the functional properties in these versatile enzymes. The grand challenge to the synthetic chemist lies in the preparation of model compounds that mimic the steric and electronic effects of the second or even third coordination sphere as defined by the polypeptide environment in the enzymes sufficiently to allow for functional biomimetic compounds with the desired catalytic properties.
Although successful strategies for the assembly of first coordination shell MMOH model complexes containing two Fe(II) ions, two bridging and one terminal carboxylates, as well as one imidazole per iron, have been developed during the last two decades (see above),42 they do not mimic environmental effects imposed by the surrounding protein. Small differences in peripheral amino acid composition are not only responsible for altered substrate access to the active site, but also influence the reactivity at the diiron center. Incorporation of some of the effects induced by the protein environment in an enzyme structure, which have been largely neglected in MMOH models mainly due to synthetic complexity, has become of increasing interest. In the following sections, new approaches to model more complex features of the MMOH active site are summarized. In particular, we cover (i) the effect of water coordinated to the diiron site, (ii) incorporation of substrates tethered to the ligand framework, (iii) synthesis of model complexes with two N-donors in syn-disposition with respect to the diiron bond, (iv) strategies for assembling complexes with a hydrophobic substrate access route, and (v) encapsulation of the diiron complexes within dendrimer ligand sheaths to mimic the protein scaffold.
3.2.1 The effect of water coordinated to the diiron site
The diiron complexes [Fe2(μ-O2CR)4(L)2] (R = ArTol or Ar4-FPh) exist in solution as an equilibrium between paddlewheel and windmill isomers. The addition of water shifts this equilibrium quantitatively to the windmill species as a result of H2O-induced carboxylate shifts.53 The use of electron-poor N-donor ligands L, such as 4-cyano- and 4-acetylpyridine, facilitates measurement of the kinetics of these water-induced conversions and subsequent oxygenations by stopped-flow electronic absorption spectroscopy utilizing the visible Fe→L charge-transfer (MLCT) band.53 The rate of oxygenation increases by approximately an order of magnitude in the presence of water when compared to the reactivity of the corresponding anhydrous diiron(II) analogs (Scheme 4).53,54 The oxygenation acceleration of the aquated windmill complex compared to the anhydrous species presumably originates from conversion to the active windmill form, which has more open access to the diiron site for O2 attack. Thus, the open windmill configuration is crucial for the O2-reactivity of carboxylate-rich diiron complexes.
Scheme 4.
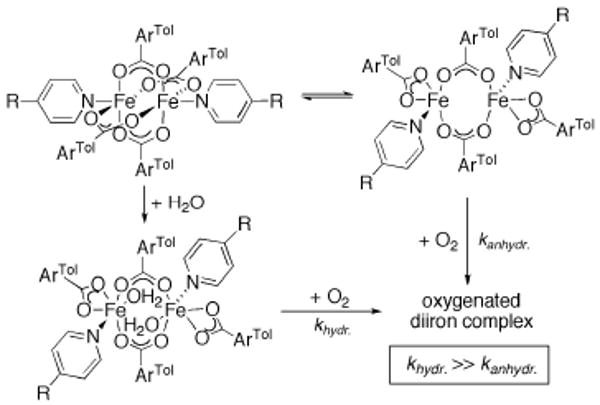
Addition of water to [Fe2(O2CR)4(4-RPy)2] (R = CN, acetyl) results in a windmill [Fe2(μ-O2CR)2(O2CR)2(H2O)2(L)2] complex, which reacts more rapidly with dioxygen than the non-aquated paddlewheel and windmill mixture.
3.2.2 Incorporation of substrates tethered to the ligand framework
The ability of an oxygenated diiron species to transfer an O–atom is often determined by examining its reaction toward external substrates. This chemistry has not yet been achieved satisfactorily with synthetic carboxylate-rich diiron complexes, possibly due to (i) restricted access of the substrate due to steric encumbrance by the ligand-framework; (ii) quenching of the reactive species by an intermolecular electron-transfer (ET) pathway; and/or (iii) slow substrate diffusion to the short-lived high-valent oxo-diiron species.
To circumvent these potential problems, substrates can be tethered to ancillary neutral donor ligands L bound to the diiron site.55 The diiron(II) complex [Fe2(μ-O2CArTol)2-(O2CArTol)2(N,N-Bn2en)2] (N,N-Bn2en = N,N-dibenzyl-ethylenediamine) reacts with O2 to afford benzaldehyde via intramolecular benzylic oxidation followed by oxidative N-dealkylation (Scheme 5A).56 A detailed investigation of the mechanism of this reaction, including a Hammett analysis and the measurement of kinetic isotope effects, suggests that it proceeds by one-electron oxidation of the amine nitrogen atom, followed by α-H atom abstraction and subsequent oxygen rebound.57 This study was extended to include benzyl- and ethyl-substituted pyridines and anilines, which upon incorporation into carboxylate-rich diiron systems and subsequent exposure to O2 yield alcohols for benzylic C–H bonds and a mixture of alcohols and ketones for the less reactive ethyl group (Scheme 5B). When steric factors are held constant, more electron-donating carboxylate and pyridine ligands increase the amount of oxidized product compared to their more electron-deficient counterparts, suggesting the need to stabilize an electrophilic intermediate to perform these transformations.45,58
Scheme 5.
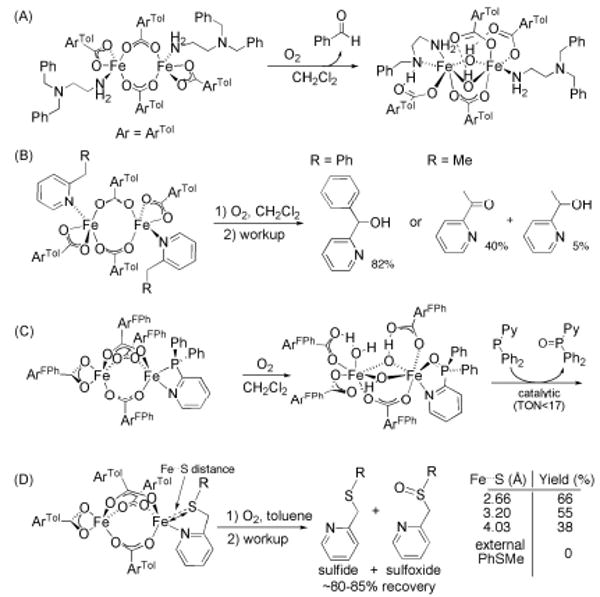
Oxidation of various substrates tethered to coordinated amine or pyridine ligands in carboxylate-rich [Fe2(O2CR)4(L)1-2] complexes. (A and B) C–H bond activation, (C) catalytic oxidation of 2-PyPPh2 and (D) Fe⃛S distance dependent sulfoxidation; R = phenyl (Fe–S distance 2.66 Å), mesityl (Fe⃛S = 3.20 Å) and 2,4,6-triisopropylphenyl (Fe⃛S = 4.03 Å).
Tethered thiol, sulfide, sulfoxide, and phosphine moieties on pyridine ligands also serve as substrates for oxidation at O2-activated carboxylate-bridged diiron(II) centers, particularly when bound in a position ortho to the N-atom of the pyridine ring (Scheme 5C and 5D).46,59 Oxidation of [Fe2(μ-O2CAr4-FPh)3(O2CAr4-FPh)(2-Ph2PPy)] in the presence of excess 2-Ph2PPy in CH2Cl2 catalytically converts the phosphine to its oxide (17 turnovers) with formation of [Fe2(μ-OH)2(μ-O2CAr4-FPh)(O2CAr4-FPh)3(OH2)(2-Ph2P(O)-Py)], which contains the biologically relevant {Fe2(μ-OH)2(μ-O2CR)}3+ core.59 The extent of substrate oxidation depends mainly on the proximity of the substrate to the diiron center. Either no or very little oxidation occurs when the substrate moiety is installed in the meta or para position of the pyridine ligand. Moreover, when the iron-sulfur distance in a series of [Fe2(μ-O2CAr)3(O2CAr)(picSR)] (picSR = ortho substituted picolyl-based thioethers) complexes was systematically elongated by increasing the steric bulk on R from phenyl to mesityl and 2,4,6-triisopropylphenyl, the sulfoxidation yield upon exposure to O2 at room temperature in toluene substantially diminished (Scheme 5D).38
3.2.3 Mimicking the protein backbone with dendrimer encapsulation of a carboxylate-bridged diiron center
Significant advances have been made in the synthesis of catalytically active dendrimer complexes as biomimetic analogs of enzymes.60 Dendrimers have highly branched and organized three-dimensional structures that facilitate the encapsulation of reactive metallocenters. Similar to the protein scaffold in a metalloenzyme, dendritic shielding creates a distinct microenvironment around the active core, which protects it from unwanted side reactions and controls its reactivity. Dendritically functionalized ligands have been explored extensively to model heme enzymes61-64 and were recently applied toward understanding non-heme diiron systems. The first dendrimer-derived mimic of a non-heme diiron enzyme contained a triazacyclononane ligand bearing poly(benzylether) dendritic substituents (L3TACN).65 The resulting mononuclear iron(II) starting material reacted upon oxygenation to form an oxo-bridged diiron(III) complex, assigned as [Fe2(μ-O)(μ-OAc)2(L3TACN)2]2+. Photoirradiation of this complex led to 2-electron reduction and subsequent oxidation to the diiron(III) complex in the presence of dioxygen.
In order to prevent deleterious intermolecular electron transfer reactions, as observed in compounds with m-terphenyl carboxylate ligands (see above), and to restrict access of solvent molecules to the active site, the basic structure of these ligands was extended with third-generation dendritic poly(benzylether) units.66 The dendrimer-appended carboxylate, −O2C-[G-3], facilitated the synthesis of doubly bridged diiron(II) complexes having the general formula [Fe2(μ-O2C-[G3])2(O2C-[G3])2(4-RPy)2] (R = cyano, pyrrolidino). The dendritic hydrophobic shield diminished gas permeability, which resulted in a 300-fold decrease in reaction rate compared with those of the unsubstituted m-terphenyl carboxylate-based complexes (Scheme 6).53
Scheme 6.
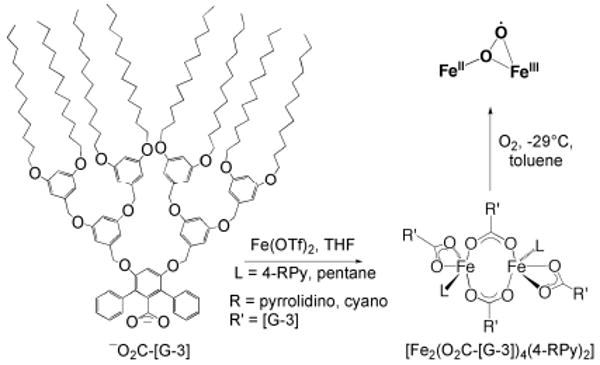
Synthesis of an encapsulated [Fe2(μ-O2CR)2(O2CR)2(L)2] complex with aid of dendritic carboxylates and formation of a superoxo intermediate after reaction with dioxygen.
Unlike the parent compounds, the dendrimer complexes stabilized a new intermediate upon oxygenation, which Mössbauer, UV-vis, EPR, and X-ray absorption spectroscopic studies suggest to be a superoxodiiron(II,III) species. This intermediate was stable at temperatures below −5 °C, a result that reflects the value of the protective shell of the dendrimer.
3.2.4 Modeling the syn-histidine disposition in MMOH
Despite the large number of model compounds, a structural characteristic that none of the aforementioned ligand motifs can rigidly enforce is the syn-disposition of the nitrogen donors with respect to the diiron vector present in these carboxylate-bridged non-heme diiron enzymes. The significance of this stereochemical feature is uncertain, but it is likely that nature did not choose this stereochemistry arbitrarily and that it plays a role in dioxygen activation. DFT studies of intermediate Q of MMOH suggest that a stereoelectronic effect is derived from this configuration, which helps to control the reactivity of this key intermediate.28 For this reason, dinucleating ligands that enforce the desired coordination mode were designed and synthesized. A requirement of such ligands is that the linker must fix the N-donor groups at the proper distance and orientation while being sufficiently flexible to accommodate different Fe–Fe distances. In addition, the resulting metal complexes should have a carboxylate-rich coordination environment and withstand bimolecular decomposition, oligomerization, or ligand oxidation by high-valent iron-oxo intermediates.
To address these challenges, a series of ligands with a 1,2-diethynylbenzene (DEB) linker connecting two heterocycles such as pyridines, quinolines, or imidazoles were prepared.67-69 The facile functionalization of the pyridine substituent allowed the synthesis of a series of ligands with the 1, 2-bis(pyridin-3-ylethynyl)benzene moiety (Figure 4A).68,69 This type of ligand has proved to be a useful template for preparing dimetallic complexes with a syn N-donor configuration. The structures of several such diiron complexes were recently determined by X-ray crystallography, and interesting structural features were noted upon inspection of dimetallic compounds with this ligand scaffold. The complexes revealed M–M distances that range from 3.11 to 5.17 Å (Chart 1), suggesting that this seemingly rigid linker is flexible enough to accommodate changes in the Fe–Fe distance upon reaction with dioxygen.67,70,71 Additionally, the diethynylbenzene backbone provides a pocket in which a bridging oxo-group can be accommodated, as may occur in intermediate Q of MMOH. Finally, functionalization of the pyridine moiety can provide additional protection from bimolecular decomposition, formation of polymers, or head-to-head ligand dimerization as observed with PIC2DET in [Fe2(μ-OTf)2(PIC2DET)2]2+ (Chart 1).
Figure 4.
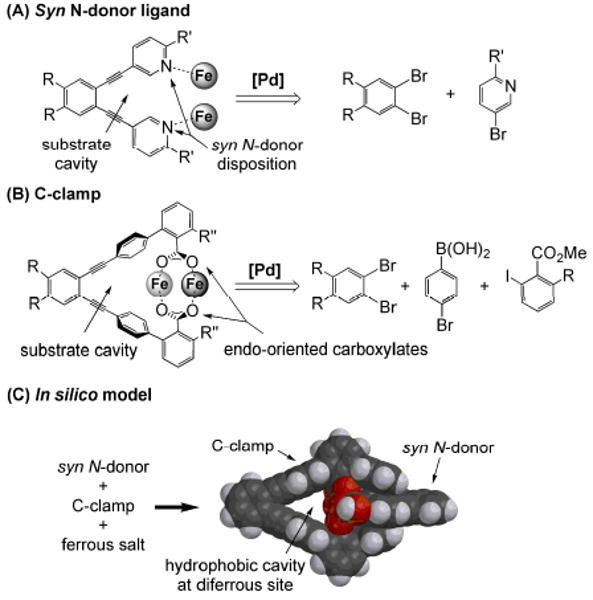
General synthetic pathway via Pd-catalyzed cross-coupling reactions to (A) syn N-donor,68 and (B) C-clamp ligands.69 (C) Energy-minimized structure of [Fe2{DEB(PICMe)2}{DEB(terphCO2)2}] displaying the substrate-access cavity (adapted from ref. 69).
Chart 1.
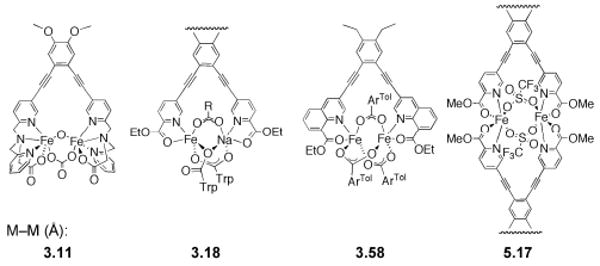
Structures of complexes (from left to right): [Fe2(μ-O)(μ-CO3)BPG2DEV], [NaFe(PIC2DET)(μ-O2CTrp)3], [Fe2(μ-O2CArTol)3-(Et2BCQEBEt)]+, [Fe2(μ-OTf)2(PIC2DET)2]2+, and comparison of the M–M distances in these compounds.
The quinoline-based ligand Et2BCQEBEt (1,2-bis(3-ethynyl-8-carboxylate-quinoline)-benzene ethyl ester), afforded a diiron(II) complex, [Fe2(Et2BCQEBEt)(μ-O2CArTol)3]+, with three bridging carboxylates.67 Another carboxylate-rich but heterodinuclear complex, [NaFe(PIC2DET)(μ-O2CTrp)3], was isolated.71 The labile sodium ion could be replaced by titration with a ferrous salt, resulting in a diiron complex. It has been observed by crystallographic analysis of various X-ray crystal structures that the position of Fe2 is more flexible and distorted than Fe1 in MMOH (Figure 2).7 Thus, this model system simulates the lability of one iron atom in MMOH (see above).
The more recently introduced BPG2DEV2− ligand affords three oxo-bridged diiron(III) complexes, [Fe2(μ-O)(H2O)2-BPG2DEV](ClO4)2, [Fe2(μ-O)(μ-O2CAriPrO)BPG2DEV]-(ClO4), and [Fe2(μ-O)(μ-CO3)BPG2DEV], which form a peroxodiiron(III) species upon reaction with hydrogen peroxide.72 The spectroscopic properties of these inter-mediates differ significantly from those of related (μ-oxo)(μ-peroxo)diiron(III) species,73 which may be due to the rigid scaffold that restrains the diiron distance to shorter values or to protonation of the peroxo species.
3.2.5 Toward modeling the hydrophobic substrate pocket with C-clamp ligands
The work on syn N-donor ligands was further expanded for the synthesis of diiron complexes having specific hydrophobic cavities to allow for substrate access to the diiron site. This feature is of special interest considering the importance of instantaneous access of a substrate to a short-lived high-valent diiron intermediate. In MMOH, methane resides in a hydrophobic cavity at the active site at the time of formation of intermediate Q. Molecular recognition plays an important rule in selective C–H activation of small molecules.74 C-clamp ligands as potential chelate hosts capable of binding a guest molecule in their endo-dicarboxylate pockets have been prepared with a flexible aromatic diamine linker75 and with a sterically more constraining ligand having a diethynyl benzene backbone (Figure 4B).69 The syn N-donor ligand provides a platform to which a C-clamp ligand, with two bridging, endo-oriented carboxylate groups, can bind in such a manner that a cavity is formed. A space-filling diagram of the energy-optimized structure of a diiron complex containing a C-clamp and syn N-donor ligand is shown in Figure 4C. This in silico model features a hydrophobic substrate-access cavity as well as two N-donors in syn-disposition.
4 Bioinspired, N-rich MMOH model complexes
The first carboxylate-bridged diiron complexes were reported in the early 1980s using nitrogen-rich capping ligands and bridging carboxylates (Chart 2) as models for met-hemerythrin.8 Although many models containing a N-rich structural motif have been published subsequently, these diiron complexes do not strictly resemble the coordination stoichiometry and environment of carboxylate-bridged non-heme diiron enzymes involved in oxidation chemistry.42 The lack of a carboxylate-rich ligand environment typically results in diiron intermediates and compounds having different electronic spin states than those in the biological systems. Thus, their UV-vis and Mössbauer spectroscopic properties often differ significantly from those in the enzymes and the oxidative strength towards substrates is greatly diminished. However, to some surprise, the only high-valent intermediate similar to intermediate Q,76 and the cleavage of strong C–H and O–H bonds in external substrates, have been achieved with such N-rich ligand systems. These bioinspired, rather then biomimetic, transformations fully justify the introduction of this alternative ligand set, without diminishing the ultimate goal of achieving the chemistry with a more relevant donor atom set. In the following sections we describe a few selected oxygenated diiron intermediates in model compounds containing a nitrogen-rich environment, which are of potential relevance to the active site of sMMO and related enzymes.
Chart 2.
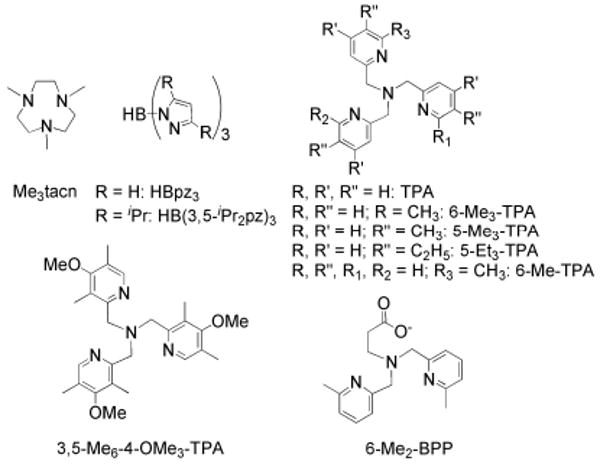
Commonly used classical N-rich capping ligands for the assembly of diiron complexes (top) and ligands used to study peroxo complexes in Scheme 7 and 8.
4.1 Superoxodiiron(II,III) model intermediates
A superoxo intermediate has been observed only in a carboxylate-rich system in which the diiron core was embedded within a dendritic ligand sheath (see above).66 Reaction of [Fe2(μ-OH)2(6-Me3-TPA)2], where 6-Me3-TPA is tris(6-methyl-2-pyridylmethyl)amine, with O2 at −80 °C gave rise to an end-on bound η1-superoxo diiron(II,III) complex and a η1-hydroperoxodiiron(III,III) species, as revealed by resonance Raman spectroscopy and kinetic studies (Scheme 7).77 These intermediates are precursors to a (μ-oxo)(μ-peroxo)diiron(III) species, which forms by warming the reaction mixture to −60 °C. In contrast to the peroxo species, which is inert toward 2,4-di-tert-butylphenol (DTBP), the superoxo intermediate readily performs a one-electron oxidation on this substrate, a result suggesting that metal-superoxo species may play a previously unanticipated role as oxidants in metalloenzymes.
Scheme 7.
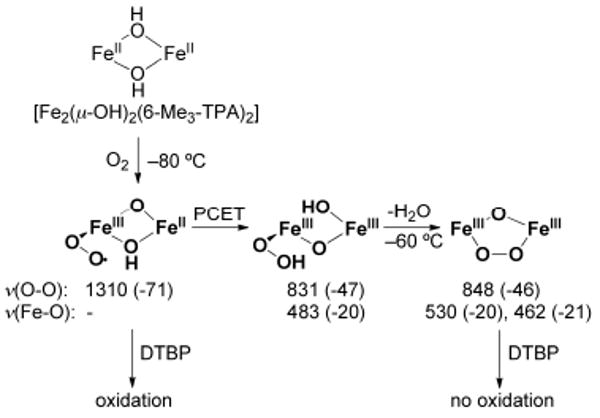
Proposed mechanism of superoxo formation of diiron complexes with 6-Me3-TPA (Chart 2) and reactivity with 2,4-di-tert-butylphenol (DTBP). Resonance Raman data (given in cm−1) for the ν(O–O) and ν(Fe–O) frequencies and the 18O-downshifts (given in parentheses) is listed below each intermediate.
4.2 Peroxodiiron(III) model intermediates
The first intermediates that are spectroscopically observed after reaction of BMMred with O2 are peroxo species, the most commonly observed intermediate in enzymatic as well as biomimetic diiron systems. Thus, detailed spectroscopic and structural information about peroxo intermediates is available. Most commonly, in synthetic models the peroxo ligand is bound in a cis-μ-1,2-fashion, with an Fe–O–O–Fe dihedral angle that depends on the nature of other bridging ligands. The value is close to 0° with a bridging phenoxide/alkoxide ligand and approximately 54° in the presence of carboxylate bridging ligands as was established by crystal structure analysis of some model compounds.8
More recently, solid state structures of peroxodiiron(III) complexes [Fe2(6-Me2-BPP)2(OH)(O2)]+ and [Fe2(6-Me2-BPP)2(O)(O2)] (6-Me2-BPP = tripodal-based ligand) have been determined (Scheme 8).78 Upon addition of acid to the (μ-oxo)(μ-peroxo)diiron(III) species, protonation occurs at the oxo-bridge to form a (μ-hydroxo)(μ-peroxo)diiron(III) intermediate. The study revealed that these two species have significantly different spectroscopic properties, with the oxo-bridged peroxo complex having a blue-shifted LMCT band in contrast to the hydroxo-bridged species. Studies of a related peroxodiiron(III) complex, [Fe2(μ-O)(μ-O2H3)(L)2]3+ (L = tris(3,5-dimethyl-4-methoxypyridyl-2-methyl)amine), suggested that protonation of oxygenated intermediates plays an important role in the formation of intermediate Q and substantially influences the reactivity. In this system, rapid conversion of the peroxo species to a diiron(IV) intermediate was observed spectroscopically;79 the latter converts into a di(μ-oxo)diiron(IV) species upon addition of acid.
Scheme 8.
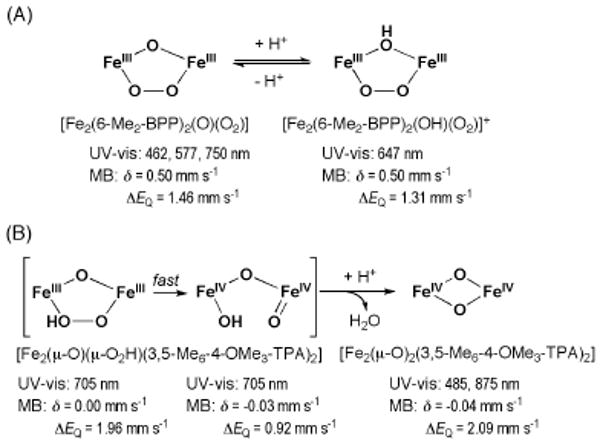
Different protonation pathways of peroxodiiron(III) complexes. Protonation of μ-oxo (A), and μ-peroxo (B) in two different model systems. For structures of 6-Me2-BPP and 3,5-Me6-4-OMe3-TPA see Chart 2.
By contrast, in the well-characterized N-rich (μ-peroxo)(μ-carboxylato)diiron(III) synthetic complex, [Fe2(μ-O2)(μ-O2Cph)(N-EtHPTB)]2+, protonation occurs at the carboxylate rather than the peroxo ligand.80 This result suggests that a carboxylate-shift can be induced by protonation, which improves the electrophilicity of the diiron unit and substrate access to the diiron center. The resulting protonated carboxylate could contribute to the activation of the diiron(III) peroxo intermediate in a number of ways. It could form a hydrogen bond to one of the peroxo oxygen atoms, rendering it more electrophilic, or contribute to the proper orientation of other nearby units, either coordinated or not.
A recent spectroscopic and computational study on a series of (μ-oxo)(μ-1,2-peroxo)diiron(III) complexes revealed a linear correlation between the ν(O–O) frequency and Fe–Fe distance.73 The metal separation can be tuned by the nature of the bridging ligand, which decreases to ∼3.05 Å in oxo-bridged peroxodiiron complexes. This correlation may facilitate use of ν(O–O) values to estimate iron-iron distances in peroxo intermediates in biological systems where no crystal structure can be obtained.
4.3 Modeling intermediate Q
The nature of the high-valent diiron(IV) intermediate Q is less well understood than those of mononuclear iron(IV)-oxo compounds.81 The first examples of oxo-bridged diiron(IV) complexes employed a tetra-anionic tetracyclic ligand (TAML) and were capable of oxidizing PPh3 and alcohols to aldehydes. However, they did not reproduce the MMOH active site ligand environment.82 A di(μ-oxo)diiron(IV) complex has been prepared by electrochemical one-electron oxidation of the mixed valent diiron(III,IV) complex [Fe2(μ-O)2(L)2]3+ (L = tris(3,5-dimethyl-4-methoxypyridyl-2-methyl)amine; Chart 2).76 Spectroscopic analysis confirmed that this species contains a [Fe2(μ-O)2] core with a short Fe–Fe distance of 2.68 Å, somewhat longer than that of ∼2.5 Å proposed for intermediate Q. The ability of this diiron(IV) species to oxidize 9,10-dihydroanthracene is about 10-fold greater than that of the diiron(III,IV) precursor, but significantly less pronounced than that of a related mononuclear oxo-iron(IV) complex (Scheme 9). This surprisingly low reactivity for the dimetallic species may be explained by its low-spin state, for it is suggested by DFT calculations that high-spin diiron complexes are more reactive than their low-spin counterparts.76
Scheme 9.
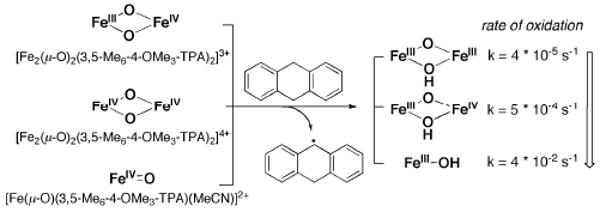
Comparison of C–H activation by mono- and diiron(IV) complexes with 3,5-Me6-OMe3-TPA ligand (adapted from ref. 76).
More recently, [FeIII2(μ-O)(L)2]2+ (L = N,N-bis-(3′,5′-dimethyl-4′-methoxypyridyl-2′-methyl)-N′-acetyl-1,2-diaminoethane), which has a (μ-oxo)bis(μ-carboxamido) diiron core and a relatively small Fe–O–Fe angle (approximately 130°), has been oxidized electrochemically to a low-spin diiron(IV) (S = 1) species that maintained the structure, as confirmed by X-ray absorption and Mössbauer spectroscopy. The latter compound has much greater reactivity toward hydrocarbons than previously reported diiron(IV) complexes.83 This species, which has the highest redox potential of the three diiron(IV) complexes reported so far, functions as a one-electron oxidant toward hydrocarbons having C–H bond activation energies as high as 100 kcal mol−1, but dehydrogenation of the substrate instead of hydroxylation occurs (Scheme 10). This high-valent diiron(IV) complex is a unique example of a complex that cleaves the strong O–H bonds of alcohols. The rate of cyclohexane oxidation for this system, however, is still orders of magnitude smaller than the rate of methane hydroxylation in intermediate Q, which in this case might be a result of the difference in iron spin states for these oxidants (see above).
Scheme 10.
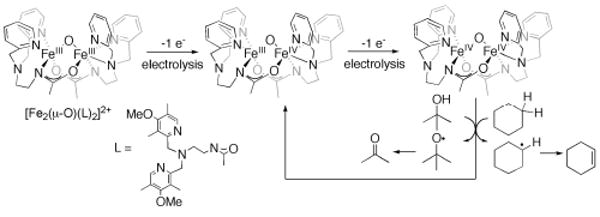
Electrochemical generation of a diiron(IV) complex and its ability for C–H and O–H bond activation. The dinucleating ligand L is shown in the inset.
5 Conclusions
Recent progress and attempts to mimic more closely the active sites and protein scaffolds of carboxylate-bridged non-heme diiron enzymes, MMOH in particular, are described in this review. Ligand design is the key factor for assembling diiron complexes with the desired steric and electronic properties. m-Terphenyl-based carboxylate ligands facilitate the synthesis of diiron complexes having the flexibility adequate to reproduce biological features such as the carboxylate shift and the proper substituents to enforce a hydrophobic ligand environment, but they cannot stabilize high-valent species at ambient temperature. Compounds with dendrimer-appended terphenyl carboxylates protect the diiron core in such a way that allows for the isolation of novel oxygenated diiron species. Although not strictly structurally biomimetic models, nitrogen-rich ligand systems have the ability to stabilize high-valent diiron species, but with the iron atoms in a low-spin rather than a high-spin state. This low-spin configuration is presumably a contributing factor for the lower reaactivity of the oxygenated species and their non-biomimetic spectral properties. Syn N-donor ligands can afford diiron complexes that mimic not only the stoichiometry but also the geometry of the enzyme active sites with respect the syn disposition of the two histidines. We await with interest the evolution of new strategies that allow access to model compounds that reproduce the geometric and electronic structural features as well as the functional dioxygen-activation chemistry of carboxylate-bridged non-heme diiron enzyme cores.
Acknowledgments
This work was supported by Grant GM032134 from the National Institute of General Medical Sciences (to S.J.L.) and EPSRC (EP/H00338X/1 to E.R.). The authors thank Ms. Christy Tinberg for providing ribbon diagrams of sMMO and helpful discussions.
Biographies

Dr Simone Friedle received her Diploma in chemistry (Dipl.-Chem.) from the University of Karlsruhe, Germany, conducting her diploma thesis research abroad in the group of Professor Richard R. Holm at Harvard University on metal dithiolene complexes. Her graduate work under the guidance of Professor Stephen J. Lippard involved modeling the actives sites of non-heme diiron enzymes. At present, she is a postdoctoral fellow in the laboratory of Professor Samuel W. Thomas III at Tufts University investigating contact electrification of electrostatically responsive materials.

Dr Erwin Reisner performed his doctorate work on electron-transfer activated metal-based anticancer complexes at the University of Vienna, Austria, and the Instituto Superior Técnico, Lisbon, Portugal. He then joined Professor Stephen J. Lippard at MIT as an Erwin Schrödinger postdoctoral fellow working on synthetic models of the diiron active site of soluble methane monooxygenase. After working with Professor Fraser A. Armstrong as a postdoctoral research assistant on solar hydrogen production with enzyme-modified nanoparticles at the University of Oxford, he moved to The University of Manchester as an EPSRC Career Acceleration Fellow (principal investigator) to develop bio-inspired systems for the solar-light driven production of green fuels.

Stephen J. Lippard is the Arthur Amos Noyes Professor of Chemistry at the Massachusetts Institute of Technology. His research activities span the fields of inorganic chemistry, biological chemistry, and neurochemistry. Included are studies to understand and improve platinum anticancer drugs, the synthesis of diiron complexes as models for carboxylate-bridged diiron metalloenzymes, structural and mechanistic investigations of bacterial multicomponent monooxygenases, and investigations of inorganic neurotransmitters and signal transducers, especially nitric oxide and zinc.
References
- 1.Lippard SJ, Berg JM. Principles of Bioinorganic Chemistry. University Science Books; Mill Valley, CA: 1994. [Google Scholar]
- 2.Liu X, Theil EC. Acc Chem Res. 2005;38:167–175. doi: 10.1021/ar0302336. [DOI] [PubMed] [Google Scholar]
- 3.Stubbe J, Nocera DG, Yee CS, Chang MCY. Chem Rev. 2003;103:2167–2201. doi: 10.1021/cr020421u. [DOI] [PubMed] [Google Scholar]
- 4.Shanklin J, Guy JE, Mishra G, Lindqvist Y. J Biol Chem. 2009;284:18559–18563. doi: 10.1074/jbc.R900009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vu VV, Emerson JP, Martinho M, Kim YS, Münck E, Park MH, Que L., Jr Proc Natl Acad Sci USA. 2009;106:14814–14819. doi: 10.1073/pnas.0904553106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sazinsky MH, Lippard SJ. Acc Chem Res. 2006;39:558–566. doi: 10.1021/ar030204v. [DOI] [PubMed] [Google Scholar]
- 7.Merkx M, Kopp DA, Sazinsky MH, Blazyk JL, Müller J, Lippard SJ. Angew Chem, Int Ed. 2001;40:2782–2807. doi: 10.1002/1521-3773(20010803)40:15<2782::AID-ANIE2782>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 8.Siewert I, Limberg C. Chem Eur J. 2009;15:10316–10328. doi: 10.1002/chem.200901910. [DOI] [PubMed] [Google Scholar]
- 9.Sazinsky MH, Bard J, Di Donato A, Lippard SJ. J Biol Chem. 2004;279:30600–30610. doi: 10.1074/jbc.M400710200. [DOI] [PubMed] [Google Scholar]
- 10.Sazinsky MH, Dunten PW, McCormick MS, Di Donato A, Lippard SJ. Biochemistry. 2006;45:15392–15404. doi: 10.1021/bi0618969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bollinger JM, Jr, Diao Y, Matthews ML, Xing G, Krebs C. Dalton Trans. 2009:905–914. doi: 10.1039/b811885j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurtz DM., Jr Dalton Trans. 2007:4115–4121. [Google Scholar]
- 13.Que L, Jr, Tolman WB. Nature. 2008;455:333–340. doi: 10.1038/nature07371. [DOI] [PubMed] [Google Scholar]
- 14.Waugh KC. Catal Today. 1992;15:51–75. [Google Scholar]
- 15.Lunsford JH. Catal Today. 2000;63:165–174. [Google Scholar]
- 16.Lange JP. Ind Eng Chem Res. 1997;36:4282–4290. [Google Scholar]
- 17.Hanson R, Hanson T. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalton H. Phil Trans R Soc B. 2005;360:1207–1222. doi: 10.1098/rstb.2005.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovaleva EG, Neibergall MB, Chakrabarty S, Lipscomb JD. Acc Chem Res. 2007;40:475–483. doi: 10.1021/ar700052v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray LJ, Lippard SJ. Acc Chem Res. 2007;40:466–474. doi: 10.1021/ar600040e. [DOI] [PubMed] [Google Scholar]
- 21.Lieberman RL, Rosenzweig AC. Nature. 2005;434:177–182. doi: 10.1038/nature03311. [DOI] [PubMed] [Google Scholar]
- 22.Hakemian AS, Rosenzweig AC. Annu Rev Biochem. 2007;76:223–241. doi: 10.1146/annurev.biochem.76.061505.175355. [DOI] [PubMed] [Google Scholar]
- 23.Schlichting I, Berendzen J, Chu K, Stock AM, Maves SA, Benson DE, Sweet RM, Ringe D, Petsko GA, Sligar SG. Science. 2000;287:1615–1622. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- 24.Momenteau M, Reed CA. Chem Rev. 1994;94:659–698. [Google Scholar]
- 25.Tinberg CE, Lippard SJ. Biochemistry. 2009;48:12145–12158. doi: 10.1021/bi901672n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valentine AM, Stahl SS, Lippard SJ. J Am Chem Soc. 1999;121:3876–3887. [Google Scholar]
- 27.Beauvais LG, Lippard SJ. J Am Chem Soc. 2005;127:7370–7378. doi: 10.1021/ja050865i. [DOI] [PubMed] [Google Scholar]
- 28.Baik MH, Gherman BF, Friesner RA, Lippard SJ. J Am Chem Soc. 2002;124:14608–14615. doi: 10.1021/ja026794u. [DOI] [PubMed] [Google Scholar]
- 29.Bollinger JM, Jr, Krebs C. J Inorg Biochem. 2006;100:586–605. doi: 10.1016/j.jinorgbio.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 30.Shu L, Nesheim JC, Kauffmann K, Münck E, Lipscomb JD, Que LJ. Science. 1997;275:515–518. doi: 10.1126/science.275.5299.515. [DOI] [PubMed] [Google Scholar]
- 31.Lee SK, Nesheim JC, Lipscomb JD. J Biol Chem. 1993;268:21569–21577. [PubMed] [Google Scholar]
- 32.Murray LJ, Naik SG, Ortillo DO, García-Serres R, Lee JK, Huynh BH, Lippard SJ. J Am Chem Soc. 2007;129:14500–14510. doi: 10.1021/ja076121h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.V. Izzo, C. E. Tinberg, S. Naik, B. H. Huynh and S. J. Lippard Unpublished results.
- 34.Whittington DA, Rosenzweig AC, Frederick CA, Lippard SJ. Biochemistry. 2001;40:3476–3482. doi: 10.1021/bi0022487. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Nesheim JC, Lee SK, Lipscomb JD. J Biol Chem. 1995;270:24662–24665. doi: 10.1074/jbc.270.42.24662. [DOI] [PubMed] [Google Scholar]
- 36.Ookubo T, Sugimoto H, Nagayama T, Masuda H, Sato T, Tanaka K, Maeda Y, Ōkawa H, Hayashi Y, Uehara A, Suzuki M. J Am Chem Soc. 1996;118:701–702. [Google Scholar]
- 37.Marchetti F, Melai B, Pampaloni G, Zacchini S. Inorg Chem. 2007;46:3378–3384. doi: 10.1021/ic061903h. [DOI] [PubMed] [Google Scholar]
- 38.Reisner E, Telser J, Lippard SJ. Inorg Chem. 2007;46:10754–10770. doi: 10.1021/ic701663j. [DOI] [PubMed] [Google Scholar]
- 39.Hagadorn JR, Que L, Jr, Tolman WB. J Am Chem Soc. 1998;120:13531–13532. doi: 10.1021/ja983333t. [DOI] [PubMed] [Google Scholar]
- 40.Lee D, Lippard SJ. J Am Chem Soc. 1998;120:12153–12154. [Google Scholar]
- 41.Tolman WB, Que L., Jr J Chem Soc, Dalton Trans. 2002:653–660. [Google Scholar]
- 42.Tshuva EY, Lippard SJ. Chem Rev. 2004;104:987–1012. doi: 10.1021/cr020622y. [DOI] [PubMed] [Google Scholar]
- 43.Lee D, Lippard SJ. Inorg Chem. 2002;41:2704–2719. doi: 10.1021/ic020186y. [DOI] [PubMed] [Google Scholar]
- 44.Lee D, Lippard SJ. Inorg Chim Acta. 2002;341:1–11. [Google Scholar]
- 45.Carson EC, Lippard SJ. J Inorg Biochem. 2006;100:1109–1117. doi: 10.1016/j.jinorgbio.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 46.Reisner E, Abikoff TC, Lippard SJ. Inorg Chem. 2007;46:10229–10240. doi: 10.1021/ic7014176. [DOI] [PubMed] [Google Scholar]
- 47.Friedle S, Kodanko JJ, Fornace KL, Lippard SJ. J Mol Struct. 2008;890:317–327. doi: 10.1016/j.molstruc.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee D, Du Bois J, Petasis D, Hendrich MP, Krebs C, Huynh BH, Lippard SJ. J Am Chem Soc. 1999;121:9893–9894. [Google Scholar]
- 49.Lee D, Krebs C, Huynh BH, Hendrich MP, Lippard SJ. J Am Chem Soc. 2000;122:5000–5001. doi: 10.1021/ja012251t. [DOI] [PubMed] [Google Scholar]
- 50.Lee D, Pierce B, Krebs C, Hendrich MP, Huynh BH, Lippard SJ. J Am Chem Soc. 2002;124:3993–4007. doi: 10.1021/ja012251t. [DOI] [PubMed] [Google Scholar]
- 51.Stubbe J, van der Donk WA. Chem Rev. 1998;98:705–762. doi: 10.1021/cr9400875. [DOI] [PubMed] [Google Scholar]
- 52.Chavez FA, Ho RYN, Pink M, Young VG, Jr, Kryatov SV, Rybak-Akimova EV, Andres H, Münck E, Que L, Jr, Tolman WB. Angew Chem Int Ed. 2002;41:149–152. doi: 10.1002/1521-3773(20020104)41:1<149::aid-anie149>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 53.Yoon S, Lippard SJ. J Am Chem Soc. 2005;127:8386–8397. doi: 10.1021/ja0512531. [DOI] [PubMed] [Google Scholar]
- 54.Zhao M, Song D, Lippard SJ. Inorg Chem. 2006;45:6323–6330. doi: 10.1021/ic0602906. [DOI] [PubMed] [Google Scholar]
- 55.Ménage S, Galey JB, Dumats J, Hussler G, Seité M, Luneau IG, Chottard G, Fontecave M. J Am Chem Soc. 1998;120:13370–13382. [Google Scholar]
- 56.Lee D, Lippard SJ. J Am Chem Soc. 2001;123:4611–4612. doi: 10.1021/ja0100930. [DOI] [PubMed] [Google Scholar]
- 57.Yoon S, Lippard SJ. Inorg Chem. 2006;45:5438–5446. doi: 10.1021/ic060307k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carson EC, Lippard SJ. Inorg Chem. 2006;45:828–836. doi: 10.1021/ic051471v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carson EC, Lippard SJ. Inorg Chem. 2006;45:837–848. doi: 10.1021/ic051476s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hecht S, Fréchet JMJ. Angew Chem Int Ed. 2001;40:74–91. doi: 10.1002/1521-3773(20010105)40:1<74::aid-anie74>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 61.Jiang DL, Aida T. Chem Commun. 1996:1523–1524. [Google Scholar]
- 62.Jiang DL, Aida T. J Macromol Sci, Pure Appl Chem. 1997;A34:2047–2055. [Google Scholar]
- 63.Collman JP, Fu L, Zingg A, Diederich F. Chem Commun. 1997:193–194. [Google Scholar]
- 64.Zingg A, Felber B, Gramlich V, Fu L, Collman JP, Diederich F. Helv Chim Acta. 2002;85:333–351. [Google Scholar]
- 65.Enomoto M, Aida T. J Am Chem Soc. 2002;124:6099–6108. doi: 10.1021/ja012589n. [DOI] [PubMed] [Google Scholar]
- 66.Zhao M, Helms B, Slonkina E, Friedle S, Lee D, DuBois J, Hedman B, Hodgson KO, Frechet JMJ, Lippard SJ. J Am Chem Soc. 2008;130:4352–4363. doi: 10.1021/ja076817a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuzelka J, Farrell JR, Lippard SJ. Inorg Chem. 2003;42:8652–8662. doi: 10.1021/ic034928e. [DOI] [PubMed] [Google Scholar]
- 68.Kodanko JJ, Morys AJ, Lippard SJ. Org Lett. 2005;7:4585–4588. doi: 10.1021/ol051700h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reisner E, Lippard SJ. Eur J Org Chem. 2008;2008:156–163. [Google Scholar]
- 70.Kodanko JJ, Lippard SJ. Inorg Chim Acta. 2008;361:894–900. doi: 10.1016/j.ica.2007.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kodanko JJ, Xu D, Song DT, Lippard SJ. J Am Chem Soc. 2005;127:16004–16005. doi: 10.1021/ja054943d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Friedle S, Kodanko JJ, Morys AJ, Hayashi T, Moënne-Loccoz P, Lippard SJ. J Am Chem Soc. 2009;131:14508–14520. doi: 10.1021/ja906137y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fiedler AT, Shan X, Mehn MP, Kaizer J, Torelli S, Frisch JR, Kodera M, Que L., Jr J Phys Chem A. 2008;112:13037–13044. doi: 10.1021/jp8038225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Das S, Incarvito CD, Crabtree RH, Brudvig GW. Science. 2006;312:1941–1943. doi: 10.1126/science.1127899. [DOI] [PubMed] [Google Scholar]
- 75.Farrell JR, Stiles D, Bu W, Lippard SJ. Tetrahedron. 2003;59:2463–2469. [Google Scholar]
- 76.Xue G, Wang D, De Hont R, Fiedler AT, Shan X, Münck E, Que L., Jr Proc Natl Acad Sci USA. 2007;104:20713–20718. doi: 10.1073/pnas.0708516105. and references therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shan X, Que L., Jr Proc Natl Acad Sci USA. 2005;102:5340–5345. doi: 10.1073/pnas.0409640102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang X, Furutachi H, Fujinami S, Nagatomo S, Maeda Y, Watanabe Y, Kitagawa T, Suzuki M. J Am Chem Soc. 2005;127:826–827. doi: 10.1021/ja045594a. [DOI] [PubMed] [Google Scholar]
- 79.Xue G, Fiedler AT, Martinho M, Münck E, Que L., Jr Proc Natl Acad Sci USA. 2008;105:20615–20620. [Google Scholar]
- 80.Do LH, Hayashi T, Moënne-Loccoz P, Lippard SJ. J Am Chem Soc. 132:1273–1275. doi: 10.1021/ja909718f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nam W. Acc Chem Res. 2007;40:522–531. doi: 10.1021/ar700027f. [DOI] [PubMed] [Google Scholar]
- 82.Ghosh A, Tiago de Oliveira F, Yano T, Nishioka T, Beach ES, Kinoshita I, Münck E, Ryabov AD, Horwitz CP, Collins TJ. J Am Chem Soc. 2005;127:2505–2513. doi: 10.1021/ja0460458. [DOI] [PubMed] [Google Scholar]
- 83.Wang D, Farquhar ER, Stubna A, Münck E, Que L., Jr Nature Chem. 2009;1:145–150. doi: 10.1038/nchem.162. [DOI] [PMC free article] [PubMed] [Google Scholar]


