Abstract
Natural killer (NK) cells are critical to the immune response to viral infections. Their functions are controlled by receptors for major histocompatibility complex (MHC) class I, including NKG2A and killer-cell immunoglobulin-like receptors (KIR). In order to evaluate the role of MHC class I receptors in the immune response to hepatitis C virus infection we have studied patients with chronic HCV infection by multi-parameter flow cytometry directly ex vivo. This has permitted evaluation of combinatorial expression of activating and inhibitory receptors on single NK cells. Individuals with chronic HCV infection had fewer CD56dim NK cells than healthy controls (4·9 ± 3·4% versus 9·0 ± 5·9%, P < 0·05). Expression levels of the inhibitory receptor NKG2A was up-regulated on NK cells from individuals with chronic hepatitis C virus (HCV) (NKG2A mean fluorescence intensity 5692 ± 2032 versus 4525 ± 1646, P < 0·05). Twelve individuals were treated with pegylated interferon and ribavirin. This resulted in a down-regulation of NKG2A expression on CD56dim NK cells. Individuals with a sustained virological response (SVR) had greater numbers of NKG2A-positive, KIR-negative NK cells than those without SVR (27·6 ± 9·6% NK cells versus 17·6 ± 5·7, P < 0·02). Our data show that NKG2A expression is dysregulated in chronic HCV infection and that NKG2A-positive NK cells are associated with a beneficial response to pegylated interferon and ribavirin therapy.
Keywords: hepatitis C, interferon-alpha, killer cell immunoglobulin-like receptors, natural killer cells, NKG2A
Introduction
Hepatitis C is a common chronic viral infection. The virus poses a significant challenge to the immune system, as the majority of individuals exposed to hepatitis C virus (HCV) are unable to clear HCV spontaneously, develop a chronic infection and are predisposed to cirrhosis and hepatocellular carcinoma. This failure to mount a successful immune response is multi-factorial in nature and includes abnormalities in T, B and dendritic cell responses.
Natural killer (NK) cells are lymphocytes that can interact directly with virus-infected host cells, activate dendritic cells and also secrete T helper type 1 (Th1) cytokines to augment cytotoxic T cell responses [1]. Their functions are controlled by multiple activating and inhibitory receptors, and they integrate signals derived from these receptors to determine whether or not they become activated. NK cells are enriched in the liver in comparison to peripheral blood [2] and a number of studies have documented abnormalities of NK cells in chronic HCV infection. These include changes in NK cell frequency, activity and subpopulation [3–8].
Key activating receptors on NK cells are the natural cytotoxicity receptors (NKp46, NKp44 and NKp30) and NKG2D. Key inhibitory receptors are the killer-cell immunoglobulin-like receptors (KIR) and NKG2A. These inhibitory receptors have major histocompatibility complex (MHC) class I ligands, and in particular the KIR exhibit substantial population diversity. The KIR bind polymorphic MHC-A, -B and -C class I molecules [9] and NKG2A binds human leucocyte antigen (HLA)-E [10]. The inhibitory NK receptor KIR2DL3 in combination with its group 1 HLA-C ligands is associated with spontaneous resolution of HCV infection [11,12].
Natural killer cell receptors are expressed abnormally in HCV infection. In chronic HCV infection both up- and down-regulation has been reported of the natural cytotoxicity receptors NKp30 and NKp46 on peripheral NK cells [13,14]. More recently, low levels of NKG2D on peripheral blood NK cells in HCV infection have been demonstrated [15], which may be related to the finding that HCV can down-regulate MHC class I-related chain A (MICA), a ligand for NKG2D [16]. Additionally, individuals with chronic HCV infection have higher frequencies of NK cells expressing the inhibitory receptor NKG2A [13,17], and expression of this receptor on CD56dim NK cells correlates inversely with HCV viral load [18]. Conversely, inhibitory KIR expression is normal or low in chronic HCV infection [15].
A number of abnormalities of NK cell function in HCV infection have been reported. Initial studies showed that NK cells had lower levels of cytotoxicity which normalized during successful interferon (IFN)-α therapy [3,4,6]. However, a more recent study did not replicate these findings [7] and in vitro, NK cells have been shown to degranulate more readily to both cytokines and receptor cross-linking [15]. Additionally they have an abnormal cytokine profile, secreting low levels of IFN-γ, and relatively higher levels of interleukin (IL)-10 [14,15]. HCV also expresses a peptide that stabilizes HLA-E on the cell surface [19]. As HLA-E is the ligand for NKG2A, this could lead to increased inhibition of NK cells. However, the frequency of NKG2A expressing NK cells correlates inversely with HCV RNA, suggesting that this subset is protective in HCV infection [18]. These conflicted findings may be caused by differences in the protocols used to stimulate NK cells, the use of fresh or frozen NK cells and the genetic heterogeneity of the populations studied.
IFN-α is a cytokine that, in combination with ribavirin, forms the cornerstone of current anti-HCV therapies. IFN-α acts both on infected cells to reduce viral replication and on cells of the immune system. It is a potent stimulus to NK cell IFN-γ production and cytolytic function [20,21]. Successful treatment with IFN-α may restore low levels of NK cell cytotoxicity and result in an increase in intrahepatic NK cells [4,16]. However, the effects of this cytokine on NK cell receptor expression in vivo has not yet been determined. Natural killer cells express MHC class I receptors stochastically, and this generates a complex repertoire of NK cells expressing different combinations of receptors within a single individual. This may account for heterogeneity in the response to HCV infection [22]. The aim of this study was therefore to determine the role of the KIR and NKG2A receptors in chronic HCV infection and in the outcome of treatment for HCV.
Materials and methods
Patients
The study was approved by the Local Research Ethics Committee. Thirty-five patients (21 male, mean age 44 years) infected chronically with HCV and 15 healthy controls (nine male, mean age 36 years) were studied. All HCV-positive individuals were positive for anti-HCV by second-generation enzyme-linked immunosorbent assay (ELISA) and were confirmed viraemic using HCV COBAS Amplicor Monitor version 2·0 (Roche, Burgess Hill, Sussex, UK). The mean viral load was 3·83 × 106 iu/ml. HCV genotyping was performed using quantitative polymerase chain reaction (PCR) (iQur® Ltd, Southampton, UK). Thirteen individuals had genotype 1 infection, and the remainder had either genotypes 2 or 3 infection (the iQUR® assay does not distinguish genotype 2 from genotype 3). Two individuals had biopsy-confirmed cirrhosis. Individuals were treated with a combination of pegylated interferon-α2a 180 µg once weekly in combination with 1200 mg or 1000 mg ribavirin daily, according to weight, for a total of 48 weeks if they had genotype 1 infection or 24 weeks for genotypes 2/3 infection.
HLA genotyping
Genomic DNA was extracted from peripheral blood lymphocytes using the QIAamp™ blood kit (Qiagen, Crawley, UK). HLA typing was performed by direct sequencing of PCR products [23]. HLA types which were not resolved by sequencing or which gave unusual results were also tested by sequence specific oligonucleotide probe typing [PCR–sequence-specific oligonucleotide probe (PCR–SSOP)], using a commercial kit (Dynal, RELI SSO™, Wirral, UK).
Flow cytometry
NK cell phenotyping was performed directly ex vivo. One hundred µl whole blood was incubated for 15 min with the following antibodies: CD158b,j-fluorescein isothiocyanate (FITC) (KIR2DL2/2DL3/2DS2), CD56-phycoerythrin (PE)-Cy7, CD3-allophycocyanin (APC)-Cy7 (BD Biosciences-Pharmingen, Oxford, UK), CD158e1-biotin (3DL1) and streptavidin–peridinin chlorophyll (PerCP) (Abcam, Cambridge, UK), CD158a,h-APC (KIR2DL1/2DS1), NKG2A-PE, NKp30-PE, NKp46-PE (Beckman Coulter, High Wycombe, UK) and NKG2D-PE (R&D Systems, Inc., Minneapolis, MN, USA). Red cells were lysed using FACS Lysing Solution™ (BD Biosciences) and the lymphocytes washed three times in phosphate-buffered saline (PBS) prior to analysis on a FACSCanto flow cytometer (Becton Dickinson) using diva software (Becton Dickinson).
Statistical analysis
Phenotyping analysis between healthy and HCV-infected individuals was performed using GraphPad Prism version 5 (GraphPad, La Jolla, CA, USA). Statistical comparisons were made using the Mann–Whitney U-test unless stated otherwise.
Results
Low numbers of NK cells and changes in receptor frequency are associated with chronic HCV infection
In order to enumerate accurately the NK cells from individuals with chronic HCV we performed a direct ex-vivo analysis of fresh CD3-CD56+ NK cells from 35 HCV-infected donors and 15 uninfected controls. Individuals with chronic HCV infection had significantly fewer CD56dim NK cells than healthy controls (4·9 ± 3·4% versus 9·0 ± 5·9%, P < 0·05) (Fig. 1a and b). No difference was found in the frequencies of CD56bright NK cells between the two populations. Analysis of MHC class I receptor expression on CD56dim NK cells indicated that individuals with chronic HCV had fewer CD158a,h-positive NK cells than healthy controls (21·7 ± 11·2% versus 31·1 ± 10·4%, P < 0·025) (Fig. 1c). The frequencies of other MHC class I receptors, including NKG2A, was similar among both populations. The lower frequency of CD158a,h was not associated with a specific subpopulation of NK cells that expressed other MHC class I receptors. As recent work [24] has shown that subpopulations of NK cells in the mouse can undergo expansion in a receptor specific pattern, and that these ‘adaptive’ NK cells have enhanced responses, we determined the HLA type of the individuals and the frequencies of their NK cells expressing cognate receptors for MHC class I. However, there was no expansion of either NK cells expressing a KIR cognate for the HLA class I alleles of that individual or those expressing NKG2A, which interacts with HLA-E (Fig. 1d). Interestingly, we noted that overall individuals with group 1 HLA-C allotypes had more NK cells expressing a cognate receptor for HLA-C than those without a group 1 HLA-C allotype. This was present in both individuals with chronic HCV (41·8% versus 20·2%, P < 0·005) and healthy controls (47·7% versus 28·0% P < 0·03) (Fig. 1e). Thus the difference in expression of a cognate HLA-C receptor between individuals with group 1 and group 2 HLA-C allotypes is generalizable to uninfected individuals. This implies that healthy individuals who acquire HCV and have group 1 HLA-C allotypes will have more NK cells expressing a cognate HLA-C receptor than those with group 2 HLA-C allotypes. As spontaneous resolution of HCV infection is associated with group 1 HLA-C allotypes, then resolution of infection may correlate with the number of NK cells expressing a receptor cognate for HLA-C.
Fig. 1.
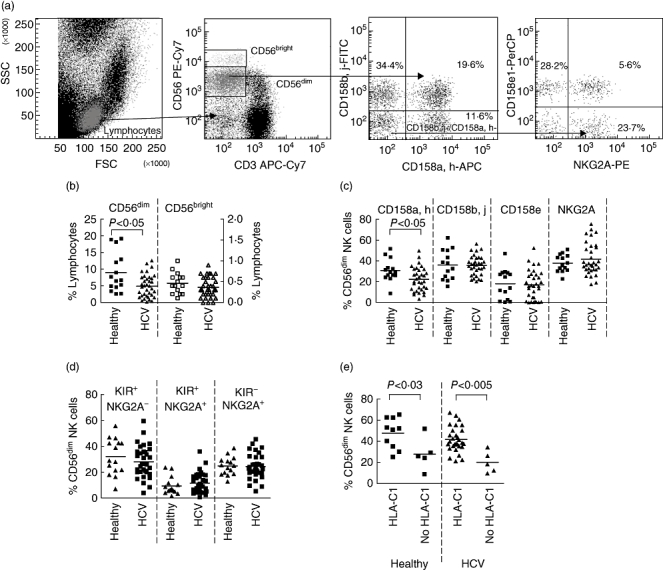
A comparison of CD3-CD56+ natural killer (NK) cells between hepatitis C virus (HCV)-infected individuals (squares) with healthy individuals (triangles) as determined by flow cytometry. (a) Strategy for the six-colour flow cytometry, gating on CD56dim, CD3- NK cells to determine killer-cell immunoglobulin-like receptors (KIR) (CD158a,h, CD158b,j and CD158e) and NKG2A expression. Shown are three dot plots demonstrating the expression of CD158e and NKG2A on CD158a,h-,CD158b,j-, CD3-CD56dim NK cells. (b) Percentage of CD56dim and CD56bright NK cells as a fraction of the total lymphocyte population. The CD56dim NK cells are recorded on the left axis (closed symbols) and CD56bright NK cells on the right axis (open symbols). (c) Frequencies of NK cells expressing individual major histocompatibility complex (MHC) class I receptors as determined by anti-CD158a,h (KIR2DL1/2DS1), anti-CD158b,j (KIR2DL2/2DL3/2DS2), anti-CD158e (KIR3DL1) and Z199 (NKG2A). The data shown are the percentage of total CD56dim NK cells expressing the indicated receptor. (d) Comparison of the overall frequencies of NK cells expressing receptors for which they have an MHC class I ligand (an HLA-B or HLA-C allele for the KIR, or HLA-E for NKG2A). (e) Frequencies of NK cells expressing a cognate HLA-C receptor in individuals with and without an HLA-C group 1 allele. Comparisons between the healthy donors and HCV infected individuals were performed using the Mann–Whitney U-test. P-values of 0·05 or less are indicated.
Levels of activating and inhibitory receptor expression is elevated in HCV infection
Previous work [13,17,18] has implicated NKG2A+ NK cells in the pathology of HCV, and as we observed no abnormalities in the frequency of NKG2A-expressing NK cells we determined the level of NKG2A as indicated by the mean fluorescence intensity (MFI) of surface NKG2A on the NK cells (Fig. 2a). The MFI of NKG2A was increased significantly on NKG2A-positive NK cells within both the total CD56dim and CD56bright subsets of NK cells in individuals with chronic HCV infection (5692 ± 2032 versus 4525 ± 1646, P < 0·05; 11 307 ± 3509 versus 8549 ± 3495, P < 0·025, respectively) (Fig. 2b). It was not increased on NKG2A-positive T cells (8785 ± 2978 versus 8051 ± 2354), indicating that these differences are specific to NK cells, and are also not caused by an artefact of the flow cytometry. Our previous work has shown that KIR2DL3 (recognized by the CD158b,j antibody) is associated with protection from chronic HCV infection [11]. In order to determine if the KIR have an influence on the over-expression of NKG2A in our patient population we determined the levels of NKG2A on the subpopulations of CD158a,h and CD158b,j NK cells that co-expressed NKG2A. The increase in NKG2A was most significant in the CD158a,h+/CD158b,j- (4827 ± 1880 versus 3600 ± 1425, P < 0·025) and the CD158a,h-/CD158b,j- (6527 ± 2303 versus 5081 ± 2082 P < 0·05) subpopulations (Fig. 2c).
Fig. 2.
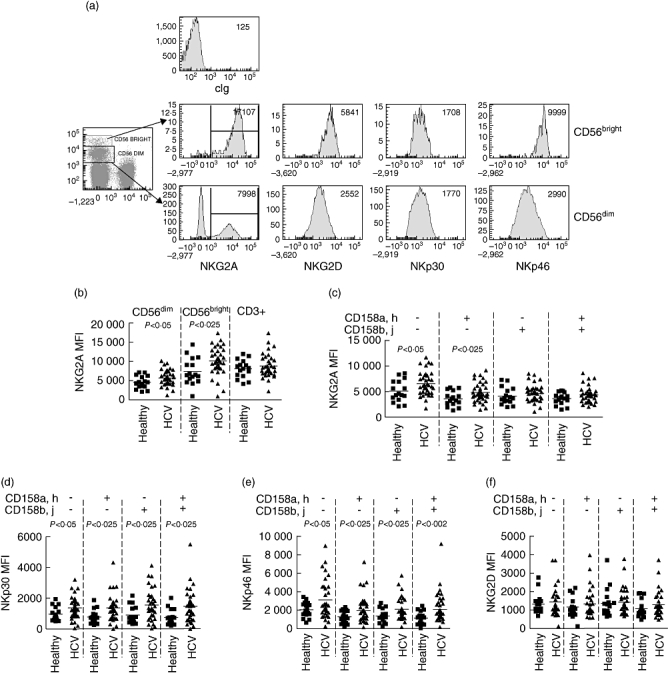
A comparison of the expression levels of inhibitory and activating receptors on natural killer (NK) cells from hepatitis C virus (HCV)-infected individuals with healthy individuals as determined by flow cytometry. (a) Flow cytometry plots indicating the expression of NKG2A, NKG2D, NKp46 and NKp30 on CD3-CD56bright and CD3-CD56dim NK cells. The mean fluorescence intensity of the positive population for NKG2A and of the total NK cell population for the activating receptors is indicated. (b,c) Levels of NKG2A expression on NKG2A+CD56dim NK cells, NKG2A-CD56bright NK cells and on NKG2A+CD3+ T cells (b); and on the subsets of CD56dim NK cells expressing specific combinations of killer-cell immunoglobulin-like receptors (KIR) as determined by antibodies to CD158a,h and CD158b,j (c). (d–f) Levels of the activating receptors on the same subpopulations of NK cells are shown for NKp30 (d), NKp46 (e) and NKG2D (f). Comparisons were performed using the Mann–Whitney U-test and P-values of 0·05 or less are indicated.
The natural cytotoxicity receptors NKG2D, NKp30 and NKp46 are expressed on all mature NK cells [25] and are responsible for recognition of some tumour targets and virally infected cells [26,27]. In order to determine if the abnormalities of NKG2A expression were specific to this inhibitory receptor or reflective of more general changes in the CD158a,h-positive population, we compared the levels of expression of these receptors on the different subpopulations of NK cells. We found that in chronic HCV infection the levels of expression of both NKp30 and NKp46 were elevated across all subpopulations of CD56dim NK cells, but those of NKG2D were unaltered (Fig. 2d–f).
Pegylated IFN-α and ribavirin normalizes NKG2A, but not natural cytotoxicity receptors (NCR) expression
The effect of pegylated IFN and ribavirin (PEG/Rib) on NK cell receptor expression was determined in 12 patients undergoing treatment for HCV. Flow cytometry analysis was performed during the first 12 weeks of treatment (Fig. 3a). In our treatment protocol this is the time-frame within which individuals who have a sustained virological response (SVR) have a significant decline in viral load [28]. Overall NKG2A expression declined rapidly by week 2 on the CD56dim NK cells to an average of 0·58 times the pretreatment levels. This decrease was less significant on the CD56bright NK cells (mean fold change compared to baseline 0·84 ± 0·02 versus 0·59 ± 0·02 (CD56dim NK cells), P = 0·001) and also on T cells (mean fold change = 0·88 ± 0·21, P < 0·002 versus CD56dim NK cells (Fig. 3b). Furthermore, this was not specific to subpopulations of NK cells that expressed specific KIR, as it was also found on NK cells that expressed no KIR (Fig. 3c). By contrast, the expression of the activating receptors remained relatively unchanged on NK cells (Fig. 3d–f). However, there was a slight decline in expression of the activating receptor NKG2D on T cells which reached significance by week 12 (mean fold change 0·42 ± 0·18 compared to the baseline level (P < 0·05) (Fig. 3g). This implies that in the early phase of PEG/Rib therapy there is a change in the relative balance between activating and inhibitory receptors, which favours a loss of the inhibitory signal from the HLA-E : NKG2A system on NK cells, but not on T cells.
Fig. 3.
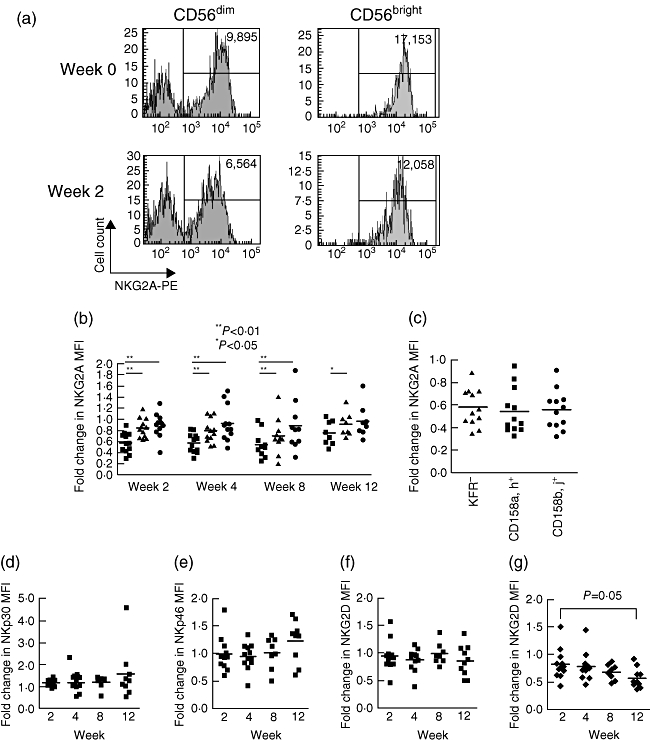
The effect of pegylated interferon and ribavirin on expression of inhibitory and activating receptors. (a) Flow cytometry plot of natual killer (NK)G2A expression pretreatment and at week 2 on CD3-CD56bright and CD3-CD56dim NK cells, showing the change in mean fluorescence intensity. (b) Fold change in mean fluorescence intensity of NKG2A on all NKG2A-positive CD56dim NK cells (squares), CD56bright NK cells (triangles) and CD3+NKG2A+ T cells (circles) during the first 12 weeks of therapy. The chart shows the comparison of the expression level of NKG2A at the indicated weeks compared to the baseline level. Expression of NKG2A declined most rapidly on the CD56dim NK cells compared to the CD56bright NK cells and CD3+ T cells. Statistical analysis was performed using the Wilcoxon matched-pairs test. (c) Fold change in NKG2A expression compared to baseline on the indicated subsets of NKG2A-positive CD56dim NK cells at week 2. (d–f) Fold change compared to baseline in the mean fluorescence intensity of the activating receptors NKp30 (d), NKp46 (e) and NKG2D (f) on CD56dim NK cells and of NKG2D on CD3+ T cells (g) during treatment.
Sustained virological response is associated with the baseline frequency of NKG2A+KIR- NK cells
Successful treatment with pegylated-IFN/Rib is associated with an SVR in which HCV RNA remains undetectable for at least 6 months following cessation of therapy, and usually continues into the long term [29]. Individuals with and without SVR had similar levels of decline in NKG2A expression (not shown) and so we therefore studied the association of the baseline NK cell phenotype with treatment outcome. Twenty-nine of the 35 individuals completed treatment and 23 had an SVR. There was a non-significant trend towards individuals with SVR having a higher frequency of CD56dim NKG2A+ NK cells (Fig. 4a). Subset analysis indicated that this was due to KIR-NKG2A+ NK cells (27·6 ± 9·6% (SVR) versus 17·6 ± 5·7 (no SVR), P < 0·02), i.e. cells inhibited only by HLA-E, and not those inhibited by MHC-B and -C (Fig. 4b). No other baseline phenotypic parameter was associated significantly with outcome.
Fig. 4.
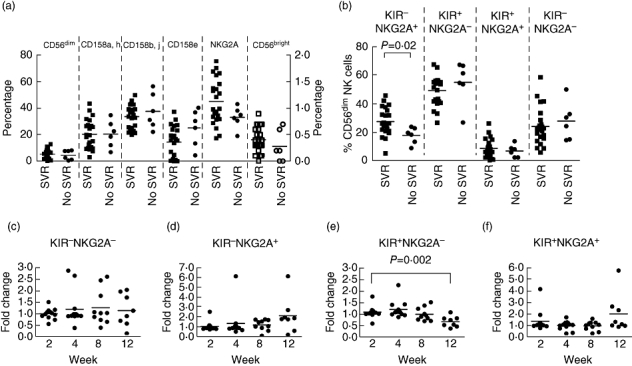
The association of sustained virological response to pegylated interferon and ribavirin with the frequency of specific subpopulations of natural killer (NK) cells. (a) Pretreatment frequencies of CD56dim NK cells expressing the indicated receptors in individuals with (filled squares) and without (filled circles) sustained virological response (SVR) plotted on the left Y-axis. Also shown are the frequencies of the CD56bright NK cells from the same individuals plotted on the right Y-axis (open squares denote individuals with SVR and open circles those without SVR). (b) Pretreatment frequency of CD56dim NK cells expressing the indicated receptors on individuals with (squares) and without (circles) SVR. Comparisons between those with and without SVR were made using the Mann–Whitney U-test. (c–f) changes in the indicated subpopulations of NK cells during treatment. Statistical comparisons were made using the analysis of variance (anova) test. For all panels P-values of less 0·05 are indicated.
As KIR-NKG2A+ NK cells are associated with SVR, we determined if these cells increase in frequency during the early phase of PEG/Rib therapy (Fig. 4c–e). There was a trend towards an increase in KIR-NKG2A+ NK cells (P > 0·1), and a significant decline in KIR+NKG2A- NK cells (P = 0·005). However, these changes occurred later during treatment at weeks 8–12, and were not associated with SVR. Consideration of the HLA type of the individuals demonstrated no association with HLA-C and no expansions of NK cells that expressed a KIR specific for the HLA-C allele of that individual.
Discussion
The role of MHC class I receptors and NK cells in HCV infection relates to both frequency of specific subpopulations of NK cells and also receptor expression levels. Previous work has yielded conflicting results, and this may be due in part to differences in preparation such as freezing or in cytokine stimulation [13,14]. We have therefore analysed NK cell receptor expression on NK cells in chronic HCV infection directly ex vivo in order to avoid such bias in receptor subpopulations. The use of multi-parameter flow cytometry has allowed the simultaneous evaluation of multiple receptors on single NK cells in order to provide insights into the relative contribution of receptors with different MHC class I ligands. Overall, we found that NKG2A-positive NK cells were associated with a beneficial response to treatment. In vitro this subset of NK cells can lyse vaccinia-infected targets in the context of autologous MHC class I [30]. Thus this specific subset of NK cells has been shown to have anti-viral activity.
The most marked changes were noted in levels of NKG2A expression, which occurred on both CD56bright and CD56dim NK cells. This increase in NKG2A expression on NK cells may be representative of chronic activation of NK cells, which can be induced by cytokines such as IL-12 or IL-21 [31,32]. In studies using NK cell clones the inhibition mediated by NKG2A was weaker and more variable than that mediated by KIR [33]. Thus our observed changes may have functional consequences for the inhibition of NK cells. Conversely, during treatment with PEG/Rib, the decline in NKG2A levels may reverse this increased inhibition and hence allow a more ready activation of the NKG2A-positive NK cells. This change is more likely to be important on NK cells that are not inhibited by other inhibitory receptors, and hence is consistent with our observation that the frequency of KIR-negative, NKG2A-positive NK cells are associated with a successful outcome of PEG/Rib therapy.
We noted a rapid decline in expression of NKG2A, but not activating receptors, following the administration of PEG/Rib. However, this change did not correlate with outcome. This is not unexpected, as many factors influence the outcome of PEG/Rib therapy for HCV, the most notable of which is the viral genotype. This change in expression preceded alterations in NK cell subsets, which did not occur until weeks 8–12 following therapy; thus it is most probably a direct effect on NKG2A receptor expression. NK cells express multiple inhibitory receptors for MHC class I, and so NK cells expressing NKG2A as their sole inhibitory receptor are the ones that will have their activating/inhibitory receptor balance affected the most by a reduction in NKG2A expression. As we observed no change in activating receptor expression, this implies that these cells have this balance altered in favour of activation. Consistent with the hypothesis that these are a protective subset of NK cells we found that individuals with SVR have more KIR-NKG2A+ NK cells than those without SVR, a quantitative model of NK cells in viral infections as has been observed for murine cytomegalovirus (MCMV) [34]. Recent work has demonstrated that NK cells have a number of adaptive properties, including antigen specificity, clonal expansion, long-lived progeny and recall responses [35]. We saw a trend towards an increase in the number of KIR-NKG2A+ NK cells during PEG/Rib therapy, which is consistent with a role in the anti-HCV response.
From our data it is not clear as to the mechanism by which interferon and/or ribavirin affects expression of NKG2A on NK cells. It may be a direct effect on NK cells due to changes in the cytokine milieu, or to interactions with other cells of the immune system that can subsequently effect NK cell receptor expression. In chronic HCV, but not HBV, infection NKG2A expression correlates with necroinflammatory score [18], which is consistent with our observations. However, longitudinal analysis of individuals with chronic HBV infection treated with IFN and in vitro culture experiments may be helpful in determining the disease specificity and mechanisms behind our in vivo observations of NKG2A expression.
Genetic studies have focused upon the KIR system in HCV infection, and in chronic HCV infection we have noted specific changes in the frequency of NK cells expressing CD158a,h, but not CD158b,j. These markers are not specific for inhibitory KIR and currently there are no reagents available to distinguish the activating from the inhibitory forms of these receptors. The majority of these cells express the inhibitory receptor, as determined by biochemical studies and the analysis of NK cell clones [36–38]. In particular, a comprehensive analysis of NK cell clones demonstrated that all cells expressing CD158a,h were inhibited by group 2 HLA-C allotypes and similarly for CD158b,j and group 1 HLA-C allotypes [33]. Thus it appears that in our analysis the KIR2DL1 subset of NK cells is perturbed in chronic HCV infection relative to the KIR2DL2/KIR2DL3 subset. Differences in our data compared to that of Oliviero et al. [15], in which changes in KIR3DL1 were most prominent, are related most probably to genetic differences among the patient populations studied, especially as allelic diversity of KIR3DL1 is associated with marked differences in the level of expression [39]. However, in their study there was also a lower frequency of CD158a,h-positive NK cells in individuals with chronic HCV compared to healthy donors, although this did not reach statistical significance. Similarly, Bonorino et al. noted non-significant lower levels of CD158a,h but not CD158b,j in individuals with chronic HCV [18].
Overall, we did not observe an influence of HLA-C on the frequency of NK cells in chronic HCV infection compared to healthy controls, although we did observe that individuals with a group 1 HLA-C ligand had more NK cells expressing a KIR cognate for HLA-C allele than those without. This was found both in healthy donors and in HCV-infected individuals. This is a quantitative correlate of the protective effect of NK cells in spontaneously resolving HCV infection. The diversity of the KIR system provides one rationale for differences in response among individuals, but this does not preclude the NKG2A system from being important. Indeed, it has been shown that individuals with spontaneously resolved HCV infection have lower numbers of NKG2A/C/E-positive NK cells than those with chronic HCV infection [17]. Furthermore, the mature NK cell repertoire is influenced both by KIR and MHC class I genotype [40]. Thus the interplay between the two receptor systems is complex and likely to synergize in infections, such that both receptors systems are important in the anti-HCV response. Our data, however, demonstrate the importance of the NKG2A: HLA-E system in the response to pegylated interferon and ribavirin, and thus highlights the anti-viral role of this evolutionarily conserved receptor : ligand system.
Acknowledgments
We would like to thank the Hepatology Specialist Nurses at Southampton General Hospital for their assistance in collecting these samples.
Disclosure
Competing interests
None.
Funding
This work was funded by The Wellcome Trust.
References
- 1.Moretta L, Ferlazzo G, Bottino C, et al. Effector and regulatory events during natural killer-dendritic cell interactions. Immunol Rev. 2006;214:219–28. doi: 10.1111/j.1600-065X.2006.00450.x. [DOI] [PubMed] [Google Scholar]
- 2.Doherty DG, Norris S, Madrigal-Estebas L, et al. The human liver contains multiple populations of NK cells, T cells, and CD3+CD56+ natural T cells with distinct cytotoxic activities and Th1, Th2, and Th0 cytokine secretion patterns. J Immunol. 1999;163:2314–21. [PubMed] [Google Scholar]
- 3.Corado J, Toro F, Rivera H, Bianco NE, Deibis L, De Sanctis JB. Impairment of natural killer (NK) cytotoxic activity in hepatitis C virus (HCV) infection. Clin Exp Immunol. 1997;109:451–7. doi: 10.1046/j.1365-2249.1997.4581355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonavita MS, Franco A, Paroli M, et al. Normalization of depressed natural killer activity after interferon-alpha therapy is associated with a low frequency of relapse in patients with chronic hepatitis C. Int J Tissue React. 1993;15:11–16. [PubMed] [Google Scholar]
- 5.Deignan T, Curry MP, Doherty DG, et al. Decrease in hepatic CD56(+) T cells and V alpha 24(+) natural killer T cells in chronic hepatitis C viral infection. J Hepatol. 2002;37:101–8. doi: 10.1016/s0168-8278(02)00072-7. [DOI] [PubMed] [Google Scholar]
- 6.Par G, Rukavina D, Podack ER, et al. Decrease in CD3-negative-CD8dim(+) and Vdelta2/Vgamma9 TcR+ peripheral blood lymphocyte counts, low perforin expression and the impairment of natural killer cell activity is associated with chronic hepatitis C virus infection. J Hepatol. 2002;37:514–22. doi: 10.1016/s0168-8278(02)00218-0. [DOI] [PubMed] [Google Scholar]
- 7.Morishima C, Paschal DM, Wang CC, et al. Decreased NK cell frequency in chronic hepatitis C does not affect ex vivo cytolytic killing. Hepatology. 2006;43:573–80. doi: 10.1002/hep.21073. [DOI] [PubMed] [Google Scholar]
- 8.Meier UC, Owen RE, Taylor E, et al. Shared alterations in NK cell frequency, phenotype, and function in chronic human immunodeficiency virus and hepatitis C virus infections. J Virol. 2005;79:12365–74. doi: 10.1128/JVI.79.19.12365-12374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–51. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 10.Braud VM, Allan DS, O'Callaghan CA, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–9. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 11.Khakoo SI, Thio CL, Martin MP, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–4. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 12.Romero V, Azocar J, Zuniga J, et al. Interaction of NK inhibitory receptor genes with HLA-C and MHC class II alleles in hepatitis C virus infection outcome. Mol Immunol. 2008;45:2429–36. doi: 10.1016/j.molimm.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nattermann J, Feldmann G, Ahlenstiel G, Langhans B, Sauerbruch T, Spengler U. Surface expression and cytolytic function of natural killer cell receptors is altered in chronic hepatitis C. Gut. 2006;55:869–77. doi: 10.1136/gut.2005.076463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Maria A, Fogli M, Mazza S, et al. Increased natural cytotoxicity receptor expression and relevant IL-10 production in NK cells from chronically infected viremic HCV patients. Eur J Immunol. 2007;37:445–55. doi: 10.1002/eji.200635989. [DOI] [PubMed] [Google Scholar]
- 15.Oliviero B, Varchetta S, Paudice E, et al. Natural killer cell functional dichotomy in chronic hepatitis B and chronic hepatitis C virus infections. Gastroenterology. 2009;137:1151–60. doi: 10.1053/j.gastro.2009.05.047. [DOI] [PubMed] [Google Scholar]
- 16.Wen C, He X, Ma H, et al. Hepatitis C virus infection downregulates the ligands of the activating receptor NKG2D. Cell Mol Immunol. 2008;5:475–8. doi: 10.1038/cmi.2008.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golden-Mason L, Madrigal-Estebas L, McGrath E, et al. Altered natural killer cell subset distributions in resolved and persistent hepatitis C virus infection following single source exposure. Gut. 2008;57:1121–8. doi: 10.1136/gut.2007.130963. [DOI] [PubMed] [Google Scholar]
- 18.Bonorino P, Ramzan M, Camous X, et al. Fine characterization of intrahepatic NK cells expressing natural killer receptors in chronic hepatitis B and C. J Hepatol. 2009;51:458–67. doi: 10.1016/j.jhep.2009.05.030. [DOI] [PubMed] [Google Scholar]
- 19.Nattermann J, Nischalke HD, Hofmeister V, et al. The HLA-A2 restricted T cell epitope HCV core 35–44 stabilizes HLA-E expression and inhibits cytolysis mediated by natural killer cells. Am J Pathol. 2005;166:443–53. doi: 10.1016/S0002-9440(10)62267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- 21.Matikainen S, Paananen A, Miettinen M, et al. N-alpha and IL-18 synergistically enhance IFN-gamma production in human NK cells: differential regulation of Stat4 activation and IFN-gamma gene expression by IFN-alpha and IL-12. Eur J Immunol. 2001;31:2236–45. [PubMed] [Google Scholar]
- 22.Yawata M, Yawata N, Draghi M, Partheniou F, Little AM, Parham P. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 2008;112:2369–80. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunn PP, Cox ST, Little AM. Sequencing protocols for detection of HLA class I polymorphism. Methods Mol Biol. 2003;210:191–222. doi: 10.1385/1-59259-291-0:191. [DOI] [PubMed] [Google Scholar]
- 24.Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457:557–61. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moretta A, Bottino C, Vitale M, et al. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol. 2001;19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- 26.Arnon TI, Markel G, Mandelboim O. Tumor and viral recognition by natural killer cells receptors. Semin Cancer Biol. 2006;16:348–58. doi: 10.1016/j.semcancer.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 27.Hamerman JA, Ogasawara K, Lanier LL. NK cells in innate immunity. Curr Opin Immunol. 2005;17:29–35. doi: 10.1016/j.coi.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Zeuzem S, Berg T, Moeller B, et al. Expert opinion on the treatment of patients with chronic hepatitis C. J Viral Hepatol. 2009;16:75–90. doi: 10.1111/j.1365-2893.2008.01012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maylin S, Martinot-Peignoux M, Moucari R, et al. Eradication of hepatitis C virus in patients successfully treated for chronic hepatitis C. Gastroenterology. 2008;135:821–9. doi: 10.1053/j.gastro.2008.05.044. [DOI] [PubMed] [Google Scholar]
- 30.Brooks CR, Elliott T, Parham P, Khakoo SI. The inhibitory receptor NKG2A determines lysis of vaccinia virus-infected autologous targets by NK cells. J Immunol. 2006;176:1141–7. doi: 10.4049/jimmunol.176.2.1141. [DOI] [PubMed] [Google Scholar]
- 31.Draghi M, Yawata N, Gleimer M, Yawata M, Valiante NM, Parham P. Single-cell analysis of the human NK cell response to missing self and its inhibition by HLA class I. Blood. 2005;105:2028–35. doi: 10.1182/blood-2004-08-3174. [DOI] [PubMed] [Google Scholar]
- 32.Brady J, Hayakawa Y, Smyth MJ, Nutt SL. IL-21 induces the functional maturation of murine NK cells. J Immunol. 2004;172:2048–58. doi: 10.4049/jimmunol.172.4.2048. [DOI] [PubMed] [Google Scholar]
- 33.Valiante NM, Uhrberg M, Shilling HG, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–51. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 34.Scalzo AA, Fitzgerald NA, Simmons A, La Vista AB, Shellam GR. Cmv-1, a genetic locus that controls murine cytomegalovirus replication in the spleen. J Exp Med. 1990;171:1469–83. doi: 10.1084/jem.171.5.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun JC, Lanier LL. Natural killer cells remember: an evolutionary bridge between innate and adaptive immunity? Eur J Immunol. 2009;39:2059–64. doi: 10.1002/eji.200939435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moretta A, Tambussi G, Bottino C, et al. A novel surface antigen expressed by a subset of human CD3– CD16+ natural killer cells. Role in cell activation and regulation of cytolytic function. J Exp Med. 1990;171:695–714. doi: 10.1084/jem.171.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moretta A, Vitale M, Bottino C, et al. P58 molecules as putative receptors for major histocompatibility complex (MHC) class I molecules in human natural killer (NK) cells. Anti-p58 antibodies reconstitute lysis of MHC class I-protected cells in NK clones displaying different specificities. J Exp Med. 1993;178:597–604. doi: 10.1084/jem.178.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Litwin V, Gumperz J, Parham P, Phillips JH, Lanier LL. Specificity HLA class I antigen recognition by human NK clones: evidence for clonal heterogeneity, protection by self and non-self alleles, and influence of the target cell type. J Exp Med. 1993;178:1321–36. doi: 10.1084/jem.178.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gardiner CM, Guethlein LA, Shilling HG, et al. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol. 2001;166:2992–3001. doi: 10.4049/jimmunol.166.5.2992. [DOI] [PubMed] [Google Scholar]
- 40.Shilling HG, Guethlein LA, Cheng NW, et al. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol. 2002;168:2307–15. doi: 10.4049/jimmunol.168.5.2307. [DOI] [PubMed] [Google Scholar]


