Abstract
The intestinal microbiota includes a diverse group of functional microorganisms, including candidate probiotics or viable microorganisms that benefit the host. Beneficial effects of probiotics include enhancing intestinal epithelial cell function, protecting against physiologic stress, modulating cytokine secretion profiles, influencing T-lymphocyte populations, and enhancing antibody secretion. Probiotics have demonstrated significant potential as therapeutic options for a variety of diseases, but the mechanisms responsible for these effects remain to be fully elucidated. Accumulating evidence demonstrates that probiotics communicate with the host by modulating key signaling pathways, such as NFκB and MAPK, to either enhance or suppress activation and influence downstream pathways. Beneficial microbes can profoundly alter the physiology of the gastrointestinal tract, and understanding these mechanisms may result in new diagnostic and therapeutic strategies.
Key words: probiotic, commensal, signaling, macrophage, dendritic cell, intestinal epithelial cell, innate immune system, intestine
Introduction: Probiotic Modulation of Host Signaling Pathways
Commensal microbes in the gastrointestinal tract play an essential role in nutrition and food digestion.1 These microbes also impact metabolism, endocrinology, proper gut development and regulation of the immune system.2–4 Probiotics provide opportunities to apply beneficial microbes in specific human health and clinical applications. By modifying the microbial community within the gut, we may be able to prevent or treat gut disorders such as inflammatory bowel disease (IBD) and irritable bowel syndrome (IBS) as well as systemic disorders like eczema, allergies, asthma and diabetes.3–10 According to the Food and Agricultural Organization of the United Nations and the World Health Organization, probiotics are “Live microorganisms, which, when consumed in adequate amounts, confer a health benefit on the host.”11 Numerous studies in mice and humans have already shown how a single probiotic strain or combinations of strains may modulate gut function and ameliorate disease. Many of these studies have yielded promising results regarding the use of probiotics in the treatment of acute gastroenteritis, Clostridium difficile-associated diarrhea or colitis, irritable bowel syndrome, necrotizing enterocolitis, and others.5 At the same time, other studies have shown either no effect or minimal effects by probiotics.12,13 Such negative data could be due to lack of efficacy of probiotics-based strategies, poor study design, or inappropriate selection of model systems or probiotic strains for specific applications. Probiotic strains may be improperly selected for study without clear scientific rationale for strain selection. The compilation of studies yielding evidence of disease amelioration and immunomodulation suggest probiotics communicate with the host in multiple ways. However, the mechanisms by which probiotics actually exert their beneficial effects have not been clearly defined. Clear understanding of these mechanisms will allow for appropriate probiotic strain selection for specific applications and may uncover novel probiotic functions.
Probiotics include exogenous and indigenous bacterial species that interact with various cellular components within the intestinal environment. Intact, viable bacteria may be essential for probiotic effects, or these effects may be mediated by a cell wall component or structurally diverse secreted molecules. Several mechanistic studies show that key biological signaling pathways like NFκB, MAPK, Akt/PI3K and PPARγ are targets for probiotics or their products. These pathways can be modified in different ways by individual probiotic strains, including strains of the same species. For example, one Lactobacillus reuteri strain, ATCC PTA 6475, can inhibit LPS-induced TNF production from myeloid cells through suppression of the activator protein-1 (AP-1) pathway, while another L. reuteri strain, DSM 17938, does not inhibit LPS-induced TNF production.14
The goal of this review is to explore probiotics-host communication via signaling pathway modulation within the intestine. While it is equally important to consider how the host influences the gut microbiota, this review focuses on how gut microbes influence the host.
Probiotic-Induced Changes in Intestinal Epithelial Cell Signaling Pathways Modulate Cell Survival and Cytokine Secretion
Intestinal epithelial cells (IECs) are an initial point of contact between the host and intestinal microbes. IECs are the first line of defense against pathogenic bacteria, and they communicate extensively with commensal microbes and probiotics. Probiotics can affect IECs in multiple ways, some of which are enhancing barrier function,15–19 increasing mucin production,20,21 inducing antimicrobial and heat shock protein production,22,23 interfering with pathogenic organisms,24–26 and modulating signaling pathways (Table 1) and cell survival (Fig. 1).
Table 1.
Probiotic modulation of signaling pathways in intestinal epithelial cells and macrophages
| Probiotic species | Model system | Signaling pathway | Probiotic effect(s) | Reference(s) |
| Bacillus subtilis JH642 | IECs | hsp | Induces hsp27, hsp25 and hsp70 | Fujiya, et al. 2007,30 |
| Bacillus subtilis JH642 | IECs | MAPKs | Increases p38 phosphorylation | Fujiya, et al. 2007,30 |
| Bacteroides fragilis ATCC 23745 | IECs | hsp | Induces hsp25 and hsp72 | Kojima, et al. 2003,29 |
| Bacteroides thetaiotaomicron ATCC 29184 | IECs | MAPKs | Activation of ERK1/2 and p38 | Resta-Lenert, et al. 2006,46 |
| Bacteroides thetaiotaomicron | IECs | NFκB | Enhances RelA nuclear export via PPARγ | Kelly, et al. 2004,49 |
| Bacteroides vulgatus | IECs | NFκB | Increases IκBα phosphorylation | Haller, et al. 2002,44 |
| Bifidobacterium adolescentis ATCC 15703 | Macrophages | NFκB | Decreases IκBα phosphorylation, increases SOCS | Okada, et al. 2009,67 |
| Bifidobacterium bifidum B536 | Macrophages | NFκB | Decreases LPS binding to CD14 | Menard, et al. 2004,62 |
| Bifidobacterium breve BbC50 | Macrophages | NFκB | Decreases LPS binding to CD14 | Menard, et al. 2004,62 |
| Bifidobacterium lactis BB12 | IECs | NFκB | Activates RelA | Ruiz, et al. 2005,43 |
| Bifidobacterium lactis BB12 | IECs | MAPKs | Increases p38 phosphorylation | Ruiz, et al. 2005,43 |
| Bifidobacterium longum | IECs | NFκB | Decreases p65 translocation | Bai, et al. 2004;39 Bai, et al. 2006,40 |
| Enterococcus faecalis EC1/EC3/EC15/EC16 | IECs | PPARγ | Induced phosphorylation of PPARγ1 | Are, et al. 2008,50 |
| Escherichia coli M17 | Macrophages | NFκB | Inhibits p65 nuclear binding | Fitzpatrick, et al. 2008,58 |
| Faecalibacterium prausnitzii DSM 17677 | IECs | NFκB | Inhibits NFκB activation | Sokol, et al. 2008,41 |
| Lactobacillus acidophilus ATCC 4356 | IECs | MAPKs | Activation of ERK1/2 and p38 | Resta-Lenert, et al. 2006,46 |
| Lactobacillus acidophilus ATCC 4356 | IECs | NFκB | Decreases IκBα phosphorylation | Resta-Lenert, et al. 2006,46 |
| Lactobacillus bulgaricus | IECs | NFκB | Decreases p65 translocation | Bai, et al. 2004,39 |
| Lactobacillus casei DN-114 001 | IECs | NFκB | Prevents IκBα degradation | Tien, et al. 2005,35 |
| Lactobacillus casei | IECs | PPARγ | Increases PPARγ mRNA | Eun, et al. 2007,52 |
| Lactobacillus casei Shirota | Macrophages | NFκB | Inhibits IκBα phosphorylation | Watanabe, et al. 2009,59 |
| Lactobacillus casei Shirota | Macrophages | MAPKs | Inhibits ERK1/2 phosphorylation | Watanabe, et al. 2009,59 |
| Lactobacillus casei YIT 9029 | Macrophages | NFκB | Activation of NFκB | Matsuguchi, et al. 2003,65 |
| Lactobacillus crispatus M247 | IECs | PPARγ | Increased activation and transcriptional activity | Voltan, et al. 2008,47 |
| Lactobacillus crispatus | Macrophages | NFκB | Activation of NFκB | Klebanoff, et al. 1999,69 |
| Lactobacillus fermentum DSMZ 20052 | IECs | NFκB | Blocks NFκB activation | Frick, et al. 2007,38 |
| Lactobacillus fermentum DSMZ 20052 | IECs | MAPKs | Decreases p38 activation | Frick, et al. 2007,38 |
| Lactobacillus fermentum YIT 0159 | Macrophages | NFκB | Activation of NFκB | Matsuguchi, et al. 2003,65 |
| Lactobacillus fermentum YIT 0159 | Macrophages | MAPKs | Activation of JNK | Matsuguchi, et al. 2003,65 |
| Lactobacillus plantarum | IECs | NFκB | Inhibits proteasome | Petrof, et al. 2009,37 |
| Lactobacillus plantarum K8 | Macrophages | NFκB | Inhibits IκBα degradation | Kim, et al. 2008;63 Kim, et al. 2008,64 |
| Lactobacillus plantarum K8 | Macrophages | MAPKs | Decreases p38, JNK, ERK1/2 phosphorylation | Kim, et al. 2008;63 Kim, et al. 2008, 64 |
| Lactobacillus plantarum S1, DB22, & DS41 | PBMCs | apoptosis | Increases TRAIL production and secretion | Horinaka, et al. 2010,73 |
| Lactobacillus reuteri | IECs | NFκB | Prevents IκBα | degradation Ma, et al. 2004,36 |
| Lactobacillus reuteri ATCC PTA 6475 | Macrophages | apoptosis | Suppress Bcl-2 and Bcl-xL | Iyer, et al. 2008,71 |
| Lactobacillus reuteri ATCC PTA 6475 | Macrophages | NFκB | Decreases IκBα ubiquitination | Iyer, et al. 2008,71 |
| Lactobacillus reuteri ATCC PTA 6475 | Macrophages | MAPKs | Increases JNK and p38 phosphorylation | Iyer, et al. 2008,71 |
| Lactobacillus reuteri ATCC PTA 6475 | Macrophages | MAPKs | Decreases ERK1/2 phosphorylation | Iyer, et al. 2008,71 |
| Lactobacillus reuteri ATCC PTA 6475 | Macrophages | AP-1 | Decreases c-Jun phosphorylation | Lin, et al. 2008,14 |
| Lactobacillus rhamnosus GG ATCC 53103 | IECs | hsp | Induces hsp25 and hsp72, heat shock TF1 | Tao, et al. 2006,27 |
| Lactobacillus rhamnosus GG ATCC 53103 | IECs | MAPKs | Increased phosphorylation of p38 and JNK | Tao, et al. 2006,27 |
| Lactobacillus rhamnosus GG ATCC 53103 | IECs | NFκB | Prevents IκBα ubiquitination and degradation | Kumar, et al. 2007;32 Lin, et al. 2009;33 Zhang, et al. 2005,34 |
| Lactobacillus rhamnosus GG ATCC 53103 | IECs | MAPKs | Upregulation of MAPK related genes | Di Caro, et al.54 2005; Lin, et al. 2008,55 |
| Lactobacillus rhamnosus GG ATCC 53103 | IECs | apoptosis | Activates Akt/PI3K | Yan, et al. 2002;56 Yan, et al. 2007,57 |
| Lactobacillus rhamnosus GG ATCC 53103 | Macrophages | NFκB | Induces NFκB binding activity | Miettinen, et al. 2000,70 |
| Lactobacillus rhamnosus Lcr | Macrophages | apoptosis | Increases pro-apoptotic Bax, releases cytochrome-c, activates caspase-9 & 3 | Chiu, et al. 2009,72 |
| Lactobacillus rhamnosus GR-1 | Macrophages | MAPKs | Activates JAK2-STAT3 to inhibit JNK activation | Kim, et al. 2006,68 |
| Ruminococcus gnavus FRE1 | Macrophages | NFκB | Decreases LPS binding to CD14 | Menard, et al. 2004,62 |
| “Saccharomyces boulardii”§ | IECs | apoptosis | Prevents caspase-3 activation | Czerucka, et al. 2000,17 |
| “Saccharomyces boulardii”§ | Macrophages | NFκB | Prevents IκBα degradation | Sougioultzis, et al. 2006,60 |
| Salmonella pullorum | IECs | NFκB | Inhibits IκBα ubiquitination | Neish, et al. 2000,31 |
| Salmonella typhimurium PhoP | IECs | NFκB | Inhibits IκBα ubiquitination | Neish, et al. 2000,31 |
| Streptococcus thermophilus ATCC 19258 | IECs | MAPKs | Activation of ERK1/2 and p38 | Resta-Lenert, et al. 2006,46 |
| Streptococcus thermophilus ATCC 19258 | IECs | NFκB | Decreases IκBα phosphorylation | Resta-Lenert, et al. 2006,46 |
| Streptococcus thermophilus St065 | Macrophages | NFκB | Decreases LPS binding to CD14 | Menard, et al. 2004,62 |
| VSL#3* | IECs | hsp | Induces hsp25 and hsp72, heat shock TF1 | Petrof, et al. 2004,28 |
| VSL#3* | IECs | MAPKs | Decreases p38 phosphorylation | Jijon, et al. 2004,45 |
| VSL#3* | IECs | NFκB | Inhibits proteasome | Petrof, et al. 2004,28 |
| VSL#3* | IECs | PPARγ | Enhanced expression of PPARγ | Ewaschuk, et al. 2006,51 |
The proper taxonomic designation for “Saccharomyces boulardii” is Saccharomyces cerevisiae subsp boulardii.
VSL#3 is a cocktail of several bacterial species including Streptococcus salivarius subsp thermophilus, Lactobacillus casei, L. plantarum, L. acidophilus, L. delbrueckii subsp bulgaricus, Bifidobacterium longum, B. infantis and B. breve.
Abbreviations: ERK, extracellular signal-regulated kinases; hsp, heat shock protein; IEC, intestinal epithelial cell; IκBα, inhibitor of NFκB; JNK, c-Jun N-terminal kinase; LPS, lipopolysaccharide; MAPK, mitogen-activated protein kinase; NFκB, nuclear factor-kappaB; PPARγ, peroxisome proliferator activated receptor-gamma; SOCS, suppressor of cytokine signaling; STAT, signal transducer and activator of transcription.
Figure 1.
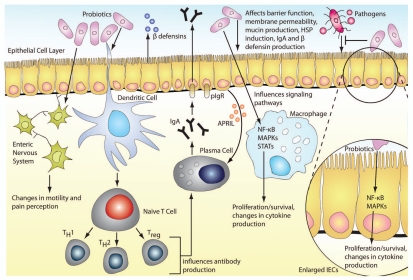
Probiotics benefit the host by communicating with a variety of cell types. Intestinal epithelial cell (IEC) barrier function is enhanced through probiotic modulation of tight junctions as well as enhanced mucin production. Probiotics interfere with pathogens by increasing β defensin secretion from IECs and IgA from plasma cells and by directly blocking the signaling pathways hijacked by pathogens. Cytokine secretion by IECs, macrophages and dendritic cells is regulated by probiotics through modulation of key signaling pathways such as NFκB and MAPKs. Changes in these pathways can also affect proliferation and survival of target cells. Through interactions with dendritic cells, probiotics can influence T cell subpopulations and skew them towards a Th1, Th2 or Treg response. Probiotics can also cause changes in gut motility and pain perception by modulating pain receptor expression and secreting potential neurotransmitter molecules. APRIL, a proliferation-inducing ligand; hsp, heat shock protein; IEC, intestinal epithelial cell; Ig, immunoglobulin; MAPK, mitogen-activated protein kinase; NFκB, nuclear factor-kappaB; pIgR, polymeric immunoglobulin receptor; STAT, signal transducers and activator of transcription; Treg, T regulatory cell.
Induction of cytoprotective heat shock proteins.
Cells exhibit “stress tolerance” when they encounter thermal, osmotic, oxidative and other stressors. Cellular heat shock proteins (hsp) are induced in response to these stressors. These highly conserved proteins confer protection against insults and prevent cell death from occurring.27,28 In the intestine, the induced heat shock proteins primarily include hsp25 and hsp72, which help maintain tight junctions between IECs and promote barrier function.27–29 For example, hsp 72 prevents cellular proteins from denaturing and hsp25 stabilizes actin.27
Commensal or probiotic bacteria induce production of cytoprotective heat shock proteins in the intestine. Even transient exposure of IECs to Lactobacillus rhamnosus GG ATCC 53103 (LGG) cell-free conditioned media induced expression of hsp25 and hsp72. Evidence indicates that hsp induction by probiotics is transcriptionally regulated. For example, LGG activated MAPKs p38 and JNK to induce heat shock transcription factor 1 and increase mRNA levels of hsp25 and hsp72. Selective inhibitors of these MAPKs prevent hsp72 induction by LGG but not hsp25, suggesting that multiple mechanisms may be involved.27 Induction of hsp25 and hsp72 in IECs also occurred when cells were treated with conditioned media from VSL#3, a cocktail of four Lactobacillus species (not including LGG), three Bifidobacterium species, and one Streptococcus strain. Similar to LGG, VSL#3 regulates hsp gene expression by induction of heat shock transcription factor 1.28 A third probiotic, Bacillus subtilis JH642, produces a quorum-sensing pentapeptide known as competence and sporulation factor (CSF) that induces hsp27 in an IEC culture model, and hsp25 and hsp70 in an ex vivo intestinal preparation. CSF can also stimulate cell survival by phosphorylation of p38 and protein kinase B (Akt).30 Finally, a study looking at Bacteroides fragilis ATCC 23745 demonstrated that this bacteria induces expression of hsp25 and hsp72 in IECs by a yet undefined mechanism.29 Induction of heat shock proteins by these various probiotic strains may allow host IECs to overcome insults and stressors that would be detrimental in the absence of this mutualistic relationship.
Modulation of inflammatory signaling pathways in IECs.
The NFκB pathway is one key signaling channel for activation of immune responses secondary to a variety of stimuli, and this pathway may represent a key “conversation node” between probiotics or beneficial microbes and IECs. Under non-stimulatory conditions, NFκB is present in its inactive form in the cytoplasm, bound to the inhibitor molecule, IκB. When pro-inflammatory stimuli trigger signaling pathways, IκB is phosphorylated by IKK, targeting it for ubiquitination by E3-SCFβ-TrCP and subsequent proteasomal degradation. Once freed from IκB, NFκB is able to migrate into the nucleus, bind target promoters and activate transcription of effector genes.31 Many nodes in this pathway present opportunities for probiotics to prevent the activation of NFκB and influence downstream cytokine secretion. For example, direct contact with avirulent Salmonella typhimurium PhoP and Salmonella pullorum decrease IL-8 and TNFα secretion from polarized T84 epithelial cells by inhibiting IκBα polyubiquitination and subsequent proteasomal degradation (Fig. 2).31 NFκB cannot enter the nucleus and activate transcription when bound to its inhibitor molecule, IκB. LGG ATCC 53103 modulates ubiquitin-mediated degradation of IκBα through the generation of reactive oxygen species (ROS). Increased quantities of ROS inactivate the Ubc12 enzyme, which is responsible for neddylation of the cullin-1 subunit of the E3 ligase, E3-SCFβ-TrCP. In the absence of cullin-1 neddylation, the E3 ligase cannot contribute to polyubiquitination of IκBα and NFκB is not released.32,33
Figure 2.
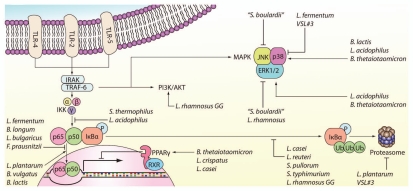
Probiotics modulate key signaling pathways in intestinal epithelial cells. Various probiotics prevent NFκB activation by inhibiting IκBα phosphorylation, ubiquitination, proteasomal degradation, or translocation of NFκB into the nucleus (suppression is indicated by a block sign “??”). Probiotics can also enhance RelA export from the nucleus via PPARγ. Other probiotics increase NFκB activation through enhanced translocation into the nucleus (activation is indicated by an arrow sign “→”). Apoptosis of intestinal epithelial cells can be prevented by probiotic modulation of the PI3K/Akt pathway. Probiotic-induced changes in phosphorylation levels of p38, JNK, and ERK1/2 MAPKs can affect cytokine secretion and apoptosis. ERK, extracellular signal-regulated kinases; IκBα, inhibitor of NFκB α; IKK, IκB kinase; IRAK, interleukin-1 receptor-associated kinase; JNK, c-Jun N-terminal kinase; P, phosphorylation; PPARγ, peroxisome proliferator activated receptor-γ; RXR, retinoid X receptor; TLR, Toll-like receptor; Ub, ubiquitin.
Several other probiotic strains can also prevent degradation of IκB. A study by Zhang et al. investigating the effects of both viable and heat-killed LGG in an epithelial cell model demonstrated the probiotic's ability to decrease IκB degradation and subsequent NFκB translocation into the nucleus, resulting in decreased TNF-induced IL-8 production.34 Others have shown that pretreatment of epithelial cells with L. casei DN-114 001 decreased Shigella flexneri-induced NFκB activation due to inhibition of IκBα degradation. Subsequent gene expression studies showed that L. casei modulates several genes involved in ubiquitination and proteasomal processes.35 Ma et al. demonstrated that preincubation of viable, whole cell L. reuteri with IECs inhibited TNF-induced and Salmonella enterica serovar Typhimurium-induced IL-8 transcription by preventing NFκB activation. Mechanistic studies indicate that L. reuteri can block TNF-induced p65 nuclear translocation secondary to a decrease in IκB degradation.36 Whether inhibition of IκB degradation is due to a block in ubiquitination or in the proteasome itself is not known, but different probiotic species are known to inhibit both of these components of the NFκB activation pathway. An example of a known proteasome inhibitor is Lactobacillus plantarum. Conditioned media from L. plantarum inhibits NFκB binding activity and IκB degradation in an IEC model system. L. plantarum does not affect ubiquitination of IκB, but instead impedes the chymotrypsin-like activity of the proteasome, thus preventing IκB degradation. Even though treatment with L. plantarum inhibits the proteasome, it does not cause cell death and toxicity like other proteasome inhibitors.37 The probiotic mixture, VSL#3, is also capable of inhibiting the chymotrypsinlike activity of the proteasome to block IκB degradation but not ubiquitination (Fig. 2).28
Many other studies demonstrate that probiotics have inhibitory effects on the NFκB pathway in IECs, but fail to specifically define how NFκB activation is prevented. For example, secreted factors from Lactobacillus fermentum DSMZ 20052 can decrease Yersinia enterocolitica-induced IL-8 secretion from IECs by suppressing NFκB activation. MAPKs may also play a role in inhibiting IL-8 in this model system since p38 activation was also decreased.38 Both Bifidobacterium longum and Lactobacillus bulgaricus can modestly reduce TNF-induced IL-8 secretion from a HT-29 cell model system by decreasing NFκB p65 migration into the nucleus.39 Incubation of B. longum with colonic biopsy explants from patients with ulcerative colitis caused a modest decrease in TNFα and IL-8 due to inhibition of NFκB.40 Supernatant from a newly identified probiotic, Faecalibacterium prausnitzii DSM 17677, is able to suppress IL-1β-induced NFκB activation.41 Other studies showed that treatment of Caco-2 cells with butyrate, a short-chain fatty acid produced by anaerobic bacterial fermentation of dietary fiber, can inhibit IL-1β-induced IL-8 mRNA expression and suppress nuclear translocation and DNA binding of NFκB. Butyrate can also induce higher expression levels of IκB-β, causing increased cytoplasmic sequestering of NFκB, preventing its activation (Fig. 2).42
Not all probiotic bacteria inhibit NFκB activation. Rather some stimulate NFκB to cause increased cytokine secretion. A study investigating Bifidobacterium lactis BB12 showed this strain can transiently induce activation of RelA, a transcriptionally active subunit of NFκB, to increase IL-6 secretion. In addition to RelA activation, p38 MAPK is phosphorylated by B. lactis BB12. Specific inhibitors to both RelA and p38 showed that both pathways are needed to induce IL-6 production. In addition, B. lactis BB12-induced IL-6 expression is dependent on the TLR2 signaling pathway since no effect was seen in TLR2 deficient mouse embryogenic fibroblasts.43 The commensal, non-pathogenic Bacteroides vulgatus activates NFκB via the IL-1R/TLR4 pathway. Incubation with IECs induced interleukin-1 receptor-associated kinase-1 (IRAK1) degradation, IκBα phosphorylation and degradation, RelA phosphorylation, and NFκB DNA binding with increased transcriptional activity. Interestingly, the effects of B. vulgatus are modified in a co-culture model system consisting of Caco-2 cells and peripheral blood mononuclear cells (PBMCs). In the presence of PBMCs, IκBα degradation and NFκB activation are inhibited despite the fact that IRAK-1 degradation and IκBα and RelA phosphorylation still occur.44 These findings show that other cell types, particularly cells of the innate and adaptive immune system, modulate interactions between IECs and probiotic bacteria.
Probiotics can also affect MAPK signaling pathways independent of NFκB signaling. Suppression of p38 phosphorylation has been associated with inhibition of IL-8 secretion without impacting IL-8 mRNA levels or the activation of NFκB. This effect has been observed in IECs treated with VSL#3 DNA.45 Resta-Lenert et al. treated IECs with IFNγ either after pretreatment or simultaneously with a probiotic combination of Streptococcus thermophilus ATCC 19258 and Lactobacillus acidophilus ATCC 4356 or the commensal bacteria Bacteroides thetaiotaomicron ATCC 29184. Both the probiotic combination and commensal bacteria caused changes in MAPK signaling cascades, including sustained activation of ERK1/2 and increased p38 phosphorylation, which prevented IFNγ-induced changes in ion transport. The probiotic combination also caused a decrease in IκBα phosphorylation, an effect that was not observed in the commensal treated cells.46
PPARγ, a nuclear hormone receptor that can regulate intestinal inflammation and homeostasis, is another target for probiotic modulation.47 PPARγ may play a role in attenuating colitis as evidenced by PPARγ−/+ heterozygous mice being more susceptible to developing colitis. PPARγ may diminish colitis by inhibiting NFκB activity.48 Inquiries into PPARγ as a target for modulation demonstrated that Bacteroides thetaiotaomicron decreases phorbol 12-myristate 13-acetate (PMA)-, flagellin- and E. coli 0157:H7-induced IL-8 secretion via enhanced nuclear export of RelA through a PPARγ dependent pathway. Only viable, whole bacteria are able to influence RelA and IL-8 secretion, indicating a cell contact dependent mechanism. RNAi of PPARγ as well as dominant negative PPARγ can prevent B. thetaiotaomicron-induced nuclear export of RelA.49 Another commensal that regulates PPARγ is Enterococcus faecalis, a microbe that colonizes the human gut early in life. Four strains of E. faecalis, EC1, EC3, EC15 and EC16, isolated from newborns were capable of inducing PPARγ1 phosphorylation and enhancing DNA binding and activation of downstream effector genes such as IL-10.50 Subsequent studies demonstrate that PPARγ is also a target for modulation by probiotics. Culturing VSL#3 in the presence of linoleic acid produces conjugated linoleic acids, which possess anti-inflammatory and anticarcinogenic properties. Treatment of IECs with conditioned media containing VSL#3-produced conjugated linoleic acids causes enhanced PPARγ expression compared to VSL#3 or linoleic acids alone.51 Another probiotic study showed Lactobacillus casei decreases LPS-induced levels of COX-2, TLR4 and IL-8, possibly by increasing PPARγ mRNA expression and activating a peroxisome proliferator response element.52 Lactobacillus crispatus M247 uses hydrogen peroxide as a signal-transducing molecule to increase PPARγ activation and transcriptional activity.47 PPARγ mRNA and protein expression is reduced in the colonic epithelial cells of patients with ulcerative colitis compared to healthy individuals and patients with Crohn disease.48 All of these studies suggest that treatment with a specific commensal or probiotic strain may increase expression of PPARγ and help ameliorate ulcerative colitis-associated inflammation.
In vivo studies examining gene expression changes induced by probiotics demonstrate that probiotics can affect a multitude of genes. In one such study, gene expression profiling was performed on healthy human duodenal samples 6 hours post oral ingestion of Lactobacillus plantarum WCFS1. L. plantarum was administered at various growth phases, including midlog and stationary phase, and as dead bacteria. Host gene expression patterns differed by growth phase of the administered L. plantarum. Stationary-phase and dead L. plantarum induced a gene expression pattern involving upregulation of NFκB-, JUN- and TNF-dependent pathways. For example, upregulation of several NFκB subunits as well as three NFκB inhibitor molecules, BCL3, A20 and IκB was observed. In contrast, the gene expression pattern induced by midlog-phase L. plantarum involved MYC- and cyclin D1-dependent pathways, positive regulators of proliferation.53 Other studies indicate that LGG may affect mammalian gene expression by modulating genes involved in MAPK pathways. In a study by Di Caro et al. duodenal mucosal biopsies were taken from male patients ingesting LGG for one month and gene expression changes were compared to patients taking placebo. Microarray analysis showed that 334 genes were upregulated and 92 genes were downregulated compared to the placebo control. Diverse pathways were affected, including genes involved in cell adhesion, apoptosis, immunity and the MAPK signaling cascade.54 In a mouse small intestine model, LGG ATCC 53103 upregulated several genes involved in cytoprotection including MAPK-related, anti-apoptotic, and proliferation- and migration-related genes.55 The above studies demonstrate that proper selection of probiotics for a specific application include the appropriate species/strain, the growth parameters or physiologic state of the bacteria, and whether key factors or signals are produced by probiotics in vivo.
Current probiotic mechanistic studies demonstrate that beneficial bacteria of the gut are capable of changing host signaling pathways in IECs in a variety of ways. The NFκB pathway is very important in immune responses, and its modulation can have potent pro- or anti-inflammatory effects. NFκB activation can be blocked by probiotics at the key regulatory points of IκB phosphorylation and ubiquitination. NFκB subunits can be prevented from entering the nucleus or can undergo enhanced export from the nucleus via PPARγ. Even the proteasome itself can be inhibited to diminish degradation of NFκB-inhibitory molecules. In contrast, other probiotic strains enhance transport of NFκB subunits into the nucleus to increase activation and upregulate transcription of target genes. Other pathways regulated by probiotics include JNK, p38 and ERK1/2 MAPKs. These probiotic effects are strain specific, and as such it should not be assumed that these effects are shared by all strains of a particular species. The effectors of probiotic strains that allow for target specific modulation of NFκB, MAPK, and other signaling pathways remain unknown, but this information may be helpful in identifying bacterial strains that possess overlapping effects. Modulating multiple signaling pathways simultaneously with a combination of functionally non-redundant probiotic strains could have a profound synergistic effect on IEC function and produce potent pro- or anti-inflammatory effects.
Regulation of apoptosis in IECs.
While less well studied than probiotic modulation of IEC signaling pathways, certain probiotics can regulate apoptosis in IECs. Lactobacillus rhamnosus GG ATCC 53103 can activate anti-apoptotic Akt/protein kinase B and inhibit pro-apoptotic p38 MAPK in TNF-, IL-1α or IFNγ stimulated IECs.56 Subsequent experiments indicated that LGG secretes two proteins, p75 and p40. These proteins promote cell proliferation and activate anti-apoptotic Akt in a PI3K-dependent manner to protect human and mouse IECs from cytokineinduced apoptosis.57 The ability of probiotics to regulate apoptosis may be a useful strategy to minimize the deleterious effects of enteric infection. For example, during EPEC infection, apoptosis is induced via caspase-3 activation, which can be prevented by the probiotic yeast, “Saccharomyces boulardii”.17
Probiotic Modulation of Macrophage Signaling Pathways
Probiotics modulate various signaling pathways in macrophages and have corresponding effects on mucosal immunity (Table 1). Select probiotic strains suppress inflammation by inhibiting NFκB signaling and reducing pro-inflammatory cytokine secretion. For example, treatment of RAW 264.7 mouse macrophages with Escherichia coli strain M17 decreased LPS-induced proinflammatory cytokines TNFα, IL-1β and IL-6. In addition, E. coli M17 inhibited TNFα-induced NFκB and p65 nuclear binding (Fig. 3). In vivo studies of E. coli M17 treatment in a DSS-induced mouse model of colitis demonstrated decreased secretion of IL-12, IL-6, IL-1β and IFNγ due to an inhibitory effect on NFκB signaling.58
Figure 3.
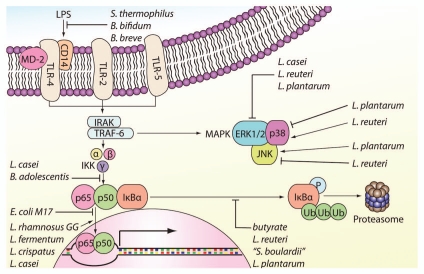
Probiotics modulate inflammatory signaling pathways in macrophages. Select probiotics can block binding of LPS to the CD14 receptor, interfering with LPS signal transduction. Various probiotics prevent activation of NFκB by decreasing phosphorylation or ubiquitination of IκBα or blocking NFκB translocation into the nucleus (suppression is indicated by a block sign “??”). NFκB activation is enhanced by other probiotics via increased nuclear translocation of transcriptionally active NFκB subunits (activation is indicated by an arrow sign “→”). MAPK proteins p38, JNK and ERK1/2 are also targets of probiotic modulation in macrophages. ERK, extracellular signal-regulated kinases; IκBa, inhibitor of NFκBα; IKK, IκB kinase; IRAK, interleukin-1 receptor-associated kinase; JNK, c-Jun N-terminal kinase; LPS, lipopolysaccharide; MD-2, myeloid differentiation 2; P, phosphorylation; TLR, Toll-like receptor; Ub, ubiquitin.
Certain probiotics and commensal microbes secrete soluble factors that suppress NFκB signaling in macrophages. Suppression of NFκB signaling may also be associated with changes in pattern recognition receptors (PRRs) and MAPK signaling pathways. This can be seen with culture supernatant from Lactobacillus casei strain Shirota (LcS), which can inhibit TNFα protein production and mRNA from LPS-stimulated human monocytoid THP-1 cells. Mechanistic studies showed that secreted factors from LcS suppress LPS-induced IκBα phosphorylation and subsequent degradation of this NFκB inhibitor. LPS-induced ERK1/2 phosphorylation is also diminished, indicating that MAPKs may also play a role in TNFα suppression by LcS. L-lactate is a candidate immunomodulatory factor produced in copious quantities by some Lactobacillus species, including LcS. L-lactate inhibits TNFα gene expression, in addition to suppressing IκBα phosphorylation and degradation. However, L-lactate does not affect ERK1/2 phosphorylation. These results suggest that LcS may secrete multiple factors, including L-lactate, that are necessary for TNFα inhibition or changes in MAPK pathways are not essential for TNFα inhibition in this model system.59 “Saccharomyces boulardii,” a strain of Saccharomyces cerevisiae, produces a small, heat stable, water-soluble factor that has anti-inflammatory properties. In THP-1 cells, this secreted factor prevented NFκB reporter gene activation and IκBα degradation, resulting in decreased p65 nuclear translocation and NFκB DNA binding.60 Another factor, butyrate, is secondarily enhanced by probiotic-mediated changes in intestinal microbial composition and function, and it seems to have anti-inflammatory affects in macrophages in addition to the anti-inflammatory affects observed in IECs. In both PBMCs and THP-1 cells, butyrate prevented LPS-induced TNFα and IL-6 secretion and inhibited IL-1β by blocking NFκB nuclear translocation through stabilization of IκBα.61 Finally, a study comparing several probiotics and commensal bacteria found LPS-induced TNFα production from both PBMCs and THP-1 cells is diminished by both Bifidobacterium breve BbC50 and Streptococcus thermophilus St065 in a dose dependent manner. Commensal bacterial strains of the species Bifidobacterium bifidum B536 and Ruminococcus gnavus FRE1 also inhibit LPS-induced TNFα. This study suggests that bacterial products may interfere with LPS signal transduction. These secreted factors decreased LPS binding to the CD14 receptor, causing an overall reduction in NFκB activation (Fig. 3).62
Lipoteichoic acid (LTA) is a major component of the outer peptidoglycan layer of gram-positive bacteria, and it may be an important factor in septic shock associated with gram-positive bacterial infections. The LTA from select probiotics, however, appears to have a potent anti-inflammatory effect. LTA isolated from Lactobacillus plantarum K8 (pLTA) inhibits Staphylococcus aureus LTA (aLTA)-induced TNFα production by preventing signal transduction through both NFκB and MAPK pathways.63 A study by Kim et al. examined the effects of LTA from L. plantarum K8 (pLTA) and found that pretreatment of THP-1 cells inhibited LPS-induced TNFα secretion.64 Decreased degradation of both IκBα and IκBβ resulted in diminished activation of NFκB. Additionally, pLTA suppressed LPS-induced phosphorylation of ERK1/2, JNK and p38. Pattern recognition receptors and upstream pathway components involved in TNFα production are also affected by pLTA. Many pattern recognition receptors have decreased expression following pLTA pretreatment, including TLR4, NOD1 and NOD2. IRAK-M, a negative regulator of TLRs, is induced by pLTA while LITAF, a molecule involved in LPS-induced TNFα expression, is decreased, resulting in an overall reduction in TNFα.64 In contrast to these anti-inflammatory studies, other studies have demonstrated that Lactobacillus LTA has an immunostimulatory effect. LTAs purified from L. casei YIT 9029 and L. fermentum YIT 0159 (FERM P-13859) stimulate TNFα production and activation of NFκB and JNK in RAW 264.7 cells.65 Interestingly, a mutant strain of L. plantarum, NCIMB8826, with a defect in D-alanylation of techoic acids (Dlt-mutant) has an enhanced ability to induce IL-10 production and to inhibit secretion of pro-inflammatory cytokines, IL-12, IFNγ and TNFα, compared to the wild type L. plantarum strain. When the Dlt− mutant is put into a trinitrobenzene sulphonic acid (TNBS)-induced colitis model, it is significantly more protective compared to control groups receiving wild type L. plantarum or no treatment.66 These results suggest that while probiotic LTA stimulates an inflammatory response mediated by macrophages, certain strains of probiotics may harbor changes in the teichoic acid biosynthesis pathway that result in anti-inflammatory effects mediated by their LTA.
Probiotics have diverse effects on the immune system, and stimulation or suppression of innate immunity ultimately depends on the biological features of individual probiotic strains. As with IECs, probiotics regulate diverse signaling pathways in addition to NFκB signaling. Specific strains of Lactobacillus reuteri (ATCC PTA 6475) can suppress TNFα production from LPS-stimulated macrophages by inhibiting the c-Jundependent activator protein-1 (AP-1) pathway.14 Suppressor of cytokine signaling (SOCS) family proteins can inhibit TLR signal transduction, and SOCS1 inhibits LPS induction of NFκB signaling in macrophages. Treatment of RAW 264.7 cells with Bifidobacterium adolescentis ATCC 15703 causes decreased IL-1β, TNFα and IL-12p40 through inhibition of IκBα phosphorylation, while increasing mRNA levels of SOCS1 and SOCS3 compared to LPS treatment alone.67 Lactobacillus rhamnosus GG ATCC 53103 and L. rhamnosus GR-1 conditioned media can induce anti-inflammatory cytokines IL-10 and G-CSF from THP-1 cells. G-CSF is essential for mediating L. rhamnosus GR-1 suppression of E. coli- or LPS-induced TNFα production by activating the JAK2-STAT3 pathway and suppressing JNK activation (Fig. 3).68 These studies demonstrate that a variety of inflammatory pathways may be modulated by different probiotic strains in order to induce similar anti-inflammatory effects.
Some probiotic strains have an immunostimulatory phenotype, causing activation of NFκB or STAT signaling in macrophages. STATs are inactive cytoplasmic transcription factors that can become activated by cytokine or antigen stimulation to induce inflammatory response cascades. Lactobacillus crispatus induced TNFα and IL-1β production through the activation of NFκB in THP-1 cells.69 In human PBMCs, viable Lactobacillus rhamnosus GG ATCC 53103 directly induced NFκB binding activity, as well as cytokine-mediated STAT1 and STAT3 DNA binding to promote an inflammatory response.70 Unlike lipoteichoic acid from L. plantarum K8, which inhibits NFκB and MAPK signaling,63,64 lipoteichoic acid isolated from both L. casei YIT 9029 and L. fermentum YIT 0159 activates NFκB, inducing TNFα production by RAW 264.7 cells (Fig. 3).65 These immunostimulatory phenotypes may be important in promoting host defense against pathogens.
In addition to influences on cytokine production, probiotics may affect apoptosis in macrophages by modulating NFκB signaling. Secreted factors from L. reuteri ATCC PTA 6475 may promote apoptosis in TNFα-stimulated myeloid cells through the suppression of anti-apoptotic factors Bcl-2 and Bcl-xL and through the inhibition of cell proliferation mediators, Cox-2 and cyclin D1. Increased apoptosis by L. reuteri secreted factors results from suppression of NFκB activation. L. reuteri suppressed IκBα ubiquitination resulting in decreased NFκB p65 nuclear translocation. Secreted factors from L. reuteri also modulated MAPK signaling as seen by enhanced phosphorylation of JNK and p38 and suppression of ERK1/2 phosphorylation.71 In addition to L. reuteri, L. rhamnosus Lcr promotes monocyte apoptosis in a time- and dose-dependent manner. In order to induce apoptosis, L. rhamnosus disrupts the mitochondrial membrane potential, increases pro-apoptotic Bax relative to Bcl-2, causes release of cytochrome-c, and activates downstream caspases 9 and 3.72 The pro-apoptotic effects of probiotics may prove to be useful in cancer therapy. A recent study showed that three different strains of L. plantarum, S1, DB22 and DS41, increased secretion of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) from PBMCs. This cytokine stimulates apoptosis of malignant cancer cells both in vitro and in vivo and could be a promising new area of cancer therapy.73
Select probiotic strains can influence macrophage signaling pathways to affect cytokine secretion and apoptosis. As in IECs, NFκB is a key target pathway, along with MAPKs, ERK1/2 and p38. Probiotics also demonstrate effects at the cell surface by inhibiting agonist-mediated signaling through cell surface receptors. AP-1, STATs and SOCS represent additional intracellular targets of probiotic modulation unique to macrophages. Modifications of cell surface components such as LTA may account for probiotic effects, and activated immune cell populations (macrophages) may be selectively deleted by probiotics via pro-apoptotic mechanisms.
Probiotic Modulation of Dendritic Cells Influences T-Lymphocyte Populations
Dendritic cells (DCs) directly sample gut luminal contents through cellular processes that extend between intestinal epithelial cells into the lumen.74 This feature of intestinal DCs, combined with their ability to orchestrate T-lymphocyte responses, highlights the role of DCs as a bridge between microbes, innate immunity and adaptive immunity (Fig. 1). Bifidobacterium breve C50 produces a fermentation product, BbC50sn, which induces DC maturation, increases DC survival, and increases antiinflammatory IL-10 production. BbC50sn can induce PI3K/Akt phosphorylation similar to the TLR2 agonist, Zymosan and p38 phosphorylation similar to the TLR4 agonist, LPS. PI3K activation appears to regulate the prolonged survival induced by BbC50sn.75 In addition, BbC50sn may prolong DC survival by increasing anti-apoptotic Bcl-xL and inactivating pro-apoptotic Bad.76 Both PI3K and p38 induce DC maturation by proportionately increasing expression of CD83 and CD86 maturation markers. Increased IL-10 production by BbC50sn is regulated by PI3K, p38 and by ERK1/2.75 As seen in other host-microbe interactions, different species and strains of Lactobacillus may have diverse effects with respect to modulation of cytokine production, regulation of MHC class II and B7-2 surface receptors, among other effects. Lactobacillus species may counteract each other's effects on DCs, emphasizing the point that aggregate microbial composition and balance may determine the nature of mucosal immune responses. For example, L. reuteri DSM 12246 inhibits the induction of IL-12, IL-6 and TNFa by L. casei CHCC3139, and also diminishes L. casei-induced increases of B7-2 receptors.77 DCs may play a key role in maintaining immune tolerance to commensal bacteria in the gut. Intestinal DCs can harbor viable commensal bacteria and transport the bacteria to the mesenteric lymph nodes (MLNs), where it is retained and prevented from entering the systemic immune compartment. By this mechanism, DCs can selectively induce IgA to protect against mucosal invasion, while restricting immune responses to the local microbial community in the gut.78–80
DCs regularly interact with gut microbes and other stimuli to determine if tolerance or an immune response will be induced. Various microbial factors interact with different DC cell surface pattern recognition receptors to determine DC maturation and subsequent DC-regulated differentiation of naïve T cells into Th1, Th2, Th17 or T regulatory cells.81–83 Several types of regulatory T cells, including Th3, TR1, CD4+CD25+ regulatory, CD8+ suppressor, and γδ T cells may be affected by probiotics and beneficial microbes.84 Commensals may even be essential for the appropriate balance of regulatory T-cell populations. A study by Ivanov et al. demonstrated that a specific group of intestinal microbes, the cytophaga-flavobacter-bacteroidetes (CFB), was important in Th17 development in the lamina propria and in maintaining the appropriate balance of Th17 and Treg cell populations.85 This study highlights the important role of the intestinal microbiota in maintaining intestinal homeostasis and in T-cell development. Further evidence can be gathered from studies with germ-free mice showing that they possess a smaller population of antigen-experienced CD4+CD45Rblow T cells compared to conventional mice. Colonization of these germ-free mice with Bacteroides fragilis NCTC 9343 replenished the CD4+CD45Rblow T-cell population, an effect that was dependent on B. fragilis polysaccharide A (PSA). When a B. fragilis mutant strain that could not produce PSA colonized germfree mice, no increase in the CD4+CD45Rblow population was observed.86
A zealous Th1 response and its cytokine profile (IFNγ and IL-12p40) may contribute to chronic intestinal inflammation in Crohn disease,74 and diminished Th1 cell populations or corresponding cytokines may help ameliorate intestinal inflammation.87 Treatment of DCs with VSL#3 caused decreased IFNγ production by DC-stimulated T cells, along with a corresponding decrease in the number of Th 1 cells.88 Similarly, T cells co-cultured with L. paracasei B21060-treated DCs had diminished IFNγ, IL-2, IL-6 and IL-10 production, suggesting a reduced Th1 population. When L. paracasei was administered intra-gastrically to a DSS-mouse model of colitis, it demonstrated a protective effect, with reduced disease severity and delayed disease progression.13 A combination of L. paracasei 1602 and L. reuteri 6798 strains significantly reduced cecocolic inflammation induced by Helicobacter hepaticus in an IL-10-deficient mouse model of colitis. The probiotics-treated animals demonstrated diminished IL-12p40 mRNA levels, suggesting that select probiotics may suppress the Th1 immune response.9
Regulatory T cells are a subset population of T cells that can suppress the function of Th effector cells. Many functions have been proposed for Treg cells, including induction of oral tolerance, suppression of allergies and asthma, and induction of tolerance to commensal bacteria.89 Intestinal immune function relies on several lymphocyte populations, including regulatory T cells specific to the intestine. The intestinal components are charged with the task of maintaining tolerance to dietary antigens and the commensal microbiota. Since regulatory T cells play a significant role in dampening misdirected immune responses,90 probiotics may exert their anti-inflammatory effects by influencing this T-cell population. Bifidobacterium lactis W51, L. acidophilus W55, and L. plantarum W62 induce FOXP3+ Treg cell differentiation from a CD25− T cell population, with the most potent induction by L. acidophilus W55. The expression of FOXP3 is stable, and these cells display a suppressive phenotype, indicating that some probiotic species are capable of inducing regulatory T cells.91 In a TNBS mouse model of chronic colitis, VSL#3 administration decreased the severity of the colitis, an effect that is transferrable to control mice by transplanting lamina propria mononuclear cells (LPMCs) from probiotic-treated mice. A greater number of regulatory CD4+ T cells with surface TGFβ are present in probiotic-treated mice. TGFβ expressing regulatory T cells were necessary to generate the beneficial effects of VSL#3 administration since anti-TGFβ antibodies blocked the protective effect.92 Transfer of DCs pretreated with L. rhamnosus Lr32 protected against TNBS-induced colitis and reduced inflammation scores. L. rhamnosus may induce tolerogenic DCs and, consequently, stimulate regulatory T lymphocytes because the DCs lose their protective effect in mice lacking CD25+ T cells. Supporting this hypothesis is the fact that protection is associated with elevated localized expression of indoleamine 2,3 dioxygenase (IDO), an enzyme that catabolizes tryptophan and regulates T-cell function through depletion of tryptophan.81 DCs treated with one of five different probiotics, including L. casei DN-114 001, Streptococcus thermophilus DN-001 621, Bifidobacterium animalis DN-173 010, Bifidobacterium adolescentis DN-150 017, or Bacteroides thetaiotaomicron, stimulate naïve T cells to develop suppressor functions, demonstrated by the ability of differentiated T cells to diminish proliferation and IFNγ production by CD4+ effector T cells.93 MLN cells co-cultured with DCs that had encountered L. reuteri 100-23 demonstrated an increase in the number of FOXP3+ T cells along with a corresponding decrease in proliferation of the T cell population, which is consistent with an enhanced Treg population.94 Finally, oral treatment with a probiotic cocktail consisting of L. acidophilus, L. casei, L. reuteri, B. bifidium and S. thermophilus increased generation of CD4+Foxp3+ Tregs from CD4+CD25− T cells in the MLN. In addition, the suppressor activity of CD4+CD25+ Tregs was enhanced by the probiotic-induced increase in TGFβ and cytotoxic T-lymphocyte-associated-4 (CTLA-4) expression.95 Modulation of DCs and T cell populations and phenotypes by commensals and probiotics may contribute to the maintenance of gut homeostasis and immune tolerance to gut microbes.
Unlike the more abundant αβ T-cell receptor (TCR)-expressing T cells, γδ T cells express TLRs and function like APCs. γδT cells induce maturation and proliferation of DCs and directly activate CD4+ αβ T cells, providing yet another link between innate and adaptive immunity.96 One interesting study shows that γδ T-cell-deficient mice will develop spontaneous colitis, an effect that can be prevented or suppressed by γδ T cell transfer.97 These results indicate that γδ T cells likely reduce the inflammatory response. The gram-negative probiotic strain E. coli Nissle 1917 can increase the activation and expansion of γδ T cells but not αβ T cells.96,98 Once the γδ T cells have been activated, E. coli Nissle can also induce apoptosis of these activated cells through the Fas ligand-dependent pathway, suggesting the existence of a regulatory loop.96 Treatment with L. acidophilus Bar 13 and Bifidobacterium longum Bar 33 in a TNBS-induced model of acute colitis attenuated intestinal damage, possibly by decreasing the number of CD4+ T cells found in the lamina propria and intraepithelial lymphocyte populations. Further study into distinct subpopulations showed that these two probiotics can increase the number of γδ T cells present in the intraepithelial lymphocyte population, while decreasing the number of γδ in the lamina propria lymphocyte population.97 Location-specific differences in T-cell subpopulations of the gut mucosa may have implications for the nature of the immune response to the gut microbiota.
Probiotics can regulate apoptosis in different cell types of the intestinal mucosa, and may contribute to maintenance of the appropriate number and balance of lymphocyte subpopulations. Explants of inflamed ileum samples from patients with Crohn disease were cultured with Lactobacillus casei DN-114 001 and examined for cytokine secretion and markers of apoptosis. In addition to inhibiting pro-inflammatory cytokines IL-6 and TNFα, L. casei also decreased expression of the anti-apoptotic protein Bcl-2 in the lamina propria lymphocytes. More TUNEL-positive lymphocytes and fewer activated T cells were present in the explants with L. casei, indicating that the probiotic can reduce the number of activated T cells by inducing apoptosis. Similar effects were not observed when L. casei was co-cultured with non-inflamed tissue, underlining the importance of current host immune state prior to addition of probiotics.99 Finally, a study examining the effects of VSL#3 in the IL-10-deficient mouse model of colitis demonstrated upregulation of galectin 2, a protein that induces apoptosis of activated T cells, compared to untreated mice.100 These recent studies demonstrate that several probiotic strains can influence DC maturation, survival/proliferation and cytokine production as well as the induction of specific T-cell phenotypes.
Probiotics Influence Antibody Production in the Intestine
In the lamina propria of the gut, B cells differentiate into plasma cells and secrete dimeric IgA antibodies. The polymeric Ig receptor on the basolateral surface of intestinal epithelial cells complexes with IgA and transports it to the apical cell surface where it is secreted into the intestinal lumen.84,101 Secretory IgA is important in mucosal-associated immunity and helps protect the host by binding a variety of antigens from bacteria, viruses and fungi.84,101,102 Mucosal immune responses to commensal bacteria include IgA production that is independent of T lymphocytes and an organized lymphoid tissue component.103 Epithelial cells and DCs produce molecules such as a proliferation-inducing ligand (APRIL), CD40 ligand, and TGFβ that induce T cell-independent IgA class-switching.104 Commensal bacteria such as Lactobacillus plantarum NCIMB88, L. plantarum WCFS1, Bacillus subtilis JH642, and TLR-activating bacterial products can induce intestinal epithelial cells to produce APRIL, which triggers IgA class switching to IgA2, an immunoglobulin that is prevalent in the distal intestine and more resistant to bacterial proteases.53,105 In Rag−/− IgA-deficient mice, which are lacking a functional adaptive immune system, commensal Bacteroides thetaiotaomicron VPI-5482 enhances innate immunity by increasing activity of inducible nitric oxide synthase (iNOS) and upregulating innate immunity-associated genes and downstream signaling pathways such as STATs and NFκB. The observed enhancement in innate immunity was in comparison to Rag−/− mice with B. thetaiotaomicron-specific IgA or to wild-type mice, indicating IgA and the adaptive immune system play an important role in gut homeostasis and in mucosal immune responses to commensal gut microbes.104
Several studies have shown that probiotics can stimulate the production of IgA by plasma cells (Fig. 1). However, the ability to induce IgA may be strain-dependent. In a small study, seven healthy children received oral Bifidobacterium lactis Bb-12 for 21 days. Administration of this bacterial strain correlated with increased total fecal IgA and anti-poliovirus IgA, suggesting B. lactis stimulates IgA secretion in the gut.106 Oral administration of Bifidobacterium bifidum Bb-11 in mice demonstrated that this probiotic strain enhances the number of IgA-secreting cells present in the MLNs and spleen and increases systemic and intestinal IgA, while not affecting B. bifidum-specific IgA. This effect is increased substantially when B. bifidum is encapsulated, suggesting that it is dependent on intact, viable cells of B. bifidum in the intestine.107 Long-term (98 days) oral administration of fermented milk containing L. casei DN-114001, L. delbrueckii subsp bulgaricus and Streptococcus thermophilus showed increased numbers of IgA+ cells in both the small and large intestine compared to controls.102 Finally, a comparison between two Lactobacillus species, L. johnsonii NCC 533 and L. paracasei NCC 2461, in germfree mice showed that both strains can induce lymphoid follicle formation and increase the number of IgA+ plasma cells present in the lamina propria. The pattern of increased Lactobacillus-specific IgA differs between the two species. L. johnsonii stimulates a significant increase in L. johnsonii-specific IgA in both the Peyer's patches and gut lumen, while L. paracasei results in a modest increase in specific IgA by comparison. Based on these findings, one would expect the colonization of L. johnsonii to be less robust than that of L. paracasei, but the opposite is true, indicating that other components of the innate immune system play an important role in regulating bacterial load.108 The discrepancy between specific IgA production and bacterial load suggests that differences in immunogenicity of LAB strains may be independent of the strain's capacity to persist in the host.
Outside of the gut, probiotics influence immunoglobulin levels by altering systemic Ig isotype profiles. Oral administration of L. johnsonii NCC 533 skewed systemic IgG isotypes towards a greater proportion of IgG1, an isotype that is associated with IL-4 induction of B cells and a Th2 predominant immune response. In contrast, L. paracasei NCC 2461 induced a greater proportion of IgG2a, which results from IFNγ stimulation of B cells, and is associated with a Th1 predominant immune response.108 Treatment with orally administered stationary phase L. casei ATCC 393 or L. murinus CNRZ increased the IgG1/IgG2a ratio without altering the overall amount of systemic IgG. This increased IgG1 response may reflect a higher CD4+ Th2 cell activity in these mice.109 Differences in immunoglobulin induction patterns indicate that different probiotic strains can induce unique systemic T-cell responses.
Commensals Impact GALT Development and Function
Gut associated lymphoid tissue (GALT) is found in the intestine in the form of Peyer's patches and MLNs, and these areas may be in active communication with the gut microbiota. In one study, mice were given Lactobacillus plantarum Lp6 for 2 weeks, followed by jejunal Peyer's patch isolation and DNA microarray analysis of differentially expressed genes. L. plantarum treatment increased expression of 420 genes involved in immune responses, cell differentiation, cell-cell signaling, cell adhesion, and signal transcription and transduction.110 In a chicken tonsil model (part of the GALT in avian species), treatment with L. acidophilus DNA induced upregulation of several genes involved in proinflammatory signaling pathways, including STAT2, STAT4, IL-18, MyD88, IFNα and IFNγ.111 The administration of probiotic species induced a multitude of genetic changes, demonstrating how the GALT may respond to changes in the gut microbiota.
The two-way communication between the lymphoid organs of the gut and the intestinal bacteria is important for the development of GALT as well as for homeostasis. For example, Peyer's patches and lymph nodes are underdeveloped and structureless in germ-free mice.79 Polysaccharide (PSA) from Bacteroides fragilis NCTC 9343 promotes lymphoid organogenesis, a process that does not occur normally when a B. fragilis PSA mutant strain is used.112 Intestinal lymphoid tissues such as Peyer's patches, MLNs, and isolated lymphoid follicles (ILFs) produce microbe-reactive, IgA-producing B cells that can help prevent overgrowth of unwanted bacterial species. Mice with diminished IgA have an expanded bacterial community in their gut along with numerous and abnormally large ILFs. Gut bacterial communities are also significantly altered in the absence of ILFs. Interestingly, the formation of lymphoid tissues, specifically mature ILFs, requires induction by gut microbiota. Gram-negative peptidoglycan is necessary and sufficient to induce formation of ILFs by binding NOD1, activating CCR6 and inducing β-defensin 3 and CCL20-mediated signaling.113 These observations demonstrate the beneficial relationship between commensal microbes and host immunity and development. Continued investigations into this important crosstalk are necessary to fully appreciate the role of the microbiota in human health and disease.
Anatomical Distribution of the Intestinal Microbiota and the Subsequent Effects on the Host
The exact composition and distribution of the microbiota within the gastrointestinal tract is still relatively unknown. This paucity of knowledge is being aggressively addressed by the Human Microbiome Project (HMP) and the International Human Microbiome Consortium (IHMC), a global effort to characterize bacterial communities present at a variety of body sites in healthy adults. Current culture-based analysis shows that the bacterial load within the intestine is relatively low in the duodenum (50–100 cfu/mL) and increases distally through the small and large intestines, with the distal ileum and cecum possessing greater than 500,000 cfu/mL of bacteria.114 Culture-independent analyses, namely 16S rRNA gene-sequence analysis, of the colon demonstrates that the predominant phyla of healthy humans are the Bacteroidetes and Firmicutes.115 Human-associated bacteria may be members of relatively few phyla, but ample diversity at the species and strain levels is apparent within an individual and between individuals.115,116
The colonization pattern of commensals and probiotics within the intestine may dictate where the associated beneficial effects of these organisms occur. For example, the cecum and ascending colon are known to be the sites where a majority of short-chain fatty acids like butyrate are produced.117 The immunomodulatory effects of butyrate, such as inhibition of IκB degradation, may be restricted to the cells of the cecum and ascending colon. However, if probiotics only exerted their beneficial effects locally, it would be difficult to explain the mechanisms of systemic effects conferred by orally administered probiotics. A study by Yun et al. in a mouse model of type 2 diabetes demonstrated that a twice daily, high dose, oral administration of L. gasseri BNR17 for 12 weeks could reduce fasting and postprandial two hour blood glucose levels after 6 weeks of probiotic administration. In addition, L. gasseri BNR17-treated mice had improved glucose tolerance for two hours at weeks 6 and 9, and the BNR17 group showed a trend towards decreased HbA1c after 12 weeks.118 Probiotics have also demonstrated effectiveness in the prevention of allergic disorders, such as eczema. In one such study, high risk pregnant mothers were given an oral probiotic cocktail for several weeks prior to delivery and their infants were given the same oral cocktail for six months after birth. The probiotic cocktail included L. rhamnosus GG ATCC 53103, L. rhamnosus LC705 (DSM 7061), Bifidobacterium breve Bb99 (DSM 13692), and Propionibacterium freudenreichii subsp shermanii JS (DSM 7076). Infants who received the probiotic cocktail demonstrated a significant reduction in the incidence of eczema compared to placebo controls.6 Another study examined the effects of oral pretreatment with Lac-B, a mixture of Bifidobacterium infantis and B. longum, in a rat model of nasal hypersensitivity. Prolonged pretreatment with Lac-B suppressed histamine H1 receptor expression as well as the expression and activity of histidine decarboxylase, the enzyme responsible for synthesizing histamine from L-histidine. In addition, Lac-B pretreatment reduced allergic-like symptoms in the rats, including sneezing, watery rhinorrhea, and nasal swelling and redness.10 These studies plus others have demonstrated that orally administered probiotics can exert beneficial effects at sites remote to the gastrointestinal tract. These results argue that probiotic effects, at least for certain strains, are not geographically restricted.
Many probiotic studies have been performed with species and strains that are not indigenous to the human or mouse gastrointestinal tracts. It may be difficult for these non-indigenous bacterial species and strains to colonize the intestine, especially in areas of increased bacterial density or diversity such as the cecum. Even transient exposure to probiotics may be sufficient for beneficial effects to be observed. For example, transient exposure to L. rhamnosus GG ATCC 53103 induces hsp expression in IECs.27 In a mouse pup model, there is no evidence of long-term gut colonization with human-derived strains L. reuteri ATCC PTA 6475 and L. reuteri DSM 17938. These two L. reuteri strains, however, have profound effects on mouse gastrointestinal physiology and immunology despite their transient passage through the gut (Preidis G, unpublished data). The ability of probiotics to exert their beneficial effects even with transient exposure to the host suggests that colonization of the gut may not be essential for probiotic function, depending on the strain and biological effects of interest. A recent study demonstrated that the human gut microbiota approaches adult-like complexity in terms of composition by the end of an infant's first year of life.119 This limited time frame may provide a “window of opportunity” in which the microbial composition of the gut can be significantly altered. Only continued work in the HMP and the various associated projects examining healthy versus diseased states will enable us to answer these types of questions.
Conclusions and Future Directions
The complex gut microbiome is not a “silent organ” or simply a collection of passenger microorganisms; rather, intestinal microbial communities represent active participants in mammalian immunity and physiology. Probiotics include member species of indigenous gastrointestinal microbial communities, but this class of microbes also includes organisms that may be “alien” invaders of established microbiomes. Commensal microbes and microbiome research will yield new classes of beneficial microbes and probiotics. By virtue of well-documented effects on mammalian cells in vitro and in animal models, probiotics clearly can cause biologically significant changes in the gastrointestinal tract despite the presence of complex, established microbial ecosystems. Whether these microbes are classified as indigenous or allochthonous, probiotics are illuminating key signaling pathways important in mucosal immunity relevant to gut microbes. Probiotics can also serve as a tool to probe mammalian immunity and physiology of the gastrointestinal tract, and may reveal novel therapeutic approaches for alleviation of human diseases.
As summarized in this review, probiotics have been documented to modulate different signaling pathways in epithelial and immune cells. These pathways include NFκB, MAPKs, PI3K/Akt, and transcriptional regulators such as heat shock transcription factor 1 and PPARγ. Intestinal epithelial cells, macrophages, dendritic cells and lymphocytes are all affected in some way by individual probiotic strains. Future directions in this area should continue to elucidate the effects of different strains and include global mammalian gene expression profiling in order to explore all currently unknown and unappreciated pathways in the host. More gene expression, proteomics and metabolomics studies are needed to explore how bacteria affect intestinal biology in the context of in vivo model systems, laser-microdissected intestinal mucosa, and studies of defined cellular populations in vivo. New noninvasive imaging studies may help us to understand how fluctuations in the intestinal microbiota may contribute to changes in intestinal physiology in vivo.
Investigations of microbial-host signaling inevitably include changes in microbial communities and secondary alterations to functional metagenomics caused by introduction of probiotics. New sequencing technologies are behind a rapid surge in metagenomics and studies of microbial composition in the gastrointestinal tract. The recent discovery of segmented filamentous bacterium (SFB) and its association with Th17 immune responses in the mammalian intestine offers a dramatic punchline to the relevance of microbiome research for understanding mammalian immunity.120 In addition to aggregate microbial composition, the identification of key probiotic genes and strains is evolving by virtue of systems biology approaches in probiotics and “reference genomes” projects of commensal microbes. Strain specificity is an important concept that has emerged originally from studies of bacterial pathogenesis. Ultimately any reproducible biological effect depends on a stable phenotype emanating from distinct bacterial clones or strains. In this review, we have witnessed examples of the tremendous diversity of biological effects conferred by different strains of the same or different bacterial species. We will be able to make intelligent decisions on how to treat disease phenotypes with probiotics only with greater understanding of strain-specific biological features and the ways in which such strains modulate host signaling pathways. In conclusion, new cell or gene therapy-based approaches may depend on the rational selection of natural gut-derived microbes or genetically-engineered probiotics for applications in human gut health.
Acknowledgements
Special thanks to Matthew D. Burstein for the production of the figures included in this review and to Jennifer K. Spinler for her constructive feedback on the manuscript. This work was supported by National Institutes of Health (NIH) National Institute of Digestive and Kidney Disease (UH2 DK083990, R01 DK065075, P30 DK56338), NIH National Center for Complementary and Alternative Medicine (R01 AT004326 and R21 AT003482), and NIH National Human Genome Research Institute (U54 HG003272).
Abbreviations
- A20
TNFalpha-induced protein 3
- AP-1
activator protein-1
- APC
antigen-presenting cell
- APRIL
a proliferation-inducing ligand
- BCL3
B-cell CLL/lymphoma 3
- CFB
cytophaga-flavobacter-bacteroidetes
- COX-2
cyclooxygenase-2
- CSF
competence and sporulation factor
- CTLA-4
cytotoxic T-lymphocyte-associated-4
- DC
dendritic cell
- DSS
dextran sodium sulfate
- EPEC
enteropathogenic Escherichia coli
- ERK
extracellular signal-regulated kinases
- FOXP3
forkhead box P3
- GALT
gastrointestinal-associated lymphoid tissue
- G-CSF
granulocyte colony-stimulating factor
- HbA1c
hemoglobin A1c
- HMP
human microbiome project
- hsp
heat shock protein
- LAB
lactic acid bacteria
- IBD
inflammatory bowel disease
- IBS
irritable bowel syndrome
- IDO
indoleamine 2,3 dioxygenase
- IEC
intestinal epithelial cell
- IFN
interferon
- Ig
immunoglobulin
- IκBα
inhibitor of NFκB alpha
- IKK
IκB kinase
- IL
interleukin
- ILFs
isolated lymphoid follicles
- iNOS
inducible nitric oxide synthase
- IRAK
interleukin-1 receptor-associated kinase
- JAK2
janus kinase 2
- JNK
c-Jun N-terminal kinase
- LITAF
lipopolysaccharide-induced tumor necrosis factor-alpha factor
- LPMCs
lamina propria mononuclear cells
- LPS
lipopolysaccharide
- LTA
lipoteichoic acid
- MAPK
mitogen-activated protein kinase
- MHC
major histocompatibility complex
- MLN
mesenteric lymph node
- MYC
v-myc oncogene homolog
- NFκB
nuclear factor-kappaB
- NOD
nucleotide-binding oligomerization domain-containing protein
- PBMCs
peripheral blood mononuclear cells
- pIgR
polymeric immunoglobulin receptor
- PI3K
phosphoinositide 3-kinase
- PMA
phorbol 12-myristate 13-acetate
- PPARγ
peroxisome proliferator activated receptor-gamma
- PRR
pattern recognition receptor
- PSA
polysaccharide
- ROS
reactive oxygen species
- RXR
retinoid X receptor
- SFP
segmented filamentous bacterium
- SOCS
suppressor of cytokine signaling
- STAT
signal transducers and activator of transcription
- TCR
T-cell receptor
- TGFβ
transforming growth factor beta
- TLR
toll-like receptor
- TNBS
trinitrobenzene sulphonic acid
- TNF
tumor necrosis factor
- TRAIL
tumor necrosis factor-related apoptosis-inducing ligand
- Ub
ubiquitin
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/11712
Conflicts of interest
The authors disclose the following: Dr. Versalovic has received unrestricted research support from Biogaia AB, Stockholm, Sweden, and has received honoraria from Group Danone, Paris, France.
References
- 1.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 2.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 3.Cani PD, Delzenne NM. The role of the gut microbiota in energy metabolism and metabolic disease. Curr Pharm Des. 2009;15:1546–1558. doi: 10.2174/138161209788168164. [DOI] [PubMed] [Google Scholar]
- 4.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 5.Preidis GA, Versalovic J. Targeting the human microbiome with antibiotics, probiotics and prebiotics: gastroenterology enters the metagenomics era. Gastroenterology. 2009;136:2015–2031. doi: 10.1053/j.gastro.2009.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kukkonen K, Savilahti E, Haahtela T, Juntunen-Backman K, Korpela R, Poussa T, et al. Probiotics and prebiotic galacto-oligosaccharides in the prevention of allergic diseases: a randomized, double-blind, placebo-controlled trial. J Allergy Clin Immunol. 2007;119:192–198. doi: 10.1016/j.jaci.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Barbara G, Cremon C, Pallotti F, De Giorgio R, Stanghellini V, Corinaldesi R. Postinfectious irritable bowel syndrome. J Pediatr Gastroenterol Nutr. 2009;48:95–97. doi: 10.1097/MPG.0b013e3181a15e2e. [DOI] [PubMed] [Google Scholar]
- 8.Barbara G, Stanghellini V, Cremon C, De Giorgio R, Corinaldesi R. Almost all irritable bowel syndromes are post-infectious and respond to probiotics: controversial issues. Dig Dis. 2007;25:245–248. doi: 10.1159/000103894. [DOI] [PubMed] [Google Scholar]
- 9.Pena JA, Rogers AB, Ge Z, Ng V, Li SY, Fox JG, et al. Probiotic Lactobacillus spp. diminish Helicobacter hepaticus-induced inflammatory bowel disease in interleukin-10-deficient mice. Infect Immun. 2005;73:912–920. doi: 10.1128/IAI.73.2.912-920.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dev S, Mizuguchi H, Das AK, Matsushita C, Maeyama K, Umehara H, et al. Suppression of histamine signaling by probiotic Lac-B: a possible mechanism of its anti-allergic effect. J Pharmacol Sci. 2008;107:159–166. doi: 10.1254/jphs.08028fp. [DOI] [PubMed] [Google Scholar]
- 11.FAO/WHO, author. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Basel, Switzerland: World Health Organization; 2001. [Google Scholar]
- 12.Prantera C, Scribano ML, Falasco G, Andreoli A, Luzi C. Ineffectiveness of probiotics in preventing recurrence after curative resection for Crohn's disease: a randomised controlled trial with Lactobacillus GG. Gut. 2002;51:405–409. doi: 10.1136/gut.51.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mileti E, Matteoli G, Iliev ID, Rescigno M. Comparison of the immunomodulatory properties of three probiotic strains of Lactobacilli using complex culture systems: prediction for in vivo efficacy. PLoS One. 2009;4:7056. doi: 10.1371/journal.pone.0007056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin YP, Thibodeaux CH, Pena JA, Ferry GD, Versalovic J. Probiotic Lactobacillus reuteri suppress proinflammatory cytokines via c-Jun. Inflamm Bowel Dis. 2008;14:1068–1083. doi: 10.1002/ibd.20448. [DOI] [PubMed] [Google Scholar]
- 15.Zyrek AA, Cichon C, Helms S, Enders C, Sonnenborn U, Schmidt MA. Molecular mechanisms underlying the probiotic effects of Escherichia coli Nissle 1917 involve ZO-2 and PKCzeta redistribution resulting in tight junction and epithelial barrier repair. Cell Microbiol. 2007;9:804–816. doi: 10.1111/j.1462-5822.2006.00836.x. [DOI] [PubMed] [Google Scholar]
- 16.Resta-Lenert S, Barrett KE. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC) Gut. 2003;52:988–997. doi: 10.1136/gut.52.7.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Czerucka D, Dahan S, Mograbi B, Rossi B, Rampal P. Saccharomyces boulardii preserves the barrier function and modulates the signal transduction pathway induced in enteropathogenic Escherichia coli-infected T84 cells. Infect Immun. 2000;68:5998–6004. doi: 10.1128/iai.68.10.5998-6004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otte JM, Podolsky DK. Functional modulation of enterocytes by gram-positive and gram-negative microorganisms. Am J Physiol Gastrointest Liver Physiol. 2004;286:613–626. doi: 10.1152/ajpgi.00341.2003. [DOI] [PubMed] [Google Scholar]
- 19.Seth A, Yan F, Polk DB, Rao RK. Probiotics ameliorate the hydrogen peroxide-induced epithelial barrier disruption by a PKC- and MAP kinase-dependent mechanism. Am J Physiol Gastrointest Liver Physiol. 2008;294:1060–1069. doi: 10.1152/ajpgi.00202.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mack DR, Ahrne S, Hyde L, Wei S, Hollingsworth MA. Extracellular MUC3 mucin secretion follows adherence of Lactobacillus strains to intestinal epithelial cells in vitro. Gut. 2003;52:827–833. doi: 10.1136/gut.52.6.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mack DR, Michail S, Wei S, McDougall L, Hollingsworth MA. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am J Physiol. 1999;276:941–950. doi: 10.1152/ajpgi.1999.276.4.G941. [DOI] [PubMed] [Google Scholar]
- 22.Wehkamp J, Harder J, Wehkamp K, Wehkamp-von Meissner B, Schlee M, Enders C, et al. NFκB- and AP-1-mediated induction of human β defensin-2 in intestinal epithelial cells by Escherichia coli Nissle 1917: a novel effect of a probiotic bacterium. Infect Immun. 2004;72:5750–5758. doi: 10.1128/IAI.72.10.5750-5758.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlee M, Harder J, Koten B, Stange EF, Wehkamp J, Fellermann K. Probiotic lactobacilli and VSL#3 induce enterocyte β-defensin 2. Clin Exp Immunol. 2008;151:528–535. doi: 10.1111/j.1365-2249.2007.03587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X, Kokkotou EG, Mustafa N, Bhaskar KR, Sougioultzis S, O'Brien M, et al. Saccharomyces boulardii inhibits ERK1/2 mitogen-activated protein kinase activation both in vitro and in vivo and protects against Clostridium difficile toxin A-induced enteritis. J Biol Chem. 2006;281:24449–24454. doi: 10.1074/jbc.M605200200. [DOI] [PubMed] [Google Scholar]
- 25.Castagliuolo I, LaMont JT, Nikulasson ST, Pothoulakis C. Saccharomyces boulardii protease inhibits Clostridium difficile toxin A effects in the rat ileum. Infect Immun. 1996;64:5225–5232. doi: 10.1128/iai.64.12.5225-5232.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castagliuolo I, Riegler MF, Valenick L, LaMont JT, Pothoulakis C. Saccharomyces boulardii protease inhibits the effects of Clostridium difficile toxins A and B in human colonic mucosa. Infect Immun. 1999;67:302–307. doi: 10.1128/iai.67.1.302-307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao Y, Drabik KA, Waypa TS, Musch MW, Alverdy JC, Schneewind O, et al. Soluble factors from Lactobacillus GG activate MAPKs and induce cytoprotective heat shock proteins in intestinal epithelial cells. Am J Physiol Cell Physiol. 2006;290:1018–1030. doi: 10.1152/ajpcell.00131.2005. [DOI] [PubMed] [Google Scholar]
- 28.Petrof EO, Kojima K, Ropeleski MJ, Musch MW, Tao Y, De Simone C, et al. Probiotics inhibit nuclear factor-κB and induce heat shock proteins in colonic epithelial cells through proteasome inhibition. Gastroenterology. 2004;127:1474–1487. doi: 10.1053/j.gastro.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 29.Kojima K, Musch MW, Ren H, Boone DL, Hendrickson BA, Ma A, et al. Enteric flora and lymphocyte-derived cytokines determine expression of heat shock proteins in mouse colonic epithelial cells. Gastroenterology. 2003;124:1395–1407. doi: 10.1016/s0016-5085(03)00215-4. [DOI] [PubMed] [Google Scholar]
- 30.Fujiya M, Musch MW, Nakagawa Y, Hu S, Alverdy J, Kohgo Y, et al. The Bacillus subtilis quorum-sensing molecule CSF contributes to intestinal homeostasis via OCTN2, a host cell membrane transporter. Cell Host Microbe. 2007;1:299–308. doi: 10.1016/j.chom.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Neish AS, Gewirtz AT, Zeng H, Young AN, Hobert ME, Karmali V, et al. Prokaryotic regulation of epithelial responses by inhibition of IκB-α ubiquitination. Science. 2000;289:1560–1563. doi: 10.1126/science.289.5484.1560. [DOI] [PubMed] [Google Scholar]
- 32.Kumar A, Wu H, Collier-Hyams LS, Hansen JM, Li T, Yamoah K, et al. Commensal bacteria modulate cullin-dependent signaling via generation of reactive oxygen species. EMBO J. 2007;26:4457–4466. doi: 10.1038/sj.emboj.7601867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin PW, Myers LE, Ray L, Song SC, Nasr TR, Berardinelli AJ, et al. Lactobacillus rhamnosus blocks inflammatory signaling in vivo via reactive oxygen species generation. Free Radic Biol Med. 2009;47:1205–1211. doi: 10.1016/j.freeradbiomed.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Li N, Caicedo R, Neu J. Alive and dead Lactobacillus rhamnosus GG decrease tumor necrosis factor-α-induced interleukin-8 production in Caco-2 cells. J Nutr. 2005;135:1752–1756. doi: 10.1093/jn/135.7.1752. [DOI] [PubMed] [Google Scholar]
- 35.Tien MT, Girardin SE, Regnault B, Le Bourhis L, Dillies MA, Coppee JY, et al. Anti-inflammatory effect of Lactobacillus casei on Shigella-infected human intestinal epithelial cells. J Immunol. 2006;176:1228–1237. doi: 10.4049/jimmunol.176.2.1228. [DOI] [PubMed] [Google Scholar]
- 36.Ma D, Forsythe P, Bienenstock J. Live Lactobacillus reuteri is essential for the inhibitory effect on tumor necrosis factor α-induced interleukin-8 expression. Infect Immun. 2004;72:5308–5314. doi: 10.1128/IAI.72.9.5308-5314.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrof EO, Claud EC, Sun J, Abramova T, Guo Y, Waypa TS, et al. Bacteria-free solution derived from Lactobacillus plantarum inhibits multiple NFκB pathways and inhibits proteasome function. Inflamm Bowel Dis. 2009;15:1537–1547. doi: 10.1002/ibd.20930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frick JS, Schenk K, Quitadamo M, Kahl F, Koberle M, Bohn E, et al. Lactobacillus fermentum attenuates the proinflammatory effect of Yersinia enterocolitica on human epithelial cells. Inflamm Bowel Dis. 2007;13:83–90. doi: 10.1002/ibd.20009. [DOI] [PubMed] [Google Scholar]
- 39.Bai AP, Ouyang Q, Zhang W, Wang CH, Li SF. Probiotics inhibit TNFa-induced interleukin-8 secretion of HT29 cells. World J Gastroenterol. 2004;10:455–457. doi: 10.3748/wjg.v10.i3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bai AP, Ouyang Q, Xiao XR, Li SF. Probiotics modulate inflammatory cytokine secretion from inflamed mucosa in active ulcerative colitis. Int J Clin Pract. 2006;60:284–288. doi: 10.1111/j.1368-5031.2006.00833.x. [DOI] [PubMed] [Google Scholar]
- 41.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn's disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu GD, Huang N, Wen X, Keilbaugh SA, Yang H. High-level expression of IκB-β in the surface epithelium of the colon: in vitro evidence for an immunomodulatory role. J Leukoc Biol. 1999;66:1049–1056. doi: 10.1002/jlb.66.6.1049. [DOI] [PubMed] [Google Scholar]
- 43.Ruiz PA, Hoffmann M, Szcesny S, Blaut M, Haller D. Innate mechanisms for Bifidobacterium lactis to activate transient pro-inflammatory host responses in intestinal epithelial cells after the colonization of germ-free rats. Immunology. 2005;115:441–450. doi: 10.1111/j.1365-2567.2005.02176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haller D, Russo MP, Sartor RB, Jobin C. IKKβ and phosphatidylinositol 3-kinase/Akt participate in non-pathogenic Gram-negative enteric bacteria-induced RelA phosphorylation and NFκB activation in both primary and intestinal epithelial cell lines. J Biol Chem. 2002;277:38168–38178. doi: 10.1074/jbc.M205737200. [DOI] [PubMed] [Google Scholar]
- 45.Jijon H, Backer J, Diaz H, Yeung H, Thiel D, McKaigney C, et al. DNA from probiotic bacteria modulates murine and human epithelial and immune function. Gastroenterology. 2004;126:1358–1373. doi: 10.1053/j.gastro.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 46.Resta-Lenert S, Barrett KE. Probiotics and commensals reverse TNFα- and IFNγ-induced dysfunction in human intestinal epithelial cells. Gastroenterology. 2006;130:731–746. doi: 10.1053/j.gastro.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 47.Voltan S, Martines D, Elli M, Brun P, Longo S, Porzionato A, et al. Lactobacillus crispatus M247-derived H2O2 acts as a signal transducing molecule activating peroxisome proliferator activated receptor-γ in the intestinal mucosa. Gastroenterology. 2008;135:1216–1227. doi: 10.1053/j.gastro.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 48.Dubuquoy L, Jansson EA, Deeb S, Rakotobe S, Karoui M, Colombel JF, et al. Impaired expression of peroxisome proliferator-activated receptor γ in ulcerative colitis. Gastroenterology. 2003;124:1265–1276. doi: 10.1016/s0016-5085(03)00271-3. [DOI] [PubMed] [Google Scholar]
- 49.Kelly D, Campbell JI, King TP, Grant G, Jansson EA, Coutts AG, et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-γ and RelA. Nat Immunol. 2004;5:104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 50.Are A, Aronsson L, Wang S, Greicius G, Lee YK, Gustafsson JA, et al. Enterococcus faecalis from newborn babies regulate endogenous PPARγ activity and IL-10 levels in colonic epithelial cells. Proc Natl Acad Sci USA. 2008;105:1943–1948. doi: 10.1073/pnas.0711734105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ewaschuk JB, Walker JW, Diaz H, Madsen KL. Bioproduction of conjugated linoleic acid by probiotic bacteria occurs in vitro and in vivo in mice. J Nutr. 2006;136:1483–1487. doi: 10.1093/jn/136.6.1483. [DOI] [PubMed] [Google Scholar]
- 52.Eun CS, Han DS, Lee SH, Jeon YC, Sohn JH, Kim YS, et al. Probiotics may reduce inflammation by enhancing peroxisome proliferator activated receptor γ activation in HT-29 cells. Korean J Gastroenterol. 2007;49:139–146. [PubMed] [Google Scholar]
- 53.van Baarlen P, Troost FJ, van Hemert S, van der Meer C, de Vos WM, de Groot PJ, et al. Differential NFκB pathways induction by Lactobacillus plantarum in the duodenum of healthy humans correlating with immune tolerance. Proc Natl Acad Sci USA. 2009;106:2371–2376. doi: 10.1073/pnas.0809919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Di Caro S, Tao H, Grillo A, Elia C, Gasbarrini G, Sepulveda AR, et al. Effects of Lactobacillus GG on genes expression pattern in small bowel mucosa. Dig Liver Dis. 2005;37:320–329. doi: 10.1016/j.dld.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 55.Lin PW, Nasr TR, Berardinelli AJ, Kumar A, Neish AS. The probiotic Lactobacillus GG may augment intestinal host defense by regulating apoptosis and promoting cytoprotective responses in the developing murine gut. Pediatr Res. 2008;64:511–516. doi: 10.1203/PDR.0b013e3181827c0f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yan F, Polk DB. Probiotic bacterium prevents cytokine-induced apoptosis in intestinal epithelial cells. J Biol Chem. 2002;277:50959–50965. doi: 10.1074/jbc.M207050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yan F, Cao H, Cover TL, Whitehead R, Washington MK, Polk DB. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology. 2007;132:562–575. doi: 10.1053/j.gastro.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fitzpatrick LR, Small J, Hoerr RA, Bostwick EF, Maines L, Koltun WA. In vitro and in vivo effects of the probiotic Escherichia coli strain M-17: immunomodulation and attenuation of murine colitis. Br J Nutr. 2008;100:530–541. doi: 10.1017/S0007114508930373. [DOI] [PubMed] [Google Scholar]
- 59.Watanabe T, Nishio H, Tanigawa T, Yamagami H, Okazaki H, Watanabe K, et al. Probiotic Lactobacillus casei strain Shirota prevents indomethacin-induced small intestinal injury: involvement of lactic acid. Am J Physiol Gastrointest Liver Physiol. 2009;297:506–513. doi: 10.1152/ajpgi.90553.2008. [DOI] [PubMed] [Google Scholar]
- 60.Sougioultzis S, Simeonidis S, Bhaskar KR, Chen X, Anton PM, Keates S, et al. Saccharomyces boulardii produces a soluble anti-inflammatory factor that inhibits NFκB-mediated IL-8 gene expression. Biochem Biophys Res Commun. 2006;343:69–76. doi: 10.1016/j.bbrc.2006.02.080. [DOI] [PubMed] [Google Scholar]
- 61.Segain JP, Raingeard de la Bletiere D, Bourreille A, Leray V, Gervois N, Rosales C, et al. Butyrate inhibits inflammatory responses through NFκB inhibition: implications for Crohn's disease. Gut. 2000;47:397–403. doi: 10.1136/gut.47.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Menard S, Candalh C, Bambou JC, Terpend K, Cerf-Bensussan N, Heyman M. Lactic acid bacteria secrete metabolites retaining anti-inflammatory properties after intestinal transport. Gut. 2004;53:821–828. doi: 10.1136/gut.2003.026252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim HG, Lee SY, Kim NR, Ko MY, Lee JM, Yi TH, et al. Inhibitory effects of Lactobacillus plantarum lipoteichoic acid (LTA) on Staphylococcus aureus LTA-induced tumor necrosis factor-α production. J Microbiol Biotechnol. 2008;18:1191–1196. [PubMed] [Google Scholar]
- 64.Kim HG, Kim NR, Gim MG, Lee JM, Lee SY, Ko MY, et al. Lipoteichoic acid isolated from Lactobacillus plantarum inhibits lipopolysaccharide-induced TNFα production in THP-1 cells and endotoxin shock in mice. J Immunol. 2008;180:2553–2561. doi: 10.4049/jimmunol.180.4.2553. [DOI] [PubMed] [Google Scholar]
- 65.Matsuguchi T, Takagi A, Matsuzaki T, Nagaoka M, Ishikawa K, Yokokura T, et al. Lipoteichoic acids from Lactobacillus strains elicit strong tumor necrosis factor a-inducing activities in macrophages through Toll-like receptor 2. Clin Diagn Lab Immunol. 2003;10:259–266. doi: 10.1128/CDLI.10.2.259-266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grangette C, Nutten S, Palumbo E, Morath S, Hermann C, Dewulf J, et al. Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc Natl Acad Sci USA. 2005;102:10321–10326. doi: 10.1073/pnas.0504084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Okada Y, Tsuzuki Y, Hokari R, Komoto S, Kurihara C, Kawaguchi A, et al. Anti-inflammatory effects of the genus Bifidobacterium on macrophages by modification of phospho-IκB and SOCS gene expression. Int J Exp Pathol. 2009;90:131–140. doi: 10.1111/j.1365-2613.2008.00632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim SO, Sheikh HI, Ha SD, Martins A, Reid G. G-CSF-mediated inhibition of JNK is a key mechanism for Lactobacillus rhamnosus-induced suppression of TNF production in macrophages. Cell Microbiol. 2006;8:1958–1971. doi: 10.1111/j.1462-5822.2006.00763.x. [DOI] [PubMed] [Google Scholar]
- 69.Klebanoff SJ, Watts DH, Mehlin C, Headley CM. Lactobacilli and vaginal host defense: activation of the human immunodeficiency virus type 1 long terminal repeat, cytokine production, and NFκB. J Infect Dis. 1999;179:653–660. doi: 10.1086/314644. [DOI] [PubMed] [Google Scholar]
- 70.Miettinen M, Lehtonen A, Julkunen I, Matikainen S. Lactobacilli and Streptococci activate NFκB and STAT signaling pathways in human macrophages. J Immunol. 2000;164:3733–3740. doi: 10.4049/jimmunol.164.7.3733. [DOI] [PubMed] [Google Scholar]
- 71.Iyer C, Kosters A, Sethi G, Kunnumakkara AB, Aggarwal BB, Versalovic J. Probiotic Lactobacillus reuteri promotes TNF-induced apoptosis in human myeloid leukemia-derived cells by modulation of NFκB and MAPK signalling. Cell Microbiol. 2008;10:1442–1452. doi: 10.1111/j.1462-5822.2008.01137.x. [DOI] [PubMed] [Google Scholar]
- 72.Chiu YH, Hsieh YJ, Liao KW, Peng KC. Preferential promotion of apoptosis of monocytes by Lactobacillus casei rhamnosus soluble factors. Clin Nutr. 2009 doi: 10.1016/j.clnu.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 73.Horinaka M, Yoshida T, Kishi A, Akatani K, Yasuda T, Kouhara J, et al. Lactobacillus strains induce TRAIL production and facilitate natural killer activity against cancer cells. FEBS Lett. 584:577–582. doi: 10.1016/j.febslet.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 74.Macdonald TT, Monteleone G. Immunity, inflammation and allergy in the gut. Science. 2005;307:1920–1925. doi: 10.1126/science.1106442. [DOI] [PubMed] [Google Scholar]
- 75.Hoarau C, Martin L, Faugaret D, Baron C, Dauba A, Aubert-Jacquin C, et al. Supernatant from bifidobacterium differentially modulates transduction signaling pathways for biological functions of human dendritic cells. PLoS One. 2008;3:2753. doi: 10.1371/journal.pone.0002753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hoarau C, Lagaraine C, Martin L, Velge-Roussel F, Lebranchu Y. Supernatant of Bifidobacterium breve induces dendritic cell maturation, activation and survival through a Toll-like receptor 2 pathway. J Allergy Clin Immunol. 2006;117:696–702. doi: 10.1016/j.jaci.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 77.Christensen HR, Frokiaer H, Pestka JJ. Lactobacilli differentially modulate expression of cytokines and maturation surface markers in murine dendritic cells. J Immunol. 2002;168:171–178. doi: 10.4049/jimmunol.168.1.171. [DOI] [PubMed] [Google Scholar]
- 78.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 79.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 80.Macpherson AJ, Geuking MB, McCoy KD. Immune responses that adapt the intestinal mucosa to commensal intestinal bacteria. Immunology. 2005;115:153–162. doi: 10.1111/j.1365-2567.2005.02159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Foligne B, Zoumpopoulou G, Dewulf J, Ben Younes A, Chareyre F, Sirard JC, et al. A key role of dendritic cells in probiotic functionality. PLoS One. 2007;2:313. doi: 10.1371/journal.pone.0000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zeuthen LH, Fink LN, Frokiaer H. Toll-like receptor 2 and nucleotide-binding oligomerization domain-2 play divergent roles in the recognition of gut-derived lactobacilli and bifidobacteria in dendritic cells. Immunology. 2008;124:489–502. doi: 10.1111/j.1365-2567.2007.02800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kapsenberg ML. Dendritic-cell control of pathogen-driven T-cell polarization. Nat Rev Immunol. 2003;3:984–993. doi: 10.1038/nri1246. [DOI] [PubMed] [Google Scholar]
- 84.Delcenserie V, Martel D, Lamoureux M, Amiot J, Boutin Y, Roy D. Immunomodulatory effects of probiotics in the intestinal tract. Curr Issues Mol Biol. 2008;10:37–54. [PubMed] [Google Scholar]
- 85.Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 87.Sartor RB. Mechanisms of disease: pathogenesis of Crohn's disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol. 2006;3:390–407. doi: 10.1038/ncpgasthep0528. [DOI] [PubMed] [Google Scholar]
- 88.Hart AL, Lammers K, Brigidi P, Vitali B, Rizzello F, Gionchetti P, et al. Modulation of human dendritic cell phenotype and function by probiotic bacteria. Gut. 2004;53:1602–1609. doi: 10.1136/gut.2003.037325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Corthay A. How do regulatory T cells work? Scand J Immunol. 2009;70:326–336. doi: 10.1111/j.1365-3083.2009.02308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Izcue A, Coombes JL, Powrie F. Regulatory lymphocytes and intestinal inflammation. Annu Rev Immunol. 2009;27:313–338. doi: 10.1146/annurev.immunol.021908.132657. [DOI] [PubMed] [Google Scholar]
- 91.de Roock S, van Elk M, van Dijk ME, Timmerman HM, Rijkers GT, Prakken BJ, et al. Lactic acid bacteria differ in their ability to induce functional regulatory T cells in humans. Clin Exp Allergy. 2009 doi: 10.1111/j.1365-2222.2009.03344.x. [DOI] [PubMed] [Google Scholar]
- 92.Di Giacinto C, Marinaro M, Sanchez M, Strober W, Boirivant M. Probiotics ameliorate recurrent Th1-mediated murine colitis by inducing IL-10 and IL-10-dependent TGFβ-bearing regulatory cells. J Immunol. 2005;174:3237–3246. doi: 10.4049/jimmunol.174.6.3237. [DOI] [PubMed] [Google Scholar]
- 93.Baba N, Samson S, Bourdet-Sicard R, Rubio M, Sarfati M. Commensal bacteria trigger a full dendritic cell maturation program that promotes the expansion of non-Tr1 suppressor T cells. J Leukoc Biol. 2008;84:468–476. doi: 10.1189/jlb.0108017. [DOI] [PubMed] [Google Scholar]
- 94.Livingston M, Loach D, Wilson M, Tannock GW, Baird M. Gut commensal Lactobacillus reuteri 100-23 stimulates an immunoregulatory response. Immunol Cell Biol. 88:99–102. doi: 10.1038/icb.2009.71. [DOI] [PubMed] [Google Scholar]
- 95.Kwon HK, Lee CG, So JS, Chae CS, Hwang JS, Sahoo A, et al. Generation of regulatory dendritic cells and CD4+Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc Natl Acad Sci USA. 107:2159–2164. doi: 10.1073/pnas.0904055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guzy C, Paclik D, Schirbel A, Sonnenborn U, Wiedenmann B, Sturm A. The probiotic Escherichia coli strain Nissle 1917 induces κδ T cell apoptosis via caspase- and FasL-dependent pathways. Int Immunol. 2008;20:829–840. doi: 10.1093/intimm/dxn041. [DOI] [PubMed] [Google Scholar]
- 97.Roselli M, Finamore A, Nuccitelli S, Carnevali P, Brigidi P, Vitali B, et al. Prevention of TNBS-induced colitis by different Lactobacillus and Bifidobacterium strains is associated with an expansion of κδT and regulatory T cells of intestinal intraepithelial lymphocytes. Inflamm Bowel Dis. 2009;15:1526–1536. doi: 10.1002/ibd.20961. [DOI] [PubMed] [Google Scholar]
- 98.Sturm A, Rilling K, Baumgart DC, Gargas K, Abou-Ghazale T, Raupach B, et al. Escherichia coli Nissle 1917 distinctively modulates T-cell cycling and expansion via toll-like receptor 2 signaling. Infect Immun. 2005;73:1452–1465. doi: 10.1128/IAI.73.3.1452-1465.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Carol M, Borruel N, Antolin M, Llopis M, Casellas F, Guarner F, et al. Modulation of apoptosis in intestinal lymphocytes by a probiotic bacteria in Crohn's disease. J Leukoc Biol. 2006;79:917–922. doi: 10.1189/jlb.0405188. [DOI] [PubMed] [Google Scholar]
- 100.Reiff C, Delday M, Rucklidge G, Reid M, Duncan G, Wohlgemuth S, et al. Balancing inflammatory, lipid and xenobiotic signaling pathways by VSL#3, a biotherapeutic agent, in the treatment of inflammatory bowel disease. Inflamm Bowel Dis. 2009;15:1721–1736. doi: 10.1002/ibd.20999. [DOI] [PubMed] [Google Scholar]
- 101.Mazanec MB, Nedrud JG, Kaetzel CS, Lamm ME. A three-tiered view of the role of IgA in mucosal defense. Immunol Today. 1993;14:430–435. doi: 10.1016/0167-5699(93)90245-G. [DOI] [PubMed] [Google Scholar]
- 102.de Moreno de LeBlanc A, Chaves S, Carmuega E, Weill R, Antoine J, Perdigon G. Effect of long-term continuous consumption of fermented milk containing probiotic bacteria on mucosal immunity and the activity of peritoneal macrophages. Immunobiology. 2008;213:97–108. doi: 10.1016/j.imbio.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 103.Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222–2226. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 104.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 105.He B, Xu W, Santini PA, Polydorides AD, Chiu A, Estrella J, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity. 2007;26:812–826. doi: 10.1016/j.immuni.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 106.Fukushima Y, Kawata Y, Hara H, Terada A, Mitsuoka T. Effect of a probiotic formula on intestinal immunoglobulin A production in healthy children. Int J Food Microbiol. 1998;42:39–44. doi: 10.1016/s0168-1605(98)00056-7. [DOI] [PubMed] [Google Scholar]
- 107.Park JH, Um JI, Lee BJ, Goh JS, Park SY, Kim WS, et al. Encapsulated Bifidobacterium bifidum potentiates intestinal IgA production. Cell Immunol. 2002;219:22–27. doi: 10.1016/s0008-8749(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 108.Ibnou-Zekri N, Blum S, Schiffrin EJ, von der Weid T. Divergent patterns of colonization and immune response elicited from two intestinal Lactobacillus strains that display similar properties in vitro. Infect Immun. 2003;71:428–436. doi: 10.1128/IAI.71.1.428-436.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Maassen CB, Boersma WJ, van Holten-Neelen C, Claassen E, Laman JD. Growth phase of orally administered Lactobacillus strains differentially affects IgG1/IgG2a ratio for soluble antigens: implications for vaccine development. Vaccine. 2003;21:2751–2757. doi: 10.1016/s0264-410x(03)00220-2. [DOI] [PubMed] [Google Scholar]
- 110.Chang G, Shi Y, Le G, Xu Z, Sun J, Li J. Effects of Lactobacillus plant arum on genes expression pattern in mice jejunal Peyer's patches. Cell Immunol. 2009;258:1–8. doi: 10.1016/j.cellimm.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 111.Brisbin JT, Zhou H, Gong J, Sabour P, Akbari MR, Haghighi HR, et al. Gene expression profiling of chicken lymphoid cells after treatment with Lactobacillus acidophilus cellular components. Dev Comp Immunol. 2008;32:563–574. doi: 10.1016/j.dci.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 112.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 113.Bouskra D, Brezillon C, Berard M, Werts C, Varona R, Boneca IG, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 114.Gorbach SL, Plaut AG, Nahas L, Weinstein L, Spanknebel G, Levitan R. Studies of intestinal microflora II. Microorganisms of the small intestine and their relations to oral and fecal flora. Gastroenterology. 1967;53:856–867. [PubMed] [Google Scholar]
- 115.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wilson M. Bacteriology of Humans An Ecological Perspective. Malden, MA: Blackwell Publishing; 2008. pp. 266–326. [Google Scholar]
- 118.Yun SI, Park HO, Kang JH. Effect of Lactobacillus gasseri BNR17 on blood glucose levels and body weight in a mouse model of type 2 diabetes. J Appl Microbiol. 2009;107:1681–1686. doi: 10.1111/j.1365-2672.2009.04350.x. [DOI] [PubMed] [Google Scholar]
- 119.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]


