Abstract
The variability of radiation responses in ovarian tumors and tumor-derived cell lines is poorly understood. Since both DNA repair capacity and p53 status can significantly alter radiation sensitivity, we evaluated these factors along with radiation sensitivity in a panel of sporadic human ovarian carcinoma cell lines. We observed a gradation of radiation sensitivity among these sixteen lines, with a five-fold difference in the LD50 between the most radiosensitive and the most radioresistant cells. The DNA-dependent protein kinase (DNA-PK) is essential for the repair of radiation induced DNA double-strand breaks in human somatic cells. Therefore, we measured gene copy number, expression levels, protein abundance, genomic copy and kinase activity for DNA-PK in all of our cell lines. While there were detectable differences in DNA-PK between the cell lines, there was no clear correlation with any of these differences and radiation sensitivity. In contrast, p53 function as determined by two independent methods, correlated well with radiation sensitivity, indicating p53 mutant ovarian cancer cells are typically radioresistant relative to p53 wild-type lines. These data suggest that the activity of regulatory molecules such as p53 may be better indicators of radiation sensitivity than DNA repair enzymes such as DNA-PK in ovarian cancer.
Keywords: ovarian cancer, non-homologous end joining, p53, p21, DNA-PK, DNA-PKcs, Ku, Ku70, Ku80, XRCC4, Ligase IV, radioresistance, radiosensitivity, apoptosis, double-strand break repair, microarray, comparative genomic hybridization
Introduction
Epithelial ovarian cancer (EOC) remains the leading cause of death from gynecological malignancy among women in the USA (1). Since there are no methods for early detection of ovarian cancer, late detection is common with invasion of pelvic organs and the surrounding abdomen and peritoneal cavity. A large number of chemotherapeutic agents including platinum compounds, alkylants and taxane derivatives have been used to treat EOC. Platinum-based combination chemotherapy with either cisplatin or carboplatin and paclitaxel is the standard treatment of care for ovarian cancer. It is estimated that 20–30% of EOCs are resistant to platinum-based chemotherapy before treatment begins (2,3). It is also known that a substantial number of tumors, which were originally responsive to platinum-based chemotherapy develop resistance over the course of their treatment. Therefore, there is an active interest in whole abdominal radiotherapy as one component of a multimodal treatment program to increase therapeutic efficacy. Determination of the optimal radiation dose has been particularly problematic in the advanced stages of ovarian cancer as well as other cancers. Predictive markers for tumor response to radiation and chemical therapeutics would be of significant clinical value. Both DNA stress response and DNA repair pathways can directly affect radiosensitivity/ resistance and therefore make attractive candidates for predicting tumor response to radiation treatment (4).
Mutations in p53 are one of the most commonly described genetic abnormalities in ovarian cancer and is inactivated in over half of all advanced ovarian cancers (5). Seminal studies revealed a significant correlation between p53 status and aneuploidy in advanced stage disease (6). Studies have revealed that p53 status (measured by p53 protein overexpression) correlated with favorable response to taxane-platinum based therapy (7). Therefore, loss of p53 function and ablation of the DNA-damage induced p53-dependent apoptosis pathway is a strong predictor of taxane-platinum-based therapy.
The non-homologous end joining (NHEJ) repair pathway is the major pathway responsible for the repair of radiation-induced DNA double-strand breaks (DSBs) in mammalian cells. Mutations which inactivate enzymes involved in the NHEJ pathway result in severely compromised DSB repair and concomitant X-ray sensitivity. The non-homologous end-joining machinery consists minimally of the DNA-dependent protein kinase (DNA-PK) consisting of Ku70, Ku80 and the DNA-PK catalytic subunit (DNA-PKcs) and the hetero-multimeric XRCC4/Ligase IV (8). Positive correlations between Ku levels and radio- or chemo-resistance in esophageal tumors, advanced stage head and neck tumors, rectal cancer and cervical cancer have been documented (9–12). However, DNA-PK activity was measured in a panel of sporadic breast cancer cell lines and no major differences were detected (13). Thus, some relationship may exist between the NHEJ pathway and radiosensitivity/resistance in malignancies; however, this relationship does not appear to be universal and may be cell/tumor-type specific. Henkels et al first studied the relationship of DNA-PK and platinum resistance in ovarian cell lines which suggested to us that DNA-PK levels might also dictate sensitivity/resistance to X-ray treatment (14). Since the NHEJ pathway was able to help predict the response of radiation and chemotherapeutic treatments in many different cancer types, we hypothesized that the NHEJ pathway may help predict relative radiosensitivity of sporadic ovarian cancer.
To determine the relative predictive value of DSB repair (DNA-PK) and DNA damage signaling pathways (p53) for radiosensitivity of ovarian tumors, we evaluated key members of these two distinct pathways in a panel of ovarian cancer cell lines. We first determined the X-ray sensitivity profiles of sixteen human ovarian cell lines by performing colony-formation assays. Using array-based comparative genomic hybridization (CGH), we determined if genomic regions encoding DNA-PK catalytic subunit (cs) or if Ku70/80 genes were amplified or deleted in the ovarian carcinoma cell lines. Gene expression analysis by Affymetrix GeneChip® microarray was used to measure relative message levels of DNA-PK components. Our panel of cell lines was also analyzed for DNA-PKcs, Ku70 and p53 protein levels. DNA-PK-specific activity from each of the cell lines were measured by an in vitro kinase assay. Lastly, we screened genomic DNA isolated from each of the ovarian cell lines for exon-specific mutations in p53. Our results demonstrate there is a range of sensitivity/resistance among the sixteen ovarian cell lines and that p53 can act as a general predictor of relative radiosensitivity for most of the ovarian cancer cell lines tested confirming previous observations; however, we found no strong correlations between radiosensitivity and DNA-PK. These cell lines and others will be used in future studies to help identify novel chemotherapeutic targets for advanced stage ovarian cancer.
Materials and methods
Cell culture
Ovarian cancer cell lines were a generous gift from Dr Joe Gray. All ovarian cancer cell lines were cultured in 10% fetal bovine serum, with the media indicated in Table I with antibiotics/antimycotics at 37°C and 5% CO2. All cell culture media and related products were purchased from Invitrogen, Carlsbad, CA or the UCSF Cell Culture Facility (San Francisco, CA).
Table I.
Description of ovarian cancer cell lines used in this study.
| Cell line | Media | Histotype | Source |
|---|---|---|---|
| CAOV4 | L15 | Adenocarcinoma | Metastatic site |
| CAOV3 | DMEM | Adenocarcinoma | Unknown |
| A2780 | RPMI-1640 | Ovarian carcinoma | Primary tumor |
| OVCAR8 | RPMI-1640 | Adenocarcinoma | Unknown |
| OVCAR3 | RPMI-1640 | Adenocarcinoma, poorly differentiated |
Ascites |
| IOSE29 | M199:MCDB105 | Normal surface epithelial cells |
Ovary surface |
| OVCA429 | MEM | Recurrent serous carcinoma |
Ascites |
| OVCAR5 | RPMI-1640 | Adenocarcinoma | Unknown |
| OVCA432 | MEM | Recurrent serous carcinoma |
Ascites |
| ES2 | M199:MCDB105 | Clear cell carcinoma |
Primary tumor |
| TOV112D | M199:MCDB105 | Endometroid carcinoma |
Primary tumor |
| OV90 | M199:MCDB105 | Adenocarcinoma | Ascites |
| SKOV3 | RPMI-1640 | Adenocarcinoma | Ascites |
| HEY | RPMI-1640 | Cyst adenocarcinoma |
Ascites |
| OVCA420 | MEM | Recurrent serous carcinoma |
Ascites |
X-ray exposure and colony formation (survival) assays
Cells were trypsinized and counted using a Coulter counter. Appropriate number of cells were plated in triplicate and allowed to attach overnight. Plated cells were irradiated with either 0 through 7 Gy of X-rays filtered through a 0.5-mm copper plate using a Pantek® generator operating at 320 kV/110 mA. After irradiation, cells were returned to the incubator and left undisturbed for 10 to 25 days until colonies formed. The colonies were stained with a crystal violet solution consisting of 10% paraformaldehyde, 0.05% crystal violet phosphate-buffered saline solution. Colonies were counted and an average from three dishes was used to define percent survival. LD50 was determined by the use of Graph Pad software.
Genomic DNA isolation and array CGH
Relative copy numbers of the 16 ovarian cell lines were measured with three types of CGH arrays. The Hum 2.0 arrays comprise 2465 BACs selected at approximately megabase intervals along the genome and were prepared by the UCSF Comprehensive Cancer Center Array Core. The BAC DNA targets of RTK and G950 arrays were selected, DNA samples for array CGH were labeled and after hybridization, the slides were washed and imaged using a 16-bit CCD camera through CY3, CY5, and DAPI filters. The resulting images were analyzed to determine CY3/CY5 ratios for each array element using custom software (15,16).
Gene expression analysis
An Affymetrix high throughput analysis (HTA) automated GeneChip system® was used for collection of microarray data for gene expression profiling as described previously (17). Target preparation, washing and staining were performed on an Affymetrix/Caliper robotic system, scanning was performed using a CCD-based Affymetrix high throughput scanner which is a fully automated epiflourescent imaging system. Data analysis software includes GeneSpring, Bioconductor, GeneTraffic and Cluster 3.0.
DNA-PK-kinase assays and radiation-induced p21 induction
Nuclear extracts were prepared from ovarian cancer cell lines by hypotonic lysis. DNA-PK kinase assays were performed using an assay kit according to the manufacturer's protocol (Promega, Madison, WI). Multiple kinase assays were performed and a representative assay is presented.
Radiation-induced p21 induction was measured by exposing ovarian cells to 0 or 10 Gy of X-rays and harvesting cells 24 h later. Cell lysates were resolved by 12% SDS-PAGE and transferred to nitrocellulose. The membrane was probed with an antibody specifically recognizing p21 (ab14061 Ab Cam, Cambridge, MA). A summary of those experiments is provided in Table II.
Table II.
p53 status and p21 response to X-ray exposure of sixteen ovarian cancer cell lines.
| Cell line | Call | Exon | Codon | p21 induction |
|
|---|---|---|---|---|---|
| Relative radioresistance | |||||
| TOV21G | Wildtype | n/a | n/a | +++ | |
| CAOV4 | Polymorphism- C/G (aa:P/R) |
Exon 4 | 72_2 | − | |
| CAOV3 | Polymorphism- C/G (aa:P/R) |
Exon 4 | 72_2 | +++ | |
| A2780 | Polymorphism- C/G (aa:P/R) |
Exon 4 | 72_2 | +++ | |
| OVCAR8 | A/G | Intron 4 | −1_−1 | − | |
| OVCAR3 | not determined | n/a | n/a | − | |
| IOSE29 | Wildtype | n/a | n/a | SV40 transformed |
|
| OVCA429 | A/C (aa:H/P) | Exon 5 | 151_2 | − | |
| G/T (aa:V/V) | Exon 8 | 274_3 | |||
| OVCAR5 | Wildtype | n/a | n/a | +++ | |
| OVCA432 | T/G (aa:F/C) | Exon 8 | 277_2 | − | |
| ES2 | Polymorphism- C/G (aa:P/R) |
Exon 4 | 72_2 | − | |
| T/C (aa:F/S) | Exon 7 | 241_2 | |||
| TOV112D | A/G (aa:H/R) | Exon 5 | 175_2 | − | |
| OV90 | C/A (aa:R/S) | Exon 6 | 215_1 | − | |
| SKOV3 | Polymorphism- C/G (aa:P/R) |
Exon 4 | 72_2 | − | |
| −/C | Exon 4 | 88_2–89_3 | |||
| HEY | Wildtype | n/a | n/a | − | |
| OVCA420 | Polymorphism- C/G (aa:P/R) |
Exon 4 | 72_2 | − | |
| A/G (aa:H/R) | Exon 8 | 273_2 |
Cell line, p53 mutational status and X-ray induced p21 protein determination. (−) represents no observable induction of p21, (+++) indicates p21 induction in response to X-ray exposure. n/a either no mutation was found (wild-type) or not determined for that cell line (OVCAR8).
SDS-PAGE and immunochemical techniques
Total protein cellular lysates (20 µg) were electrophoretically resolved on denaturing tris-glycine (SDS)-polyacrylamide gels (4–12%), transferred to nitrocellulose and probed with specific antisera using standard techniques. DNA-PK [18-2], Ku70/80 monoclonal antibodies were a generous gift of Dr D.J. Chen. p53 (DO-1) and β-tubulin were purchased from Santa Cruz Technologies (Santa Cruz, CA).
Roche p53 AmpliChip® analysis
The Roche p53 AmpliChip is a microarray-based resequencing chip in development and is designed for the p53 gene, co-developed by Roche and Affymetrix, for the diagnosis of mutations in the p53 coding sequence (18,19). Genomic DNAs are isolated from the ovarian cancer cells as described above for CGH, and labeled and hybridized to oligo probes on the AmpliChip, which correspond to the whole p53 gene. This chip was used for the determination of the entire sequence of the tumor suppressor gene p53 in the ovarian cancer cells. The wild-type sequence was used as a reference. Details of the labeling, hybridization, scanning and data analysis are available from the authors on request.
Results
Response of ovarian cancer cell lines to X-ray exposure
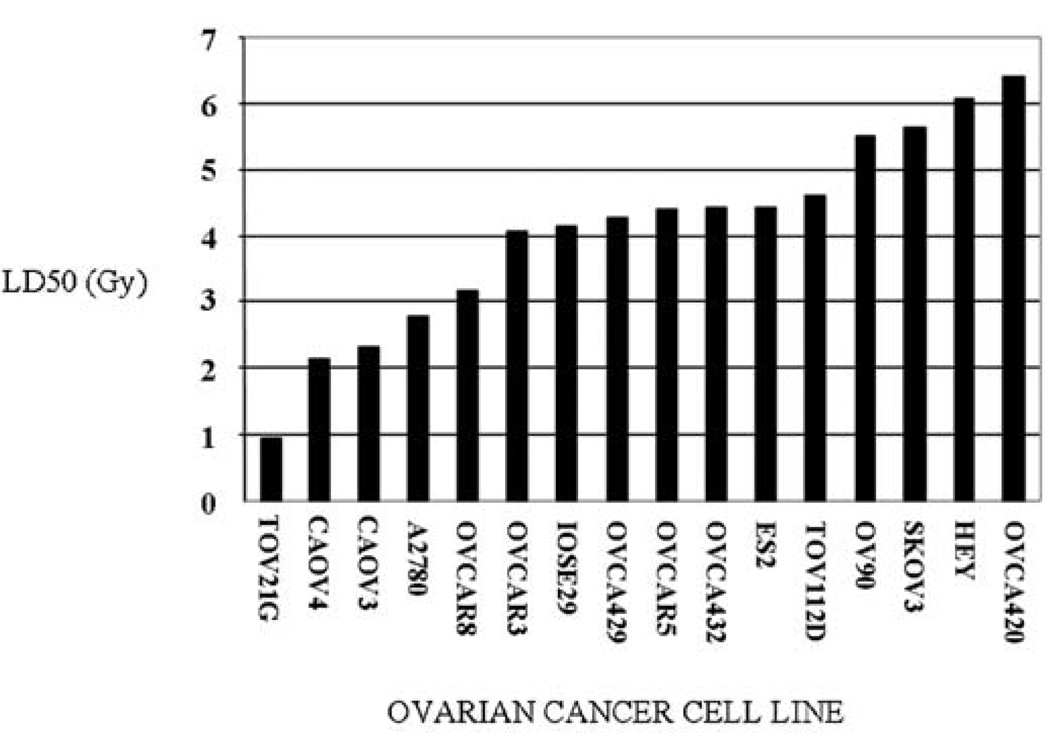
Ovarian tumors have a widely varied response to chemo- and radio-therapeutic treatments, we therefore assessed whether this was also true for our panel of 16 ovarian-derived cell lines in vitro. This specific panel of cancer cell lines was chosen because most are derived from advanced stage ovarian tumors (see Table I). Survival plots were generated from colony formation assays for each cell line at each X-ray dose and LD50 values calculated from these plots (Fig. 1). Our panel of ovarian cancer cell lines exhibit relatively smooth progression of radiation sensitivity when sorted according to their respective LD50 values (Fig. 1). It is interesting to note that there is greater than five-fold difference in radiation sensitivity across this collection of ovarian cancer cell lines.
Figure 1.
Ovarian cancer cells vary in their response to X-ray exposure. Colony formation assays were performed to measure sensitivity/resistance to increasing doses of X-ray exposure. LD50 values were determined by GraphPad analysis and plotted in Microsoft Excel. Further experimental details are presented in Materials and methods.
The TOV21G and CAOV4 lines showed the highest levels of radiation sensitivity, while HEY, and OVCA420 were the most radioresistant from the panel tested. Curiously, the control cell line IOSE29, derived from normal ovarian tissue, is intermediate in the range of radiosensitivities measured (Fig. 1). Since both DNA-PK and p53 status have been reported to correlate with radiosensitivity/resistance in various cancer lines, we evaluated these two as potential predictors for radiation treatment in our panel of ovarian cancer cell lines.
DNA-PKcs gene, message, protein and specific activity as a predictor of X-ray-sensitivity in ovarian cancer cell lines
DNA-PK is an abundant nuclear serine/threonine protein kinase consisting of a 460-kDa catalytic subunit (DNA-PKcs) and a heterodimeric DNA binding component (Ku). Cell lines and mice deficient for any DNA-PK component are extremely sensitive to agents that cause DNA double-strand breaks. Since Ku70 and DNA-PK reside on chromosomes that are frequently aberrant in ovarian cancers, CGH profiling was performed on the panel of ovarian cell lines to determine if gross amplifications or deletions in these regions correlate with relative X-ray sensitivity (Fig. 2A). CGH data of DNA-PKcs, Ku70 or Ku80 show no significant correlations with X-ray sensitivity, with Pearson correlation coefficient lower than (0.15) in all three cases. Using Affymetrix microarray analysis we measured the relative amounts of mRNA for DNA-PK genes (Fig. 2A) and determined there was no strong correlation between DNA-PK message levels and radio-sensitivity.
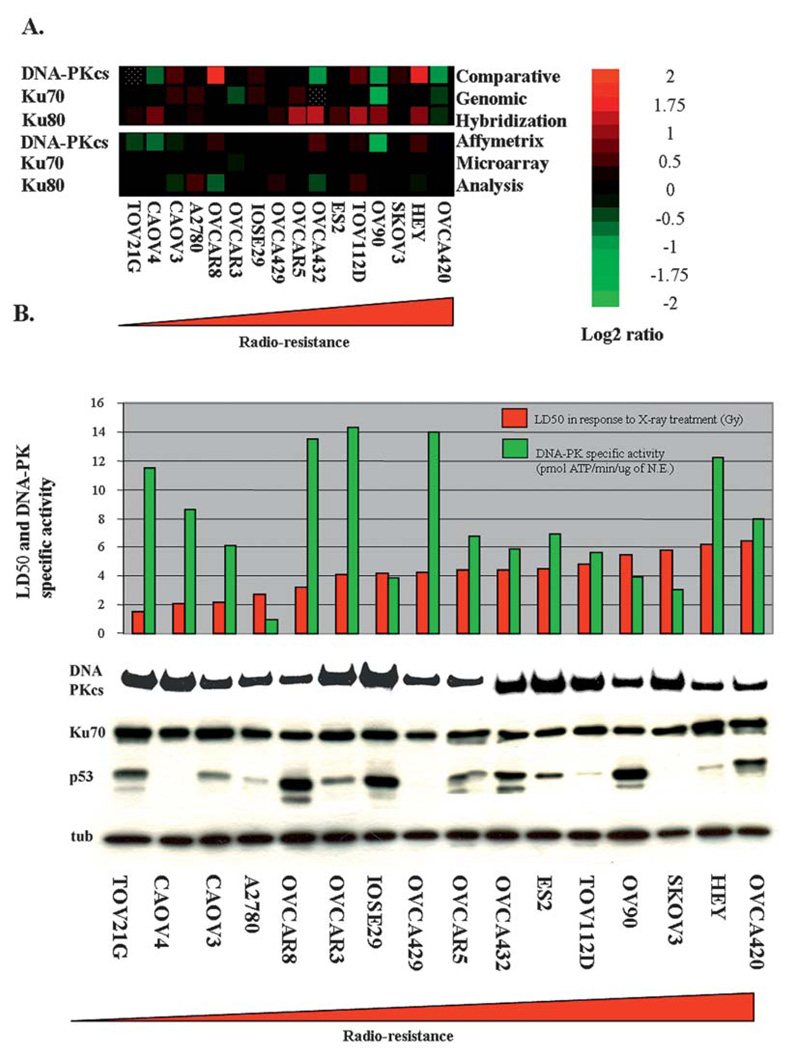
Figure 2.
Molecular and biochemical characterization of DNA-PK in ovarian cancer cell lines. (A) Heatmaps were generated from comparative genomic hybridization (CGH) and Affymetrix microarray analysis of DNA-PK (DNA-PKcs, Ku70 and Ku80 genes) in the panel of ovarian cell lines (The data are normalized and in log2 ratio format). Red levels indicate gene amplification (CGH) or gene overexpression (microarray), and green levels indicate gene deletion (CGH) or underexpression (microarray). Dotted squares indicate where data are not available. (B) Biochemical analysis of DNA-PK. Cell lysates isolated from the ovarian cancer lines were separated by SDS-PAGE (7.5% gel) and transferred to nitrocellulose membrane by wet transfer. The membrane was probed simultaneously with antibodies that specifically recognize DNA-PKcs, Ku70, p53 and α-tubulin. The Western blots were visualized using standard ECL and autoradiography. LD50s were derived from survival curves in response to increasing doses of X-rays (0–7 Gy). DNA-PK specific activity levels were determined by performing in vitro DNA-PK kinase assays.
Since many DNA repair pathways are regulated by post-translational modifications, the mRNA analysis may not be indicative of DNA-PK activity. Therefore, we characterized DNA-PK protein integrity and kinase activity from our panel of ovarian cell lines. The motilities of DNA-PKcs and Ku70 were evaluated by Western blot analysis (Fig. 2B). In all cell lines tested, similar amounts of both of DNA-PKcs and Ku70 were observed with no significant variation in protein mobility between the cell lines.
Defects in DNA-PK enzymatic activity may not necessarily be detected in protein abundance, we therefore assayed DNA-PK kinase activity from our cell lines by performing in vitro kinase assays (Fig. 2B). There was significant variation of DNA-PK activity between the different cell lines; however, there was no strong correlation (Pearson's coefficient: <0.1 but >−0.1) between radiation sensitivity (LD50) and relative DNA-PK kinase activity. Interestingly, there was a strong correlation (Pearson's coefficient: 0.72) between DNA-PK gene copy number and message expression, but no strong correlation between message levels and enzymatic activities. These data strongly suggest that both post-transcriptional and post-translational modifications regulate DNA-PK and the NHEJ pathway and more importantly suggest that DNA-PK copy number, message level, or kinase activity are not reliable predictors of radiosensitivity in the panel of ovarian cell lines tested.
Evaluation of p53 status by Roche p53 AmpliChip analysis
Although initial reports suggested that p53 mutant ovarian cell lines are radioresistant, these studies evaluated only a handful of cell lines (20,21). Therefore, we interrogated the p53 status of each of the 16 ovarian cell lines by utilizing microarray-based technology (Roche AmpliChip p53) to detect changes in the coding sequence of p53 (Table II). Interestingly, many of the cell lines contained a polymorphism in p53, and unexpectedly one of the most radioresistant cell lines is p53-wild-type. However, it must be noted that this cell did not accumulate p21 protein as measured by Western blot analysis (data summarized in Table II) in response to X-ray treatment. Moreover, many of the mutations detected are not typical of the mutational hotspots detected in p53 (Table II). We have confirmed initial reports that p53-status may be used as a general predictive marker of radiation sensitivity in ovarian cancer and that the Roche p53 AmpliChip may be used to accurately predict the p53-status of the ovarian cancer cell lines.
Discussion
In order to minimize patient radiation exposure, advanced techniques such as intraperitoneal radiocolloid therapy as well as small field external beam also known as linear accelerator-based radiotherapy are currently being studied. Another method to reduce patient exposure to radiation would be to determine which tumors would be responsive to radiation treatment and which would not. In order to help develop more individualized ovarian tumor radiotherapy treatment methods, we determined the relative X-ray sensitivity of a panel of 16 ovarian cancer cell lines, investigated the role of the NHEJ and the p53-dependent DNA damage pathways in predicting radiosensitivity. Using microarray-based CGH, we determined if there were gross amplifications or deletions at the loci that contain the NHEJ genes. There were changes in the copy number of DNA-PKcs, Ku70 and Ku80 but these changes in gene copy number did not correlate with radiosensitivity.
Next, we examined the transcript levels of NHEJ genes by Affymetrix microarray expression technology to determine if there are differences in NHEJ message levels between the ovarian cancer cell lines and if these differences correlated with radiosensitivity. While measurable differences in NHEJ gene mRNA levels were observed across the panel, these variations did not correlate well with radiosensitivity. DNA-PK activity is essential for the repair of DSBs in the cell, and other groups have reported that Ku and other NHEJ members strongly correlate with radiosensitivity/resistance in thyroid cancer. However, after investigating the relative in vitro kinase activity and abundance of DNA-PK in this panel of ovarian cancer cell lines, DNA-PK activity was detected in all of the ovarian cell lines and although there was a range of DNA-PK activity between the ovarian cell lines this did not correlate with radiation sensitivity. Western blot analysis performed on the cell lysates allowed the visualization of DNA-PKcs and Ku70 protein expression levels.
While gene copy number and mRNA levels correlated well, DNA-PK protein abundance and activity did not show a strong correlation, and the genetic and protein biochemical data did not correlate well with each other. These data clearly demonstrate that genetic amplifications of DNA-PK or Ku do not directly reflect the enzymatic activities of these proteins.
p53 protein expression significantly varied among the 16 cell lines. We evaluated p53 status by using Roche p53 AmpliChip analysis and by directly measuring X-ray induced-p21 protein accumulation. We report herein that the majority of radioresistant cell lines are usually p53 mutant and supports the earlier studies that p53 mutant ovarian lines are generally more radioresistant (20,21). Our observations suggest that DNA damage signaling pathways may be more reliable predictors than DNA repair pathways for predicting the response of ovarian malignancies to X-rays. It will be interesting to see if these observations hold true in other epithelial tumor types.
This is the first study to interrogate DNA-PK at the genomic, transcript, and protein levels as well as enzymatic activity of DNA-PK and its association with radiosensitivity/resistance in a large set of ovarian cancer cell lines. Although there were no significant correlations between DNA-PK and radioresistance, all of the ovarian cancer cell lines had detectable DNA-PK activities which make it an attractive target to use for small molecule inhibitor or siRNA-based therapeutics. Our in vitro studies further support the notion that p53 status can be a general predictor of radiation sensitivity in advanced stages of ovarian cancer. These ovarian cancer cell lines can be utilized for future studies to help better understand chemotherapeutic resistance. We also present a newly developed technology to help determine relative radiosensitivity/resistance using small amounts of genomic DNA to directly assay p53 status, which should be of great interest to radiation-oncologists treating advanced stage ovarian cancers and of other cancer types.
Acknowledgements
We would like to thank Ms. Nancy Patten and Sim Truong for their expert technical assistance. This work was supported by NIH 1F32GM071229-01 awarded to G. Langland and the National Natural Science Foundation of China, NSFC-30828010, the Zhejiang California Nanosystem Institute, DOD Breast Cancer Research Program Grant BC061995, NIH Grant R1CA95393-01, DOD BCRP BC045345 grant, UCSF Prostate Cancer SPORE award and NIH P50 Grant CA112970 awarded to either J.W. Gray or F. Chen. This work was performed under the auspices of the U.S. Department of Energy, at the University of California/Lawrence Berkeley National Laboratory under contract no. DE-AC03-76SF00098.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–76. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Trimble EL, Wright J, Christian MC. Treatment of platinum-resistant ovarian cancer. Expert Opin Pharmacother. 2001;2:1299–1306. doi: 10.1517/14656566.2.8.1299. [DOI] [PubMed] [Google Scholar]
- 3.Stuart GC. First-line treatment regimens and the role of consolidation therapy in advanced ovarian cancer. Gynecol Oncol. 2003;90:S8–S15. doi: 10.1016/s0090-8258(03)00472-4. [DOI] [PubMed] [Google Scholar]
- 4.El-Deiry WS. The role of p53 in chemosensitivity and radio-sensitivity. Oncogene. 2003;22:7486–7495. doi: 10.1038/sj.onc.1206949. [DOI] [PubMed] [Google Scholar]
- 5.Gallion HH, Pieretti M, DePriest PD, van Nagell JR., Jr The molecular basis of ovarian cancer. Cancer. 1995;76 Suppl 10:1992–1997. doi: 10.1002/1097-0142(19951115)76:10+<1992::aid-cncr2820761315>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 6.Kihana T, Tsuda H, Teshima S, Okada S, Matsuura S, Hirohashi S. High incidence of p53 gene mutation in human ovarian cancer and its association with nuclear accumulation of p53 protein and tumor DNA aneuploidy. Jpn J Cancer Res. 1992;83:978–984. doi: 10.1111/j.1349-7006.1992.tb02010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kupryjanczyk J, Kraszewska E, Ziolkowska-Seta I, Madry R, Timorek A, Markowska J, Stelmachow J, Bidzinski M, Polish Ovarian Cancer Study Group (POCSG) TP53 status and taxane-platinum versus platinum-based therapy in ovarian cancer patients: a non-randomized retrospective study. BMC Cancer. 2008;8:27–34. doi: 10.1186/1471-2407-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collis SJ, DeWeese TL, Jeggo PA, Parker AR. The life and death of DNA-PK. Oncogene. 2005;24:949–961. doi: 10.1038/sj.onc.1208332. [DOI] [PubMed] [Google Scholar]
- 9.Zhao HJ, Hosoi Y, Miyachi H, Ishii K, Yoshida M, Nemoto K, Takai Y, Yamada S, Suzuki N, Ono T. DNA-dependent protein kinase activity correlates with Ku70 expression and radiation sensitivity in esophageal cancer cell lines. Clin Cancer Res. 2000;6:1073–1078. [PubMed] [Google Scholar]
- 10.Pavon MA, Parreno M, Leon X, Sancho FJ, Cespedes MV, Casanova I, Lopez-Pousa A, Mangues MA, Quer M, Barnadas A, Mangues R. Ku70 predicts response and primary tumor recurrence after therapy in locally advanced head and neck cancer. Int J Cancer. 2008;123:1068–1079. doi: 10.1002/ijc.23635. [DOI] [PubMed] [Google Scholar]
- 11.Chang HW, Kim SY, Yi SL, Son SH, Song do Y, Moon SY, Kim JH, Choi EK, Ahn SD, Shin SS, Lee KK, Lee SW. Expression of Ku80 correlates with sensitivities to radiation in cancer cell lines of the head and neck. Oral Oncol. 2006;42:979–986. doi: 10.1016/j.oraloncology.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 12.Wilson CR, Davidson SE, Margison GP, Jackson SP, Hendry JH, West CM. Expression of Ku70 correlates with survival in carcinoma of the cervix. Br J Cancer. 2000;83:1702–1706. doi: 10.1054/bjoc.2000.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mérel P, Prieur A, Pfeiffer P, Delattre O. Absence of major defects in non-homologous DNA end joining in human breast cancer cell lines. Oncogene. 2002;21:5654–5659. doi: 10.1038/sj.onc.1205742. [DOI] [PubMed] [Google Scholar]
- 14.Henkels KM, Turchi JJ. Induction of apoptosis in cisplatin-sensitive and -resistant human ovarian cancer cell lines. Cancer Res. 1997;57:4488–4492. [PubMed] [Google Scholar]
- 15.Hodgson G, Hager JH, Volik S, Hariono S, Wernick M, Moore D, Nowak N, Albertson DG, Pinkel D, Collins C, Hanahan D, Gray JW. Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nat Genet. 2001;29:459–464. doi: 10.1038/ng771. [DOI] [PubMed] [Google Scholar]
- 16.Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, Hamilton G, Hindle AK, Huey B, Kimura K, Law S, Myambo K, Palmer J, Ylstra B, Yue JP, Gray JW, Jain AN, Pinkel D, Albertson DG. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet. 2001;29:263–264. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- 17.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.George B, Datar RH, Wu L, Cai J, Patten N, Beil SJ, Groshen S, Stein J, Skinner D, Jones PA, Cote RJ. p53 gene and protein status: the role of p53 alterations in predicting outcome in patients with bladder cancer. J Clin Oncol. 2007;25:5352–5358. doi: 10.1200/JCO.2006.10.4125. [DOI] [PubMed] [Google Scholar]
- 19.Dicker F, Herholz H, Schnittger S, Nakao A, Patten N, Wu L, Kern W, Haferlach T, Haferlach C. The detection of TP53 mutations in chronic lymphocytic leukemia independently predicts rapid disease progression and is highly correlated with a complex aberrant karyotype. Leukemia. 2009;23:117–124. doi: 10.1038/leu.2008.274. [DOI] [PubMed] [Google Scholar]
- 20.Petru E, Sevin BU, Gottlieb C. Radiosensitivity patterns of four human ovarian cancer cell lines in vitro. Gynecol Oncol. 1997;64:490–492. doi: 10.1006/gyno.1996.4574. [DOI] [PubMed] [Google Scholar]
- 21.Concin N, Zeillinger C, Stimpfel M, Schiebel I, Tong D, Wolff U, Reiner A, Leodolter S, Zeillinger R. p53-dependent radioresistance in ovarian carcinoma cell lines. Cancer Lett. 2000;150:191–199. doi: 10.1016/s0304-3835(99)00393-6. [DOI] [PubMed] [Google Scholar]