Summary
During heart morphogenesis, epicardial cells undergo an epithelial to mesenchymal transition (EMT) and migrate into the subepicardium. The cellular signals controlling this process are poorly understood. Here, we show that epicardial cells exhibit two distinct mitotic spindle orientations, directed either parallel or perpendicular to the basement membrane. Cells undergoing perpendicular cell division subsequently enter the myocardium. We found that loss of β-catenin led to a disruption in adherens junctions and a randomization of mitotic spindle orientation. Loss of adherens junctions also disrupted Numb localization within epicardial cells and disruption of Numb and Numblike expression in the epicardium led to randomized mitotic spindle orientations. Taken together, these data suggest that directed mitotic spindle orientation contributes to epicardial EMT and implicate a junctional complex of β-catenin and Numb in the regulation of spindle orientation.
Keywords: epicardium, β-catenin, Numb
Introduction
The establishment of cell polarity is important for many biological processes including asymmetric cell division and directed cell migration. Par6, aPKC and Par3 are part of the polarity complex that establishes apical-basal polarity (Kemphues et al., 1988; Muller and Wieschaus, 1996), directs asymmetric cell division (Cheng et al., 1995; Kemphues et al., 1988), and positions adherens junctions during epithelial development (Harris and Peifer, 2004). As adherens junctions are the primary sites of epithelial cell attachment, they are essential for maintaining epithelial sheet integrity and apical-basal polarity, and one necessary component of the adherens junction is β-catenin (Yap et al., 1997).
The epicardium is a single layer of mesothelial cells that attaches to and spreads over the myocardium beginning at E9.5 in the mouse. During heart morphogenesis, a subset of these cells migrates into the subepicardial space via the process of epithelial to mesenchymal transition (EMT) and subsequently differentiates into fibroblasts and vascular smooth muscle cells (VSMC) (Gittenberger-de Groot et al., 1998; Mikawa and Fischman, 1992; Poelmann et al., 1993; Wessels and Perez-Pomares, 2004). Previously, it has been demonstrated that β-catenin is essential for normal epicardial cell EMT (Zamora et al., 2007), but the cellular mechanisms behind this requirement remain unclear.
Here, we show that epicardial cells divide predominantly along two distinct axes, either parallel or perpendicular to the basement membrane. Cell divisions perpendicular to the basement membrane result in one cell remaining in the epicardium while the other cell exits the mesothelial monolayer. This process is reminiscent of an asymmetric cell division, a mechanism for cell differentiation in several epithelial cell populations (Lechler and Fuchs, 2005). The loss of β-catenin in the epicardium led to disruption of adherens junctions and randomized epicardial cell division patterns.
Furthermore, we found that Numb, an intracellular adaptor protein, associates with β-catenin in epicardial cells. Numb was originally identified in Drosophila as a cell fate determinant, where it is asymmetrically distributed in two daughter cells during neural progenitor division (Frise et al., 1996; Petersen et al., 2002; Spana and Doe, 1996; Uemura et al., 1989; Wirtz-Peitz et al., 2008). In mammals, two homologs, Numb and Numblike, exist (Verdi et al., 1996; Zhong et al., 1996), and they appear to function redundantly during development (Li et al., 2003; Petersen et al., 2004). Although the best known function of Numb is its role in asymmetric cell division (Knoblich et al., 1995; Rhyu et al., 1994; Wu et al., 2007), it also has been suggested to function in migration (Nishimura and Kaibuchi, 2007), cell death (Orgogozo et al., 2002), cell adhesion, and cell polarity (Rasin et al., 2007). We found that Numb and β-catenin co-localized to adherens junctions, and that epicardial cells with Numb and Numblike deletion displayed abnormal spindle orientation and failed to enter into the myocardium similar to epicardial cells lacking β-catenin.
Results
Epicardial cell proliferation is required for entry into myocardium
Epicardial cells arise from a small outcropping of cells called the proepicardium (Komiyama et al., 1987; Viragh and Challice, 1981). To determine how this limited number of cells expands to encompass the entire heart, we examined epicardial cell proliferation using BrdU incorporation. Because epicardial cells are a single layer of mesothelial cells surrounding the myocardium, we were able to use collagen IV staining to delineate the epicardial cell population (Fig. 1A). From the point of epicardial spreading until a few days after birth, we observed the highest rate of proliferation at E11.5 with a consistent decrease in the proliferation rate each subsequent day (Fig. 1B–C).
Figure 1. Cell proliferation is required for epicardial cell EMT.
(A) The basement membrane of the epicardium was identified by staining for collagen IV (Col IV). The layer of mesothelial cells attached to the basement membrane on the outer surface of the heart is the epicardium. (B) BrdU labeling indicates epicardial cell proliferation. Arrowheads point to epicardial cells that are BrdU+. Scale bar = 20 µm. (C) Proliferation rate of epicardial cells at indicated time points. Six midcoronal sections per heart for each age were quantified. (D–E) WT1 and GFP from PDGFRαGFP/+ mice were used to visualize epicardial cell location with ex vivo culture and aphidicolin treatment. (F) Quantification of WT1+ cells present within myocardium after treatment. (G) PDGFRαGFP/+ hearts were used to visualize epicardial cell location with ex vivo culture and cyclin D1 over-expression at 48 hours. (H) Quantification of GFP+ within the myocardium after 48 hours of culture with control adenovirus or cyclin D1 adenovirus. Six midcoronal sections per heart were quantified. Scale bar = 10 µm. See also figure S1.
To test whether epicardial cell division was required for epicardial cell entry into the myocardium, we arrested the cell cycle using a specific inhibitor of DNA polymerase, aphidicolin. E12.5 hearts were treated with aphidicolin or vehicle and then labeled with BrdU. As shown by BrdU incorporation, aphidicolin treatment led to an arrest in the cell cycle (Fig. S1A). We next determined if aphidicolin treatment resulted in a failure of epicardial cell entry into the myocardium. Epicardial cells were traced by using a PDGFRαGFP/+ knock-in line (Hamilton et al., 2003). In this line, H2b–eGFP is under the control of the PDGFRα promoter. In heterozygous PDGFRαGFP/+ mice, GFP expression faithfully recapitulates PDGFRα expression and at E12.5 is restricted to a single layer of epicardial cells surrounding the heart (Fig. S1B). A secondary means of identifying epicardial and epicardial derived cells (EPDC) was the expression of Wilm’s tumor antigen 1 (WT1) (Moore et al., 1999; Zhou et al., 2008). At time zero in the control, WT1+ and GFP+ (Fig. 1D–E) cells resided solely within the epicardial layer, but after 48 hours ex vivo culture, control hearts contained labeled cells within the subepicardium. In contrast, there were significantly fewer WT1+ or GFP+ cells in this region in aphidicolin treated hearts (Fig. 1D–F). The inability of aphidicolin treated epicardial cells to enter the subepicardial space was not permanent, because epicardial cells entered the subepicardial space after aphidicolin was removed (Fig. 1D–F). The reduced number of GFP+ cells in myocardium was not because of suppression of PDGFRα transcription by aphidicolin, as PDGFRα mRNA expression and GFP intensity was not significantly altered (Fig. S1 C – D). We observed very similar results when a second cell cycle inhibitor, doxorubicin, was used (Fig. S1E). Conversely, when we overexpressed cyclin D1 using adenoviral transduction (Ad-cyclin D1) to drive cell cycle progression, we observed an increase in the number of epicardial cells that had entered into the myocardium (Fig. 1G–H). Fig. S1F illustrates that there was a high efficiency of epicardial transduction. These results indicate that cell division is an important component of epicardial cell entry into the myocardium.
Epicardial mitotic spindles occur in two orientations
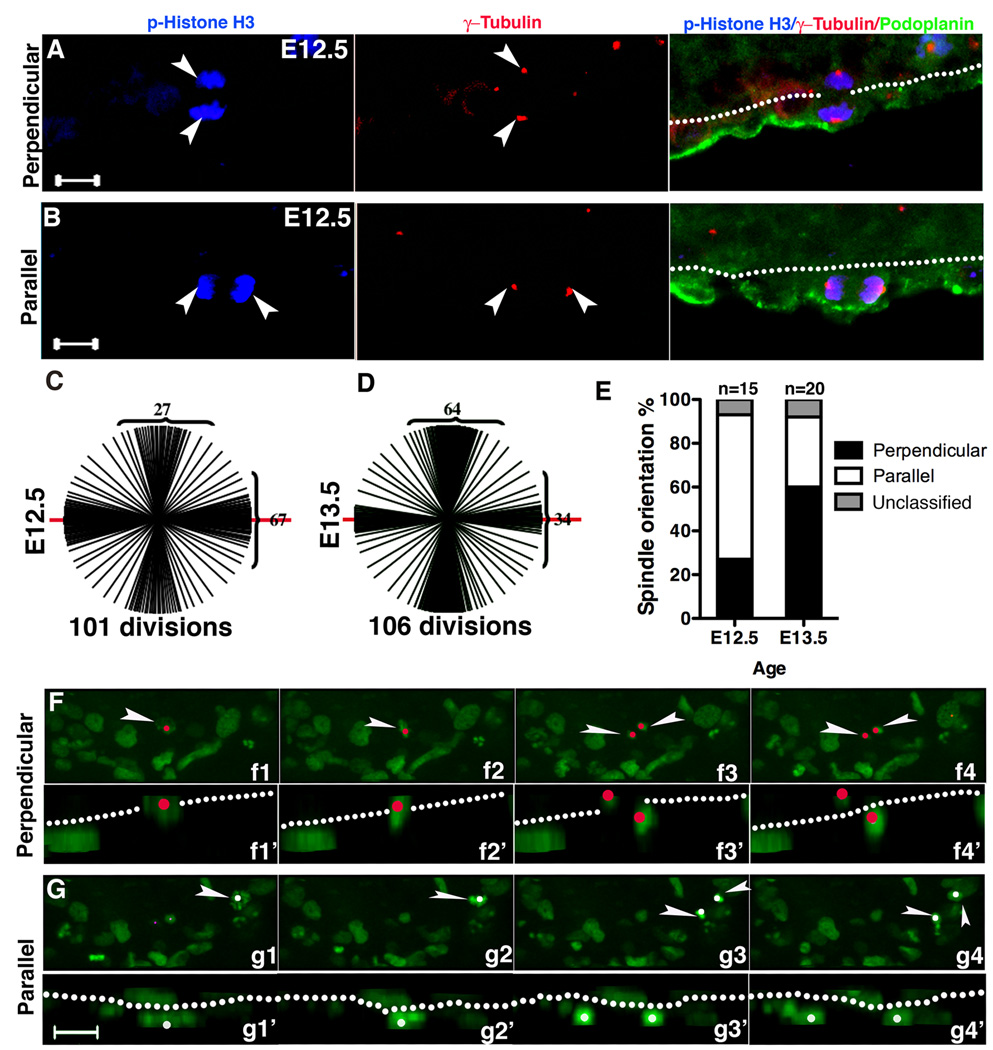
During development a subset of epicardial cells must exit the mesothelial monolayer and become mesenchymal. To determine if orientation of cell division impacts these events, we investigated spindle orientation of dividing epicardial cells identified by phosphorylated Histone H3 (p-Histone H3) and podoplanin (Mahtab et al., 2008). We observed that epicardial mitotic spindles occurred predominantly in two orientations, either perpendicular or parallel to the basement membrane. We quantified these patterns using α, β, or γ-tubulin staining. If the angle of the spindle was less than 30° from the basement membrane it was defined as a parallel division. If the angle of the mitotic spindle was 60–90° from the basement membrane it was defined as a perpendicular division (Fig. 2A–B and Fig. S2A).
Figure 2. Mitotic spindle orientation of epicardial cells.
(A–B) E12.5 hearts stained with podoplanin, p-Histone H3 and γ-tubulin indicate epicardial mitotic spindles are either (A) perpendicular or (B) parallel to the basement membrane indicated by dotted line. Arrowheads point to epicardial cells positive for p-Histone H3 labeled centrosomes. (C–E) Quantification of epicardial cell spindle orientations at E12.5 and E13.5. Spindles were visualized by staining for γ-tubulin (as in A and B) or α-tubulin (as in Fig. S2). The basement membrane was considered 0° and is represented by the red line. Each line represents the spindle orientation of a dividing epicardial cell. Numbers in brackets quantify the divisions that occur between 0–30° and 60°–90°. (E) Ratio of spindle orientations at each embryonic stage. (F,G) Time lapse and Z stack images of E12.5 hearts cultured ex vivo. Tracing GFP positive cells, we observed divisions that were either (F) perpendicular or (G) parallel to the basement membrane. f1–f4 are consecutive images that were taken every 6 minutes. Arrowhead points to a cell that starts in (f1) pro-metaphase, then moves to (f2) metaphase, (f3) telophase, and (f4) produces two daughters. g1–g4 is similar to f1–f4 except this cell completed a parallel division. Images indicated by the primes are side views, and nuclei are indicated by red and white dots, respectively. Dotted lines indicate the basement membrane. Scale bar = 10 µm. See also figure S2.
To specifically identify dividing epicardial cells, we stained for WT1 and α-Tubulin. We found WT1 expressing cells undergoing perpendicular and parallel divisions (Fig. S2B). To demonstrate that the perpendicular division results in a viable cell within the subepicardium, we used murine stem cell virus (MSCV) transduction to preferentially target dividing cells. Whole hearts were transduced with MSCV-GFP and after 48 hours were sectioned and imaged. As expected we found pairs of cells in two configurations. The first configuration was one cell in the epicardium and the other abutting it in the subepicardium; the second configuration resulted in both cells residing in the epicardium (Fig. S2C). Quantification of these events showed that 27% ± 2.5% of the pairs at E12.5 and 74% ±3.2% at E13.5 resulted from a perpendicular cell division (n=172 pairs from 6 E12.5 hearts and n=136 pairs from 3 E13.5 hearts). These results are consistent with the division ratio demonstrated by tubulin staining at E12.5 and E13.5 (Fig. 2C–E). At E12.5 parallel divisions were more prominent, while at E13.5 perpendicular divisions occurred more often.
To observe these division patterns in real time, we used time-lapse confocal microscopy to image whole hearts. Using confocal microscopy and hearts from PDGFRαGFP/+, we traced the division patterns of epicardial cells. Consistent with our above results, epicardial cells displayed two distinct division patterns. Furthermore, the orientation of cell division appeared to influence the fate of the epicardial cell. When perpendicular epicardial cell division occurred, we observed one GFP+ cell in the epicardium and one cell in the subepicardium (Fig. 2F). When a parallel division occurred the two GFP+ cells remained within the plane of the epicardium (Fig. 2G). Using GFP to track epicardial cell divisions via time-lapse imaging, we found that 33% of the epicardial cells divided perpendicularly and 67% divided in the parallel orientation (109 cell divisions from 18 E12.5 hearts). In total, we observed 38 cells entering the myocardium, and 36 cells entered via perpendicular division, while two appeared to migrate in without any apparent cell division. Time-lapse movies of a vertical and perpendicular cell division are provided in supplementary data (Movie S1a and S1b).
It has been proposed that epicardial cells undergo EMT in a stereotypic fashion where the dorsal cells at the atrio-ventricular junction undergo EMT earlier than ventral cells (Komiyama et al., 1987). Our data suggested that preferential perpendicular divisions coincide with the induction of EMT. To document this, we imaged E12.5 and E13.5 whole hearts for epicardial cell spindle orientation. The epicardial cells were identified by podoplanin staining (Mahtab et al., 2008) and spindle orientation was determined by p-Histone H3 and α-tubulin. The dominant spindle orientation of E12.5 epicardial cells was parallel, while for E13.5 epicardial cells, it was perpendicular (Fig. 3A–B and Table SI). Taken together these data suggest that two distinct orientations of cell division occur within the epicardium and that perpendicular divisions follow the pattern of EMT in the developing epicardium.
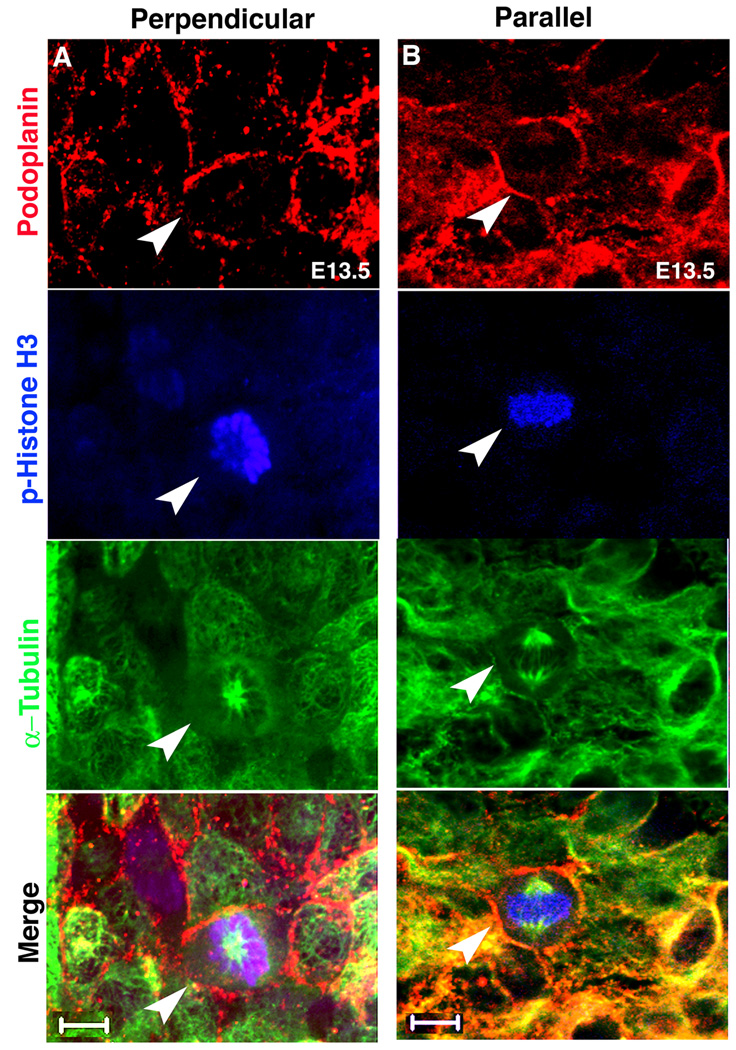
Figure 3. Whole mount staining of dividing epicardial cells.
(A) Perpendicular and (B) parallel divisions can be observed in whole mount stained hearts. Podoplanin, p-Histone H3 and α-Tubulin stained: epicardial cells, mitotic cells and spindles respectively. Arrowheads point to the mitotic cells. Scale bars = 10 µm. See Table S1 for additional information.
β-catenin is required to establish adherens junctions and cell polarity
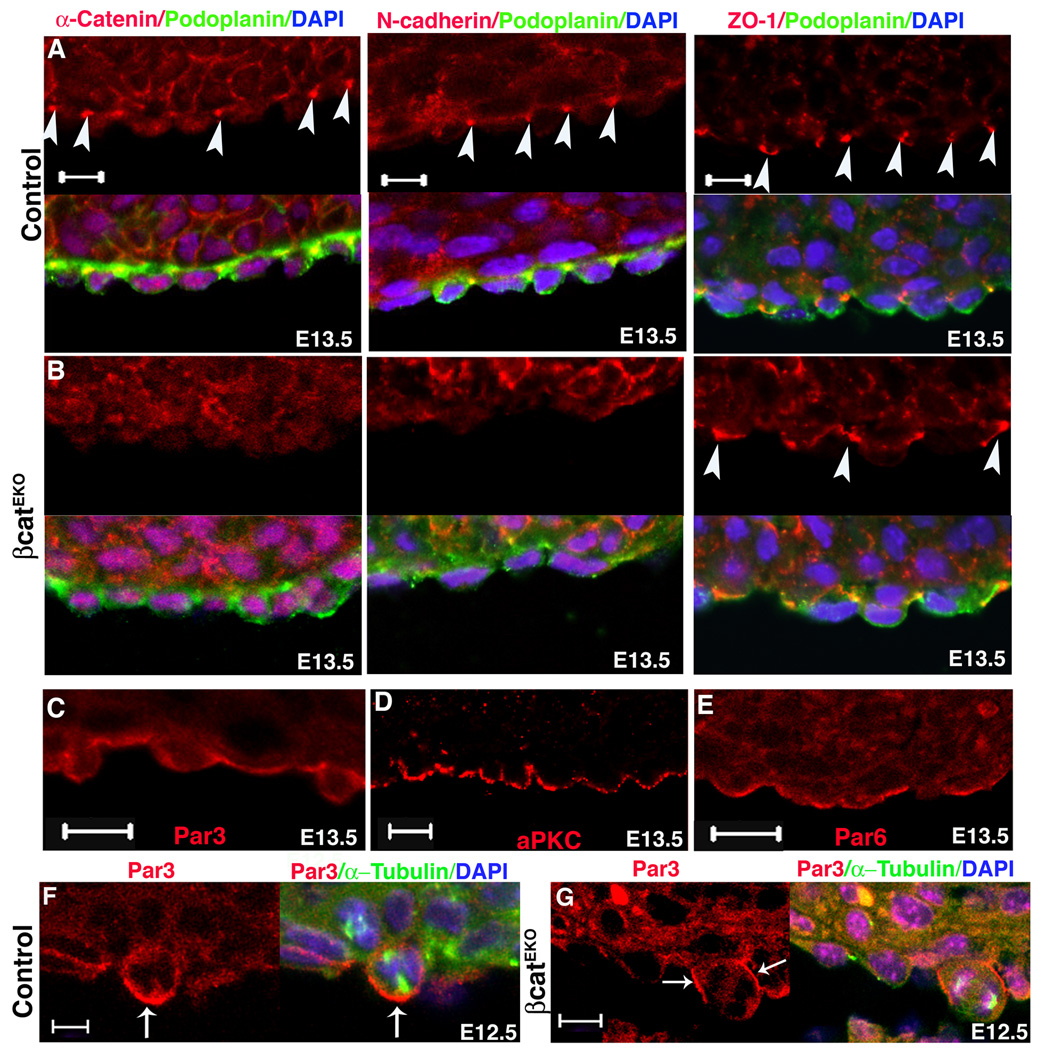
Previous work has shown that epicardial deletion of β-catenin (βcatEKO) results in failure of epicardial cell EMT in vivo, although β-catenin null epicardial cells still undergo EMT in vitro (Zamora et al., 2007). As β-catenin is an essential component of the adherens junction complex, we determined if adherens junctions were disrupted in βcatEKO epicardial cells. In vertebrates, adherens junction components include the transmembrane protein cadherins, α-catenin and β-catenin (Hartsock and Nelson, 2008; Tepass, 2002), and we investigated the localization of N-cadherin and α-catenin in epicardial cells. We found that the adherens junctions proteins, N-cadherin and α-catenin, localized to the basal lateral domains of epicardial cells in control but not in βcatEKO hearts (Fig. 4A–B), while the tight junction marker ZO-1 displayed normal localization (Fig. 4A–B) in both genotypes. Adherens junctions are also required to establish cell polarity and asymmetric cell division (Kaplan et al., 2009; Lu et al., 2001a; Marthiens and ffrench-Constant, 2009), and this notion has been supported by reports that β-catenin localization establishes cell polarity in some tissues (Cox et al., 1996; Fu et al., 2006; Morel and Arias, 2004). We then assessed whether β-catenin deficient epicardium displayed disrupted epicardial cell polarity. We first determined the distribution of cell polarity machinery proteins in wild type epicardial cells. Par3, Par6, and aPKC (PKCζ) are important components of the epithelial cell polarity complex (Knoblich, 2008). We found that all three of these proteins were localized to the apical surface of epicardial cells (Fig. 4C–E). These data demonstrate that the epicardium is a polarized epithelium. We also examined the polarity of these proteins in βcatEKO. Although we did not see abnormal localization of these polarity proteins in interphase cells (data not shown), in 33 out of 40 dividing βcatEKO cells, Par3 was no longer localized to the apical cortex (Fig. 4F–G).
Figure 4. Epicardial cell junctions and polarity.
Formation of adherens junctions (α-catenin and N-cadherin) and tight junctions (ZO-1) in (A) control and (B) βcatEKO. Arrowheads point to the junctions. Podoplanin marked epicardial cells. (C–E). Wild type hearts were stained for the indicated cell polarity proteins. (F–G) Par3 localization in (F) control and (G) βcatEKO mitotic epicardial cells. Arrow indicates accumulated Par3. Scale bar = 10 µm.
Loss of β-catenin leads to randomized spindle orientation
Previous reports have suggested that adherens junctions are important for mitotic spindle orientation (den Elzen et al., 2009; Kraut et al., 1996; Lechler and Fuchs, 2005; Lu et al., 2001a). Because we observed a disruption in epicardial cell adherens junctions, we examined the spindle orientation in βcatEKO epicardial cells. βcatEKO epicardial cells at E12.5 contained a lower percentage of perpendicular divisions than the controls. Similar results were obtained in E13.5 epicardial cells (Fig. 5A–B). In the absence of β-catenin, epicardial cell division occurred randomly, which correlated with fewer cells entering the myocardium (Zamora et al., 2007). These results suggest that β-catenin may be important for directing epicardial spindle orientation.
Figure 5. Spindle orientation in βcatEKO epicardial cells and rescue of EMT by a transactivaton domain deficient β-catenin.
(A,B) Spindle orientation of control and βcatEKO epicardial cells at E12.5 and E13.5. For illustration purposes, spindle orientation events from only 9 hearts for each genotype and age are shown in the diagrams. Bar graphs at the right indicate the summary of spindle orientations. (C) In βcatEKO hearts, adenoviruses overexpressg full length (Ad-βcat) or transactivation domain deleted β-caten (Ad-trβcat) rescue βcatEKO EMT but Ad-GFP and Ad-dnTCF4 do not. (D) EMT results from wild type hearts transduced with Ad-dnTCF4 or Ad-TCF4. See also figure S3.
To distinguish between the adherens junction function of β-catenin and its role in transcription, we rescued βcatEKO hearts with either full-length or truncated, constitutively active β-catenin (Barth et al., 1999; Reya et al., 2003). The truncated β-catenin lacks the transactivation domain (Ad-trβcat) (Hsu et al., 1998; Vleminckx et al., 1999). Expression of both the truncated β-catenin and full length β-catenin promoted epicardial cell entrance into the myocardium (Fig. 5C and Fig. S3A). In contrast, Ad-GFP or a dominant negative TCF4 (Ad-dnTCF4) (Kolligs et al., 1999) failed to rescue epicardial cell EMT (Fig. 5C). We also found that Ad-TCF4 and Ad-dnTCF4 did not alter wild type epicardial cell entry into the myocardium (Fig. 5D). We verified that the Ad-trβcat and Ad-dnTCF4 inhibited Wnt3a induced gene transcription in primary cultured epicardial cells (Fig. S3B). In primary cultured βcatEKO epicardial cells, Ad-βcat induced Wnt target gene transcription, while Ad-trβcat did not (Fig.S3C), confirming the transactivation incompetence of the truncated β–catenin.
To further demonstrate that the transactivation activity of β–catenin is not required for epicardial cell entry into the myocardium, we treated hearts with Inhibitor of Wnt Response (IWR) ENDO, which abrogates Axin degradation (Chen et al., 2009). We found that activation of β-catenin signaling was reduced by IWR ENDO (Fig. S3D–F). However, this reduction did not affect epicardial cell entry into the myocardium (Fig. S3G–H). In summary, these results suggest that β-catenin transcriptional activity was not required for epicardial cell entry into the myocardium.
Numb and β-catenin associate with each other
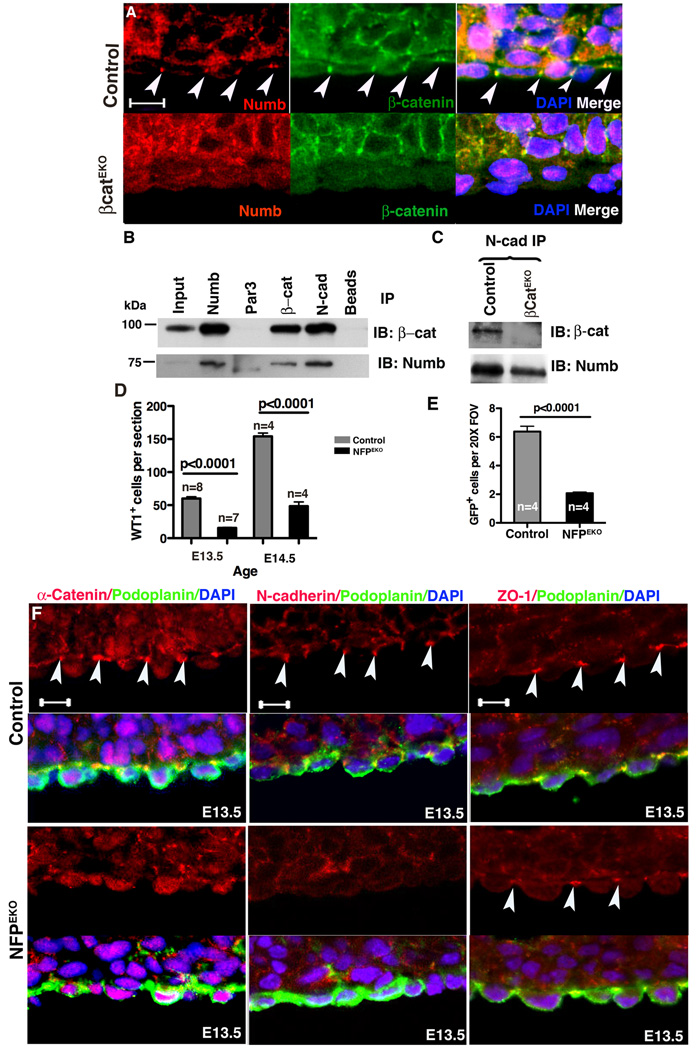
The polarization of epicardial cells caused us to further investigate proteins that are asymmetrically localized. Because Numb associates with β-catenin (Rasin et al., 2007) and has been shown to localize asymmetrically in progenitor cell populations (Zhong et al., 1996), we examined its expression in the epicardium. Numb was expressed by both cardiomyocytes and epicardial cells but in the latter localized to adherens junctions (Fig. 6A, Fig. S4A–B and Fig. S5A). We then observed that Numb localization changed in dividing cells. In late metaphase, regardless of the orientation of cell division, Numb weakly localized to the basal lateral surface in roughly 70% of the dividing epicardial cells (n=114 mitotic events in E12.5 10 hearts) (Fig. S4B and data not shown). Consistent with reports that localization of Numb and apical proteins are reciprocal (Smith et al., 2007), Par3 and Numb localized to the apical and basal domain of cells respectively, regardless of their division pattern (Fig. 4F, Fig. 6A, Fig. S4, Fig. S5A and data not shown).
Figure 6. Numb function in adherens junctions and NFPEKO epicardial phenotype.
(A) Numb localized to adherens junctions in control but not in βcatEKO epicardial cells. Arrowheads indicate adherens junctions. (B) Numb, Par3, N-cadherin (N-cad) and β-catenin (β-cat) were immunoprecipitated from primary epicardial cell lysates and blotted for the indicated proteins. (C) N-cadherin was immunoprecipitated from control and βcatEKO epicardial cell lysates and blotted for interactions with the indicated proteins. Input was 5% of the lysate. (D) Fewer WT1+ cells were present in the myocardium of NFPEKO than in control hearts. (E) Ad- GFP was used to trace epicardial cell EMT in an ex vivo culture system. (F) Adherens junctions (α-catenin and N-cadherin) and tight junctions (ZO-1) in NFPEKO epicardial cells. Arrowheads point to junctions. Podoplanin was used to identify epicardial cells. Scale bar = 10 µm. See also figure S4.
The disrupted cell polarization and adherens junctions of βcatEKO epicardial cells prompted us to examine Numb localization in these hearts. In wild type epicardium, Numb was localized to adherens junctions and co-localized with β-catenin. However, in βcatEKO epicardial cells, Numb localization in adherens junctions and at the basal surface did not occur (Fig. 6A). We then determined whether Numb, β-catenin and N-cadherin existed in a complex using primary epicardial cell lysates. We found that β-catenin and N-cadherin precipitates, but not Par3 precipitates, contained Numb. Similarly, Numb and N-cadherin, precipitates contained β-catenin (Fig. 6B). These data show that β-catenin and Numb exist in a complex similar to what had been suggested previously (Rasin et al., 2007). To determine if the interaction between Numb and N-cadherin was β-catenin dependent, we precipitated N-cadherin from βcatEKO and control epicardial cells lysates. We found that the N-cadherin and Numb interaction occurred in the absence of β-catenin (Fig. 6C). These results suggested that N-cadherin, β-catenin and Numb may exist as a complex.
Numb family proteins (NFP) are required for epicardial cell development
To study the functions of NFP in epicardial cells, we generated animals lacking Numb and Numblike in epicardial cells (NFPEKO). Fig. S5A–B demonstrates efficient loss of expression Numb in epicardial cells. Real-time PCR demonstrated a 76%±3 and 56%± 7 reduction in Numb and Numblike expression levels in primary epicardial cell cultures, respectively (data not shown). Loss of NFP in the epicardium resulted in embryonic lethality between E12.5 and E14.5. Because our data suggested an interaction with β-catenin, we examined the EMT process in these mutant hearts. We found that fewer WT1+ cells entered the myocardium in NFPEKO hearts at both E13.5 and E14.5 (Fig. 6D and Fig. S5C). We also used Ad-GFP transduction (Mellgren et al., 2008) to trace epicardial cell migration ex vivo and found that NFPEKO hearts had significantly fewer GFP+ cells in the myocardium after stimulation (Fig. 6E). Finally, we used GATA5CreTg:ROSA26RYFP lineage tracing to follow the epicardially derived cells and found fewer cells within the subepicardial space (Fig. S5D–E).
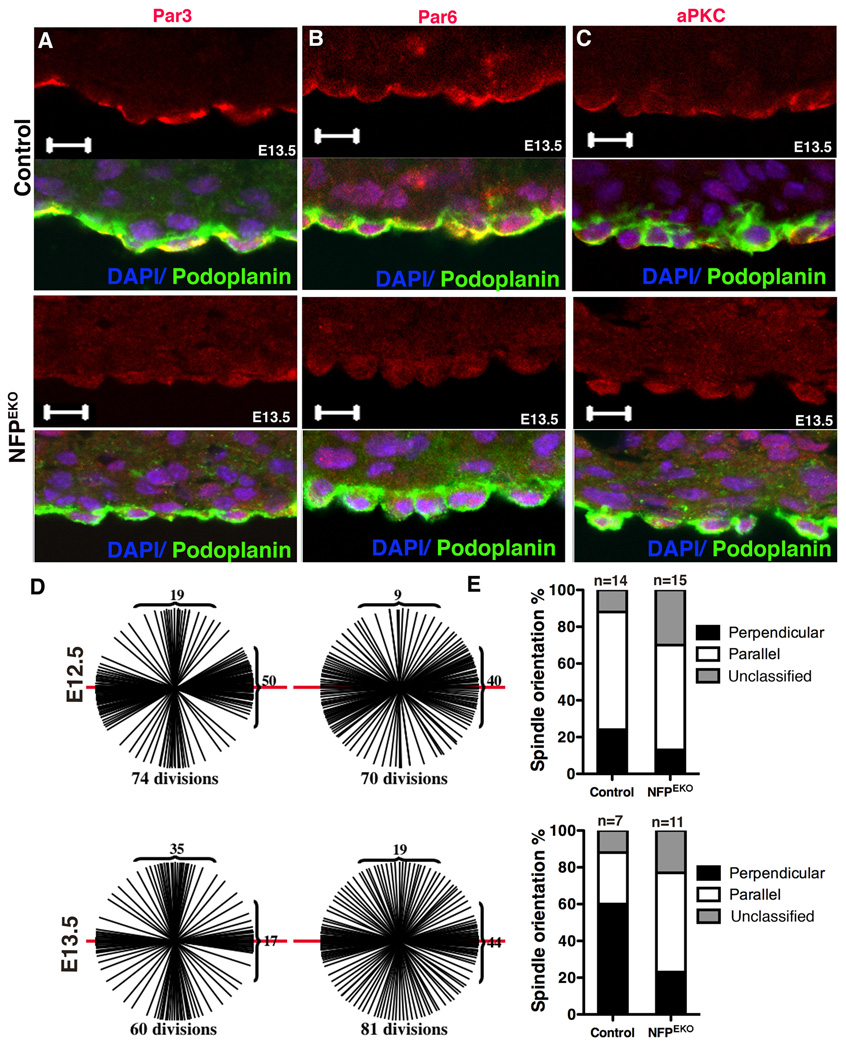
As Numb is a component of adherens junctions (Kuo et al., 2006; Wang et al., 2009), we investigated whether NFP EKO epicardial cells displayed adherens junction and cell polarity defects similar to βcatEKO hearts. Similar to βcatEKO, adherens junction proteins were mislocalized (Fig. 6F). Par3, aPKC and Par6 were also mis-localized in NFPEKO epicardial cells (Fig. 7A–C). By contrast, ZO-1 localization was not disrupted (Fig. 6F).
Figure 7. NFP mutant hearts exhibit disrupted epicardial polarity and spindle orientation.
(A–C) NFPEKO epicardial cells displayed abnormal apical basal polarity based on Par3, Par6 and aPKC localization. (D) Spindle orientation of control and NFPEKO epicardial cells. Spindle orientation events from 9 hearts for each genotype, except control at E13.5 which used 7 hearts. (E) Indicates the ratio of spindle orientations at each embryonic stage. Scale bar = 10 µm. See also figure S5.
We then investigated spindle orientation in NFPEKO epicardial cells. Compared to controls, NFPEKO epicardial cells had fewer cells undergoing perpendicular divisions, and the spindle orientation appeared to occur more in the parallel orientation (Fig. 7D–E). Taken together, these results suggest that β-catenin and NFP expression are important for directing the orientation of epicardial cell division and that this may have some impact on the ability of epicardial cells to undergo EMT.
Discussion
The development of the epicardium is a highly dynamic process that plays a critical role in producing coronary VSMC and cardiac fibroblasts. Little is known about the processes that direct a subpopulation of these cells to exit the mesothelial epicardium. A better understanding of these events may allow the manipulation of adult epicardial cells to facilitate myocardial regrowth after cardiac damage has occurred.
During EMT, epithelial cells undergo morphological changes and become mobile mesenchymal cells (Boyer and Thiery, 1993; Hay, 2005; Liebner et al., 2004). EMT is a highly regulated, yet heterogeneous, multiple-step process (Thiery and Sleeman, 2006). EMT starts with the disassembly of cell-cell junctions, loss of apical-basal polarity, and the transition to a purely mesenchymal cell type (Hay, 2005; Thiery and Sleeman, 2006). However, these cellular events are not obligatory for each EMT process. EMT is a species and cell context specific process, and each EMT uses unique initiation cues, transcription factors, and signal transduction cascades. During mouse gastrulation (an EMT dependent process), TGFβ, Wnt, and FGF pathways are important for mesoderm cell formation (Nakaya and Sheng, 2008), while TGFβ, BMP, and notch signaling are important for endocardial cushion EMT (Boyer et al., 1999; Camenisch et al., 2002; Ma et al., 2005; Timmerman et al., 2004). During neural crest EMT, cells from the dorsal neural tube undergo EMT and dissociate from the neural epithelium and migrate to their destined regions in a context dependent manner (Duband et al., 1995; Duband and Thiery, 1987; Nichols, 1981; Sauka-Spengler and Bronner-Fraser, 2008; Tucker et al., 1988). Recently, it has been demonstrated that neural crest cells leave the neural tube by diverse morphological processes (Ahlstrom and Erickson, 2009). This suggests that even within a given cell population, EMT may not be a canonical process.
Recent studies demonstrate that WT1 is an essential component of the epicardial EMT process by controlling Snail and E-cadherin expression (Martinez-Estrada et al., 2009), yet it is apparently expressed in all mesothelial epicardial cells and transiently expressed in EPDCs. Because WT1 shuttles between the nucleus and cytoplasm, it could be a candidate factor that distributes asymmetrically during epicardial EMT. However, we were unable to observe any differences in WT1 intensity or localization when we compared WT1 expression in the epicardium and in cells entering the subepicardial space (data not shown). Therefore, WT1 may be involved in an EMT function that is downstream of oriented cell division.
The process of epicardial EMT has been studied extensively in avian proepicardial explants (Dettman et al., 1998; Lu et al., 2001b), but until recently very few experiments have been performed in mice. Interestingly, we found epicardial EMT may not be a canonical EMT process. The epicardium is a mesothelial layer rather than a true epithelial layer. For example, vimentin is expressed uniformly in the mouse epicardium (Mellgren et al., 2008), which suggests that the mouse epicardium may be more mesothelial than epithelial in nature. In addition, unlike many other developmental EMT events, cell division appears to be required for epicardial cell EMT. Finally, in many EMT processes, β-catenin nuclear localization is the signal that initiates the mesenchymal transcription profiles (Hay, 2005), yet, here we show that β-catenin localization in the adherens junctions may be a critical component of epicardial EMT.
β-catenin is well known for its function as a transcriptional activator in cell proliferation and cell fate determination in response to Wnt signaling (Cadigan and Nusse, 1997), and our results do not exclude a transcriptional role later in epicardial cell differentiation. Another activity for β-catenin is connecting adherens junctions with the cytoskeleton (Huber and Weis, 2001; Nelson and Nusse, 2004; Tepass, 2002). Others have suggested a role for β-catenin in directing spindle orientation (Le Borgne et al., 2002; Walston et al., 2004), centrosome separation (Bahmanyar et al., 2008), and asymmetric cell division (Mizumoto and Sawa, 2007; Schneider and Bowerman, 2007). Thus, β-catenin function is probably context dependent and has to be examined in a cell type specific manner.
Centrosomes, part of the microtubule organizing center (MTOC), are important for directing spindle orientation and asymmetric cell division (Rusan and Peifer, 2007; Yamashita and Fuller, 2008; Yamashita et al., 2007). In epicardial cells, we observe that the centrosome localizes to the apical domain during interphase. During mitosis, centrosomes localize to the lateral domains in a parallel division and to the apical-basal domains in a perpendicular division. In the Drosophila male germline, the inheritance of the mother centrosome determines if the cell remains a stem cell (Yamashita et al., 2007). Future work will determine if a similar scenario exists in the mouse epicardium. Recently, it has been reported that Connexin 43, a gap junction protein, is required for efficient epicardial cell directed migration after EPDC formation (Rhee et al., 2009). In this model, gap junctional signaling helps organize actin stress fibers and permits directed migration. In vitro, examination of MTOC demonstrated connexin 43 mutants exhibited a random location of the MTOC compared to the leading edge organization observed in control epicardial cells. Thus, the MTOC could be involved in entry into the myocardium after an epicardial cell perpendicular division in vivo.
In summary, we show that loss of β-catenin leads to a disruption of Numb localization to adherens junctions, consistent with results reported for the retina (Fu et al., 2006). Our results suggest that Numb, β-catenin and N-cadherin exist as a complex and that loss of NFP leads to disrupted adherens junctions and cell polarity: a phenotype resembling the loss of β-catenin phenotype. Finally, we show that β-catenin and NFP were involved in mitotic spindle orientation of epicardial cells. These results are similar to the discovery that α-catenin and cadherins are involved in the control of mitotic spindle orientation in vivo (Lechler and Fuchs, 2005) and in vitro (den Elzen et al., 2009). We suggest a junctional function for β-catenin and NFP in the regulation of epicardial cell polarity and spindle orientation and that loss of these proteins leads to a failure in epicardial EMT.
Materials and methods
Experimental animals
Mouse strains used were: PDGFRαGFP/+ (Hamilton et al., 2003), β-cateninfl/fl (Brault et al., 2001), Numbfl/fl and Numblikefl/fl (Wilson et al., 2007; Zilian et al., 2001) and Gata5CreTg/0 (Merki et al., 2005) mice. Embryos whose genotype were β-cateninfl/fl;Gata5CreTg/0 were designated as βcatEKO, while embryos whose genotype were Numbfl/fl; Numblikefl/fl; Gata5CreTg/0 were designated as NFPEKO . All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Texas Southwestern Medical Center and performed according to the Guide for the Care and Use of Laboratory Animals published by the NIH.
Paraffin and frozen section immunohistochemistry (IHC)
IHC was performed as previously published (Lechler and Fuchs, 2005; Mellgren et al., 2008). The following primary antibodies were used: Numb (1:300, Abcam, ab14140); Numb (1:100, Abcam, ab4147); β-catenin (1:500, BD Pharmingen, 61053); Phosphorylated Y489 β-catenin (1:50, DSHB, University of Iowa); α-catenin (1:100, Abcam ab51032); N-Cadherin (1:250, BD Transduction, 610920); pan-Cadherin (1:200, Sigma C3678); Par3 (1:100, Upstate, 07–330); Par6 (1:100, Abcam, ab6022); aPKC (1:100, Santa Cruz Biotechnology, sc-216); BrdU(1:50, Becton Dickinson, 347583); Ki67(1:100, Novocastra Laboratories, NCL-L-Ki67-MM1); α-Tubulin (1:1000, Sigma, T6199), Collagen IV (1:1000, Chemicon, AB756P), β-tubulin (1:1000, BD Pharmingen, 556321), γ-Tubulin (1:1000, Sigma, T5326), acetylated α-Tubulin (1:1000, Sigma, T6793) and WT1 (1:100, DAKO, M3561 or 1:50, Santa Cruz Biotechnology, sc-19).
Epicardial cell proliferation
Pregnant females or postnatal pups were injected with 100 µg BrdU (Sigma) per gram of body weight for four hours before embryo or heart isolation. Hearts were fixed in 4% PFA for at least 1 hours, then frozen embedded, and sectioned. Antigen retrieval was carried out by microwave antigen retrieval as previously described (Mellgren et al., 2008). Sections were then blocked 45 minutes in 3% BSA in 0.1% TWEEN-20, and stained for 2 hours or overnight with a 1:50 dilution of anti-BrdU antibody (Becton Dickinson) and a 1:1000 dilution of anti-Collagen IV (Chemicon). Epicardial cells were designated as the cells adjacent to the basement membrane on the exterior of the heart. Proliferation index was calculated by dividing the number of BrdU positive cells in the epicardium by the number of DAPI stained epicardial nuclei and multiplying by 100. A minimum of six sections per heart from a total of at least three hearts was quantified at each stage.
Imaging
The following equipment was used: confocal imaging (LSM510META) (grant NIH 1-S10-RR019406-01), fluorescent imaging (Zeiss Axiovert 200 with a Hamamatsu ORCA-ER camera), time-lapse confocal imaging (Perkin Elmer Ultraview ERS spinning confocal microscope) and color imaging (Zeis Axiovert 200 with an Olympus DP71 camera).
MSCV virus
VSVG pseudotyped virus was produced, and viral transduction was carried out as described (Duncan et al., 2005) with some modifications. Briefly, concentrated MSCV-IRES-GFP viral solutions were incubated with E12.5 or E13.5 hearts for 20 hours. Hearts were washed and incubated in fresh medium for another 24 hours. Hearts were fixed and frozen-sectioned. Divisions where two daughter cells both localized to epicardium were counted as parallel division, while divisions where one daughter was in the subepicardium and the other daughter was in the epicardium were counted as perpendicular division.
Epicardial cell entry into myocardium
Wild type or PDGFRαGFP/+ E12.5 whole hearts were cultured as previously described (Mellgren et al., 2008) and treated with aphidicolin (Calbiochem) at a concentration of 2µg/ml or vehicle for the indicated time. Some hearts were stained with WT1. WT1 or GFP+ cells that were present within the myocardium were quantified. At least 6 entire midcoronal sections per heart were quantified. Neural crest cells were excluded. In one group, aphidicolin treated hearts were treated for 24 hours, and then aphidicolin was washed out. After an additional 24 hours, hearts were processed. A minimum of 3 hearts for each treatment was quantified.
Epicardial cell viral transduction
Full length cyclin D1 cDNA was cloned using SuperScript III reverse transcriptase (Invitrogen). The full-length cDNA was then cloned into pAd/CMV/V5-DEST adenovirus vector (Invitrogen, V493-20) via the Gateway LR system. Cyclin D1 was overexpressed in epicardial cells of E12.5 PDGFRαGFP/+ hearts for 48 hours via adenovirus-mediated transduction (Mellgren et al., 2008). GFP+ cells that were present within the myocardium were quantified. At least 6 entire midcoronal sections per heart and at least three hearts for each treatment were quantified as described above. Adenoviral transduction of the epicardium proved to be very efficient with near 100% of the epicardial cells expressing the adenoviral construct. See Fig. S1F for examples of transduction efficiency.
Primers and sequences for making Ad-βcat, -trβcat, -TCF4 and -dnTCF4 adenovirus are available in Supplemental Materials and Methods.
Immunoprecipitation and western blot
Individual control or βcatEKO E12.5 hearts were cultured to isolate primary epicardial cells as described (Mellgren et al., 2008). After 48 hours, heart explants were removed, and epicardial cells were expanded for five days. Lysates from hearts of each genotype were combined and processed for immunoprecipitation with 1 µg of primary antibody. Antibodies used for immunoprecipitation and western blots include Numb (1:1000); β-catenin (1:1000); N-Cadherin (1:500); pan-Cadherin (1:500); Par3 (1:250) and β-tubulin (1:1000).
Time-lapse imaging
E12.5 hearts from PDGFRαGFP/+ mice (Hamilton et al., 2003) were harvested and embedded in low melt agarose in glass bottomed 35 mm dishes. Ventricles were seated for two hours and then subjected to microscopy using a spinning disk confocal system (Ultraview ERS; PerkinElmer) set up on a microscope (Axiovert 200M; Carl Zeiss MicroImaging, Inc.) with an Orca Hamamatsu camera. Epicardial cell division was followed by imaging 9 planes with 2 µm per plane using a 40X lens for 8 hours with 6 minutes per time point. Cultures were maintained at 37°C, 5% CO2 and saturating levels of humidity. Images were acquired by Volocity (Version 5.0.3) and edited by Imaris (Version 6.2). Target cells were reviewed in movie replay after data concatenation into time-lapse stacks. To generate the AVI videos, data were collected over all continuous planes and projected into one image per time point.
Quantitative evaluation of the orientation of the epicardial cell division plane
Coronal, sagittal, and transverse sections of E12.5 and E13.5 hearts were stained with α, β, or γ-tubulin. Spindle orientations of left and right ventricular epicardial cells in late metaphase, anaphase or early telophase, where both centrioles or centrosomes are in the same focal plane, were quantified. The orientation of the spindle is determined by the angle between the spindle axis and a basement membrane reference line. Division planes positioned at 60–90° to the basement membrane were classified as perpendicular and those that were oriented at 0–30° were classified as parallel.
For whole mount, hearts were fixed in 4% PFA overnight, then permeabilized by 0.5% Triton-100 in PBS (PBT) for at least 2 hours. After permeabilization, hearts were blocked in 10% FBS for at least 2 hours, then incubated with primary antibody overnight. After removing primary antibody by washing with PBT, hearts were incubated with secondary antibody for at least two hours. Hearts were washed with PBT, and then cleared with glycerol:PBS. To determine the spindle orientation of left and right ventricular epicardial cells in E12.5 and E13.5 whole mount stained hearts, Z-stack images were acquired using a Zeiss LSM 510 Meta confocal laser-scanning microscope with one µm per section. Three-dimensional images were reconstructed using Imaris software. The spindle orientation was measured by the angle between spindle axis, determined by the two centrosomes, and the x-y plane of heart surface (Toyoshima and Nishida, 2007). To determine the spindle orientation of epicardial cells in the time lapse imaged hearts, Z-stack images were acquired using spinning disk confocal in a time-lapse manner as described above. Three-dimensional images were reconstructed and smashed to relative real size of the epicardial cells using Imaris. Spindle orientation was determined by angle between the spindle axis, determined by the two nuclei labeled by the H2b–GFP, and the x-y plane of heart surface (Toyoshima and Nishida, 2007). As described above, divisions whose angles are between 0–30° were classified as parallel, while 60–90° were classified as perpendicular.
Statistics
Data shown as mean ± standard deviation, and p values were determined by Student’s t test or one way ANOVA using Prism 5.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Tannishtha Reya for suggestions and reagents, and to Eric Olson, Terry Lechler, Alec Zhang, and Olav Zilian for critical review of the manuscript. We are thankful to the Tallquist lab members for scientific discussion. M.D.T. and M.W. are funded by NHLBI grants (HL074257 and HL100401), and M.W. is a recipient of an AHA postdoctoral fellowship (09POST2150026). C.L.S. was a recipient of an American Heart Association predoctoral grant (09PRE2280197) and an institutional cardiology training grant (T32 HL007360-32), and is funded by a NIH Ruth Kirschstein predoctoral fellowship F30 (1F30HL096277).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahlstrom JD, Erickson CA. The neural crest epithelial-mesenchymal transition in 4D: a ;tail' of multiple non-obligatory cellular mechanisms. Development. 2009;136:1801–1812. doi: 10.1242/dev.034785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahmanyar S, Kaplan DD, Deluca JG, Giddings TH, Jr, O'Toole ET, Winey M, Salmon ED, Casey PJ, Nelson WJ, Barth AI. beta-Catenin is a Nek2 substrate involved in centrosome separation. Genes Dev. 2008;22:91–105. doi: 10.1101/gad.1596308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth AI, Stewart DB, Nelson WJ. T cell factor-activated transcription is not sufficient to induce anchorage-independent growth of epithelial cells expressing mutant beta-catenin. Proc Natl Acad Sci U S A. 1999;96:4947–4952. doi: 10.1073/pnas.96.9.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer AS, Ayerinskas II, Vincent EB, McKinney LA, Weeks DL, Runyan RB. TGFbeta2 and TGFbeta3 have separate and sequential activities during epithelial-mesenchymal cell transformation in the embryonic heart. Dev Biol. 1999;208:530–545. doi: 10.1006/dbio.1999.9211. [DOI] [PubMed] [Google Scholar]
- Boyer B, Thiery JP. Epithelium-mesenchyme interconversion as example of epithelial plasticity. APMIS. 1993;101:257–268. doi: 10.1111/j.1699-0463.1993.tb00109.x. [DOI] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- Camenisch TD, Molin DG, Person A, Runyan RB, Gittenberger-de Groot AC, McDonald JA, Klewer SE. Temporal and distinct TGFbeta ligand requirements during mouse and avian endocardial cushion morphogenesis. Dev Biol. 2002;248:170–181. doi: 10.1006/dbio.2002.0731. [DOI] [PubMed] [Google Scholar]
- Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan CW, Wei S, Hao W, Kilgore J, Williams NS, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5:100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng NN, Kirby CM, Kemphues KJ. Control of cleavage spindle orientation in Caenorhabditis elegans: the role of the genes par-2 and par-3. Genetics. 1995;139:549–559. doi: 10.1093/genetics/139.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RT, Kirkpatrick C, Peifer M. Armadillo is required for adherens junction assembly, cell polarity, and morphogenesis during Drosophila embryogenesis. J Cell Biol. 1996;134:133–148. doi: 10.1083/jcb.134.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Elzen N, Buttery CV, Maddugoda MP, Ren G, Yap AS. Cadherin adhesion receptors orient the mitotic spindle during symmetric cell division in mammalian epithelia. Mol Biol Cell. 2009;20:3740–3750. doi: 10.1091/mbc.E09-01-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettman RW, Denetclaw W, Jr, Ordahl CP, Bristow J. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev Biol. 1998;193:169–181. doi: 10.1006/dbio.1997.8801. [DOI] [PubMed] [Google Scholar]
- Duband JL, Monier F, Delannet M, Newgreen D. Epithelium-mesenchyme transition during neural crest development. Acta Anat (Basel) 1995;154:63–78. doi: 10.1159/000147752. [DOI] [PubMed] [Google Scholar]
- Duband JL, Thiery JP. Distribution of laminin and collagens during avian neural crest development. Development. 1987;101:461–478. doi: 10.1242/dev.101.3.461. [DOI] [PubMed] [Google Scholar]
- Duncan AW, Rattis FM, DiMascio LN, Congdon KL, Pazianos G, Zhao C, Yoon K, Cook JM, Willert K, Gaiano N, et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat Immunol. 2005;6:314–322. doi: 10.1038/ni1164. [DOI] [PubMed] [Google Scholar]
- Frise E, Knoblich JA, Younger-Shepherd S, Jan LY, Jan YN. The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interaction in sensory organ lineage. Proc Natl Acad Sci U S A. 1996;93:11925–11932. doi: 10.1073/pnas.93.21.11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Sun H, Klein WH, Mu X. Beta-catenin is essential for lamination but not neurogenesis in mouse retinal development. Dev Biol. 2006;299:424–437. doi: 10.1016/j.ydbio.2006.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, Poelmann RE. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ Res. 1998;82:1043–1052. doi: 10.1161/01.res.82.10.1043. [DOI] [PubMed] [Google Scholar]
- Hamilton TG, Klinghoffer RA, Corrin PD, Soriano P. Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol Cell Biol. 2003;23:4013–4025. doi: 10.1128/MCB.23.11.4013-4025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJ, Peifer M. Adherens junction-dependent and -independent steps in the establishment of epithelial cell polarity in Drosophila. J Cell Biol. 2004;167:135–147. doi: 10.1083/jcb.200406024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsock A, Nelson WJ. Adherens and tight junctions: structure, function and connections to the actin cytoskeleton. Biochim Biophys Acta. 2008;1778:660–669. doi: 10.1016/j.bbamem.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay ED. The mesenchymal cell, its role in the embryo, and the remarkable signaling mechanisms that create it. Dev Dyn. 2005;233:706–720. doi: 10.1002/dvdy.20345. [DOI] [PubMed] [Google Scholar]
- Hsu SC, Galceran J, Grosschedl R. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with beta-catenin. Mol Cell Biol. 1998;18:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber AH, Weis WI. The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell. 2001;105:391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- Kaplan NA, Liu X, Tolwinski NS. Epithelial polarity: interactions between junctions and apical-basal machinery. Genetics. 2009;183:897–904. doi: 10.1534/genetics.109.108878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52:311–320. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Knoblich JA, Jan LY, Jan YN. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995;377:624–627. doi: 10.1038/377624a0. [DOI] [PubMed] [Google Scholar]
- Kolligs FT, Hu G, Dang CV, Fearon ER. Neoplastic transformation of RK3E by mutant beta-catenin requires deregulation of Tcf/Lef transcription but not activation of c-myc expression. Mol Cell Biol. 1999;19:5696–5706. doi: 10.1128/mcb.19.8.5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama M, Ito K, Shimada Y. Origin and development of the epicardium in the mouse embryo. Anat Embryol (Berl) 1987;176:183–189. doi: 10.1007/BF00310051. [DOI] [PubMed] [Google Scholar]
- Kraut R, Chia W, Jan LY, Jan YN, Knoblich JA. Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature. 1996;383:50–55. doi: 10.1038/383050a0. [DOI] [PubMed] [Google Scholar]
- Kuo CT, Mirzadeh Z, Soriano-Navarro M, Rasin M, Wang D, Shen J, Sestan N, Garcia-Verdugo J, Alvarez-Buylla A, Jan LY, et al. Postnatal deletion of Numb/Numblike reveals repair and remodeling capacity in the subventricular neurogenic niche. Cell. 2006;127:1253–1264. doi: 10.1016/j.cell.2006.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Borgne R, Bellaiche Y, Schweisguth F. Drosophila E-cadherin regulates the orientation of asymmetric cell division in the sensory organ lineage. Curr Biol. 2002;12:95–104. doi: 10.1016/s0960-9822(01)00648-0. [DOI] [PubMed] [Google Scholar]
- Lechler T, Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437:275–280. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HS, Wang D, Shen Q, Schonemann MD, Gorski JA, Jones KR, Temple S, Jan LY, Jan YN. Inactivation of Numb and Numblike in embryonic dorsal forebrain impairs neurogenesis and disrupts cortical morphogenesis. Neuron. 2003;40:1105–1118. doi: 10.1016/s0896-6273(03)00755-4. [DOI] [PubMed] [Google Scholar]
- Liebner S, Cattelino A, Gallini R, Rudini N, Iurlaro M, Piccolo S, Dejana E. Beta-catenin is required for endothelial-mesenchymal transformation during heart cushion development in the mouse. J Cell Biol. 2004;166:359–367. doi: 10.1083/jcb.200403050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Roegiers F, Jan LY, Jan YN. Adherens junctions inhibit asymmetric division in the Drosophila epithelium. Nature. 2001a;409:522–525. doi: 10.1038/35054077. [DOI] [PubMed] [Google Scholar]
- Lu J, Landerholm TE, Wei JS, Dong XR, Wu SP, Liu X, Nagata K, Inagaki M, Majesky MW. Coronary smooth muscle differentiation from proepicardial cells requires rhoA-mediated actin reorganization and p160 rho-kinase activity. Dev Biol. 2001b;240:404–418. doi: 10.1006/dbio.2001.0403. [DOI] [PubMed] [Google Scholar]
- Ma L, Lu MF, Schwartz RJ, Martin JF. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development. 2005;132:5601–5611. doi: 10.1242/dev.02156. [DOI] [PubMed] [Google Scholar]
- Mahtab EA, Wijffels MC, Van Den Akker NM, Hahurij ND, Lie-Venema H, Wisse LJ, Deruiter MC, Uhrin P, Zaujec J, Binder BR, et al. Cardiac malformations and myocardial abnormalities in podoplanin knockout mouse embryos: Correlation with abnormal epicardial development. Dev Dyn. 2008;237:847–857. doi: 10.1002/dvdy.21463. [DOI] [PubMed] [Google Scholar]
- Marthiens V, ffrench-Constant C. Adherens junction domains are split by asymmetric division of embryonic neural stem cells. EMBO Rep. 2009;10:515–520. doi: 10.1038/embor.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Estrada OM, Lettice LA, Essafi A, Guadix JA, Slight J, Velecela V, Hall E, Reichmann J, Devenney PS, Hohenstein P, et al. Wt1 is required for cardiovascular progenitor cell formation through transcriptional control of Snail and E-cadherin. Nat Genet. 2009 doi: 10.1038/ng.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellgren AM, Smith CL, Olsen GS, Eskiocak B, Zhou B, Kazi MN, Ruiz FR, Pu WT, Tallquist MD. Platelet-Derived Growth Factor Receptor {beta} Signaling Is Required for Efficient Epicardial Cell Migration and Development of Two Distinct Coronary Vascular Smooth Muscle Cell Populations. Circ Res. 2008;103:1393–1401. doi: 10.1161/CIRCRESAHA.108.176768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merki E, Zamora M, Raya A, Kawakami Y, Wang J, Zhang X, Burch J, Kubalak SW, Kaliman P, Belmonte JC, et al. Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc Natl Acad Sci U S A. 2005;102:18455–18460. doi: 10.1073/pnas.0504343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa T, Fischman DA. Retroviral analysis of cardiac morphogenesis: discontinuous formation of coronary vessels. Proc Natl Acad Sci U S A. 1992;89:9504–9508. doi: 10.1073/pnas.89.20.9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumoto K, Sawa H. Cortical beta-catenin and APC regulate asymmetric nuclear beta-catenin localization during asymmetric cell division in C. elegans. Dev Cell. 2007;12:287–299. doi: 10.1016/j.devcel.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Moore AW, McInnes L, Kreidberg J, Hastie ND, Schedl A. YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development. 1999;126:1845–1857. doi: 10.1242/dev.126.9.1845. [DOI] [PubMed] [Google Scholar]
- Morel V, Arias AM. Armadillo/beta-catenin-dependent Wnt signalling is required for the polarisation of epidermal cells during dorsal closure in Drosophila. Development. 2004;131:3273–3283. doi: 10.1242/dev.01217. [DOI] [PubMed] [Google Scholar]
- Muller HA, Wieschaus E. armadillo, bazooka, and stardust are critical for early stages in formation of the zonula adherens and maintenance of the polarized blastoderm epithelium in Drosophila. J Cell Biol. 1996;134:149–163. doi: 10.1083/jcb.134.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya Y, Sheng G. Epithelial to mesenchymal transition during gastrulation: an embryological view. Dev Growth Differ. 2008;50:755–766. doi: 10.1111/j.1440-169X.2008.01070.x. [DOI] [PubMed] [Google Scholar]
- Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols DH. Neural crest formation in the head of the mouse embryo as observed using a new histological technique. J Embryol Exp Morphol. 1981;64:105–120. [PubMed] [Google Scholar]
- Nishimura T, Kaibuchi K. Numb controls integrin endocytosis for directional cell migration with aPKC and PAR-3. Dev Cell. 2007;13:15–28. doi: 10.1016/j.devcel.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Orgogozo V, Schweisguth F, Bellaiche Y. Binary cell death decision regulated by unequal partitioning of Numb at mitosis. Development. 2002;129:4677–4684. doi: 10.1242/dev.129.20.4677. [DOI] [PubMed] [Google Scholar]
- Petersen PH, Zou K, Hwang JK, Jan YN, Zhong W. Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature. 2002;419:929–934. doi: 10.1038/nature01124. [DOI] [PubMed] [Google Scholar]
- Petersen PH, Zou K, Krauss S, Zhong W. Continuing role for mouse Numb and Numbl in maintaining progenitor cells during cortical neurogenesis. Nat Neurosci. 2004;7:803–811. doi: 10.1038/nn1289. [DOI] [PubMed] [Google Scholar]
- Poelmann RE, Gittenberger-de Groot AC, Mentink MM, Bokenkamp R, Hogers B. Development of the cardiac coronary vascular endothelium, studied with antiendothelial antibodies, in chicken-quail chimeras. Circ Res. 1993;73:559–568. doi: 10.1161/01.res.73.3.559. [DOI] [PubMed] [Google Scholar]
- Rasin MR, Gazula VR, Breunig JJ, Kwan KY, Johnson MB, Liu-Chen S, Li HS, Jan LY, Jan YN, Rakic P, et al. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat Neurosci. 2007;10:819–827. doi: 10.1038/nn1924. [DOI] [PubMed] [Google Scholar]
- Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, Hintz L, Nusse R, Weissman IL. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- Rhee DY, Zhao XQ, Francis RJ, Huang GY, Mably JD, Lo CW. Connexin 43 regulates epicardial cell polarity and migration in coronary vascular development. Development. 2009;136:3185–3193. doi: 10.1242/dev.032334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- Rusan NM, Peifer M. A role for a novel centrosome cycle in asymmetric cell division. J Cell Biol. 2007;177:13–20. doi: 10.1083/jcb.200612140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Schneider SQ, Bowerman B. beta-Catenin asymmetries after all animal/vegetal- oriented cell divisions in Platynereis dumerilii embryos mediate binary cell-fate specification. Dev Cell. 2007;13:73–86. doi: 10.1016/j.devcel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Smith CA, Lau KM, Rahmani Z, Dho SE, Brothers G, She YM, Berry DM, Bonneil E, Thibault P, Schweisguth F, et al. aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. EMBO J. 2007;26:468–480. doi: 10.1038/sj.emboj.7601495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana EP, Doe CQ. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron. 1996;17:21–26. doi: 10.1016/s0896-6273(00)80277-9. [DOI] [PubMed] [Google Scholar]
- Tepass U. Adherens junctions: new insight into assembly, modulation and function. Bioessays. 2002;24:690–695. doi: 10.1002/bies.10129. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- Timmerman LA, Grego-Bessa J, Raya A, Bertran E, Perez-Pomares JM, Diez J, Aranda S, Palomo S, McCormick F, Izpisua-Belmonte JC, et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18:99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima F, Nishida E. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 2007;26:1487–1498. doi: 10.1038/sj.emboj.7601599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker GC, Duband JL, Dufour S, Thiery JP. Cell-adhesion and substrate-adhesion molecules: their instructive roles in neural crest cell migration. Development. 1988;103 Suppl:81–94. doi: 10.1242/dev.103.Supplement.81. [DOI] [PubMed] [Google Scholar]
- Uemura T, Shepherd S, Ackerman L, Jan LY, Jan YN. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell. 1989;58:349–360. doi: 10.1016/0092-8674(89)90849-0. [DOI] [PubMed] [Google Scholar]
- Verdi JM, Schmandt R, Bashirullah A, Jacob S, Salvino R, Craig CG, Program AE, Lipshitz HD, McGlade CJ. Mammalian NUMB is an evolutionarily conserved signaling adapter protein that specifies cell fate. Curr Biol. 1996;6:1134–1145. doi: 10.1016/s0960-9822(02)70680-5. [DOI] [PubMed] [Google Scholar]
- Viragh S, Challice CE. The origin of the epicardium and the embryonic myocardial circulation in the mouse. Anat Rec. 1981;201:157–168. doi: 10.1002/ar.1092010117. [DOI] [PubMed] [Google Scholar]
- Vleminckx K, Kemler R, Hecht A. The C-terminal transactivation domain of beta-catenin is necessary and sufficient for signaling by the LEF-1/beta-catenin complex in Xenopus laevis. Mech Dev. 1999;81:65–74. doi: 10.1016/s0925-4773(98)00225-1. [DOI] [PubMed] [Google Scholar]
- Walston T, Tuskey C, Edgar L, Hawkins N, Ellis G, Bowerman B, Wood W, Hardin J. Multiple Wnt signaling pathways converge to orient the mitotic spindle in early C. elegans embryos. Dev Cell. 2004;7:831–841. doi: 10.1016/j.devcel.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Wang Z, Sandiford S, Wu C, Li SS. Numb regulates cell-cell adhesion and polarity in response to tyrosine kinase signalling. EMBO J. 2009;28:2360–2373. doi: 10.1038/emboj.2009.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels A, Perez-Pomares JM. The epicardium and epicardially derived cells (EPDCs) as cardiac stem cells. Anat Rec A Discov Mol Cell Evol Biol. 2004;276:43–57. doi: 10.1002/ar.a.10129. [DOI] [PubMed] [Google Scholar]
- Wilson A, Ardiet DL, Saner C, Vilain N, Beermann F, Aguet M, Macdonald HR, Zilian O. Normal hemopoiesis and lymphopoiesis in the combined absence of numb and numblike. J Immunol. 2007;178:6746–6751. doi: 10.4049/jimmunol.178.11.6746. [DOI] [PubMed] [Google Scholar]
- Wirtz-Peitz F, Nishimura T, Knoblich JA. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Kwon HY, Rattis F, Blum J, Zhao C, Ashkenazi R, Jackson TL, Gaiano N, Oliver T, Reya T. Imaging hematopoietic precursor division in real time. Cell Stem Cell. 2007;1:541–554. doi: 10.1016/j.stem.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM, Fuller MT. Asymmetric centrosome behavior and the mechanisms of stem cell division. J Cell Biol. 2008;180:261–266. doi: 10.1083/jcb.200707083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–521. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap AS, Brieher WM, Gumbiner BM. Molecular and functional analysis of cadherin-based adherens junctions. Annu Rev Cell Dev Biol. 1997;13:119–146. doi: 10.1146/annurev.cellbio.13.1.119. [DOI] [PubMed] [Google Scholar]
- Zamora M, Manner J, Ruiz-Lozano P. Epicardium-derived progenitor cells require beta-catenin for coronary artery formation. Proc Natl Acad Sci U S A. 2007;104:18109–18114. doi: 10.1073/pnas.0702415104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Feder JN, Jiang MM, Jan LY, Jan YN. Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron. 1996;17:43–53. doi: 10.1016/s0896-6273(00)80279-2. [DOI] [PubMed] [Google Scholar]
- Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilian O, Saner C, Hagedorn L, Lee HY, Sauberli E, Suter U, Sommer L, Aguet M. Multiple roles of mouse Numb in tuning developmental cell fates. Curr Biol. 2001;11:494–501. doi: 10.1016/s0960-9822(01)00149-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.