Gene transcription and long-term memory storage have been linked in experiments going back for more than 30 years, but the molecular mechanisms responsible for the regulation of gene expression during memory consolidation remain the subject of intense investigation. Much work has focused on the role of individual transcription factors, such as cAMP-response element-binding protein (CREB) or nuclear factor of κB (NF-κB), in memory storage (Kaplan and Abel 2003; Yeh et al. 2004), but it is now clear that transcriptional regulation involves the concerted action of multiple transcription factors that interact with chromatin, a protein complex that packages DNA (Olins and Olins 2003). Originally thought to be static and structural in purpose, chromatin is now known to be very dynamic, exerting precise control over gene expression (Felsenfeld and Groudine 2003). In particular, the idea that chromatin remodeling may regulate gene expression for memory processes has gained considerable attention recently (Levenson and Sweatt 2005). It is this very concept that Chwang et al. (2006) investigate in their studies of transcriptional regulation during memory storage, which are described in this issue of Learning & Memory. Chwang et al. (2006) reveal that a critical signaling pathway in the hippocampus for memory storage, the extracellular signal-regulated kinase (ERK)/mitogen-activated protein kinase (MAPK) pathway, functions to modify chromatin during memory consolidation (Fig. 1).
Figure 1.
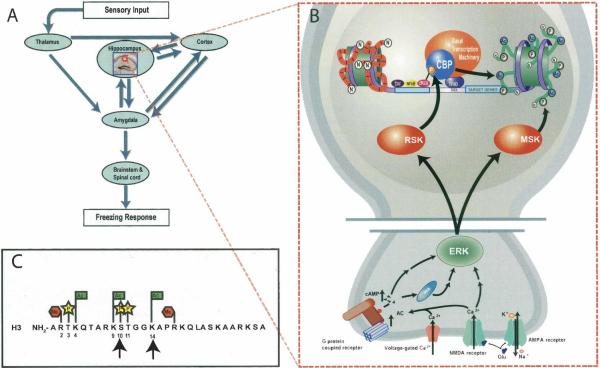
Memory formation in the hippocampus involves histone modifications. (A) Contextual fear conditioning is a behavioral paradigm used to study the neurobiology of memory and involves the activity of a complex neural circuit with many reciprocal connections. In this task, the animal is placed into a novel context and after a brief delay is given a series of footshocks. When later returned to the training context, mice display a stereotyped and species-specific anticipatory response that includes a motionless, defensive posture known as freezing, which is quantified as a measure of context-shock association. The hippocampal formation, a brain region important for encoding contextual fear conditioning (Sanders et al. 2003), receives input from both thalamus and cortex. Hippocampal output feeds back into the cortex as well as into the lateral and basolateral nuclei of the amygdala (Maren and Quirk 2004). Projections from the central nucleus of the amygdala transmit information via the periaqueductal gray and brainstem nuclei to spinal motor neurons that mediate freezing behavior. (B) The hippocampal formation is composed of multiple subregions, each of which is thought to play a distinct, yet integrative, role in hippocampal information processing. Within the CA1 subregion, changes in neuronal response properties involve activation of protein kinase A (PKA) and extracellular signal-regulated kinase (ERK). The data from Chwang et al. (2006) suggest that ERK indirectly mediates acetylation and phosphorylation of histone H3. Evidence also suggests that H3 Ser10 is a direct phosphorylation target of PKA (DeManno et al. 1999). Histone acetylation and phosphorylation are thought to have two effects: (1) to neutralize the positively charged histone tails, which reduces their affinity for DNA and thus facilitates transcription, and (2) to establish specific recognition sites for the recruitment of chromatin remodeling proteins such as histone acetyltransferases and ATP-dependent nucleosome remodeling complexes, which are associated with transcriptional activation. (C) The combinatorial patterns potentially established by different histone modifications may mediate specific gene expression profiles responsible for modulating the neuronal responses that underlie the formation of memory in the hippocampus. Black arrows in panel C indicate histone H3 residues examined by Chwang et al. (2006). Red octagons with “Me” represent methyl groups. Yellow stars with “P” represent phosphate groups. Green rectangles with “Ac” represent acetyl groups.
To understand how chromatin impacts gene expression, it is necessary to understand the nucleosome, which is the building block of chromatin. A nucleosome consists of a histone protein core (comprised of histones H2A, H2B, H3, and H4) and is the first level of packaging of genomic DNA. The amino-terminal “tails” of these histone proteins extend beyond the globular core and are sites for post-translational modifications, including acetylation, phosphorylation, methylation, ubiquitination, and sumoylation (Peterson and Laniel 2004). These histone modifications orchestrate the recruitment of specific chromatin remodeling protein complexes to mediate cell- and promoter-specific gene expression. Further, there is a dynamic interplay between histone modifications and DNA modifications (such as DNA methylation), thus creating staggering combinatorial possibilities for gene regulation. Chromatin structure can be modified in three different but related ways: First, nucleosomes may be repositioned by ATP-dependent protein complexes; second, histone variants may replace core histones; and third, histone tails may be covalently modified (Felsenfeld and Groudine 2003). Site-specific covalent modifications of histone tails can yield distinct transcriptional states. For example, the combination of histone H4 Lys8 acetylation, histone H3 Lys14 acetylation, and histone H3 Ser10 phosphorylation is often associated with transcriptional activation (Fig. 1). In contrast, tri-methylation of histone H3 Lys9 and the lack of histone H3 and H4 acetylation is associated with transcriptional repression (Peterson and Laniel 2004). Recent studies have revealed that histone modifications are especially relevant to mechanisms of transcriptional regulation during memory consolidation (Levenson and Sweatt 2005). Increasing histone acetylation at sites that correspond with transcriptional activation enhances memory and synaptic plasticity (Levenson et al. 2004), and the transcriptional coactivator and histone acetyltransferase CREB-binding protein (CBP) is critical for long-term memory and synaptic plasticity (Oike et al. 1999; Bourtchouladze et al. 2003; Alarcon et al. 2004; Korzus et al. 2004; Wood et al. 2005).
Now, Chwang et al. (2006) implicate histone H3 Ser10 phosphorylation and histone H3 Lys14 acetylation, modifications that correlate with transcriptional activation, in transcriptional regulation during memory consolidation. As an initial approach, the investigators took advantage of the well-established roles of protein kinase A (PKA) and protein kinase C (PKC) in hippocampus-dependent long-term memory formation. By pharmacological activation of these kinases in tissue slices, they found that both PKA and PKC induced transient increases in histone H3 Ser10 phosphorylation and histone H3 Lys14 acetylation in area CA1 of the hippocampus. Activation of either PKA or PKC also stimulated phosphorylation of ERK, which has been shown to be involved in H3 phosphorylation in cell culture (Soloaga et al. 2003). Furthermore, an inhibitor of MAPK/ERK kinase (MEK), the kinase primarily responsible for ERK phosphorylation, inhibited both phosphorylation and acetylation of histone H3. To examine the effect of these histone modifications on memory storage, the investigators turned to contextual fear conditioning (Fig. 1). The investigators observed a transient increase in both histone H3 Ser10 phosphorylation and histone H3 Lys14 acetylation that was paralleled by an increase in ERK phosphorylation. The peak level of these modifications was observed at 1 h post-conditioning, when each modification was >50% above its baseline level. In support of the hypothesis that these changes are associated with memory consolidation, both ERK phosphorylation and H3 modification were blocked by impairment of memory formation with repeated pre-exposure to the context in a latent inhibition paradigm or blockade of N-methyl-D-aspartate (NMDA) receptors. Finally, the investigators showed that the process that leads to modification of histone H3 requires MEK activity, suggesting a central role for MEK/ERK signaling in H3 modifications. Together, these experiments suggest that changes in histone H3 phosphorylation and acetylation in area CA1 of the hippocampus are regulated by ERK/MAPK during memory consolidation (Fig. 1). Currently, almost a dozen kinases are implicated in phosphorylating histone H3 at Ser 10 in either a stimulation-dependent manner or a mitosis-dependent manner (Bode and Dong 2005). During transcription, histone H3 phosphorylation is thought to neutralize the positive charge on histone tails by introducing a negatively charged phosphate group, thus relaxing chromatin and facilitating transcription (Grant 2001). H3 phosphorylation may also be involved in recruiting histone acetyltransferase complexes and ATP-dependent chromatin remodeling complexes to facilitate transcription (Lo et al. 2000). Interestingly, histone H3 phosphorylation and acetylation are proposed to be partly interdependent, which is the focus of much current investigation.
An intriguing hypothesis presented by Chwang et al. (2006) is that there exists a “histone code” that regulates specific gene expression profiles for distinct memory formation processes. The histone code is a theory popularized by C. David Allis and colleagues who proposed that distinct histone modification patterns, on one or more tails, can form recruitment sites for specific chromatin remodeling complexes to drive gene expression profiles required for particular cellular events (Turner 1993; Strahl and Allis 2000). Thus, the histone code extends the information potential of genomic DNA by providing an epigenetic system to regulate imprinted gene expression, cell fate, development, and now, as proposed by Chwang et al. (2006), memory formation. Indeed, the histone code and epigenetic marking of chromatin are thought to underlie stable and inherited patterns of gene expression during cellular differentiation, a process termed cellular memory (Turner 2002). Analysis using molecular biology and bioinformatics techniques will be necessary to demonstrate that specific combinations of histone modifications recruit distinct chromatin remodeling protein complexes for gene expression required for unique events.
In the meantime, Chwang et al. (2006) have set the stage by presenting compelling data showing that histone H3 is phosphorylated following contextual fear conditioning. Levenson et al. (2004) previously showed that histone H3 is acetylated following contextual fear conditioning whereas histone H4 is acetylated following a latent inhibition protocol for contextual fear conditioning. Although this dissociation between histone H3 and H4 acetylation may be a type of histone code for memory formation, these results are also consistent with the net charge model recently revisited by Dion et al. (2005). The net charge model posits that histone modifications form a simple code based on the cumulative charge of histone proteins, rather than the combinatorial complexity that could be generated by histone modification patterns, to drive different transcription profiles (for review, see Henikoff 2005). Thus, histone H3 acetylation generates one state of net charge difference, whereas histone H4 acetylation generates a second and different state, each with an associated effect on transcription. In any case, the combinatorial nature of the histone code is an attractive mechanism for gene regulation during memory formation.
If histone modifications, cellular memory, and behavioral memory all represent aspects of memory formation, what mechanisms are responsible for storing the memory? Could histone modifications be a component of the memory? We know that altering histone modifications (Alarcon et al. 2004; Korzus et al. 2004; Levenson et al. 2004) or altering activity of the enzymes that modify histones, such as CBP (Oike et al. 1999; Bourtchouladze et al. 2003; Alarcon et al. 2004; Korzus et al. 2004; Wood et al. 2005; for review, see Josselyn 2005), affects memory storage. But how can histone modifications affect memory? It may be that they are involved in a transient fashion to regulate gene expression for memory consolidation, as shown by Chwang et al. (2006). In this case, histone modifications gate a burst of transcription for a specific set of plasticity effector and regulator genes that then change the response properties of individual neurons in a network. Histone modifications may also mediate persistent changes in the expression of key plasticity effector or regulator genes required for maintenance of changes in neuronal behavior. These two possibilities are not mutually exclusive. Transient histone modifications may act downstream of signaling cascades to integrate multiple signals and ensure that a cascade of gene expression is activated only after a particular stimulus pattern (either spatially or temporally) is generated (Schreiber and Bernstein 2002). Furthermore, these histone modifications may act to integrate information about the activation and recruitment of several individual transcription factors. These histone modification patterns would alter the structure of chromatin as well as provide an interaction interface for transcriptional coactivators or corepressors that bind to modified histone tails to initiate transcription.
In addition, stable long-lasting histone modifications may maintain gene expression. For example, work by Taubenfeld et al. (2001) demonstrates that induction of the transcription factor CCAAT enhancer-binding protein (C/EBP) is observed in the hippocampus 9–20 h post-conditioning in an inhibitory avoidance task, suggesting that long-term memory involves a cascade of gene expression. Interestingly, C/EBP in Aplysia has been shown to be regulated by histone acetylation during synaptic plasticity (Guan et al. 2002), suggesting that these expression cascades are regulated by histone modification. Histone modifications are well-suited to regulate time-dependent gene expression in such cascades. In the yeast Saccharomyces cerevisiae, where ground-breaking research has elucidated much of what we currently know about the enzymes and protein complexes involved in chromatin regulation, histone modifications have been shown to be retained after transcription has subsided, suggesting that long-lasting modifications may provide a mark of recent transcription and possibly facilitate future gene expression (Turner 2003). The characterization of additional histone modifications, such as lysine methylation, during memory formation will determine whether such long-lasting changes occur with long-term memory formation. Identification of effector genes involved in long-lasting forms of memory and understanding the relationship of histone modifications to the expression of these genes will be essential to studying the role of stable long-lasting histone modifications in memory storage.
Although much of our discussion here has focused on the modifications of chromatin following learning, it is striking that researchers are able to see such changes in the acetylation and phosphorylation of “bulk” histones in hippocampal CA1 extracts at all. Indeed, one might expect to have to look at the modifications of histones in particular regulatory regions of subsets of neurons to see specific changes. The fact that changes can be observed in many neuronal properties, including synaptic transmission (McKernan and Shinnick-Gallagher 1997), GluR1 insertion (Rumpel et al. 2005), Arc expression (Guzowski et al. 1999, 2006), and changes in the slow afterhyperpolarization (AHP) (Wu et al. 2004), suggests that acquisition alters the properties of a large number of neurons. Together these studies suggest that 20%–40% of the neurons in a specific brain region may be activated by learning. The involvement of such a large percentage of hippocampal neurons during establishment of a memory suggests that initial representation may be distributed, rather than sparse. A sparse representation in which only a few neurons represent stored information maximizes the total number of possible engrams stored in the network, whereas a distributed network in which many neurons represent information sacrifices storage capacity for increased complexity and robustness (Rolls and Treves 1998). Because biochemical measures of neuronal activation, such as histone modification, integrate activity over a large window of time relative to individual neuronal activity, it is possible that the apparent network identified by these measures is a conjunction of many truly sparse networks. The final representation involved in the association may involve only a few of these individual networks, instead of the sum of networks activated during acquisition. Perhaps an important part of consolidation is the post-acquisition focusing of the network on certain gene targets in a subset of neurons.
It is becoming increasingly clear that histone modifications and chromatin remodeling are critical for gene expression during memory formation. The role of promoter-specific histone modifications has also become central to other areas of neuroscience, including research in epilepsy (Huang et al. 2002; Tsankova et al. 2004), drug addiction (Kumar et al. 2005; Levine et al. 2005), depression (Tsankova et al. 2006), and neurodegenerative diseases (Steffan et al. 2001). In addition to histone modifications, chromatin structure can be modified by ATP-dependent chromatin remodeling complexes, as well as the incorporation of histone variants into actively transcribed areas. Investigating each of these areas promises to enrich our understanding of the role of chromatin regulation in gene expression required for memory processes and perhaps enable the development of novel drugs to treat memory deficits that accompany many neurological and psychiatric disorders. By identifying H3 phosphorylation as a histone modification involved in memory storage, Chwang et al. (2006) have brought the field one step closer to understanding the complex interplay between chromatin and memory.
Acknowledgments
We would like to thank K. Matthew Lattal, Leslie Thompson, Noreen O'Connor-Abel, and John Guzowski for their comments on this manuscript. We would also like to thank Michele P. Kelly for the Nissl stain image in Figure 1.
References
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: A model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Bode AM, Dong Z. Inducible covalent posttranslational modification of histone H3. Sci. STKE. 2005;2005:re4. doi: 10.1126/stke.2812005re4. [DOI] [PubMed] [Google Scholar]
- Bourtchouladze R, Lidge R, Catapano R, Stanley J, Gossweiler S, Romashko D, Scott R, Tully T. A mouse model of Rubinstein-Taybi syndrome: Defective long-term memory is ameliorated by inhibitors of phosphodiesterase 4. Proc. Natl. Acad. Sci. 2003;100:10518–10522. doi: 10.1073/pnas.1834280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chwang WB, O'Riordan KJ, Levenson JM, Sweatt JD. ERK/MAPK regulates hippocampal histone phosphorylation following contextual fear conditioning. Learn. Mem. 2006 doi: 10.1101/lm.152906. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeManno DA, Cottom JE, Kline MP, Peters CA, Maizels ET, Hunzicker-Dunn M. Follicle-stimulating hormone promotes histone H3 phosphorylation on serine-10. Mol. Endocrinol. 1999;13:91–105. doi: 10.1210/mend.13.1.0222. [DOI] [PubMed] [Google Scholar]
- Dion MF, Altschuler SJ, Wu LF, Rando OJ. Genomic characterization reveals a simple histone H4 acetylation code. Proc. Natl. Acad. Sci. 2005;102:5501–5506. doi: 10.1073/pnas.0500136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- Grant PA. A tale of histone modifications. Genome Biol. 2001;2:reviews0003. doi: 10.1186/gb-2001-2-4-reviews0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Giustetto M, Lomvardas S, Kim JH, Miniaci MC, Schwartz JH, Thanos D, Kandel ER. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell. 2002;111:483–493. doi: 10.1016/s0092-8674(02)01074-7. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat. Neurosci. 1999;2:1120–1124. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Miyashita T, Chawla MK, Sanderson J, Maes LI, Houston FP, Lipa P, McNaughton BL, Worley PF, Barnes CA. Recent behavioral history modifies coupling between cell activity and Arc gene transcription in hippocampal CA1 neurons. Proc. Natl. Acad. Sci. 2006;103:1077–1082. doi: 10.1073/pnas.0505519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Histone modifications: Combinatorial complexity or cumulative simplicity? Proc. Natl. Acad. Sci. 2005;102:5308–5309. doi: 10.1073/pnas.0501853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Doherty JJ, Dingledine R. Altered histone acetylation at glutamate receptor 2 and brain-derived neurotrophic factor genes is an early event triggered by status epilepticus. J. Neurosci. 2002;22:8422–8428. doi: 10.1523/JNEUROSCI.22-19-08422.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josselyn SA. What's right with my mouse model? New insights into the molecular and cellular basis of cognition from mouse models of Rubinstein-Taybi syndrome. Learn. Mem. 2005;12:80–83. doi: 10.1101/lm.93505. [DOI] [PubMed] [Google Scholar]
- Kaplan MP, Abel T. Genetic approaches to the study of synaptic plasticity and memory storage. CNS Spectr. 2003;8:597–610. doi: 10.1017/s1092852900018873. [DOI] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, Russo SJ, Laplant Q, Sasaki TS, Whistler KN, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat. Rev. Neurosci. 2005;6:108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- Levenson JM, O'Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J. Biol. Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Levine AA, Guan Z, Barco A, Xu S, Kandel ER, Schwartz JH. CREB-binding protein controls response to cocaine by acetylating histones at the fosB promoter in the mouse striatum. Proc. Natl. Acad. Sci. 2005;102:19186–19191. doi: 10.1073/pnas.0509735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, Marmorstein R, Berger SL. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell. 2000;5:917–926. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- Maren S, Quirk GJ. Neuronal signalling of fear memory. Nat. Rev. Neurosci. 2004;5:844–852. doi: 10.1038/nrn1535. [DOI] [PubMed] [Google Scholar]
- McKernan MG, Shinnick-Gallagher P. Fear conditioning induces a lasting potentiation of synaptic currents in vitro. Nature. 1997;390:607–611. doi: 10.1038/37605. [DOI] [PubMed] [Google Scholar]
- Oike Y, Hata A, Mamiya T, Kaname T, Noda Y, Suzuki M, Yasue H, Nabeshima T, Araki K, Yamamura K. Truncated CBP protein leads to classical Rubinstein-Taybi syndrome phenotypes in mice: Implications for a dominant-negative mechanism. Hum. Mol. Genet. 1999;8:387–396. doi: 10.1093/hmg/8.3.387. [DOI] [PubMed] [Google Scholar]
- Olins DE, Olins AL. Chromatin history: our view from the bridge. Nat. Rev. Mol. Cell Biol. 2003;4:809–814. doi: 10.1038/nrm1225. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Laniel MA. Histones and histone modifications. Curr. Biol. 2004;4:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Rolls ET, Treves A. Neural networks and brain function. Oxford University Press; Oxford, UK: 1998. [Google Scholar]
- Rumpel S, LeDoux J, Zador A, Malinow R. Postsynaptic receptor trafficking underlying a form of associative learning. Science. 2005;308:83–88. doi: 10.1126/science.1103944. [DOI] [PubMed] [Google Scholar]
- Sanders MJ, Wiltgen BJ, Fanselow MS. The place of the hippocampus in fear conditioning. Eur. J. Pharmacol. 2003;463:217–223. doi: 10.1016/s0014-2999(03)01283-4. [DOI] [PubMed] [Google Scholar]
- Schreiber SL, Bernstein BE. Signaling network model of chromatin. Cell. 2002;111:771–778. doi: 10.1016/s0092-8674(02)01196-0. [DOI] [PubMed] [Google Scholar]
- Soloaga A, Thomson S, Wiggin GR, Rampersaud N, Dyson MH, Hazzalin CA, Mahadevan LC, Arthur JS. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 2003;22:2788–2797. doi: 10.1093/emboj/cdg273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL, Kazantsev A, Schmidt E, Zhu YZ, Greenwald M, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in. Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Taubenfeld SM, Wiig KA, Monti B, Dolan B, Pollonini G, Alberini CM. Fornix-dependent induction of hippocampal CCAAT enhancer-binding protein β and δ co-localizes with phosphorylated cAMP response element-binding protein and accompanies long-term memory consolidation. J. Neurosci. 2001;21:84–91. doi: 10.1523/JNEUROSCI.21-01-00084.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova NM, Kumar A, Nestler EJ. Histone modifications at gene promoter regions in rat hippocampus after acute and chronic electroconvulsive seizures. J. Neurosci. 2004;24:5603–5610. doi: 10.1523/JNEUROSCI.0589-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat. Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- Turner BM. Decoding the nucleosome. Cell. 1993;75:5–8. [PubMed] [Google Scholar]
- Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- Turner BM. Memorable transcription. Nat. Cell Biol. 2003;5:390–393. doi: 10.1038/ncb0503-390. [DOI] [PubMed] [Google Scholar]
- Wood MA, Kaplan MP, Park A, Blanchard EJ, Oliveira AM, Lombardi TL, Abel T. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn. Mem. 2005;12:111–119. doi: 10.1101/lm.86605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WW, Chan CS, Disterhoft JF. Slow afterhyperpolarization governs the development of NMDA receptor-dependent afterdepolarization in CA1 pyramidal neurons during synaptic stimulation. J. Neurophysiol. 2004;92:2346–2356. doi: 10.1152/jn.00977.2003. [DOI] [PubMed] [Google Scholar]
- Yeh SH, Lin CH, Gean PW. Acetylation of nuclear factor-κB in rat amygdala improves long-term but not short-term retention of fear memory. Mol. Pharmacol. 2004;65:1286–1292. doi: 10.1124/mol.65.5.1286. [DOI] [PubMed] [Google Scholar]