Summary
The SecY/Sec61 translocon complex, located in the endoplasmic reticulum membrane of eukaryotes (Sec61) or the plasma membrane of prokaryotes (SecY), mediates the transmembrane secretion or insertion of nascent proteins. Mutations that permit the secretion of nascent proteins with defective signal sequences (Prl-phenotype), or interfere with the transmembrane orientation of newly synthesized protein segments, can affect protein topogenesis. The crystallographic structure of SecYEβ from Methanococcus jannaschii revealed widespread distribution of mutations causing topogenesis defects, but not their molecular mechanisms. Based upon prolonged molecular dynamics simulations of wild-type M. jannaschii SecYEβ and an extensive sequence-conservation analysis, we show that the closed-state of the translocon is stabilized by hydrogen-bonding interactions of numerous highly conserved amino acids. Perturbations induced by mutation at various locations are rapidly relayed to the plug segment that seals the wild-type closed-state translocon, leading to displacement and increased hydration of the plug.
Keywords: protein biosynthesis, SecY/Sec61 translocon, hydrogen bonding, molecular dynamics simulations
Introduction
Proteins destined for secretion across or insertion into the endoplasmic reticulum of eukaryotes must pass from the ribosome through the Sec61 translocon complex (Rapoport, 2007) where the decision to secrete or insert is made, based largely on biophysical principles (White and Von Heijne, 2008; Jaud et al., 2009). The equivalent and highly homologous SecY complex serves the same function in the plasma membrane of prokaryotes. But, unlike eukaryotes, nascent proteins in bacteria follow two pathways. Membrane proteins are inserted co-translationally as for eukaryotes, but most secreted proteins are transported through SecY post-translationally by the SecA ATPase, which receives unfolded proteins from the versatile SecB chaperone (Randall and Hardy, 2000). Targeting of proteins to the SecY pathway is generally encoded either by a cleavable N-terminal signal sequence on secreted proteins or by the first hydrophobic transmembrane (TM) segment (signal anchor) of membrane proteins. Prl (protein localization) mutations destabilize SecY to allow secretion of proteins with defective or absent signal sequences (Emr and Hanley-Way, 1981; Mori et al., 2003; Smith et al., 2005). The recently discovered correspondence between mutations that cause prl phenotypes in yeast, and mutations that cause an increased translocation of the more positively-charged end of signal anchors, were interpreted to suggest a role for the timely displacement of the plug segment that seals the translocon in its closed state (Junne et al., 2007). In order to understand the mechanism by which specific mutations can cause translocation defects, we have carried out extensive molecular dynamics simulations of the wild-type and mutant SecYEβ translocons from Methanococcus jannaschii (Van den Berg et al., 2004) embedded in lipid bilayers.
Although an exact understanding of how the translocon opens is lacking, it is believed that the transmembrane segments TM2 and TM7 of SecY (Figure 1A,B) act as a gate that opens laterally to release hydrophobic protein segments into the lipid membrane (Plath et al., 1998; Van den Berg et al., 2004; Du Plessis et al., 2009) and that the small plug domain (Figures 1A–E) is displaced (Tam et al., 2005). Protein flexibility and significant conformational changes must be critical for the functioning of the translocon, because locking the translocon in particular conformational states has significant effects on function (Tam et al., 2005; Du Plessis et al., 2009). Changes in the preferred conformation of SecY (Mori et al., 2003; Smith et al., 2005) and a relaxed SecYEβ association (Duong and Wickner, 1999) have been proposed to explain why prl mutations broaden the specificity of the translocon by allowing translocation of secretory proteins with defective or absent signal peptides (Emr and Hanley-Way, 1981; Mori et al., 2003; Smith et al., 2005). Structural instability (Gumbart and Schulten, 2008) of the plug region (Figure 1E) correlates with an increased flexibility of the hydrophobic ring located above the plug.
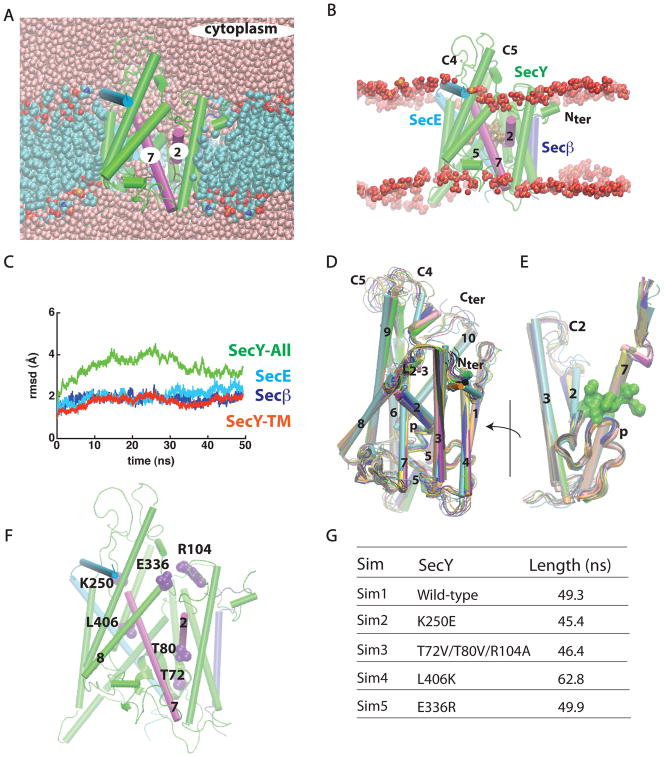
Figure 1.
SecYEβ in a POPC lipid bilayer. Loops C2, C4, and C5 correspond to loops connecting, respectively, TM2/TM3, TM6/TM7, and TM8/TM9. (A, B) Cut away images of SecYEβ viewed along the membrane plane. Color scheme: SecY, green; SecE, light blue; Secβ, purple; gate helices TM2 and TM7, magenta; water oxygen atoms, pink; lipid phosphorous atoms, orange; lipid nitrogen, blue; lipid oxygen, red; and lipid carbon atoms, cyan. Sidechains of the hydrophobic pore are shown as brown surfaces (the probe radius used was 1.4 Å). In (B), only lipid phosphate groups of a ~50 Å-thick slab containing the membrane are shown. In all figures of the M. jannaschii translocon, the protein is oriented to align the z-axis parallel to the membrane normal. (C) The root-mean-squared deviation (rmsd; Å) of the SecYEβ Cα atoms relative to the starting crystallographic structure. For simplicity, the rmsd is shown at intervals of 10ps. (D,E) Overlap of 10 snapshots from the last 10ns of Sim1 taken at every 1ns (ending conformation, green; starting conformation, cyan). In (E), the hydrophobic ring at the end of Sim1 is shown as a green surface. Flexibility of the cytoplasmic loops and the C-terminus is largely responsible for rmsd values of SecY (green curve in panel D). The plug segment (p) of wild-type SecY (Sim1) samples a relatively restricted conformational space. (F) Location of amino acids whose mutations were investigated in Sim2 - Sim5. (G) Summary of the simulations performed (Sim1 - Sim5). The lengths of the simulations correspond to the unconstrained molecular dynamics trajectories. The average values (± s.d.) of the Cα rmsd of the SecY TM region in the last 10ns segments of Sim1-Sim5 are 1.9 ± 0.2, 1.7 ± 0.3, 1.6 ± 0.1, 2.0 ± 0.1, and 2.0 ± 0.1 Å for Sim1, Sim2, Sim3, Sim4, and Sim5, respectively (see Figure S1 for rmsd plots of the mutant simulations).
TM segments of multi-spanning membrane proteins move through the Sec61 translocon in a highly controlled fashion (Sadlish et al., 2005) while achieving their proper orientation (Sadlish et al., 2005; Junne et al., 2007). The crystal structures of the putatively open structures of bacterial SecY from Thermotoga maritima with bound SecA (Zimmer et al., 2008) and from Thermus thermophilus with a bound Fab fragment (Tsukazaki et al., 2008) indicate significant conformational changes relative to the closed state of SecY from M. jannaschii (Van den Berg et al., 2004). The crystal structures, however, provide little information about the molecular mechanism that unlocks the translocon or how the translocon and the surrounding lipid membrane couple during membrane protein insertion.
Knowledge of the structure and dynamics of prl mutants should provide fundamental information on how the translocon opens, because these mutants are believed to open prematurely without the binding of a signal peptide (Mori et al., 2003; Smith et al., 2005; Junne et al., 2007). The premature displacement of the plug in the mutants perturbs the electrostatic field that in the wild-type translocon contributes to orienting the incoming signal peptide according to its flanking charges; as a consequence, defects in the orientation of the signal peptide may occur (Junne et al., 2007). For example, the K284E mutation in the Saccharomyces cerevisiae Sec61p causes a prl phenotype, and an increased translocation of the more positively charged end of signal anchors (Junne et al., 2007). How the K284E mutation could cause premature displacement of the plug is intriguing, because in the closed-state of the M. jannaschii translocon (Van den Berg et al., 2004) the distance between the corresponding amino acid K250 (located on the cytoplasmic side of TM7) and the plug is >20Å. The mechanism by which the perturbation induced by the mutation is relayed to the plug, and the structural events that accompany translocon opening in the mutants, remain elusive in the absence of structural information.
To begin to understand the mechanics of the opening of the translocon in its lipid membrane environment, we combined prolonged molecular dynamics simulations (Sims) with bioinformatics analyses to examine wild-type and translocation-defective SecY mutants. The dynamics and hydrogen bonding (H bonding) of wild-type M. jannaschii were investigated in Sim1. The effects of mutations were examined in Sims 2 – 5 (Figure 1F). Sim2 was concerned with the effect of the K250E mutation. K250, located at the cytoplasmic tip of TM7/L6-7, is highly conserved. Replacing the corresponding Lys by Glu in yeast causes a prl phenotype and enhances translocation of the more positively charged ends of signal anchors (Junne et al., 2007). In Escherichia coli, the Lys corresponding to the M. jannaschii K250 is in the center of the SecA-binding hotspot (Robson et al., 2007). Because of the crucial role of the gate helix TM2 in the insertion of TM helices into the bilayer membrane, we examined in Sim3 the effect of switching off H bonds that we hypothesized to be important in controlling the dynamics of TM2 using the in silico triple mutant T72V/T80V/R104A. The L406K mutant was studied in Sim4 in order to examine the effect of introducing an H-bonding amino acid into the hydrophobic pore domain. Replacing L406 with Lys in yeast reduces the translocation of the C terminal of signal-anchor peptides (Junne et al., 2007). The E336R mutation (Sim5) increases the translocation of the more positively charged end of signal-anchor peptides in yeast (Junne et al., 2007). Such an effect on the translocation of the positively-charged peptide end was also observed in the case of K250E, except that E336R does not cause a prl phenotype (Junne et al., 2007).
We identified an extensive H-bond network that inter-connects different segments of the translocon and stabilizes its closed state. Most H bonds are highly conserved in all branches of life; for some amino acids, including those mediating the interaction between the TM2/TM7 gate helices, the conservation depends on the organism. The network of H bonds apparently allows rapid long-distance relay of structural perturbations. Perturbations caused by mutations in remote regions of the translocon are consequently relayed rapidly to the plug, causing its displacement and increased hydration relative to the wild-type closed state. Although the plug responds to all mutations assessed here, the exact structural and dynamical changes depend on the mutation.
Results
The five independent sets of molecular dynamics simulations (Sim1 - Sim5, Figure 1G) consisted of the SecYEβ heterotrimer, 475 palmitoyloleoyl phosphatidylcholine (POPC) lipids, and 48,568 water molecules, for a total of 217,820 atoms (Figure 1A). The dimensions of the unit cell reached plateau values after ~15ns of unconstrained simulations. Coordinate sets were saved for analysis every 1ps. All histograms and average values were computed using the 10,000 coordinate sets of the last 10ns of the simulations. In all five simulations we used for the starting protein coordinates the native SecYEβ crystal structure from Van den Berg et al. (2004). Although the limited timescale of our simulations may not have captured the full extent of the structural changes induced by mutations, Sim2 - Sim5 are valuable for illuminating how structural perturbations may be relayed throughout the translocon.
Dynamics of wild-type SecY in a hydrated lipid bilayer
To mediate the secretion of soluble proteins and the membrane incorporation of membrane proteins, the translocon must communicate with the aqueous and membrane environments, respectively. The thickness of the lipid bilayer surrounding the translocon could be important for the incorporation and orientation of the transmembrane segments of nascent membrane proteins (Jaud et al., 2009), but wild-type SecYEβ did not perturb the lipid bilayer significantly in our simulation. The largest bilayer perturbations were observed in the vicinity of TM2 (which dips into the bilayer) and TM5, which extends into the aqueous bulk (Figure 1B). We found that the cytoplasmic parts of TM2/TM7 are well hydrated (Figure 1A). The loop connecting TM8 and TM9, denoted as C5 by Taura et al. (1994), partially occludes the translocon on the cytoplasmic side (Figures 1B). The plug segment sampled a relatively restricted conformational space (Figure 1E).
The TM region of SecY is rather rigid, with relatively small root-mean-square deviations of the Cα atoms relative to the crystal structure (rmsd; Figure 1C). In contrast, the C-terminus and the cytoplasmic loops C4 (connecting TM6/TM7) and C5 are very dynamic relative to the transmembrane region of SecY, and sample a large range of conformations (Figures 1C,D), largely contributing to the rmsd of the SecY (Figure 1C). The pronounced flexibility of these cytoplasmic loops and of the C-terminus may be required for function. C4 and C5 contain amino acids important for co-translational translocation in yeast (Raden et al., 2000; Cheng et al., 2005), and for binding of SecA to the E. Coli SecY (Mori and Ito, 2006); C5 is required for high-affinity binding of the ribosome (Raden et al., 2000). In the SecA/SecY complex from T. maritima, loops C4 and C5 make extensive contacts with SecA (Zimmer et al., 2008). The C-terminus of SecY interacts with SecA in E. coli (Mori and Ito, 2006), and with the ribosome in yeast (Raden et al., 2000). The flexibility suggested by our simulations may allow the C-terminus and loops C4 and C5 to sample conformations that facilitate productive interactions with their cytosolic partners. C4 and C5 flexibility may also facilitate the opening of a path for substrate entry into the cytoplasmic vestibule of the translocon.
A network of conserved hydrogen bonds stabilizes the closed state of the translocon
We identified more than seventy H bonds in the wild-type translocon that we grouped into 10 clusters, 6 on the cytoplasmic (CP-1 to CP-6) and 4 on the extracellular side of SecY (EC-1 to EC-4) (Figures 2A–C, S2, and Tables S1–S2). The H bonds inter-connect different structural elements, and one TM segment can be part of more than one H-bonding cluster (Figure 2D). The detailed analysis of the dynamics of all H-bonding clusters is summarized in Tables S1–S2. Systematic studies of the conservation of H bonds of translocons from archaea, bacteria, and eukarya, discussed briefly below, are given in Supplementary Information.
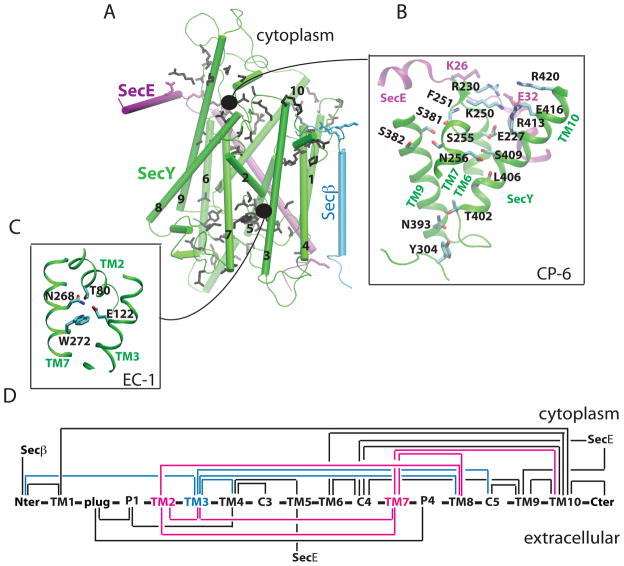
Figure 2.
H-bonding clusters in wild-type SecY. As the H-bond criterion, we used a distance of less than 3.5 Å between the donor and acceptor heavy atoms. (A) H-bonding sidechains whose dynamics were investigated here are depicted in black, purple, or cyan for the SecY, SecE, and Secβ subunits of the translocon, respectively. (B) The H-bonding cluster EC-1 connects TM2, TM3, and TM7. (C) The crowded cytoplasmic cluster CP-6 that includes TM7. (D) H-bonding connectivity in wild-type SecY (for details see Figure S2, Table S1, and Table S2). Connections between two structural elements indicates that at least one H bond between them is sampled during Sim1. Note that Taura et al. (1994) denote the N- and C-terminus of SecY as C1 and C6, respectively. Examples of H bonding interactions in the T. maritima and T. thermophilus SecY translocons are described in Figures S3 and S4, respectively. Known mutation effects of H-bonding amino acids are given in Table S3.
Of the 10 H-bonding clusters investigated, CP-6 (Figure 2B) and EC-1 (Figure 2C) appear particularly important, because they involve the lateral gate helices TM2 and TM7 to which the signal peptide is thought to bind (Plath et al., 1998; Van den Berg et al., 2004). CP-6 is a remarkably crowded cluster that extends from the cytoplasm to the central part of the translocon and connects the TM7 lateral-gate helix to TM9 and TM10 (Figure 2B); H bonds mediated by TM6-R230 further connect CP-6 to SecE (Figures 2B & S1B). EC-1 is the only H-bonding cluster that bundles TM2 and TM7 together (Figure 2C,D).
TM2 and TM7 connect with each other and with TM3 (Figures 2C,D). In the crystal structure, the carboxyl group of TM3-E122 is within H-bonding distance from the sidechains of TM2-T80 (2.7 Å) and TM7-N268 (3.0 Å), and N268 H bonds to T80 (3.2 Å). In Sim1 and in another independent set of simulations on the wild-type translocon, we observed H bonding of T80 to N268; E122 H bonded to T80 and N268, but also to W272, which in the starting crystal structure is somewhat too far from E122 for a H bond (4.4 Å).
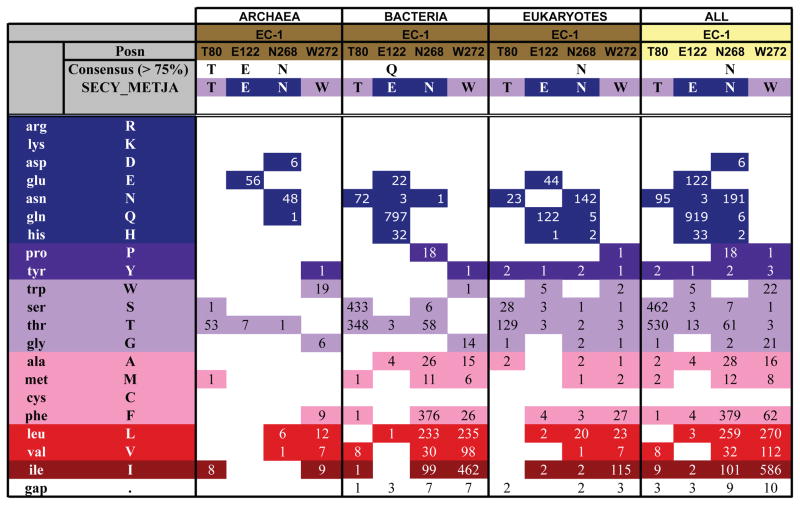
The sequence analysis indicates that the H-bonding connecting the gate helices TM2/TM7 and TM3 (cluster EC-1, Figure 2C) may be specific to M. jannaschii and some other archaeal translocons. Importantly, however, an interconnectivity of TM2, TM3, and TM7 appears to be a general characteristic of the translocons from archaea and eukarya; bacteria share an interconnectivity of TM2 and TM3. E122 is present either as Glu (56 sequences) or Thr (7 sequences) in the archaeal translocons, as Gln in bacteria, and as Gln or Glu in eukarya (Figures 3, 4, S5–S8). T80 is highly conserved in all organisms; N268 is significantly conserved in archaea and eukaryotes, while W272 has moderate conservation only in archaea (Figure 3).
Figure 3.
Conservation of H bonding in the EC-1 cluster. In the table, ‘Posn’ gives the amino acid in the M. jannaschii sequence. ‘Consensus’ indicates the amino acid present in at least 75% of the sequences, compared to the sequence of the M. jannaschii SecY (‘SECY_METJA’). ’Gap’ indicates insertion/deletion mutations. The numbers indicate how many sequences have a specific amino acid at each of the positions investigated. For example, in archaea T80 is found as Thr in 53 sequences, and replaced with Ser, Met, or Ile in 1, 1, and 8 sequences, respectively. Sequence alignments for several pertinent SecY/Sec61p sequences are shown in Figure S5. Conservation of H-bonding residues for other H-bond clusters are shown in Figures S6-S11. Complete sequence alignments for archaea are shown in Figure S12. The conservation analysis was generated according to the Kyte and Doolittle scheme (Kyte and Doolittle, 1982), in which amino acids are colored from red (most hydrophobic) to blue (most hydrophilic). We analyzed the conservation of H-bonding amino acids in archaea, bacteria, and eukarya (78, 865, and 187 sequences, respectively). The initial list of SecY sequences was compiled using the Pfam database (Finn et al., 2008). The complete alignments for all sequences are available on our web site: http://blanco.biomol.uci.edu/download/Bondar_SecYE_align.zip.
Figure 4.
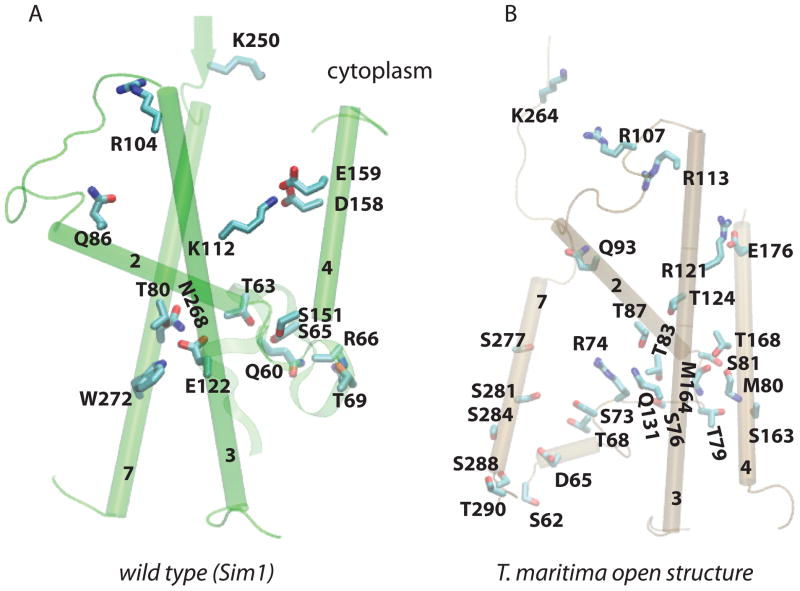
Selected H-bonding amino acids of the TM2/TM7 gate helices and of the plug in the closed state of the M. jannaschii and in the open structure of the SecA-bound SecY from T. maritima. (A) Snapshot at the end of Sim1 (wild-type M. jannaschii translocon). (B) The translocon from T. maritima. In the crystal structure of the closed state of the M. jannaschii translocon (Van den Berg et al., 2004) and in Sim1 (wild-type), helices TM2, TM3, and TM7 are interconnected via H bonding (panel A, and Figure 2C). In the open structure of the SecA-bound SecY (Zimmer et al., 2008), TM2 and TM3 H bond via T83 and Q131, which correspond to M. jannaschii T80 and E122, respectively. In both translocons, TM4 can H bond to the extracellular tip of TM2 to the plug via a Ser/Thr amino acid.
The sequences of translocons from archaea differ significantly from those of bacterial and eukaryotic SecY in the TM7 segment of the M. jannaschii sequence containing N268 and W272 (Figures 3–4, S6–S8). This region of the sequence is, however, critical for the proper function of the bacterial translocons: in E. coli, the segment of the sequence corresponding to the M. jannaschii W272 is rich in amino acids (V274G, I278S/N/T, S282R, I290T, P287L) whose mutation affects translocon function (Emr and Hanley-Way, 1981; Sako and Ino, 1988; Osborne and Silhavy, 1993; Smith et al., 2005) (see Figure S5 for an alignment of SecYs from E. coli and S. cerevisiae, and of SecYs whose crystal structure is known). The crystal structure of SecY from T. maritima (Zimmer et al., 2008) reveals an interesting array of Ser/Thr amino acids along TM7 (Figure 4B). The hydroxyl group of each of these Thr/Ser is involved in H bonding with backbone carbonyl groups of other TM7 amino acids.
The crowded CP-6 cluster contains H-bonding amino acids whose mutations affect translocon function (Figure 2B, Table S3) (Emr and Hanley-Way, 1981; Osborne and Silhavy, 1993; Smith et al., 2005; Junne et al., 2007). H bonding of S255 with S381 and S382 bridges the cytoplasmic tips of TM7 and TM9. K250 and E416 H bond with each other and have additional H bonds with E227 and R413/R420, respectively (Figure 2B and Table S1). Mutation of K250 and E416 to Glu and Lys, respectively, cause prl phenotypes in yeast (Emr and Hanley-Way, 1981; Osborne and Silhavy, 1993; Smith et al., 2005; Junne et al., 2007). K250 is highly conserved as Lys/Arg in all organisms, but the majority of sequences have Lys at this position (Figures S6–S8). The presence in most archaea and eukarya translocons of a Glu at the sequence position corresponding to M. jannaschii E416 supports an important role of the charge and length of this amino acid sidechain, underscored by the prl phenotype and translocation defects caused by its mutation to Lys in yeast (Emr and Hanley-Way, 1981; Osborne and Silhavy, 1993; Smith et al., 2005; Junne et al., 2007).
Long-distance perturbations are rapidly relayed to the plug
Cysteine crosslink experiments indicate that the plug moves towards SecE during peptide translocation, and that the movement of the plug is a rate-limiting step of the translocation reaction (Tam et al., 2005). Work with synthetic mimics of the plug segment led to the proposal of a second periplasmic site to which the plug would relocate upon translocon opening (Robson et al., 2009). Locking the plug in the open state by cross-linking to SecE, or in the F67C E. coli mutant, increased peptide translocation (Tam et al., 2005). Site-directed mutagenesis experiments were interpreted to suggest that the premature displacement of the plug in prl mutants interferes with the orientation of the signal peptide inside the translocon (Emr and Hanley-Way, 1981; Osborne and Silhavy, 1993; Smith et al., 2005; Junne et al., 2007). The proper location of the positively-charged Arg amino acid(s) of the plug segment could be particularly important in orienting the signal peptide according to its flanking charges (Emr and Hanley-Way, 1981; Osborne and Silhavy, 1993; Smith et al., 2005; Junne et al., 2007). The molecular picture of how the plug opens and the structural rearrangements associated with its displacement are, however, not clear. Another key question is how structural perturbations on the cytoplasmic side arising from the binding of the signal peptide and ribosome (or SecA) are relayed to the plug on the extracellular side. Our computations suggest that tight coupling between the plug domain and other regions of the translocon ensures that translocon perturbations are rapidly sensed by the plug.
In wild-type SecY, there is H bonding between amino acids of the plug, and between amino acids of the plug and the rest of the protein (Figures 4, S2H,J, Table S2). For example, the hydroxyl group of T69 H bonds with the peptide carbonyl group of R66, whereas the amide and carbonyl groups of T69 H bond to T72 (Figures 4, S2J). T72 also H bonds with W59, and with the L70 carbonyl group. The H bond between S151 and the carbonyl group of G74 connects TM4 to the segment that links the plug to TM2 (Figure S2J). The plug also contacts the hydrophobic pore through A64 (Figure 5A). These interactions of the plug likely explain its sampling of a relatively restricted conformational space in Sim1 (Figure 1E), and its rapid response to remote perturbations.
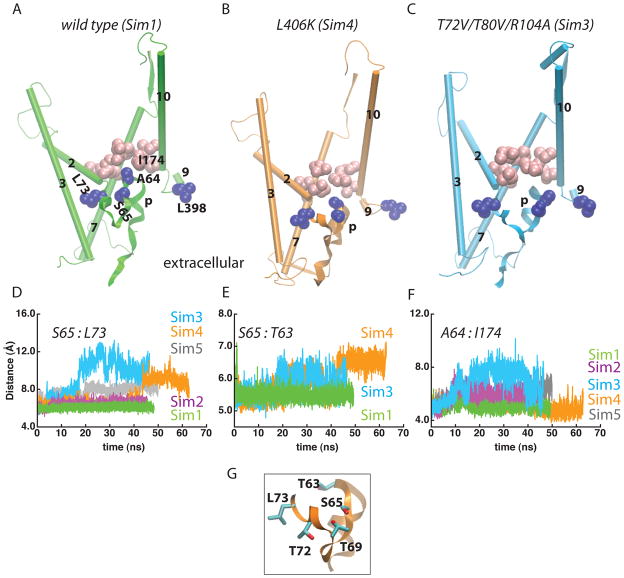
Figure 5.
Mutations perturb the structure and interactions of the plug. TM2, TM3, TM7, TM10 the hydrophobic pore, and the plug are depicted for the wild-type (panel A; Sim1), the L406K mutant (panel B; Sim4), and the in silico mutant T72V/T80V/R104A (panel C; Sim3). Amino acids of the hydrophobic ring are shown as pink van der Waals spheres, and other selected amino acids as blue. (D–F) Time series of the distance between the Cα atoms of S65 and L73 (panel D), S65 and T63 (panel E), and between A64 and I174 (panel F), illustrate the perturbation of the plug in the mutants. The average Cα distances for S65:L73, S65:T63, and A64:I174 in Sim1 are, respectively, 6.1±0.2 Å, 5.5±0.1 Å, and 5.0±0.3 Å, as compared to 5.5 Å, 5.1 Å and, 4.8 Å in the crystal structure of (Van den Berg et al., 2004). The average S65:T63 distances in the K250E (Sim2) and E336R (Sim5) mutants are 5.6±0.2 Å and 5.4±0.2 Å, respectively. (G) Close view of T63, S65, and L73 in L406K (Sim4). T63 and S65 are also depicted in Figure S2J for the wild type (Sim1).
The structure and interactions of the plug are perturbed in all mutants (Figures 5–7). Although the details depend on the mutant, the mutations perturb the location of the plug relative to the segment that connects to TM2 (Figures 5A,D) and to the pore ring (Figure 5F). In the T72V/T80V/R104A (Sim3) and L406K (Sim4) mutants, we also observe perturbation of the local structure of the cytoplasmic tip of the plug, monitored as an increased distance between the Cα atoms of T63 and S65 (Figures 5A–C, 5E, 5G). That is, although the details depend on the mutation, the reaction coordinate for the movement of the plug can be described as a loosening of the plug structure, detachment of the plug from the interactions with amino acids upstream in the sequence towards TM2, and displacement towards the extracellular side.
Figure 7.
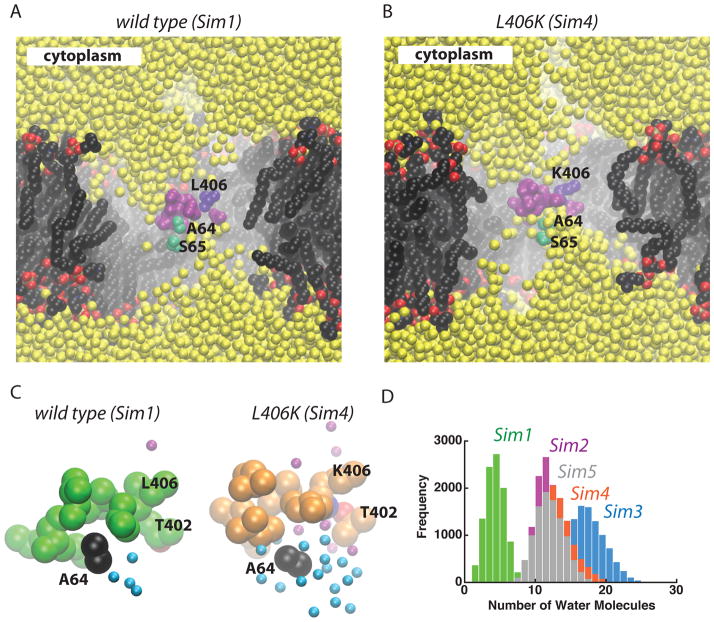
Increased hydration in the extracellular half of the translocon upon displacement of the plug. The wild-type translocon (panel A; Sim1) is compared to the L406K mutant (panel B; Sim4). Color code used for van der Waals spheres: sidechains of the amino acids forming the hydrophobic pore - mauve, except for L406 which is depicted in violet; sidechains of S65 and A64 - green; water oxygen atoms – yellow; lipid nitrogen, oxygen, phosphorous, and carbon atoms are depicted in blue, red, orange, and black, respectively. For simplicity, hydrogen atoms are not shown. (C) Close-up view of the hydrophobic pore in the wild-type (Sim1; green) and L406K (orange; Sim4). Sidechains of pore ring amino acids and A64 are shown as van der Waals spheres; water molecules within 6 Å of A64 and within 6 Å of L/K406 are shown as cyan and purple spheres, respectively. (D) Histograms of the number of water molecules within 6 Å of A64 in the wild-type (Sim1; green), K250E (Sim2; purple), L406K (orange, Sim4), and E336R (Sim5). The histograms were computed with a bin size of 1.
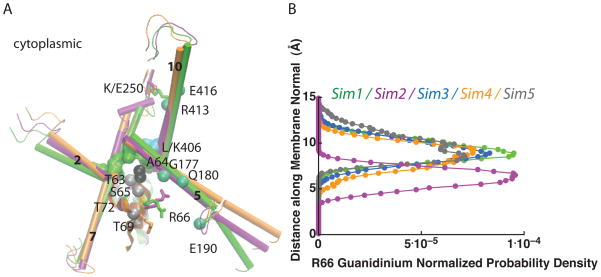
Perturbation of the plug in Sim3-Sim5 is associated with increased dynamics of the R66 sidechain: the distribution of the location along the membrane normal of the R66 guanidinium group is broader in Sim3-Sim5 than in Sim1 (Figure 6). In Sim2 (K250E), the R66 guanidinium establishes new interactions with TM5-Q180 and G177 (Figure 6).
Figure 6.
Coupling of TM segments, plug, and hydrophobic-pore amino acids. (A) Overlap of snapshots from Sim1 (wild-type), Sim2 (K250E) and Sim4 (L406K) depicted in green, purple, and orange, respectively. The sidechains of the pore amino acids in Sim1 are shown as a green surface, except for L406 which is colored cyan. The sidechain and Cα atom of A64 are shown as black van der Waals spheres, and other Cα atoms in Sim1 as silver or green van der Waals spheres. H atoms are not shown. (B) The change in the plug location and hydration (see Figures 5 and 7) is associated with changes in the dynamics of R66. The normalized number density of the guanidinium group of R66 along the membrane normal is depicted in green, purple, cyan, orange, and gray for the wild type (Sim1), K250E (Sim2), T72V/T80V/R104A (Sim3), L406K (Sim4), and E336R (Sim5), respectively. Note that relative to the wild type, in Sim3-Sim5 the distribution of the R66 guanidinium group location broadens, and in Sim2 R66 samples preferentially a more cytoplasmic orientation.
Relocation of the plug away from the hydrophobic pore in the mutants (Figure 5) is associated with an enhanced solvation of the plug (Figure 7), which allows water molecules to enter the extracellular half of the plug up to the pore amino acids. Unlike in the wild type (Sim1), the sidechain of A64 at the cytoplasmic tip of the plug is surrounded by water molecules in the four mutants (Figures 5A, 7).
A direct comparison between the structure and dynamics of the plug domains from translocons of different organisms is difficult due to significant variability in this region of the sequence (Figures 4, S5, S12). Nevertheless, the detailed analysis of the sequences (Figures S5–S8) and the site-directed mutagenesis experiments summarized in Table S3 support an important functional role of the plug H bonds. T69 is highly conserved in all organisms, and so is H bonding at position Q60 (Figure 4A, S6–S8); mutation of the amino acids corresponding to the M. jannaschii Q60 or T69 impairs translocon function (Table S3) (Emr and Hanley-Way, 1981; Osborne and Silhavy, 1993; Taura et al., 1994; Smith et al., 2005; Junne et al., 2007). H bonding at T72 is highly conserved in archaea and eukarya (Figure 4A, S6–S8, Table S3). In yeast, T63 is replaced by a Leu whose mutation to Asn causes a prl phenotype and increases translocation of the more positively charged end of signal anchors (Junne et al., 2007). S65 is highly conserved in eukarya, and T63 in bacteria (Tables S6–S8).
The importance of the interactions mediated by TM4-S151 appears to be organism-dependent. Although in yeast the conservative mutation of S151 to Thr causes a prl phenotype and increased translocation of the more positively charged end of signal anchors (Junne et al., 2007), S151 is only moderately conserved in archaea and eukarya, and is present as Thr in most bacterial sequences (Figure S6–S8). In the T. maritima structure of the SecA-bound translocon (Zimmer et al., 2008), TM4-T168 (corresponding to M. jannaschii S151) is located close to the extracellular tip of TM2 and H bonds to the carbonyl groups of M80 and M164 (Figures 4B, S3B).
SecY:SecE and SecYE:lipid H bonds participate in the relay of structural perturbations
SecE stabilizes SecY (Matsuyama et al., 1990), and the two proteins form a complex that can be purified chromatographically in detergent (Brundage et al., 1990). Prl mutations of SecY, but also of SecE, influence the way SecY and SecE interact with each other (Flower et al., 1995; Duong and Wickner, 1999; Mori et al., 2003). A loosened association between the SecY, SecE, and Secβ subunits of the translocon in prl mutants enhances conformational flexibility and makes the initiation of translocation more efficient (Duong and Wickner, 1999). The existence of a strong coupling between SecY and SecE is also supported by the observation that a single mutation in the TM region of the E. coli SecE (S105P) can compensate for several SecY mutation defects (Mori et al., 2003).
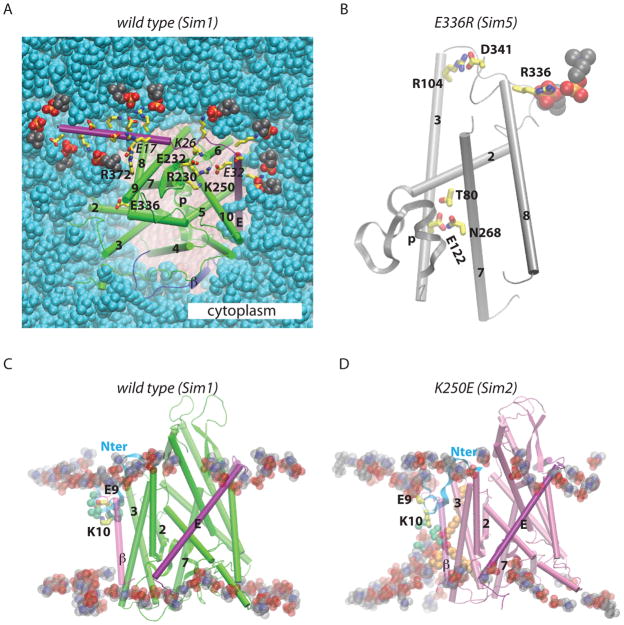
The analysis of H bonds in our simulations indicate that H bonding likely contributes to the strong coupling between SecY and SecE observed in experiments (Mori et al., 2003). SecE amino acids K26 and E32 H bond with SecY (Figures 2B, 8A, S2B,I); this segment of SecE is in the proximity of K250 (Figure 2B, 8A), and in the center of the SecA-binding hotspot in E. coli (Robson et al., 2007). The importance of the H bonding of SecE-E32 is supported by the significant conservation of this amino acid in all organisms (Figures S9–S11). On the extracellular side, SecE amino acids Y60 and K62 are in a location where they can H bond with SecY (Figure S2I).
Figure 8.
Coupling between the SecYE translocon and the lipid membrane. The wild-type translocon structure (green, Sim1; panels A and C) is compared to E336R (gray, Sim4; panel B) and K250E (pink, Sim2; panel D). (A) Snapshot from the end of Sim1 showing the SecE:lipid and SecE:SecY interactions. Charged amino acids on the cytoplasmic side of SecE, E336, and SecY amino acids that H bond to SecE on the cytoplasmic side are depicted as bonds with carbon in yellow, oxygen red, and nitrogen in blue. Selected SecE amino acids are labeled with italics. Lipid headgroups (shown as van der Waals spheres) located within 3.5 Å from cytoplasmic charged amino acids of SecE are depicted with the carbon atoms in black, oxygen red, nitrogen blue, and phosphorous atoms in orange; all other lipid heavy atoms are in cyan. Note that E335 does not interact closely with a lipid headgroup. (B) In the E336R mutant, R336 H bonds to a lipid headgroup. (C,D) Lipid and protein interactions of the N terminus in the wild type (panel C; Sim1) and K250E (panel D; Sim2). In the wild-type and in Sim3-Sim5, K10 snorkels towards the cytoplasmic side and salt-bridges with E9; in Sim2, K10 interacts with a solvated lipid headgroup. Water molecules within 9 Å of the Nζ atom of K10 are shown as green van der Waals spheres. The alkyl chains of a POPC lipid molecule whose headgroup interacts with K10 is depicted in brown.
The increased translocation of the more positively charged end of signal anchors by the yeast E382R mutant (corresponding to E336 in M. jannaschii) was explained as originating from the disturbance of the attractive interaction between the Glu and the positively-charged end of the signal anchors (Junne et al., 2007). Our computations suggest that the E336R mutation also affects the interactions between SecY and lipids and between the plug and the remainder of the protein (Figures 5, 6, 8B). In the wild-type, TM8-E336 is part of an H-bonded cluster that includes the cytoplasmic ends of TM2, TM3, and TM8 (Figure S2A). During Sim1 we observed H bonding of TM3-R104 to D341 and E336 (Table S1). In the mutant, R336 H bonds to a lipid headgroup, and R104 salt-bridges with D341 (Figure 8B). Because TM3 and TM8 are part of other H-bonding clusters (Figures 2C, S2A–D,F,G), the rearrangements of H bonds of E336 can perturb the plug and be propagated throughout the translocon.
The N terminus of SecY contains a helical segment flanked by charged amino acids (Figures 1A, S2C, S5). The crystal structure shows K10 pointing somewhat towards the extracellular side, the distance between the E9-Cδ and K10-Nζ atoms being 10.8 Å (Van den Berg et al., 2004). Within ~20ns of Sim1 and Sim3-Sim5, K10 snorkeled towards the cytoplasmic side and salt-bridged to E9 (Figure 8C). In Sim2 (K250E), K10 remained pointing towards the extracellular side and formed a stable interaction with a hydrated lipid headgroup (Figure 8D). Given the limited timescale of our simulations, we cannot definitely identify a specific effect of the K250E mutation on the lipid interactions of the SecY N terminus. It is important to note that the structure and the orientation of the long N terminus relative to SecY in the starting crystal structure (Van den Berg et al., 2004) may be affected by the crystal packing conditions.
Discussion
We have investigated the dynamics of wild-type and four mutant translocons in fluid, hydrated POPC lipid bilayers. The TM region and the plug segment are rather rigid in the closed state of the wild-type translocon (Figure 1C–E). In contrast, the cytoplasmic loops C4 and C5, and the C-terminal segment of SecY, which interact with SecY’s cytoplasmic partners, are very flexible (Figure 1C,D). The flexibility of these structural elements may be necessary to facilitate productive interactions with SecA or the ribosome, and the entry of newly synthesized peptides into the cytoplasmic channel of SecY.
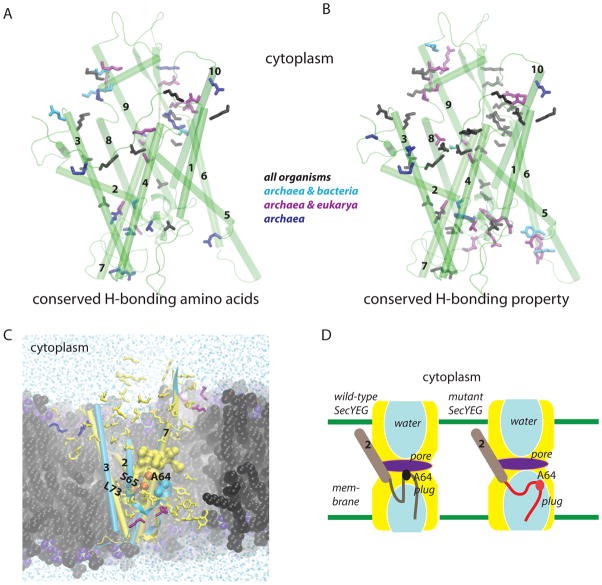
The rigidity of the TM region of the closed-state of the wild-type translocon seems to be due to the many H bonds that inter-connect various segments of the translocon (Figures 2, S1). Most of the H bonding amino acids of SecY are highly conserved; for some of the amino acids, the pattern of conservation depends on the organism (Figures 9A,B).
Figure 9.
Conserved H bonds and the relay of structural perturbations in the translocon. (A, B) Conservation of H-bonding amino acids (panel A) and of the H-bonding property (panel B). H-bonding amino acids whose dynamics were studied here are depicted as bonds colored in black, purple, cyan, or blue if they are conserved (at least 40%) in all organisms, in archaea and bacteria (cyan), archaea and eukarya (purple) or only in archaea (blue). See Figures S6–S8 for the detailed frequency analysis. (C) The many H bonds of SecY likely contribute to the fast relay of structural perturbation to the plug. H-bonding amino acids of SecY are colored in yellow, SecE – purple, and Secβ – blue. For simplicity, only helices TM2, TM3, TM7, and the plug are shown for the wild-type (yellow; Sim1) and the T72V/T80V/R104A mutant (cyan; Sim3). The hydrophobic pore amino acids of the wild type structure are shown as yellow surface (probe radius is 1.4 Å). The sidechains of A64, S65, and L73 are shown as van der Waals spheres depicted in orange for the wild type, and cyan for T72V/T80V/R104A. Notice the displacement of the plug in T72V/T80V/R104A relative to the wild type as indicated by A64 and S65. Lipid heavy atoms (cutaway view) are shown with carbon in black, phosphorous lime, oxygen violet, and nitrogen atoms in blue. Water oxygen atoms (cutaway view) are shown as blue dots. H atoms are not depicted. (D) Cartoon illustrating the displacement of the plug and enhanced solvation of the plug in the mutants.
In the M. jannaschii translocon, the gate helices TM2 and TM7, to which the signal peptide is thought to bind (Plath et al., 1998; Van den Berg et al., 2004), interconnect with each other and with TM3 (Figures 2C, 4A). Our sequence analysis suggests that the interconnectivity between TM2, TM3, and TM7 is a general characteristic of the translocons from archaea and eukarya (Figures 9A,B). Translocons from bacteria, which lack the amino acid corresponding to M. jannaschii TM7-N268 (Figures 3,4, S7), could share an interhelical H bond between TM2 and TM3 only (Figure 4B). In the crystal structure of the SecA-bound open translocon from T. maritima (Zimmer et al., 2008), TM7 is remote from the TM2-TM3 H bond (Figure 4B), but TM2 and TM3 remain H bonded via TM2-T83 and TM3-Q131 (T. maritima T83 and Q131 correspond to M. jannaschii T80 and E122, respectively; Figure S5). Our observation that TM3 mediates conserved H bonding with TM2 and TM7 in archaea and eukarya, and with TM2 in bacteria, is consistent with the important functional role of TM3 documented by experiments on the E. coli SecYEG (Shimokawa et al., 2003).
The plug segment of SecY seals the translocon in the closed state (Van den Berg et al., 2004), and displaces towards the extracellular side when the translocon opens (Tam et al., 2005; Robson et al., 2009). Our computations on the wild-type and mutant translocons suggest a relay mechanism based on H bonding and hydrophobic interactions that ensure that the plug rapidly senses perturbations, even at remote sites in the protein. In the wild-type protein, the structure and location of the plug are controlled by H bonding between amino acids of the plug, and between the plug and nearby amino acids (Figures 4, 5G, S2I,J). Through A64, the plug further connects to the hydrophobic pore (Figures 5A, 6A, 7A,C). TM segments 2, 5, 7, and 10, which contribute to the hydrophobic pore, participate in inter-helical H bonding with other structural elements of the translocon (Figures 6A, 2D). This extensive inter-connectivity likely explains the rapid propagation of perturbations from various sites of the translocon to the plug observed on the timescale of our simulations. Perturbation causes dislocation of the plug from the segment that links it to TM2, movement of the plug towards the extracellular side, and a markedly enhanced solvation of the plug amino acids (Figures 5–7, 9C–D). On the timescale of our simulations, details of the plug displacement and water interactions depend somewhat on the mutant. For example, the largest dislocation away from the linker to TM2 is observed in T72V/T80V/R104A (Sim3) and L406K (Sim4) (see the S65:L73 markers in Figure 5).
Our proposal that conserved inter-helical H bonds stabilize the closed state of the translocon and contribute to long-distance propagation of conformational changes, places the translocon among the membrane proteins in which the controlled breaking and forming of inter-helical H bonds is associated with conformational flexibility. For example, disruption of a highly conserved inter-helical salt bridge is associated with activation in the rhodopsin family of the G-protein coupled receptors (Kobilka and Deupi, 2007).
Lipid-protein H bonds have been shown to influence the local structure and dynamics of the GlpG rhomboid protease (Bondar et al., 2009). Consistent with the importance of translocon interactions with lipids, the composition of the lipid membrane surrounding the translocon affects the binding of SecA to SecYEG (Alami et al., 2007). The computations on the E336R mutant (Sim5) suggest that changes in the translocon-lipid interactions (Figures 8A,B) can contribute to the effect of specific mutations. It is also plausible that lipids influence the SecA-SecYEG complex not only via the binding of SecA to lipids (Breukink et al., 1993), but also via changes in the SecYEG local structure and dynamics: SecE H bonds to the both SecY and lipid headgroups (Figures 2B, 8A, S2B,I).
The computations presented here give a glimpse into the complex structural and dynamical response of the translocon to perturbations that interfere with H bonding. Further changes in the structure and dynamics of the mutant translocons are likely to occur on a longer timescale. A systematic study of the long-timescale dynamics of mutant translocons is necessary to derive a set of parameters that describe fully the essential structural features of the different classes of SecYEG phenotypes. Combined with the bioinformatics analyses, the detailed assessment of the translocon H bonds can also be used to select candidates for further investigation with site-directed mutagenesis experiments.
Experimental procedures
We used the crystal structure from (Van den Berg et al., 2004) for the starting SecYEβ protein coordinates in Sim1-Sim5. Hydrogen atoms were constructed using HBUILD in CHARMM (Brooks et al., 1983). The principal axes of SecY were aligned along the x, y, z directions using the VMD software (Humphrey et al., 1996). The SecYEβ complex was then embedded in the centre of a POPC lipid bilayer (475 lipid molecules) hydrated by 48,568 water molecules; chloride ions were added for charge neutrality (11, 9, 10, 12, and 13 ions in Sim1, Sim2, Sim3, Sim4, and Sim5, respectively). All Asp/Glu amino acids were considered negatively charged, and the Lys/Arg amino acids positively charged; the His amino acids, whose environment can be influenced by the local electrostatic environment (Baran et al., 2008), were modeled in the Nδ1 tautomeric state. The lengths of the bonds involving hydrogen atoms were constrained using the SHAKE algorithm (Ryckaert et al., 1977). The short-range real-space interactions were cut off at 12 Å using a switching function between 8 Å and 12 Å. Coulomb interactions were computed using the smooth particle mesh Ewald summation (Darden et al., 1993; Essmann et al., 1995). We used a Langevin dynamics scheme to maintain a constant temperature of 300K, and a Nosé-Hoover Langevin piston (Martyna et al., 1994; Feller et al., 1995) to maintain the pressure at 1 bar.
The NAMD software (Kalé et al., 1999; Phillips et al., 2005) with the CHARMM force field was used to perform MD simulations. We used the CHARMM22 (MacKerell et al., 1998) and CHARMM27 (Feller and MacKerell, 2000) parameters for the protein and lipid atoms, respectively; water molecules were TIP3P (Jorgensen et al., 1983). The reversible multiple time-step algorithm (Grubmüller et al., 1991; Tuckerman and Berne, 1992) was used to integrate the equations of motion with time-steps of 1 fs for the bonded forces, 2 fs for the short-range non-bonded forces, and 4 fs for the long-range electrostatic forces. During minimization, heating, and the first ~1 ns of equilibration, we used harmonic constraints of 5 kcal mol−1Å−2 for the protein, and 2 kcal mol−1Å−2 water, ions, and lipid molecules farther that ≈15 Å from the protein; the harmonic constraints on the protein atoms were lowered to 2 kcal mol−1Å−2 for the next ~1ns of equilibration, and switched of for a subsequent ~1 ns equilibration with harmonic constraints of 2 kcal mol−1Å−2 on the water molecules and ions. All harmonic constraints were then switched off. We used VMD (Humphrey et al., 1996) for trajectory analysis and molecular graphics.
Supplementary Material
Acknowledgments
This research was supported in part by grants from the National Institute of General Medical Sciences (GM-74637 to S.H.W) as well as the National Institute of Neurological Diseases and Stroke (GM86685 to S.H.W. and D.J.T.) and the National Science Foundation (CHE-0750175 to D.J.T.). An allocation of computer time from the National Science Foundation through the TeraGrid resources was essential for this work. C.M.D.V. is supported by ‘Programa de Retorno de Investigatores’ from the Junta de Andalucia and the Consejeria de Innovacion (TIC-02788). A.-N.B. thanks Dr. Maria Luiza Bondar for valuable discussions. The computations were performed on Ranger at the Texas Advanced Computing Center, and at the University of California, Irvine, High-Performance Computing Beowulf Cluster. We thank Joseph Farran of the University of California, Irvine, for excellent technical support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alami M, Dalal K, Lalj-Garolla B, Sligar SG, Duong F. Nanodiscs unravel the interaction between the secYEG channel and its cytosolic partner SecA. EMBO Journal. 2007;26:1995–2004. doi: 10.1038/sj.emboj.7601661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondar AN, del Val C, White SH. Rhomboid protease dynamics and lipid interactions. Structure. 2009;17:395–405. doi: 10.1016/j.str.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breukink E, Keller RC, de Kruijff B. Nucleotide and negatively charged lipid-dependent vesicle aggregation caused by SecA. Evidence that SecA contains two lipid-binding sites. FEBS Lett. 1993;331:19–24. doi: 10.1016/0014-5793(93)80289-7. [DOI] [PubMed] [Google Scholar]
- Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics. J Comput Chem. 1983;4:187–217. [Google Scholar]
- Brundage L, Hendrick JP, Schiebel E, Driessen AJM, Wickner W. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell. 1990;62:649–657. doi: 10.1016/0092-8674(90)90111-q. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Jiang Y, Mandon EC, Gilmore R. Identification of cytoplasmic residues of Sec61p involved in ribosome binding and cotranslational translocation. J Cell Biol. 2005;168:67–77. doi: 10.1083/jcb.200408188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T, York D, Pedersen L. Particle mesh Ewald: An N log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- Du Plessis DJF, Berrelkamp G, Nouwen N, Driessen AJM. The lateral gate of SecYEG opens during protein translocation. J Biol Chem. 2009;284:15805–15814. doi: 10.1074/jbc.M901855200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong F, Wickner W. The PrlA and PrlG phenotypes are caused by a loosened association among the translocase SecYEG subunits. EMBO J. 1999;18:3263–3270. doi: 10.1093/emboj/18.12.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emr SD, Hanley-Way S. Suppressor mutations that restore export of a protein with a defective signal sequence. Cell. 1981;23:79–88. doi: 10.1016/0092-8674(81)90272-5. [DOI] [PubMed] [Google Scholar]
- Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: The Langevin piston method. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- Feller SE, MacKerell AD., Jr An improved empirical potential energy function for molecular simulations of phospholipids. J Phys Chem B. 2000;104:7510–7515. [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut JS, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer ELL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower AM, Osborne RS, Silhavy TJ. The allele-specific synthetic lethality of prlA-prlG double mutants predicts interactive domains of SecY and SecE. EMBO J. 1995;14:884–893. doi: 10.1002/j.1460-2075.1995.tb07070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubmüller H, Heller H, Windemuth A, Schulten K. Generalized Verlet algorithm for efficient molecular dynamics simulations with long-range interactions. Mol Simul. 1991;6:121–142. [Google Scholar]
- Gumbart J, Schulten K. The roles of pore ring and plug in the SecY protein-conducting channel. J Gen Physiol. 2008;132:709–719. doi: 10.1085/jgp.200810062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke W, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jaud S, Fernández-Vidal M, Nillson I, Meindl-Beinker NM, Hübner NC, Tobias DJ, von Heijne G, White SH. Insertion of short transmembrane helices by the Sec61 translocon. Proc Natl Acad Sci USA. 2009;106:11588–11593. doi: 10.1073/pnas.0900638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- Junne T, Schwede T, Goder V, Spiess M. Mutations in the Sec61p channel affecting signal sequence recognition and membrane protein topology. J Biol Chem. 2007;282:33201–33209. doi: 10.1074/jbc.M707219200. [DOI] [PubMed] [Google Scholar]
- Kalé L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K. NAMD2: Greater scalability for parallel molecular dynamics. Journal of Computational Physics. 1999;151:283–312. [Google Scholar]
- Kobilka BK, Deupi X. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol Sci. 2007;28:397–406. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- Martyna GJ, Tobias DJ, Klein ML. Constant-pressure molecular-dynamics algorithms. J Chem Phys. 1994;101:4177–4189. [Google Scholar]
- Matsuyama S, Akimaru J, Mizushima H. SecE-dependent overproduction of SecY in Escherichia coli. FEBS Lett. 1990;269:96–100. doi: 10.1016/0014-5793(90)81128-b. [DOI] [PubMed] [Google Scholar]
- Mori A, Akiyama Y, Ito K. A SecE mutation that modulates SecY-SecE translocase assembly, identified as a specific suppressor of SecY defects. Journal of Bacteriology. 2003;185:948–956. doi: 10.1128/JB.185.3.948-956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Ito K. Different modes of SecY-SecA interactions revealed by site-directed in vivo photo-cross-linking. Proc Natl Acad Sci USA. 2006;103:16159–16164. doi: 10.1073/pnas.0606390103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne RS, Silhavy TJ. PrlA suppressor mutations cluster in regions corresponding to three distinct topological domains. EMBO J. 1993;12:3391–3398. doi: 10.1002/j.1460-2075.1993.tb06013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Braun B, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- Raden D, Song W, Gilmore R. Role of the cytoplasmic segments of Sec61a in the ribosome-binding and translocation-promoting activities fo the Sec61 complex. J Cell Biol. 2000;150:53–64. doi: 10.1083/jcb.150.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall LL, Hardy SJS. The promiscuous and specific sides of SecB. Nat Struct Biol. 2000;7:1077–1079. doi: 10.1038/81907. [DOI] [PubMed] [Google Scholar]
- Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- Robson A, Booth AEG, Gold VAM, Clarke AR, Collinson I. A large conformational change couples the ATP binding site of SecA to the SecY protein channel. J Mol Biol. 2007;374:965–976. doi: 10.1016/j.jmb.2007.09.086. [DOI] [PubMed] [Google Scholar]
- Robson A, Carr B, Sessions RB, Collinson I. Synthetic peptides identify a second periplasmic site for the plug of the SecYEG protein translocation complex. FEBS Lett. 2009;583:207–212. doi: 10.1016/j.febslet.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. Journal of Computational Physics. 1977;23:327–341. [Google Scholar]
- Sadlish H, Pitonzo D, Johnson AE, Skach WR. Sequential triage of transmembrane segments by Sec61α during biogenesis of a native multispanning membrane protein. Nature Struct Mol Biol. 2005;12:870–878. doi: 10.1038/nsmb994. [DOI] [PubMed] [Google Scholar]
- Sako T, Ino T. Distinct mutations in prl suppressor mutant strains of Escherichia coli respond either to suppression of signal peptide mutations or to blockage of staphylokinase processing. Journal of Bacteriology. 1988;170:5389–5391. doi: 10.1128/jb.170.11.5389-5391.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa N, Mori H, Ito K. Importance of transmembrane segments in Escherichia coli SecY. Mol Gen Genomics. 2003;269:180–187. doi: 10.1007/s00438-003-0804-8. [DOI] [PubMed] [Google Scholar]
- Smith MA, Clemons WM, Jr, DeMars CJ, Flower AM. Modeling the effects of prl mutations on the Escherichia coli SecY complex. J Bacteriol. 2005;187:6454–6465. doi: 10.1128/JB.187.18.6454-6465.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam PCK, Maillard AP, Chan KKY, Duong F. Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J. 2005;24:3380–3388. doi: 10.1038/sj.emboj.7600804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taura T, Akiyama Y, Ito K. Genetic analysis of SecY: additional export-defective mutations and factors affecting their phenotypes. Mol Gen Genet. 1994;243:261–269. doi: 10.1007/BF00301061. [DOI] [PubMed] [Google Scholar]
- Tsukazaki T, Mori H, Fukai S, Ishitani R, Mori T, Dohmae N, Perederina A, Sugita Y, Vassylyev DG, Ito K, Nureki O. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature. 2008;455:988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckerman M, Berne BJ. Reversible multiple time scale molecular dynamics. J Chem Phys. 1992;97:1990–2001. [Google Scholar]
- Van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- White SH, Von Heijne G. How translocons select transmembrane helices. Ann Rev Biophys Biophys Chem. 2008;37:23–42. doi: 10.1146/annurev.biophys.37.032807.125904. [DOI] [PubMed] [Google Scholar]
- Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.