Abstract
No consensus has been reached on how to determine if an immune response has been detected based on raw data from an ELISPOT assay. The goal of this paper is to enable investigators to understand and readily implement currently available methods for response determination. We describe empirical and statistical approaches, identifying the strengths and limitations of each approach to allow readers to rationally select and apply a scientifically sound method appropriate to their specific laboratory setting. Five representative approaches were applied to data sets from the CIMT Immunoguiding Program and the response detection and false positive rates were compared. Simulation studies were also performed to compare empirical and statistical approaches. Based on these, we recommend the use of a non-parametric statistical test. Further, we recommend that six medium control wells or four wells each for both medium control and experimental conditions be performed to increase the sensitivity in detecting a response, that replicates with large variation in spot counts be filtered out, and that positive responses arising from experimental spot counts below the estimated limit of detection be interpreted with caution. Moreover, a web-based user interface was developed to allow easy access to the recommended statistical methods. This interface allows the user to upload data from an ELISPOT assay and obtain an output file of the binary responses.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-010-0875-4) contains supplementary material, which is available to authorized users.
Keywords: ELISPOT assay, Replicate variation, Background spot production, Positive response criteria, Harmonization
Introduction
Background
The main goal of monitoring antigen-specific T cell responses in immunotherapy trials is to determine if a treated patient has mounted a response following immune intervention and if a detected response is associated with a clinical event. Here, we address the definition of immune responses, a prerequisite for determining their clinical relevance. The IFN-γ ELISPOT assay is widely used to quantify antigen-specific immunity on a single-cell level. The assay results in raw data that need to be interpreted by the investigator to determine if an immune response has been detected. To date, no commonly accepted consensus exists as to what rule to use for this determination. This complicates comparability of results across institutions and explains the urgent need to establish objective rules with which to make a response determination. Validation of ELISPOT standard operating procedures is a critical aspect but is outside the scope of this paper. A number of statistical methods have been proposed for immune response determination [e.g., 20, 21] but are not often used by investigators. The goal of this paper is to enable broader access by describing currently available statistical methods to a non-statistical audience (this section), providing real and simulated data examples to illustrate their use and to provide the reader with recommendations with respect to the interpretation of ELISPOT results (“Results”), and pointing the reader to a newly created web-based interface where the recommended statistical methods can be readily applied to investigators’ data (“Discussion”).
This paper addresses one part of a larger, recently initiated effort to harmonize T cell monitoring assays across institutions [1–6] and to introduce thorough quality control and extensive validation [7–9]. Our ultimate goal is to establish T cell immunomonitoring as a precise tool to guide clinical development and thereby accelerate the evaluation of new vaccines and immunological therapeutics [10].
Approaches for response definition
There are two main approaches that are employed to establish criteria for detecting a positive response for the ELISPOT assay. The first is empirical and the second is statistical.
Empirical rules
An empirical rule (“ER”) is usually based on observations from a specific study and provides an ad hoc tool to determine if a positive signal is detected. However, there is no theoretical basis for this rule. Several empirical approaches have been proposed in the literature for determining an ELISPOT response [11–14]. An illustration of a clear and rational method for deriving an ER to decide whether an individual is an immunological responder or not is given by Dubey et al. [14]. Using samples from 72 HIV-negative donors, a comparison was made between spot counts detected in media with HIV peptide, compared to peptide-free mock control wells, but matching DMSO content. The authors then considered the inherent background of each sample (mock control) and the magnitude of the antigen-stimulated response.
They used three components to determine their positivity rule:
A minimum threshold “x” for spot counts per 106 PBMCs above which would be considered a positive response if condition 2 below is satisfied.
A minimum threshold limit “y” for the ratio of antigen to mock above which would be considered as a positive response.
Based on the generated data with control donors, the above two thresholds (x and y) were chosen so that the false positive rate was limited to <1% by analyzing the responses against HIV-derived control peptides in HIV-negative donors.
For each of the three control peptide pools tested, they determined the thresholds that would satisfy these three criteria. They then applied the rules derived to the data generated by testing HIV-positive donors and compared the positivity rates of the different peptide pools. The resulting definition for a positive response was more than 55 spots per 1 × 106 cells and at least fourfold background. Dubey et al. clearly state that the rules they developed are only valid for the ELISPOT procedures and reagents that were used to validate them, namely the protocol they used. This is because it is unknown what the false positive rate would be in any different setting. Different rules would, therefore, be necessary for each laboratory using other ELISPOT protocols or patient populations. The goal of their paper was to advocate a method for developing an ER but explicitly not to recommend the specific cutoff values (x and y) observed in their experiments, as any laboratory would have to identify these themselves, based on their own testing results.
The three data sets from the CIP proficiency panel program that are used in “Results” to illustrate the various methods contain data from a heterogeneous group of ELISPOT protocols and hence the approach proposed by Dubey et al. to determine an ER is not appropriate. Therefore, we decided to examine two ERs that are used by many of the participating laboratories. The first ER declares a positive response based on a threshold minimum of 5 spots per 100,000 PBMCs in the experimental wells and at least a twofold increase of spot number over background. The second ER declares a positive response based only on more than a twofold difference between the spot counts in the experimental versus background wells. No minimum spot number is required in the latter rule.
Statistical tests
A statistical test (“ST”) for response determination is based on statistical hypothesis testing. This is done by constructing a null and an alternative hypotheses and then using the data to test the evidence against the null hypothesis as outlined in the following three steps:
Decide on the appropriate null and alternative hypotheses. A common null hypothesis in the ELISPOT response determination setting is that there is no difference between the average (mean) spot counts in the experimental and control wells. One commonly used alternative hypothesis is that the mean spot count in the experimental wells is greater than that of the background or control wells (a one-sided alternative hypothesis).
Decide on an appropriate test statistic. This depends on the hypotheses and the characteristics of the data.
Set the alpha level or type I error of the test. This alpha level is used to judge when there is strong evidence against the null hypothesis. In our setting, responses will be declared positive if the p value is less than or equal to alpha. The alpha level is typically set at 0.05 and represents the probability of rejecting the null hypothesis given the data when in fact the null hypothesis is true.
The p value is calculated from the assumed distribution of the test statistic under the null hypothesis so assumptions about this are needed. If the sample sizes are large (n ≥ 30 is a typical rule of thumb) or the data are known to follow a normal distribution and the null hypothesis is that the means of each group are the same, the T statistic (T = difference in means/pooled standard deviation) can be chosen as it can be assumed that the T statistic follows a Student’s t distribution under the null hypothesis. However, if the sample size is small (e.g., triplicates), or when it is difficult to estimate the distribution of the population from which the samples are taken, one cannot assume that the means follow a normal distribution by the central limit theorem. In this situation, the T statistic might still be used but with a non-parametric test (e.g., permutation or bootstrap) to calculate the p value as this avoids distributional assumptions.
In the ELISPOT setting, it is often of interest to test more than one antigen (be it peptide, peptide pool, protein, or gene) per donor. Therefore, several comparisons will be made for an individual donor (spot counts from each antigen versus control). When a ST is used to determine response, many STs will be performed per donor. This leads to the problem of multiple comparisons, namely an inflation of the false positive rate. When one ST is performed and a false positive threshold of 0.05 is selected, the probability of rejecting the null hypothesis when it is true would be 5%. However, if we perform two independent STs with the 0.05 false positive threshold, the probability that at least one test will be a false positive is 10%. This probability of at least one false positive among the multiple hypotheses tested, known as the family-wise error rate, increases with the number of simultaneous tests performed and can be calculated as 1 − (1 − α)k, where α is the false positive threshold for each test and k is the number of independent comparisons. For three, four or five concurrent tests, the probability of at least one false positive is 14, 19, or 23%, respectively.
It is of interest to control the family-wise error rate to ensure that the probability of at least one false positive for all the STs is at an acceptable level. A classical way to control the family-wise error rate is to employ a Bonferroni correction [15]. If there are k planned comparisons and the desired family-wise error rate is 0.05, the Bonferroni correction would be to set the type I error threshold for an individual test to be 0.05/k. The Bonferroni correction is most appropriate to use when the individual tests are independent. However, in the ELISPOT setting, the comparisons are not independent as all experimental conditions are compared to the same control wells and responses to antigens may not be independent due to cross-reactivity across antigens. Therefore, the Bonferroni correction will be quite conservative. Many approaches to handle the problem of multiple comparisons have been developed both in the independent and dependent settings [15–17]. It is advisable to use one of these approaches when many antigens will be tested for response so as to appropriately control the family-wise error rate.
Several STs have been proposed in the literature for ELISPOT response determination. A commonly used method for ELISPOT response determination is the t test [18] due to the ease of computation of a p value (in Excel and other programs) and common basic knowledge of the method and how to apply it. However, the t test assumes that the sample size is large enough to assume that the test statistic follows a Student’s t distribution or that the data are normally distributed. ELISPOT data are not expected to satisfy these assumptions. Typically, triplicate wells (n = 3; sometimes even less) are analyzed for each experimental condition and the responses are count data that are not generally normally distributed. This has led others to propose using the Wilcoxon rank sum test [19] or the binomial test [13] both of which do not assume the data to be normally distributed.
Hudgens et al. [20] evaluated the t test, Wilcoxon rank sum test, exact binomial test and the Severini test (an extension of the binomial test) as they would be applied in the typical ELISPOT setting. They also propose two STs based on a bootstrap and permutation resampling approach where the data are pooled across all antigens. These tests do not assume that the data are normally distributed and hence are attractive for application to ELISPOT data. Hudgens et al. also examined several approaches for handling the problem of multiple comparisons. They perform a series of simulation studies under a variety of scenarios and examine the family-wise error rate (overall false positive rate) and the overall sensitivity (positive to at least one antigen) for each test under each condition. They showed that the permutation resampling approach with the Westfall–Young adjustment for multiple comparisons generates the desired false positive rate while remaining competitive with the other methods in terms of overall sensitivity. The authors also applied all of the statistical methods to a real data set and confirmed some of their simulation results.
Moodie et al. [21] noted that in permuting the data points across all antigens as proposed by Hudgens et al., the results for one antigen could affect the response detection for another antigen. This is particularly the case in the setting where one antigen has a strong signal and the other a weak one, the weak signal may not be detected by the permutation resampling method. Moodie et al., therefore, proposed a different method that does not pool data across all antigens when permuting, rather the permutations are done separately for each antigen with the negative control (background) wells. The authors called this method distribution free resampling (DFR). For each antigen considered, the test statistic, the difference in means, is computed for all possible permutations of the antigen and negative control well data (e.g., 84 possible test statistics with 3 experimental wells and 6 negative control wells). If the null hypothesis is true, then the spot counts in the experimental wells should resemble those in the negative control wells and permuting or shuffling the data across the experimental and negative control wells should have little effect on the test statistic. Repeated permutation/shuffling and calculation of the test statistic based on the permuted data then provides an estimate of the distribution of the test statistic under the null hypothesis that does not rely on parametric assumptions (e.g., normality). The test statistic based on the observed data is then compared to those based on the permuted data to determine how extreme the observed test statistic is compared to what might be seen if the null hypothesis was true. Westfall–Young’s step-down max T approach is used to calculate p values adjusted for the multiple comparisons. Moodie et al. then compared an ER, the permutation resampling approach and their proposed DFR(eq) method, using real and simulated data. They demonstrated that in some settings their method outperformed the permutation resampling method in terms of sensitivity in detecting responses at the antigen level.
A disadvantage of the DFR method is that it should only be applied in a setting where at least three replicates were performed for both the control and experimental conditions. In contrast, the permutation resampling method can be used to make a response determination when there are only duplicates for either the control or experimental conditions provided multiple antigens are tested.
The authors have also adapted the DFR(eq) approach described in [23] for situations in which one wants to test a stricter null hypothesis and/or control the false positive rate at a lower level, e.g., 0.01. With the DFR(eq) method, the minimum p value when comparing triplicate antigen wells to triplicate control wells will always be above 0.01. Further, when background levels are high, a larger background-corrected difference may be needed for convincing evidence of a positive response. For example, a background-corrected mean of 20 per 106 PBMCs may be less compelling when the mean background is 100 per 106 PBMCs and the experimental mean is 120 per 106 PBMCs than when the mean background is 2 and the experimental mean is 22 per 106 PBMCs. The basic approach of the method is similar to what was previously proposed but with modification to the null and alternative hypotheses. The null hypothesis is that the mean of the experimental well is less than or equal to twice the mean of the negative control wells; the alternative is that it exceeds this. The method uses a slightly different non-parametric test (bootstrap test instead of the permutation test) due to the statistical hypotheses under consideration. The data are log-transformed with negative controls first multiplied by the factor specified in the null hypothesis (e.g., twofold) to reflect the data under the null. The experimental and negative control well data are then sampled with replacement a large number of times (≥1,000) and the test statistic (difference in means) computed for each. The step-down max T adjustment is used to calculate adjusted p values to account for the multiple hypotheses tested. The selection of a twofold difference was based on investigators’ biological interest although other hypotheses can be tested in the same manner. The DFR(2x) method requires data from at least three experimental wells with at least three negative control wells or at least two experimental wells with at least four negative control wells.
In the next section, we compare the following three STs for ELISPOT response determination on real data:
t test: A one-sided t test (without assuming equal variance in both groups) comparing the spot counts in the control wells versus the experimental wells.
DFR method with a null hypothesis of equal background and experimental means proposed by Moodie et al. (DFR(eq)).
DFR method with a null hypothesis of less than or equal to twofold difference between background and experimental means proposed by Moodie (DFR(2x)).
For all three statistical rules, data that result in p values less than or equal to 0.05 were considered a positive response.
Results
In order to evaluate the performance of the methods described in the previous section, two ERs (>5 spots/100,000 PBMCs & >2-fold background, >2-fold background) and the three STs (one-sided t test, DFR(eq), and DFR(2x)) were applied to results from large data sets that were generated in three consecutive interlaboratory testing projects organized by the CIP [22, 23]; data are available upon request. In the referred studies, groups of 11, 13 and 16 laboratories (phases I, II and III, respectively) quantified the number of CD8 T cells specific for two model antigens within PBMC samples that were centrally prepared and then distributed to the participating laboratories. All participants were allowed to use their preferred ELISPOT protocol. Therefore, the data sets generated in these studies can be considered representative of results generated by a wide range of different protocols commonly applied within Europe. Each participating center was asked to test in triplicate 18 preselected donors (5 in the first phase, 8 in the second phase and 5 in the third phase) with two synthetic peptides (HLA-A*0201 restricted epitopes of CMV and Influenza) as well as PBMCs in medium alone for background determination. The donors were selected so that 21 donor/antigen combinations (6 in the first phase, 8 in the second phase and 7 in the third phase) were expected to demonstrate a positive response with the remaining 15 donor/antigen combinations not expected to demonstrate a positive response. Pretesting of potential donor samples for the proficiency panels was routinely done at two time points in two independent labs. Only samples from donors that had consistent results in all four performed experiments were finally selected for distribution to the participating centers.
Statistical test versus empirical criteria
Table 1 outlines the response detection rate for each center based on the empirical and statistical response criteria. The overall response detection rate from all 19 centers across all three phases of testing was 59% based on the first ER (>5 spots/100,000 PBMCs & >2-fold background), 74% based on the second ER (>2-fold background), 76% based on the t test, 75% based on the DFR(eq) method (equal means), and 61% based on the DFR(2x) method (>2-fold difference). Table 2 details the false positive response rate for each center based on the empirical and statistical response criteria. The overall false positive rate from all 19 centers across all three phases of testing was 3% based on the first ER (>5 spots/100,000 PBMCs & >2-fold background), 17% based on the second ER (>2-fold background), 10% based on the t test, 11% based on the DFR(eq) method (equal means), and 2% based on the DFR(2x) method (>2-fold difference).
Table 1.
Detection rates per lab based on two empirical rules and three statistical tests (CIP proficiency panel phases I–III)
| LabID | # Expected responses | Detected based on empirical rule 1 | Detected based on empirical rule 2 | Detected based on t test | Detected based on DFR(eq) test | Detected based on DFR(2x) test | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | N | % | ||
| Overall | 282 | 165 | 59 | 210 | 74 | 214 | 76 | 212 | 75 | 172 | 61 |
| 1 | 21 | 13 | 62 | 17 | 81 | 16 | 76 | 17 | 81 | 13 | 62 |
| 2 | 21 | 12 | 57 | 13 | 62 | 13 | 62 | 11 | 52 | 8 | 38 |
| 3 | 21 | 11 | 52 | 17 | 81 | 17 | 81 | 17 | 81 | 14 | 67 |
| 4 | 21 | 13 | 62 | 15 | 71 | 14 | 67 | 11 | 52 | 10 | 48 |
| 5 | 21 | 5 | 24 | 5 | 24 | 8 | 38 | 8 | 38 | 4 | 19 |
| 6 | 14 | 9 | 64 | 9 | 64 | 12 | 86 | 12 | 86 | 9 | 64 |
| 7 | 21 | 14 | 67 | 18 | 86 | 19 | 90 | 19 | 90 | 16 | 76 |
| 8 | 21 | 10 | 48 | 14 | 67 | 17 | 81 | 16 | 76 | 13 | 62 |
| 9 | 21 | 16 | 76 | 21 | 100 | 20 | 95 | 20 | 95 | 18 | 86 |
| 10 | 8 | 6 | 75 | 6 | 75 | 6 | 75 | 6 | 75 | 6 | 75 |
| 11 | 21 | 11 | 52 | 15 | 71 | 16 | 76 | 16 | 76 | 14 | 67 |
| 12 | 14 | 7 | 50 | 11 | 79 | 8 | 57 | 9 | 64 | 8 | 57 |
| 13 | 15 | 9 | 60 | 14 | 93 | 13 | 87 | 13 | 87 | 10 | 67 |
| 15 | 7 | 4 | 57 | 7 | 100 | 7 | 100 | 7 | 100 | 6 | 86 |
| 16 | 7 | 5 | 71 | 5 | 71 | 5 | 71 | 6 | 86 | 4 | 57 |
| 19 | 7 | 7 | 100 | 7 | 100 | 7 | 100 | 7 | 100 | 7 | 100 |
| 21 | 7 | 4 | 57 | 7 | 100 | 5 | 71 | 6 | 86 | 5 | 71 |
| 23 | 7 | 5 | 71 | 5 | 71 | 6 | 86 | 6 | 86 | 3 | 43 |
| 24 | 7 | 4 | 57 | 4 | 57 | 5 | 71 | 5 | 71 | 4 | 57 |
The first line reports the overall results for the whole group. The following rows report the results for the 19 individual centers that participated in the three phases of the CIP proficiency panel program. The first column indicates the laboratory IDs, the second column indicates the number of positive donor-antigen combinations (=responses) that could have been detected under optimal conditions
Table 2.
False positive rates per laboratory based on two empirical rules and three statistical tests (CIP proficiency panel phases I–III)
| LabID | # Expected non-responses | False positive based on empirical rule 1 | False positive based on empirical rule 2 | False positive based on t test | Detected based on DFR(eq) test | Detected based on DFR(2x) test | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | N | % | ||
| Overall | 196 | 5 | 3 | 33 | 17 | 20 | 10 | 21 | 11 | 4 | 2 |
| 1 | 15 | 0 | 0 | 4 | 27 | 3 | 20 | 2 | 13 | 0 | 0 |
| 2 | 15 | 1 | 7 | 1 | 7 | 1 | 7 | 0 | 0 | 0 | 0 |
| 3 | 15 | 0 | 0 | 1 | 7 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 15 | 0 | 0 | 2 | 13 | 2 | 13 | 1 | 7 | 0 | 0 |
| 5 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | 12 | 0 | 0 | 3 | 25 | 1 | 8 | 2 | 17 | 1 | 8 |
| 7 | 15 | 0 | 0 | 1 | 7 | 0 | 0 | 1 | 7 | 0 | 0 |
| 8 | 15 | 0 | 0 | 2 | 13 | 2 | 13 | 2 | 13 | 0 | 0 |
| 9 | 15 | 1 | 7 | 3 | 20 | 2 | 13 | 1 | 7 | 1 | 7 |
| 10 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 15 | 1 | 7 | 5 | 33 | 4 | 27 | 5 | 33 | 1 | 7 |
| 12 | 12 | 0 | 0 | 4 | 33 | 2 | 17 | 3 | 25 | 1 | 8 |
| 13 | 11 | 0 | 0 | 3 | 27 | 0 | 0 | 1 | 9 | 0 | 0 |
| 15 | 3 | 0 | 0 | 1 | 33 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 21 | 3 | 0 | 0 | 1 | 33 | 1 | 33 | 1 | 33 | 0 | 0 |
| 23 | 3 | 2 | 67 | 2 | 67 | 2 | 67 | 2 | 67 | 0 | 0 |
| 24 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
The first line reports the overall results for the whole group. The following rows report the results for the 19 individual centers that participated in the three phases of the CIP proficiency panel program. The first column indicates the laboratory IDs, the second column indicates the number of negative donor-antigen combinations (=negative control donors)
The first ER yielded response detection rates that were lower than those derived from the t test, the DFR(eq) method and the second ER (>2-fold background). However, the false positive rates with the first ER were similar to the false positive rate found for DFR(2x), lower than the false positive rates with the t test or DFR(eq) method and much lower than the false positive rate of the second ER. The DFR(eq) method had similar response detection rates as the t test—only in 17 of 478 comparisons did the conclusion of the STs differ. The DFR(eq) method with a null hypothesis of equal means had higher detection rates compared to the DFR(2x) method where the null hypothesis was less than or equal to a twofold difference of the experimental counts over the background. However, the DFR(eq) also resulted in a higher false-positive rate than the DFR(2x) method.
There were 478 comparisons made: 282 donor/antigen combinations versus control expected to demonstrate a positive response and 196 donor/antigen combinations versus control expected not to demonstrate a positive response. There were 20 instances where a response designation was not possible with both the DFR methods due to some laboratories having only performed duplicates for a control or experimental condition. Comparing the DFR(eq) response determination rule to the first ER, there was disagreement for 76 of the 478 comparisons; for 74 comparisons, the DFR(eq) test declared the triplicate a positive response while the ER did not while for two comparisons the reverse was true. Comparing the DFR(eq) response determination to the second ER (>2-fold background), there were 50 disagreements: 25 times the DFR(eq) test declared the triplicate a positive response while the ER did not and 25 times the ER declared the triplicate a positive response while the DFR(eq) test did not. Comparing the DFR(2x) response determination rule to the first ER (>5 spots/100,000 PBMCs & >2-fold background), there was disagreement for 43 of the 478 comparisons; for 29 comparisons, the DFR(2x) test declared the triplicate a positive response while the ER did not while for 14 comparisons the reverse was true. Comparing the DFR(2x) response determination to the second ER (>2-fold background), there were 58 disagreements: the ER declared the triplicate a positive response while the DFR(2x) test did not.
This led us to investigate under what conditions the ST differs in response determination from the ER and under what conditions the two statistical DFR tests differ.
Simulation study to compare response determination with STs and ERs
A simulation study was conducted to assess under what conditions a ST would differ in response determination from an ER (Supplementary Figures 1a and 1b). One thousand hypothetical donors with triplicate wells for background and experimental conditions were generated. Spot count data were randomly generated by assuming that the counts follow a Poisson distribution. The mean spot count for the background wells was set at 10 per 100,000 PBMCs, reflective of the mean in our example data set. The mean spot count for the experimental wells was varied over 40 values from a mean of 10 to 50 per 100,000 PBMCs. The signal-to-noise ratio for each experimental condition was calculated as the mean of the triplicate in the experimental well divided by the mean of the triplicate in the background well for a given donor. A signal-to-noise ratio greater than two would be considered a positive response based on the first ER. A one-sided t test was also performed comparing each experimental condition to its corresponding background. The intra-replicate variation was calculated as the sample variance of the triplicate/(median of the triplicate + 1). The reason for expressing the variability in this way was to normalize the variation so as to make it comparable across replicates with large differences in their spot counts. In the setting where there is a large outlier in one of the experimental wells compared to the other two wells, e.g., 50, 2, 6 spots, the median reflects the central tendency of the data but, unlike the mean, is not influenced by the outlier (i.e. 50 spots). Hence, we consider the ratio of the variance to median to identify cases that have large variability in the experimental well replicates but have a small median. Since the median response may in some cases be 0 spots, a 1 is added to the denominator to avoid division by 0. The response determination based on the empirical (>2 signal-to-noise ratio) and the statistical rule (one-sided t test p value ≤ 0.05) is the same for most of the experimental triplicates (Supplemental Figures 1a and 1b). However, when the intra-replicate variation is large, the ER would sometimes consider the triplicate a response while the ST would not. Conversely, when the intra-replicate variation was small, the ST would sometimes consider the triplicate a response while the ER would not. This simulation clearly showed that ERs should only be applied in settings where the variation within replicates is known and can be reliably consistent across experiments. It also demonstrates that STs account for the variation within reported triplicates. Conversely, the ST may not declare a large signal-to-noise ratio a positive response if there is very high variability between replicates. This may indicate that the declaration of a positive response requires more compelling evidence for that sample.
Simulation study to compare response determination with DFR(eq) and DFR(2x) statistical methods
A simulation study was conducted to evaluate the overall false positive rate and the overall true positive rate (sensitivity) of each DFR method under a variety of conditions. An overall positive response is declared if at least one antigen is declared positive. To calculate the overall false positive rate, background and experimental spot counts for each donor were generated under the same model. Hence, for these donors, no response should be detected. Five thousand donors with triplicate wells for background and experimental conditions were generated. Spot count data were randomly generated by assuming that the counts follow a Poisson distribution. The mean spot count for the background and experimental (i.e., antigen-containing) wells was 2, 5, 10, 15, 20, 25, 30, 35, 40, 45, or 50 (per 100,000 PBMCs). This was examined in the setting when testing with two or ten antigen preparations (k = 2, 10). To assess the overall true positive rate, background spot counts for each donor were again generated from a Poisson distribution with background mean spot counts of 2, 5, 10, 15, 20, 25, 30, 35, 40, 45, 50; however, the experimental means were shifted by 6 (small difference), 20 (moderate difference), or 50 (large difference) relative to the background means. All other conditions were the same as in the simulations for assessing the overall false positive rate.
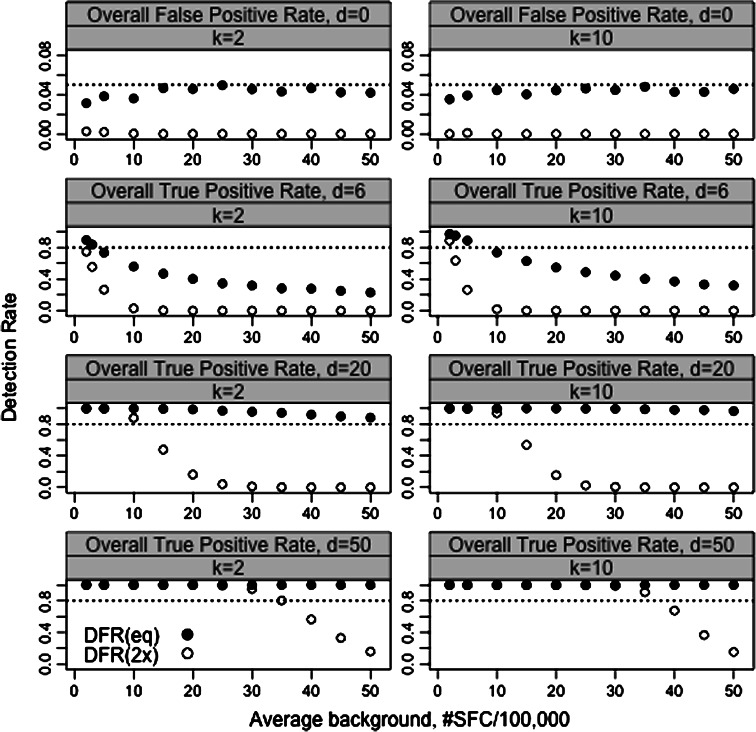
Figure 1 illustrates the response detection rates versus the mean background spot counts for both the DFR(eq) method (closed circle) and the DFR(2x) method (open circle). The two graphs on the upper row display the response detection rates in the setting where the mean number of spots was the same for both the experimental and background wells (d = 0). In this setting, the response detection rates for the two methods are expected to be low. In fact, in 5,000 simulated data sets, the average response detection rates for at least one of the antigens (k = 2 or 10) were <5% with the DFR(eq) method and <1% with the DFR(2x) method across a variety of mean background and experimental spot counts (2 to 50).
Fig. 1.
Simulation study comparing response determination using DFR(eq) and DFR(2x) statistical rules. The figure displays the response detection rate on the y-axis versus the average background spot count on the x-axis (ranging from 2 to 50 spots/100,000 PBMCs) from 5,000 simulations. In the top row, the expected mean difference between the experimental and control wells is zero; hence, responses detected are false positives. The bottom three rows have an expected difference of 6, 20 and 50 spots per 100,000 PBMC over background. Solid circles indicate response detection rates obtained by DFR(eq); open circles indicate response detection rate using the DFR(2x) test. The first column shows the results for k = 2 antigens; the second column shows the results for k = 10 antigens
The graphs in the rows 2, 3 and 4 of Fig. 1 display the overall response detection rates for small (d = 6), moderate (d = 20), or large (d = 50) mean differences. The response detection rates are high (>80%) in the DFR(eq) method for the large differences in background and experimental wells (d = 20 or 50) for a wide range of background levels (2–50). However, the response detection rates for the DFR(2x) method are much lower for higher background levels. This is not surprising given that the null hypothesis for the DFR(2x) method is less than or equal to a twofold difference over the background and therefore background levels that exceed d/2 would generally fail to reject the null hypothesis and not be considered a positive response. For small differences in background and experimental wells (d = 6), response detection rates were high only for low background levels for both DFR(eq) and DFR(2x) methods although the DFR(eq) method had higher sensitivity.
These simulations suggest that the DFR(eq) method can be used in situations where a 5% false positive rate is acceptable and an experimental mean larger than background implies a positive response regardless of the level of that background. The DFR(2x) method is appropriate in settings where one wants to control the false positive rate at a lower level, e.g., 1%, or when a fold difference in the means of experimental versus control well is more of interest than inequality of means in determining positivity, e.g., when high background is present.
Intra-replicate variation
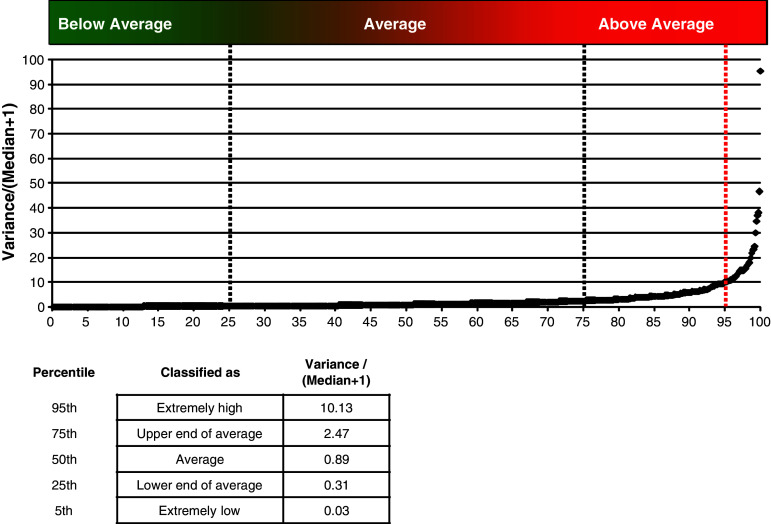
Even if a ST declares a positive response, it does not automatically imply that this result is biologically meaningful. When the spot counts found in the replicates of an experimental condition are highly variable, the experimental results are suspect and therefore response detection results for these replicates would not be believable even when declared statistically significant. However, ‘highly variable’ is a subjective term that may differ from laboratory to laboratory. We sought to quantify the typical range of intra-replicate variation found across a broad variety of different ELISPOT protocols in order to determine a variability cutoff for recommending that those replicates should be re-run. Data from the three CIP proficiency panel phases were used to analyze the intra-replicate variability of experimental results in ELISPOT assays. Nineteen different laboratories participated in at least one of the three phases and they reported a total of 717 triplicate experiments (this includes control and experimental wells). The intra-replicate variation was calculated as the sample variance of the replicates/(median of the replicates + 1) as explained in “Simulation study to compare response determination with STs and ERs”.
Figure 2 displays the intra-replicate variation of all 717 experiments reported on the vertical axis with its corresponding rank (percentile) plotted on the horizontal axis. The minimum intra-replicate variation was zero and the maximum was 95.4 with the 25th and 75th percentiles (the middle 50% of reported results) between 0.31 and 2.47 (Fig. 2, inserted table). To determine a filter for results that have ‘very high’ variability, we looked at the variance value at the 95th percentile, 10.13. Based on this finding, we would recommend that triplicates with variability greater than 10 should be considered unreliable data. Supplementary Table 1a shows the number of replicates with extremely high variation for each of the 19 participating laboratories. In depth analysis of the 36 replicates above the 95th percentile revealed that 7 of the 19 laboratories reported 3 or more triplicates with very large variation for a total of 28 replicates. The remaining eight highly variable replicates were reported by six laboratories, implying that replicates with extremely high variation do not occur randomly across all participating laboratories but rather accumulate in a few centers.
Fig. 2.
Variation of triplicates expressed as variance/(median + 1). Summary of variation found for 717 replicates that have been analyzed during three phases of the CIP ELISPOT proficiency panel program. All results were ordered in ascending order. The x-axis shows the percentile rank and the y-axis indicates the variance/(median + 1). Percentile ranks 5, 25, 50, 75 and 95 are indicated in the inserted table
Revisiting the data from the three phases (summarized in Tables 1 and 2), there were only 7 experimental replicates in the 282 positive donor/antigen combinations that had a large variability (>10). Removing these replicates with large variability, the response rate was 59% (n = 161/275) for the first ER, 75% (n = 206/275) for the second ER, 77% (n = 211/275) for the t test, 76% (n = 208/275) for the DFR(eq) ST, and 61% (n = 169/275) for the DFR(2x) ST. There were 14 experimental replicates in the 196 donor/antigen combinations not expected to demonstrate a positive response that had large variability. Removing the replicates with large variability, the false positive rate was 2% (n = 3/182) for the first ER, 17% (n = 31/182) for the second ER, 10% (n = 20/182) for the t test, 10% (n = 19/182) for the DFR(eq) ST, and 2% (n = 3/182) for the DFR(2x) ST. Hence, the response detection rates did not change after removing the replicates with large variability. This is not surprising due to the small number of replicates with large variability that were removed from the total data set.
Estimation of the limit of detection in ELISPOT assays
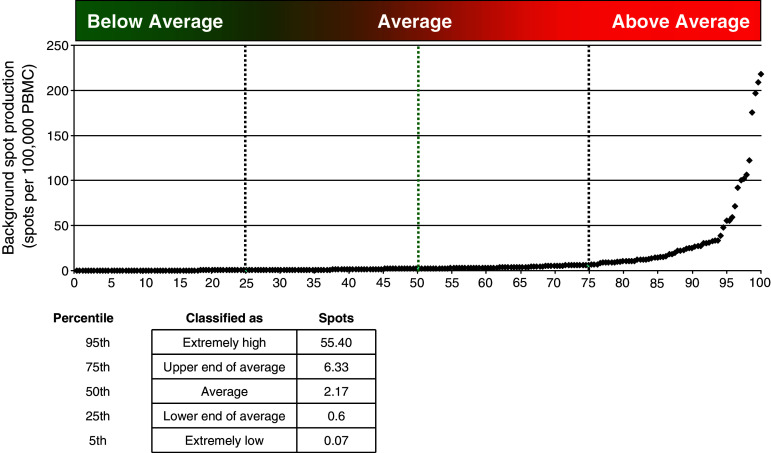
A second factor to consider when deciding on the relevance of a positive response is the limit of detection of the ELISPOT assay. The international conference on harmonization of technical requirements for registration of pharmaceuticals for human use (ICH) produced a guideline on the validation of analytical procedures (http://www.ich.org/LOB/media/MEDIA417.pdf). In this guideline (named Q2R1), the limit of detection is defined as the lowest amount of analyte in a sample which can be detected but not necessarily quantified as an exact value. The guideline describes three approaches to estimate the limit of detection for an analytical test: visual evaluation, signal-to-noise, and response based on standard deviation and slope. Visual evaluation and response based on standard deviation and slope are not applicable to the ELISPOT setting. The signal-to-noise approach compares spot counts in the experimental wells (signal) to spot counts from the medium control wells (noise). A signal-to-noise ratio between 2:1 and 3:1 is generally considered acceptable for estimating the detection limit. We applied this guideline to estimate the limit of detection of the ELISPOT assay for a broad range of protocols.
There were 239 triplicate medium control experiments reported from all three CIP proficiency panel phases. The mean of these triplicates ranged from 0 to 218 spots per 100,000 PBMCs. The median of the triplicate background means was 2.1 spots/100,000 PBMCs with the 25th and 75th percentiles, 0.6 spots/100,000 PBMCs and 6.5 spots/100,000 PBMCs, respectively. This is illustrated in Fig. 3 where the mean medium spot count for all reported replicates is plotted on the vertical axis with its corresponding rank displayed on the horizontal axis. Using an acceptable signal-to-noise ratio of 2:1 or 3:1 and choosing as the noise the median of the average background spot counts (50th percentile in Fig. 3), we estimate a typical detection limit for the ELISPOT assay to be 4 spots/100,000 PBMCs or 6 spots/100,000 PBMCs, respectively. For a heterogeneous group of laboratories that participate in a proficiency panel program, we recommend to use a threshold of 6 spots per 100,000 PBMCs (a signal-to-noise ratio of 3:1) as the typical limit of detection for an ELISPOT assay. Hence, we would recommend that even if the results of the ST lead to the rejection of the null hypothesis, if the mean of the experimental wells is less than 6 spots/100,000 PBMCs this finding should regarded with caution since it is likely that it is at the limit of detection of the ELISPOT assay, at least for laboratories with similar average performance as those included in our proficiency panel program.
Fig. 3.
Background spots production per 100,000 PBMCs. Estimation of the limit of detection based on 239 reported replicates from three phases of the CIP ELISPOT proficiency panel program. All results were ordered in ascending order. The x-axis shows the percentile rank and the y-axis indicates the reported mean spot number. Percentile ranks 5, 25, 50, 75 and 95 are indicated in the inserted table. Values between the 25th and 75th percentile were considered as being average for a typical ELISPOT protocol
This limit of detection is close to the threshold selected for the first ER (5 spots/100,000 PBMCs) and would provide further justification for applying a threshold in the ER. However, it is important to note that the limit of detection is based on the average background from all the laboratories. This means that laboratories with lower background spot counts than the average of the panel will likely have a limit of detection that is lower than 6 spots/100,000 PBMCs. Similarly, laboratories with larger background spot counts than the average of the panel will likely have a limit of detection that is higher than 6 spots/100,000 PBMCs. Therefore, the threshold or limit of detection might be too strict for some laboratories in declaring a response positive and not strict enough for others. This is clearly illustrated in supplementary Table 1B that shows the mean number of spots 100,000 PBMCs in the medium control as reported by each of the 19 participating laboratories. The mean background spot production observed in individual laboratories across all tested donors differed significantly between the participating laboratories and could be very low (0.2 spots per 100,000 PBMCs) to very high (58.1 spots per 100,000 PBMCs).
Discussion
The goal of this paper was to describe and compare objective methods that distinguish between positive and negative responses in the ELISPOT assay and to facilitate their widespread application. Two approaches, empirical and statistical, were described and applied to large data sets obtained from several proficiency panels including many laboratories operating with their own ELISPOT reagents and protocols. Simulation studies were also conducted to compare the empirical and statistical approaches. The first ER yielded lower response detection rates but had very low false positive rates in contrast to the STs and the second ER which had higher response rates but also had a larger number of false positives. The main advantage of an ER such as the one proposed by Dubey et al. is that it is generally intuitive and easy to apply. The main drawback is that it is not clear how the false positive rate is being controlled when multiple antigens are considered, as is common practice. To appropriately justify the thresholds selected for the ER, a laboratory would need to determine the false positive rates of various thresholds for a given protocol. This would require testing a large number of samples of negative controls and positive donors, which would be costly and time consuming. Additionally, the variability of the replicates is not taken into account in the determination of the rule when the average of the replicates is used. Therefore, as demonstrated in the first simulation study (Supplementary Figures 1a and 1b), if one of the replicate values is much larger than the others in that replicate, the resulting average would be large and might cause an incorrect classification of response (false positive). Also the reverse can occur where there is low variability in replicates but the average is just below the specified positivity threshold value. In this setting, a response may be missed (false negative). Furthermore, there is no formal way to adjust for multiple antigen comparisons and so the underlying false positive rate is unknown even if the thresholds for the rule were set based on a specific false positive rate.
The main advantage of a ST is that it can be applied without prior knowledge of the performance criteria of the test, provided the assumptions of the ST are valid. In addition, STs allow control of the false positive rate by the setting of the threshold for acceptance (alpha). As the t test is not necessarily appropriate due to the required assumption of a parametric distribution of the test statistic, we favor the use of non-parametric tests. Both DFR methods (DFR(eq) and DFR(2x)) control the overall false positive rate when testing multiple antigens and avoid parametric assumptions about the data. These methods can now be readily implemented in freely available software or via a web interface (http://www.scharp.org/zoe/runDFR/).
Response determination is made based on a comparison of the medium control and experimental wells. The background values for spot production in ELISPOT can differ between several donors and between different time points for one and the same donor within the observation period. Any increase in the background spot production will directly impact on the sensitivity of the method as the acceptance of a result as a positive response is directly linked to the background. To increase the power of the test, it would be ideal to have more replicate wells for both control and experimental conditions. Even an increased number of replicates for the control wells alone would already increase the power to detect a response [20, 21]. This is particularly true in the setting where many antigens are tested since the control wells are used multiple times for comparison (once with each antigen). Intuitively, this makes sense because with more control wells one can be more certain about the underlying background which is being used as the basis for comparison to the antigen responses. In the setting where duplicate or triplicate experimental wells will be performed, Hudgens et al. demonstrate that having six background wells increased the sensitivity of the test from 0.61 to 0.75 for a (2,2) versus a (6,2) format. To increase the power to detect differences between spot counts in experimental versus medium control wells, we therefore recommend using six medium control wells for each sample, whenever possible. Alternatively, utilizing four wells for both, medium control and experimental conditions, would give similar power to detect differences [21].
In addition to applying a ST, we further recommend that replicates with variability greater than 10, defined as the sample variance of the replicates/(median of the replicates + 1), should be excluded and/or re-run prior to response determination as replicates with such high variation are likely to be artefacts and should not be considered reliable to use for response determination (Fig. 2). In the setting where the responses are expected to be large, a less strict variability cutoff can be used. For example, the HIV Vaccine Trials Network uses a variability cutoff of 25 [21] based on their laboratory experience.
Our data also suggest that experimental replicates demonstrated to be a response by one of the response detection rules but having a mean below 6 spots per 100,000 PBMCs should be viewed with caution (Fig. 3). Depending on the specific study design, some investigators might have valid reasons for introducing a threshold spot count below which results are not considered of interest.
Both the filters for large variability and estimated limit of detection are based on data generated by a variety of representative ELISPOT protocols. An individual laboratory may have different thresholds that are applicable only to them. Laboratories that have very low intra-replicate variability may set a lower threshold for filtering out replicates that are experimentally problematic. Laboratories that consistently observe absent or minimal background spot production may assume a lower limit of detection than the one proposed in this paper. Similarly, individual laboratories that regularly observe background spot production above 2 spots per 100,000 PBMC, which is very common (Supplemental Table 1), should consider limits of detection that are above our estimated value. In all instances, it is important that investigators critically review their own test performance to determine their laboratory-specific estimates for high variability and limit of detection. Any threshold proposed should be justified based on experimental results.
Another strategy to increase the validity of generated data sets could be the regular use of positive control replicate which may either (a) consist of a non-specific positive control stimulus that is added to a donor’s PBMCs or (b) PBMC from a control donor known to be reactive against a given antigen. Provided the positive control replicate does not pass the positivity call of the applied response determination rule or statistical test, the results from the corresponding ELISPOT plate should be regarded with caution as they could contain false negative results.
In summary, both ERs and STs may serve as appropriate response definition criteria; however, ERs need to be justified using data sets from control populations and are only valid for the test protocol used to define the thresholds for acceptance. In contrast, STs maintain validity independent of the test protocol applied provided the assumptions of the statistical test are met. As such we would recommend that a non-parametric ST should be used to determine if a response is detected. To increase the sensitivity in detecting a response using a ST, six replicates (instead of only three) should be performed for the medium control wells or four wells each for both medium control and experimental conditions. Further, to factor in biological plausibility and relevance, we recommend filtering out replicates with high variation and viewing with caution experimental replicates below the estimated limit of detection. The DFR(eq) method would be preferred in the setting where a 5% false positive rate is acceptable and it is of interest to detect even low to moderate positive responses regardless of the level of that background. The DFR(2x) method is appropriate in settings where one wants to control the false positive rate at a lower level, e.g., 1%, or when a fold difference in the means of experimental versus control wells is more of interest than inequality of means in determining positivity.
To enable broad use of the recommended non-parametric STs described in this paper, a web-based interface was created: http://www.scharp.org/zoe/runDFR/. Instructions are provided in the electronic supplemental material. The original R code is available for download and a sample scenario for illustrative purposes is provided on the website. This tool provides two methods for objective response determination that can be easily implemented in any lab, potentially leading to greater comparability of results across institutions. In addition to the web-based tool, we developed an Excel macro for user-friendly implementation of the DFR method for investigators preferring the use of Excel. This macro will be made freely available upon request.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors gratefully acknowledge the help of Craig Magaret in helping to create the web interface and Shane Coultas for website development. C.M.B., C.O., S.H.v.d.B. and C.G. received funding from a combined research grant of the Wallace Coulter Foundation (Florida, USA). C.O. and AM are supported by the Experimental Cancer Medicine Centre, Southampton.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Z. Moodie and L. Price contributed equally to this work.
References
- 1.Britten CM, Janetzki S, van der Burg SH, Gouttefangeas C, Hoos A. Toward the harmonization of immune monitoring in clinical trials: Quo vadis? Cancer Immunol Immunother. 2007;57:285–288. doi: 10.1007/s00262-007-0379-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butterfield LH, Disis ML, Fox BA, Lee PP, Khleif SN, Thurin M, Trinchieri G, Wang E, Wigginton J, Chaussabel D, Coukos G, Dhodapkar M, Hakansson L, Janetzki S, Kleen TO, Kirkwood JM, Maccalli C, Maecker H, Maio M, Malyguine A, Masucci G, Palucka AK, Potter DM, Ribas A, Rivoltini L, Schendel D, Seliger B, Selvan S, Slingluff CL, Jr, Stroncek DF, Streicher H, Wu X, Zeskind B, Zhao Y, Zocca MB, Zwierzina H, Marincola FM. A systematic approach to biomarker discovery; preamble to “the iSBTc-FDA taskforce on immunotherapy biomarkers”. J Transl Med. 2008;6:81. doi: 10.1186/1479-5876-6-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janetzki S, Panageas KS, Ben-Porat L, Boyer J, Britten CM, Clay TM, Kalos M, Maecker HT, Romero P, Yuan J, Kast WM, Hoos A. Results and harmonization guidelines from two large-scale international Elispot proficiency panels conducted by the Cancer Vaccine Consortium (CVC/SVI) Cancer Immunol Immunother. 2008;57:303–315. doi: 10.1007/s00262-007-0380-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janetzki S, Britten CM, Kalos M, Levitsky HI, Maecker HT, Melief CJ, Old LJ, Romero P, Hoos A, Davis MM. “MIATA”-minimal information about T cell assays. Immunity. 2009;31:527–528. doi: 10.1016/j.immuni.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanekom WA, Dockrell HM, Ottenhoff TH, Doherty TM, Fletcher H, McShane H, Weichold FF, Hoft DF, Parida SK, Fruth UJ. Immunological outcomes of new tuberculosis vaccine trials: WHO panel recommendations. PLoS Med. 2008;5:e145. doi: 10.1371/journal.pmed.0050145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith SG, Joosten SA, Verscheure V, Pathan AA, McShane H, Ottenhoff TH, Dockrell HM, Mascart F. Identification of major factors influencing ELISpot-based monitoring of cellular responses to antigens from Mycobacterium tuberculosis . PLoS One. 2009;4:e7972. doi: 10.1371/journal.pone.0007972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelley M (2008) Considerations while setting up cell-based assays. Validation of cell based assays in the GLP setting. A practical guide, chapter 1. Wiley, Chichester, pp 1–9
- 8.Mander A, Chowdhury F, Low L, Ottensmeier CH. Fit for purpose? A case study: validation of immunological endpoint assays for the detection of cellular and humoral responses to anti-tumour DNA fusion vaccines. Cancer Immunol Immunother. 2009;58:789–800. doi: 10.1007/s00262-008-0633-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maecker HT, Hassler J, Payne JK, Summers A, Comatas K, Ghanayem M, Morse MA, Clay TM, Lyerly HK, Bhatia S, Ghanekar SA, Maino VC, Delarosa C, Disis ML. Precision and linearity targets for validation of an IFNgamma ELISPOT, cytokine flow cytometry, and tetramer assay using CMV peptides. BMC Immunol. 2008;9:1–9. doi: 10.1186/1471-2172-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Burg SH. Therapeutic vaccines in cancer: moving from immunomonitoring to immunoguiding. Expert Rev Vaccines. 2008;7:1–5. doi: 10.1586/14760584.7.1.1. [DOI] [PubMed] [Google Scholar]
- 11.Lewis JJ, Janetzki S, Schaed S, Panageas KS, Wang S, Williams L, Meyers M, Butterworth L, Livingston PO, Chapman PB, Houghton AN. Evaluation of CD8(+) T-cell frequencies by the Elispot assay in healthy individuals and in patients with metastatic melanoma immunized with tyrosinase peptide. Int J Cancer. 2000;87:391–398. doi: 10.1002/1097-0215(20000801)87:3<391::AID-IJC13>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 12.Janetzki S, Cox JH, Oden N, Ferrari G. Standardization and validation issues of the ELISPOT assay. Methods Mol Biol. 2005;302:51–86. doi: 10.1385/1-59259-903-6:051. [DOI] [PubMed] [Google Scholar]
- 13.Cox JH, Ferrari G, Kalams SA, Lopaczynski W, Oden N, D’souza MP. Results of an ELISPOT proficiency panel conducted in 11 laboratories participating in international human immunodeficiency virus type 1 vaccine trials. AIDS Res Hum Retroviruses. 2005;21:68–81. doi: 10.1089/aid.2005.21.68. [DOI] [PubMed] [Google Scholar]
- 14.Dubey S, Clair J, Fu TM, Guan L, Long R, Mogg R, Anderson K, Collins KB, Gaunt C, Fernandez VR, Zhu L, Kierstead L, Thaler S, Gupta SB, Straus W, Mehrotra D, Tobery TW, Casimiro DR, Shiver JW. Detection of HIV vaccine-induced cell-mediated immunity in HIV-seronegative clinical trial participants using an optimized and validated enzyme-linked immunospot assay. J Acquir Immune Defic Syndr. 2007;45:20–27. doi: 10.1097/QAI.0b013e3180377b5b. [DOI] [PubMed] [Google Scholar]
- 15.Hsu JC. Multiple comparisons. Theory and methods. London: Chapman and Hall; 1996. [Google Scholar]
- 16.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 17.Westfall PH, Young SS. Resampling-based multiple testing: examples and methods for P-value adjustment. New York: Wiley; 1993. [Google Scholar]
- 18.Herr W, Protzer U, Lohse AW, Gerken G, Meyer zum Buschenfelde KH, Wolfel T. Quantification of CD8+ T lymphocytes responsive to human immunodeficiency virus (HIV) peptide antigens in HIV-infected patients and seronegative persons at high risk for recent HIV exposure. J Infect Dis. 1998;178:260–265. doi: 10.1086/517449. [DOI] [PubMed] [Google Scholar]
- 19.McCutcheon M, Wehner N, Wensky A, Kushner M, Doan S, Hsiao L, Calabresi P, Ha T, Tran TV, Tate KM, Winkelhake J, Spack EG. A sensitive ELISPOT assay to detect low-frequency human T lymphocytes. J Immunol Methods. 1997;210:149–166. doi: 10.1016/S0022-1759(97)00182-8. [DOI] [PubMed] [Google Scholar]
- 20.Hudgens MG, Self SG, Chiu YL, Russell ND, Horton H, McElrath MJ. Statistical considerations for the design and analysis of the ELISpot assay in HIV-1 vaccine trials. J Immunol Methods. 2004;288:19–34. doi: 10.1016/j.jim.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 21.Moodie Z, Huang Y, Gu L, Hural J, Self SG. Statistical positivity criteria for the analysis of ELISpot assay data in HIV-1 vaccine trials. J Immunol Methods. 2006;315:121–132. doi: 10.1016/j.jim.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 22.Britten CM, Gouttefangeas C, Welters MJ, Pawelec G, Koch S, Ottensmeier C, Mander A, Walter S, Paschen A, Muller-Berghaus J, Haas I, Mackensen A, Kollgaard T, Thor SP, Schmitt M, Giannopoulos K, Maier R, Veelken H, Bertinetti C, Konur A, Huber C, Stevanovic S, Wolfel T, Burg SH. The CIMT-monitoring panel: a two-step approach to harmonize the enumeration of antigen-specific CD8+ T lymphocytes by structural and functional assays. Cancer Immunol Immunother. 2008;57:289–302. doi: 10.1007/s00262-007-0378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mander A, Gouttefangeas C, Ottensmeier C, Welters MJ, Low L, van der Burg SH, Britten CM. Serum is not required for ex vivo IFN-gamma ELISPOT: a collaborative study of different protocols from the European CIMT Immunoguiding Program. Cancer Immunol Immunother. 2010;59:619–627. doi: 10.1007/s00262-009-0814-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.