Abstract
Background
The plurality of genetic risk for developing type 1 diabetes mellitus (T1DM) lies within the genes that code for the human leukocyte antigens (HLAs). Many T1DM studies use HLA genetic risk assessment to identify higher risk individuals, and they often conduct these tests on dried blood spots (DBSs) like those used for newborn bloodspot screening. One such study is The Environmental Determinants of Diabetes in the Young (TEDDY), a long-term prospective study of environmental risk factors. To provide quality assurance for T1DM studies that employ HLA genetic risk assessment, the Centers for Disease Control and Prevention (CDC) conducts both a voluntary quarterly proficiency testing (VQPT) program available to any laboratory and a mandatory annual proficiency testing (PT) challenge for TEDDY laboratories.
Methods
Whole blood and DBS samples with a wide range of validated HLA-DR and HLA-DQ genotypes were sent to the participating laboratories. Results were evaluated on the basis of both the reported haplotypes and the HLA genetic risk assessment.
Results
Of the reported results from 24 panels sent out over six years in the VQPT, 94.7% (857/905) were correctly identified with respect to the relevant HLA-DR or HLA-DQ alleles, and 96.4% (241/250) were correctly categorized for risk assessment. Significant improvement was seen over the duration of this program, usually reaching 100% correct categorization during the last three years. Of 1154 reported results in four TEDDY PT challenges, 1153 (99.9%) were correctly identified for TEDDY eligibility.
Conclusions
The different analytical methods used by T1DM research centers all provided accurate (>99%) results for genetic risk assessment. The two CDC PT programs documented the validity of the various approaches to screening and contributed to overall quality assurance.
Keywords: newborn screening, quality assurance, quality control, The Environmental Determinants of Diabetes in the Young
Introduction
Children at higher risk for type 1 diabetes mellitus (T1DM) can be identified by newborn bloodspot screening (NBS).1 Type 1 diabetes mellitus risk assess-ment is primarily based on characterizing genes of the major histocompatibility complex (MHC), specifically the human leukocyte antigen (HLA) class II region HLA-D. This region includes the highly polymorphic genes HLA-DRB1, HLA-DQA1, and HLA-DQB1. Because of linkage disequilibrium within the HLA-D region, alleles on the same chromosome are usually inherited as a group (haplotype). Most genotyping methods do not discern the phase of the alleles, but extended HLA-D haplotypes may usually be inferred using haplotype frequency databases (see Anthropology/Allele Frequencies at http://www.ncbi.nlm.nih.gov/projects/gv/mhc/). To maximize cost-effectiveness, T1DM studies often further infer haplotypes from limited genotyping among HLA-DRB1, HLA-DQA1, and HLA-DQB1 alleles. For each individual, the two inferred HLA haplotypes can be combined to obtain a haplogenotype, the genotype combination of two multilocus haplotypes.2 The HLA-D haplogenotype constitutes the plurality of T1DM genetic risk. Since laboratories use different analytical methods and genotyping strategies, a proficiency testing (PT) program in which all laboratories test the same samples is essential to ensuring comparability of risk assessment.
The Centers for Disease Control and Prevention (CDC) conducts two concurrent PT programs for T1DM screening. The first program, initiated in 2003, is a voluntary collaboration that includes several T1DM research centers and one manufacturer of NBS reagents. It is conducted on a quarterly basis in the United States and Europe. The second program, initiated in 2004, is part of The Environmental Determinants of Diabetes in the Young (TEDDY) study1,3,–5 (see www.teddystudy.org). This study includes six clinical centers (three in the United States and three in Europe), five laboratories conducting the screening tests, and a centralized data center. The Environmental Determinants of Diabetes in the Young has enrolled more than 8400 children in a prospective longitudinal cohort that will be monitored for environmental factors, islet cell autoimmunity, and clinical status through the entire period of risk for childhood diabetes. The CDC conducts annual PT challenges for TEDDY screening laboratories.
For reference materials to use in either the voluntary quarterly proficiency testing (VQPT) or the TEDDY PT exercises, the CDC assembled a collection of dried blood spots (DBSs) that have been independently characterized for relevant HLA-D alleles by at least two different laboratories in advance of their distribution to participants. The complexity of the HLA-D region, as well as the different methods used to detect risk alleles, presented challenges in comparing and evaluating the data submitted. This article describes the methods used to conduct PT, the protocols used to assess results, and the overall performance of participating laboratories.
Materials and Methods
Samples and Reference Haplogenotypes
Anonymous residual cord blood samples were obtained from a public health newborn screening program,6 and anonymous venous blood samples from adults were obtained from residual clinical samples. Dried blood spots were prepared from 50–75 µl blood as described previously6 using methods from a Clinical and Laboratory Standards Institute consensus standard for newborn screening.7 Dried blood spots were stored until use at -20 ˚C in low-permeability plastic resealable bags (Fisher Scientific, Norcross, GA; catalog #19240127) with 8–10 1-g desiccant packs (Poly Lam Products, Corp., Williamsville, NY; catalog # 39AG37) and a relative humidity indicator card (Poly Lam Products, Corp., Williamsville, NY; catalog #MS20003-2) to ensure low relative humidity (<30%). Since one participating laboratory used whole blood rather than DBSs, some of the original blood from each sample was aliquoted into 1-ml CryoTubes and stored at -20 ˚C.
Before samples were included in either a VQPT or a TEDDY PT survey, reference haplogenotypes for HLA-DQA1, HLA-DQB1, and HLA-DRB1 were determined in multiple laboratories using different methods, including reverse line-blot hybridization,8,9 real-time fluorescence hybridization,6,10 and sequence-based typing.3 Presumed haplogenotypes were assigned based on the linkage disequilibrium of alleles in the HLA-D region only if results from all laboratories were concordant. The distribution of reference haplogenotypes in samples used in VQPT and TEDDY PT are summarized in Tables 1 and 2, respectively. For haplogenotypes included in VQPT, risk levels were divided into three categories compared to the general population risk for T1DM based on previous studies.8,11
Table 1.
HLA haplogenotypes of DBS samples distributed for the VQ-PT program.a
| -------------- Chromosome 1 ---------------- | -------------- Chromosome 2 ---------------- | RISK GROUP | ||||||
| DRB1 | DQA1 | DQB1 | DRB1 | DQA1 | DQB1 | |||
| Qt 1 2003 | 040101 | 03 | 0302 | 010101 /07 | 0101 | 050101 | 2 | |
| 030101 | 05 | 0201 | 0402 /04 | 03 | 0302 | 3 | ||
| 070101 | 02 | 0201 | 010101 /07 | 0101 | 050101 | 1 | ||
| 0408 | 03 | 0302 | 070101 | 0201 | 030101 | 1 | ||
| 0411 | 03 | 0302 | 010201 | 01 | 050101 | 1 | ||
| Qt 2 2003 | 030101 | 050101 | 0202 | 040101 | 0302 /03 | 0302 | 3 | |
| 030101 | 050101 | 0201 | 040101 | 0302 /03 | 0302 | 3 | ||
| 010101 /07 | 01 | 050101 | 040101 | 0302 /03 | 030101 | 1 | ||
| 030101 | 050101 | 0201 | 040101 | 0302 /03 | 0302 | 3 | ||
| 110101 /14 | 0505 | 030101 | 130201 /05 | 010201 | 0609 | 1 | ||
| Qt 3 2003 | 040301 | 0302 /03 | 0302 | 070101 | 0201 | 0202 | NE | |
| 0404 | 0302 /03 | 0302 | 070101 | 0201 | 0202 | NE | ||
| 0103 | 01 | 050101 | 080101 | 0401 | 0402 | NE | ||
| 090102 | 0302 /03 | 030302 | 160201 | 010202 | 0502 | NE | ||
| 110101 | 0505 | 030101 | 130201 | 010201 | 0609 | 1 | ||
| Qt 4 2003 | 040101 | 0302 /03 | 030101 | 040101 | 0302 /03 | 030101 | 1 | |
| 030101 | 050101 | 0201 | 040101 | 030101 | 0302 | 3 | ||
| 030101 | 050101 | 0201 | 040101 | 030101 | 0302 | 3 | ||
| 030101 | 050101 | 0201 | 040101 | 030101 | 0302 | 3 | ||
| 120101 /06 | 0505 | 030101 | 130101 | 0103 | 0603 | 1 | ||
| Qt 1 2004 | 030101 | 050101 | 0201 | 040101 | 0302 /03 | 0302 | 3 | |
| 030101 | 050101 | 0201 | 0404 | 030101 | 030101 | 3 | ||
| 030101 | 050101 | 0201 | 150101 | 010201 | 0602 | 1 | ||
| 010101 /07 | 01 | 050101 | 150101 | 010201 | 0602 | 1 | ||
| 040101 | 030101 | 0302 | 130201 | 010201 | 060401 | 2 | ||
| Qt 2 2004 | 080101 | 0401 | 0402 | 160101 | 010202 | 0502 | 1 | |
| 0103 | 01 | 050101 | 080101 | 0401 | 0402 | 1 | ||
| Qt 2 2004 | 030101 | 050101 | 0201 | 040101 | 030101 | 0302 | 3 | |
| 1405 | 01 | 050301 | 150101 | 010201 | 0502 | 1 | ||
| 030101 | 050101 | 0201 | 0402 | 030101 | 0302 | 3 | ||
| Qt 1 2005 | 030101 | 050101 | 0201 | 040101 | 030101 | 0302 | 3 | |
| 030101 | 050101 | 0201 | 130201 | 010201 | 060401 | NE | ||
| 040301 | 030101 | 0302 | 070101 | 0201 | 0202 | 1 | ||
| 010101 /07 | 01 | 050101 | 040101 | 030101 | 0302 | NE | ||
| 130101 | 0103 | 0602 | 150201 | 0103 | 060101 /03 | 1 | ||
| Qt 2 2005 | 030101 | 050101 | 0201 | 040101 | 0302 /03 | 0302 | 3 | |
| 040101 | 0302 /03 | 030101 | 120201 | 060101 | 030101 | 1 | ||
| 010201 | 01 | 050101 | 0404 | 030101 | 0302 | 2 | ||
| 040101 | 030101 | 0302 | 130201 | 010201 | 060401 | 2 | ||
| 070101 | 0201 | 0202 | 150101 | 010201 | 0602 | 1 | ||
| Qt 3 2005 | 030101 | 050101 | 0201 | 090102 | 0302 /03 | 030302 | NE | |
| 040101 | 030101 | 0302 | 0402 | 030101 | 0302 | NE | ||
| 040101 | 030101 | 0302 | 070101 | 0201 | 0202 | NE | ||
| 030101 | 050101 | 0201 | 150101 | 010201 | 0602 | 1 | ||
| 030101 | 050101 | 0201 | 040101 | 030101 | 0302 | 3 | ||
| Qt 4 2005 | 030101 | 050101 | 0201 | 0404 | 030101 | 0302 | 3 | |
| 030101 | 050101 | 0201 | 040101 | 0302 /03 | 030101 | 1 | ||
| 040101 | 0302 /03 | 030101 | 070101 | 0201 | 0202 | 1 | ||
| 040101 | 030101 | 0302 | 130201 | 010201 | 060401 | 2 | ||
| 040101 | 0302 /03 | 030101 | 130201 | 010201 | 060401 | 1 | ||
| Qt 1 2006 | 040701 | 0302 /03 | 030101 | 090102 | 0302 /03 | 0202 | 1 | |
| 070101 | 0201 | 030302 | 110101 | 0505 | 030101 | 1 | ||
| 070101 | 0201 | 0202 | 130201 | 010201 | 060401 | 1 | ||
| 010101 /07 | 01 | 050101 | 0408 | 0302 /03 | 0304 | NE | ||
| 030101 | 050101 | 0201 | 040101 | 030101 | 0302 | 3 | ||
| Qt 2 2006 | 150201 | 01 | 050101 | 150201 | 010201 | 0502 | 1 | |
| 030101 | 050101 | 0201 | 040101 | 030101 | 0302 | 3 | ||
| 070101 | 0201 | 0202 | 070101 | 0201 | 030302 | 1 | ||
| 110101 | 0505 | 030101 | 130201 | 010201 | 0609 | 1 | ||
| 130201 | 010201 | 060101 | 150201 | 0103 | 060101 | 1 | ||
| Qt 3 2006 | 030101 | 050101 | 0201 | 0402 | 030101 | 0302 | 3 | |
| 0103 | 01 | 050101 | 080101 | 0401 | 0402 | 1 | ||
| 010101 /07 | 01 | 050101 | 040101 | 0302 /03 | 030101 | 1 | ||
| 080101 | 0401 | 0402 | 160101 | 010202 | 0502 | NE | ||
| Qt 3 2006 | 0402 | 030101 | 0302 | 070101 | 0201 | 0202 | 2/3 | |
| Qt 4 2006 | 030101 | 050101 | 0201 | 040101 | 030101 | 0302 | 3 | |
| 030101 | 050101 | 0201 | 040101 | 0302 /03 | 030101 | 1 | ||
| 040101 | 0302 /03 | 030101 | 110101 | 0505 | 030101 | 1 | ||
| 030101 | 050101 | 0201 | 0404 | 030101 | 0302 | 3 | ||
| 130101 | 0103 | 0603 | 130201 | 010201 | 060401 | 1 | ||
| Qt 1 2007 | 1405 | 01 | 050301 | 150101 | 010201 | 0502 | 1 | |
| 040101 | 0302 /03 | 030101 | 040101 | 0302 /03 | 030101 | 1 | ||
| 030101 | 050101 | 0201 | 040101 | 0302 /03 | 0302 | 3 | ||
| 030101 | 050101 | 0201 | 0402 | 030101 | 0302 | 3 | ||
| 040101 | 0302 /03 | 030101 | 130201 | 010201 | 060401 | 1 | ||
| Qt 2 2007 | 040301 | 030101 | 0302 | 070101 | 0201 | 0202 | 1 | |
| 0103 | 01 | 050101 | 080101 | 0401 | 0402 | 1 | ||
| 040101 | 0302 /03 | 030101 | 120201 | 060101 | 030101 | 1 | ||
| 0404 | 030101 | 0302 | 070101 | 0201 | 0202 | NE | ||
| 030101 | 050101 | 0201 | 040101 | 030101 | 0302 | 3 | ||
| Qt 3 2007 | 030101 | 050101 | 0201 | 040101 | 0302 /03 | 030101 | 1 | |
| 030101 | 050101 | 0201 | 0404 | 030101 | 0302 | 3 | ||
| 0402 | 030101 | 0302 | 070101 | 0201 | 0202 | 2/3 | ||
| 040101 | 030101 | 0302 | 040501 | 0302 /03 | 0202 | 2/3 | ||
| 070101 | 0201 | 0202 | 140101 | 01 | 050301 | 1 | ||
| Qt 4 2007 | 030101 | 050101 | 0201 | 040101 | 030101 | 0302 | 3 | |
| 030101 | 050101 | 0201 | 040101 | 030101 | 0302 | 3 | ||
| 070101 | 0201 | 0202 | 110101 | 0505 | 030101 | 1 | ||
| 130101 | 0103 | 0603 | 130201 | 010201 | 060401 | 1 | ||
| 030101 | 050101 | 0201 | 090102 | 0302 /03 | 030302 | NE | ||
| Qt 1 2008 | 010101 /07 | 01 | 050101 | 150101 | 010201 | 0602 | 1 | |
| 130101 | 0103 | 0602 | 150201 | 0103 | 060101 /03 | 1 | ||
| 030101 | 050101 | 0201 | 0404 | 030101 | 0302 | 3 | ||
| 030101 | 050101 | 0201 | 0404 | 030101 | 0302 | 3 | ||
| 010201 | 01 | 050101 | 0411 | 030101 | 0302 | NE | ||
| Qt 2 2008 | 030101 | 050101 | 0201 | 0402 | 030101 | 0302 | 3 | |
| 030101 | 050101 | 0201 | 080101 | 0401 | 0402 | 1/2 | ||
| 120101 /06 | 0505 | 030101 | 130201 | 010201 | 060401 | 1 | ||
| 110101 | 0505 | 030101 | 130202 | 010201 | 0609 | 1 | ||
| 070101 | 0201 | 0202 | 150101 | 010201 | 0602 | 1 | ||
| Qt 3 2008 | 0103 | 01 | 050101 | 030101 | 050101 | 0201 | 1 | |
| Qt 3 2008 | 040101 | 0302 /03 | 030101 | 040101 | 0302 /03 | 030101 | 1 | |
| 010101 /07 | 01 | 050101 | 040101 | 0302 /03 | 030101 | 1 | ||
| 030101 | 050101 | 0201 | 040101 | 030101 | 0302 | 3 | ||
| 040701 | 0302 /03 | 030101 | 090102 | 0302 /03 | 0202 | 1 | ||
| Qt 4 2008 | 030101 | 050101 | 0201 | 040101 | 0302 /03 | 0302 | 3 | |
| 030101 | 050101 | 0201 | 040101 | 030101 | 0302 | 3 | ||
| 160101 | 010202 | 0502 | 090102 | 0302 /03 | 030302 | 1 | ||
| 110101 | 0505 | 030101 | 070101 | 0201 | 0202 | 1 | ||
| 040101 | 030101 | 0302 | 040501 | 0302 /03 | 0202 | 3 | ||
Each row represents the haplogenotype of one DBS sample. The first set of shaded columns show the HLA DR, DQA, and DQB alleles on one chromosome, and the second set shows the alleles on the other chromosome. Each allele is designated by an alphanumeric string with two, four or six characters. Ambiguous alleles are indicated by an alternate two-character numeric string following a forward slash (e.g. 0302 /03, meaning the allele is either 0302 or 0303). The final column designates the relative risk for T1DM imparted by the haplogenotype, where 1 indicates the lowest relative risk and 3 represents the highest. NE indicates that the haplogenotype has not been evaluated for T1DM risk.
Table 2.
Human Leukocyte Antigen Haplogenotypes of Dried Blood Spot Samples Distributed for the TEDDY Proficiency Testing Programa
| -------------- Chromosome 1 ---------------- | -------------- Chromosome 2 ---------------- | Eligible | |||||
| DRB1 | DQA1 | DQB1 | DRB1 | DQA1 | DQB1 | ||
| 0101 /05 | 0101 /04 /05 | 0501 | 0101 /05 /09 /10 | 0101 /04 /05 | 0501 | No | |
| 0101 /05 | 0101 /04 /05 | 0501 | 0401 | 0301 /02 /03 | 0301 | No | |
| 0101 /05 | 0101 /04 /05 | 0501 | 0401 | 0301 /02 /03 | 0302 | F | |
| 0101 /05 | 0101 /04 /05 | 0501 | 07 | 0201 | 02010 /02 | No | |
| 0101 /05 | 0101 /04 /05 | 0501 | 1101 | 0501 /03 /05 | 0301 | No | |
| 0101 /05 | 0101 /04 /05 | 0501 | 1201 | 0501 /03 /05 | 0301 | No | |
| 0101/ 05 | 0101 /04 /05 | 0501 | 1406 | 0501 /03 /05 | 0301 | No | |
| 0101 /05 | 0102 | 0504 | 1501 | 0102 | 0602 | No | |
| 0102 | 0101 /04 /05 | 0501 | 07 | 0201 | 0303 | No | |
| 0102 | 0101/ 04 /05 | 0501 | 0301 | 0501 /03 /05 | 0201 /02 | No | |
| 0102 | 0101/ 04 /05 | 0501 | 0302 | 0401/ 02/ 03 | 0402 | No | |
| 0103 | 0101/ 04 /05 | 0501 | 0301 | 0501/ 03/ 05 | 0201 /02 | No | |
| 03 | 0501 | 0201 | 09 | 0301 | 0303 | J | |
| 0301 | 0201 | 0201 /02 | 07 | 0501 /03 /05 | 0201 /02 | No | |
| 0301 | 0501 /03 /05 | 0201 /02 | 0301 | 0501 /03 /05 | 0201 /02 | D | |
| 0302 | 0401 /02 /03N | 0402 | 0301 | 0501 /03/ 05 | 0201 /02 | No | |
| 0302 | 0401 /02 /03N | 0402 | 1101 | 0102 | 0501 | No | |
| 04 | 0301 | 0302 | 04 | 0301 | 0302 | B | |
| 04 | 0301 | 0302 | 03 | 0501 | 0201 | A | |
| 04 | 0301 | 0302 | 08 | 0401 | 0402 | C | |
| 04 | 0301 | 0302 | 13 | 0102 | 0604 | G | |
| 0401 | 0301 /02 /03 | 0301 | 0103 | 0501 /03 /05 | 0301 | No | |
| 0401 | 0301 /02 /03 | 0301 | 0401 | 0301 /02 /03 | 0301 | No | |
| 0401 | 0301 /02 /03 | 0301 | 0402 | 0301 /02 /03 | 0302 | No | |
| 0401 | 0301 /02 /03 | 0302 | 1104 | 0501 /03 /05 | 0301 | No | |
| 0401 | 0301 /02 /03 | 0302 | 1501 | 0102 | 0602 | No | |
| 0401 | 0301 /02 /03 | 0302 | 1101 | 0501 /03 /05 | 0301 | No | |
| 0401 | 0301 /02 /03 | 0302 | 0403 | 0301 /02 /03 | 0304 | H | |
| 0401 | 0301 /02 /03 | 0302 | 1201 | 0501 /03 /05 | 0301 | No | |
| 0402 | 0301 /02 /03 | 0302 | 1101 | 0501 /03 /05 | 0301 | No | |
| 0402 | 0301 /02 /03 | 0302 | 1104 | 0501 /03 /05 | 0301 | No | |
| 0404 | 0301 /02 /03 | 0302 | 0403 | 0301 /02 /03 | 0304 | H | |
| 0404 | 0301 /02 /03 | 0302 | 1201 | 0501 /03 /05 | 0301 | No | |
| 0407 | 0301 /02 /03 | 0302 | 1602 | 0501 /03 /05 | 0303 | No | |
| 07 | 0201 | 0201 /02 | 0301 | 0501 /03 /05 | 0201 /02 | No | |
| 07 | 0201 | 0201 /02 | 0401 | 0301 /02 /03 | 0301 | No | |
| 07 | 0201 | 0201 /02 | 0401 | 0301 /02 /03 | 0302 | No | |
| 07 | 0201 | 0201 /02 | 0404 | 0301 /02 /03 | 0302 | No | |
| 07 | 0201 | 0201 /02 | 0405 | 0301 /02 /03 | 0302 | No | |
| 07 | 0201 | 0201 /02 | 07 | 0201 | 0201 /02 | No | |
| 07 | 0201 | 0201 /02 | 1303 | 0501 /03 /05 | 0301 | No | |
| 07 | 0201 | 0303 | 0301 | 0501 /03 /05 | 0201 /02 | No | |
| 07 | 0201 | 0303 | 0401 | 0301 /02 /03 | 0301 | No | |
| 07 | 0201 | 0303 | 0404 | 0301 /02 /03 | 0302 | No | |
| 07 | 0201 | 0201 /02 | 1302 | 0102 | 0609 | No | |
| 07 | 0201 | 0201 /02 | 1303 | 0501 /03 /05 | 0301 | No | |
| 07 | 0201 | 0201 /02 | 1304 | 0501 /03 /05 | 0301 | No | |
| 09 | 0301 /02 /03 | 0303 | 1304 | 0501 /03 /05 | 0301 | No | |
| 10 | 0101 /04 /05 | 0501 | 07 | 0201 | 0201 /02 | No | |
| 0803 | 0103 | 0601 | 07 | 0201 | 0201 /02 | No | |
| 0804 | 0102 | 0602 | 1503 | 0401 /02 /03N | 0301 | No | |
| 1101 | 0301 /02 /03 | 0501 | 09 | 0102 | 0201 /02 | No | |
| 1101 | 0501 /03 /05 | 0301 | 0301 | 0501 /03 /05 | 0201 /02 | No | |
| 1101 | 0501 /03 /05 | 0301 | 1104 | 0501 /03 /05 | 0301 | No | |
| 1101 | 0501 /03 /05 | 0301 | 1401 | 0101 /04 /05 | 0602 | No | |
| 1102 | 0501 /03 /05 | 0301 | 1301 | 0102 | 0501 | No | |
| 1104 | 0501 /03 /05 | 0501 | 0101/ 05 | 0101 /04 /05 | 0301 | No | |
| 1201 | 0101 /04 /05 | 0501 | 1501 | 0102 | 0602 | No | |
| 1301 | 0103 | 0603 | 0301 | 0501 /03 /05 | 0201 /02 | No | |
| 1301 | 0103 | 0603 | 0401 | 0301 /02 /03 | 0301 | No | |
| 1301 | 0103 | 0603 | 0401 | 0301 /02 /03 | 0302 | No | |
| 1301 | 0103 | 0603 | 07 | 0201 | 0303 | No | |
| 1302 | 0102 | 0604 | 0401 | 0301 /02 /03 | 0301 | No | |
| 1302 | 0102 | 0604 | 1302 | 0102 | 0609 | No | |
| 1303 | 0501 /03 /05 | 0301 | 1302 | 0102 | 0609 | No | |
| 1304 | 0501 /03 /05 | 0301 | 1101 | 0501 /03 /05 | 0301 | No | |
| 1401 | 0101 /04 /05 | 0503 | 0301 | 0501 /03 /05 | 0201 /02 | No | |
| 1401 | 0101 /04 /05 | 0503 | 0401 | 0301 /02 /03 | 0301 | No | |
| 1401 | 0101 /04 /05 | 0503 | 0403 | 0301 /02 /03 | 0302 | No | |
| 1401 | 0101 /04 /05 | 0503 | 0803 | 0103 | 0601 | No | |
| 1501 | 0101/ 04/ 05 | 0501 | 0101 /05 | 0102 | 0602 | No | |
| 1501 | 0102 | 0602 | 0301 | 0501 /03 /05 | 0201 /02 | No | |
| 1501 | 0102 | 0602 | 0401 | 0301 /02 /03 | 0301 | No | |
| 1501 | 0102 | 0602 | 0404 | 0301 /02 /03 | 0302 | No | |
| 1501 | 0102 | 0602 | 0405 | 0301 /02 /03 | 0302 | No | |
| 1501 | 0102 | 0602 | 07 | 0201 | 0201 /02 | No | |
| 1501 | 0102 | 0602 | 07 | 0201 | 0303 | No | |
| 1501 | 0102 | 0602 | 0801 | 0401 /02 /03N | 0402 | No | |
| 1501 | 0102 | 0602 | 09 | 0301 /02 /03 | 0303 | No | |
| 1501 | 0102 | 0606 | 09 | 0301 /02 /03 | 0303 | No | |
| 1501 | 0102 | 0602 | 1303 | 0501 /03 /05 | 0301 | No | |
| 1501 | 0102 | 0602 | 1501 | 0102 | 0602 | No | |
| 1502 | 0102 | 0502 | 07 | 0201 | 0201 /02 | No | |
| 1502 | 0103 | 0601 | 0401 | 0301 /02 /03 | 0302 | No | |
| 1503 | 0102 | 0602 | 0301 | 0501 /03 /05 | 0201 /02 | No | |
| 1503 | 0102 | 0602 | 07 | 0201 | 0201 /02 | No | |
| 1503 | 0102 | 0602 | 09 | 0301 /02 /03 | 0201 /02 | No | |
| 1503 | 0102 | 0602 | 0302 | 0401 /02 /03N | 0402 | No | |
| 1503 | 0102 | 0602 | 1503 | 0102 | 0602 | No | |
| 1503 | 0102 | 0602 | 1503 | 0102 | 0602 | No | |
| 1503 | 0102 | 0602 | 1102 or 1114 | 0501 /03 /05 | 0301 | No | |
| 1601 | 0102 | 0502 | 0401 | 0301 /02 /03 | 0302 | No | |
| 1602 | 0102 | 0502 | 0401 | 0301 /02 /03 | 0301 | No | |
The layout is the same as Table 1. The final column designates the TEDDY eligibility code (A–J) if the haplogenotype is eligible for study participation either in the general population (code A–D) or as a FDR (E–J) or “No” if the haplogenotype is not eligible. Haplogenotypes are listed in ascending order on columns 1 (DRB1 chromosome 1) through 4 (DRB1 chromosome 2).
Specimen Panels
Resealable bags containing the stored DBS filter paper cards (each originally spotted with up to 15 DBSs) were removed from the freezer and allowed to come to room temperature before opening to avoid damage caused by moisture condensation. Sufficient DBSs for the PT panel were then cut from the filter paper card. Each DBS was labeled with a panel identification number and placed into a glassine envelope that was also labeled with the same number. For VQPT panels, the samples were sealed in a Mylar bag along with three or four desiccant packs. For the TEDDY PT panel, envelopes were bound into sets, each in numerical order, and each complete sample set was sealed in a Mylar bag along with three or four desiccant packs. With both panels, the Mylar bags were then placed into Tyvek envelopes and stored at -20 ˚C until shipping. To ensure an equivalent and timely delivery to laboratories in both the United States and Europe, DBS samples sets were shipped on a Friday by an express courier. Whole blood aliquots frozen in CryoTubes were shipped on dry ice to the one participating laboratory that did not use DBS as a screening sample.
For each VQPT, a panel of 5 samples were sent to each of the participating laboratories. Haplogenotypes representing at least two different risk levels were included in each sample set (Table 1). For each TEDDY PT, a panel of 55 samples were sent to each laboratory. Only 8 of the 10 TEDDY-eligible haplogenotypes were available in sufficient quantity to include in the panels. Samples for each panel were selected, including all available TEDDY-eligible HLA-D haplogenotypes as well as the largest possible variety of ineligible haplogenotypes. Of the 10 TEDDY-eligible haplogenotypes, 6 conferred eligibility only if the candidate had a first-degree relative (FDR) with T1DM. In each panel, 10 samples were designated as having FDR status, only some of which met TEDDY eligibility criteria. Samples that were eligible only with FDR status were also included in samples that were not designated as FDR to determine whether participating laboratories would correctly identify them as ineligible (Table 2).
Each laboratory received a spreadsheet inventory of the sample set in hardcopy enclosed with the panel (VQPT) or as an electronic file attached to an email (TEDDY PT, see Figures 1 and 2). Laboratories were asked to analyze the panel samples using their routine screening method and not to conduct specialized analyses that were not part of their routine screening method. They were given three weeks to complete their analysis.
Figure 1.
First tab of the electronic spreadsheet for the TEDDY PT challenge with instructions and haplogenotype codes.
Figure 2.
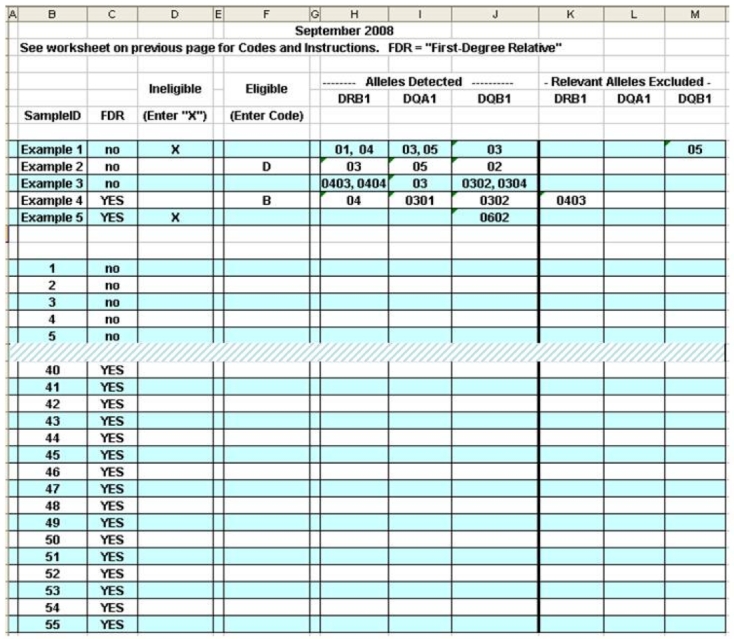
Second tab of the electronic spreadsheet for the TEDDY PT challenge showing a truncated answer sheet with examples near the top of the page.
Reporting and Evaluation
For VQPT, laboratories reported results via a hardcopy form, and their evaluation (Figure 3) was returned by mail. Participants had to correctly identify the risk level in 4 out of 5 samples for a satisfactory evaluation.
Figure 3.
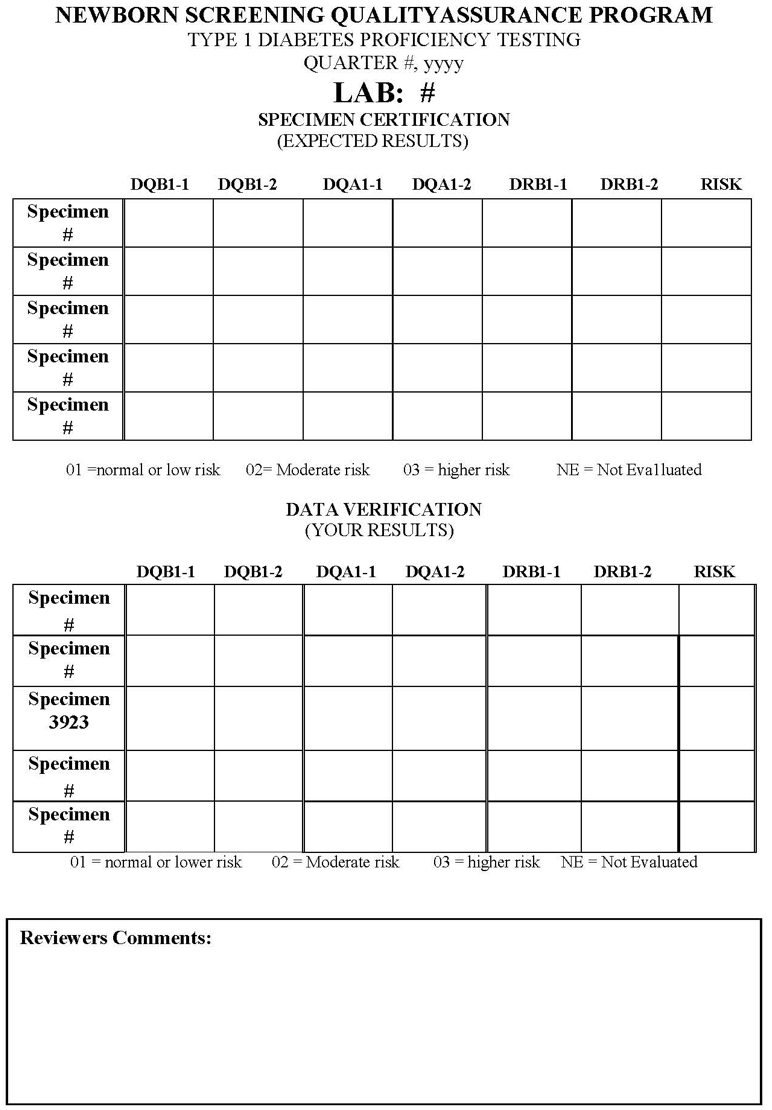
Evaluation form returned by laboratories participating in the VQPT.
For TEDDY PT, laboratories reported their results on the preformatted electronic spreadsheet (Figure 2) by email to the CDC. Results were evaluated for the correct identification of eligibility for each sample and for the correct haplogenotype code assigned to each eligible haplogenotype. Information about alleles determined to be present or absent was not evaluated unless the eligibility status was misclassified. Each laboratory was notified of its overall score within three business days of reporting. Participants had to correctly identify eligibility in at least 98% of the samples to meet the TEDDY acceptability criteria.
Results
For the VQPT, four laboratories have participated since the program began in 2003. As of 2008, a total of 560 VQPT samples had been distributed on a quarterly basis. Of the reported results, 94.7% (857/905) were correctly analyzed with respect to the haplogenotype, and 96.4% (241/250) were correctly categorized for risk level. The rate of correct responses increased significantly from the earliest to the most recent surveys (Figure 4).
Figure 4.
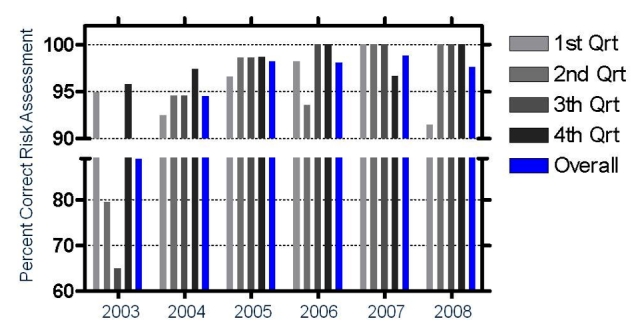
Composite results from all laboratories in the VQPT program over six years. Each bar shows the percentage of samples for which relative risk was correctly assessed. The last bar in each year shows the average percent correct of the four quarterly challenges for that year.
For the TEDDY PT, a total of five laboratories participated in the first three PT surveys from 2004–2006, and six participated in the fourth survey in 2008. A total of 1155 PT samples were distributed over four challenges, and results were received for all samples from all laboratories in every panel. Of these, 1154 (99.9%) were correctly categorized with respect to TEDDY eligibility. For all TEDDY laboratories combined, the analytical sensitivity was 99.4% (186/187), and the analytical specificity was 100% (748/748). The haplogenotypes for all 186 samples reported as eligible were correctly identified. The one sample that was misclassified as ineligible by one laboratory was correctly identified as eligible in a repeat blinded sample set analyzed by that laboratory.
Discussion
The increasing incidence of T1DM12 has been recognized as an urgent public health issue.13 The most important genetic locus, HLA-D, works in conjunction with other genes and nongenetic factors to influence T1DM risk. Thus most people with even the highest risk HLA-D haplogenotypes do not develop T1DM, while some people with lower risk haplogenotypes do. Most diabetologists believe that environmental factors play a major role in triggering T1DM, and many believe that exposures early in life (even in utero) are important. However, the exact nature of such exposures remains unknown. In addition to specific exposures, other environmental influences may be important, including birth by C-section,14 maternal stress during pregnancy,15 and vitamin D.16
Type 1 diabetes mellitus is generally caused by the autoimmune destruction of the insulin-producing beta cells in pancreatic islets.17 About half of the risk for T1DM can be attributed to genetic predisposition, and much of that risk resides in the MHC. The MHC contains genes that code for transplantation antigens, which, in humans, are known as the HLAs. The HLA genes are highly polymorphic, and certain combinations of HLA alleles are associated with increased or decreased risk for T1DM. While other genetic loci that impart increased risk have been identified,18,19 the HLA locus is by far the largest single contributor.20 The difference between susceptible and protective HLA haplogenotypes can result in as much as a 20-fold difference in the risk for T1DM.21 Moreover, the highest risk HLA haplogenotypes are those associated with an earlier age at onset, which increases the chance of diabetic ketoacidosis (DKA) and hospitalization.22 Identification of increased risk lowers the probability of hospitalization and DKA23 and therefore provides benefit even in the absence of a preventative treatment.
From a research perspective, early recognition of higher T1DM risk is required for natural history studies1 and for clinical trials of therapies that could prevent or delay autoimmune destruction of islet cells. From a public health perspective, early recognition can reduce the chance of DKA in incident cases, a major contributor to morbidity and health care expenses. The most effective public health program for early recognition of congenital risk is NBS. A number of T1DM research efforts have used NBS to identify higher risk individuals for recruitment into natural history studies and early intervention trials. Results suggest that early identification and intervention can preserve insulin production for up to five years.24,25 If the onset of T1DM can be prevented or significantly delayed, NBS for T1DM genetic risk could become a standard public health practice.
Determining the exact haplogenotypes in the HLA complex, which is required to match tissues for organ transplantation, is complex and expensive. The extensive polymorphism in the HLA results in a vast array of individual haplogenotypes, as illustrated in Tables 1 and 2. This allows identification of higher risk haplogenotypes without the necessity of identifying every allele. Still, the complexity of the HLA loci demands careful attention to specificity of allele identification. For instance, the haplogenotypes in the second and third rows of Table 2 differ only by the DQB1 alleles on the second chromosome (0301 versus 0302), but this difference in closely related alleles discriminates a protective haplogenotype from a higher risk haplogeno-type. Conversely, the two haplogenotypes designated as eligibility code “H,” differ only in the DRB1 locus of chromosome 1 (0401 versus 0404), a difference that does not affect the risk category as defined by the TEDDY study.
The T1DM research programs that use HLA-based risk assessment algorithms have identified technical approaches that make use of the known allelic associations within the HLA complex. In addition, protective alleles are dominant, so a tiered approach that first culls the general population to remove those with protective alleles can provide more cost-effective selection of higher risk individuals. Since TEDDY protocol includes only a small set of eligible haplogenotypes, screening algorithms can be designed to exclude individuals in whom any ineligible allele is detected.1
Proficiency testing is an effective method for laboratories to ensure quality control and quality assurance. It provides laboratory personnel with an objective benchmark with which to measure their accuracy and also to compare their score with other laboratories conducting similar testing. While the two PT programs discussed here use different methodologies, they both achieve the goal of objectively measuring the performance of the laboratories. To do so, the challenge panels had to include a wide variety of lower risk haplogenotypes as well has the higher risk (or TEDDY-eligible) haplogenotypes.
The difference in algorithms used to assign T1DM genetic risk presents challenges for PT. Laboratories will not necessarily test for the same genetic markers, and the tests that they employ may differ in the extent to which they can resolve closely related HLA alleles. The complex and inconsistent use of nomenclature to identify these alleles presents additional problems. Assignment of risk levels can also vary, depending on the population being tested and the reference data used by the laboratory. Taken together, these factors hamper the ability to make valid comparisons between laboratories.
The TEDDY study circumvents the problem of assessing laboratory comparability by specifying a small set of higher risk HLA haplogenotypes for study eligibility. Even though screening laboratories can use their own methods, they must all identify the eligible HLA haplogenotypes properly. All participants screened as eligible are confirmed by higher resolution testing in a single HLA reference laboratory to assure accurate identification.3 The TEDDY screening laboratories have, to date, exceeded the required accuracy of 98%, a remarkable achievement given the demand for low-cost analysis, the large sample volume, and need for rapid turn-around time.
Results from the VQPT also support the technical feasibility of newborn DBS screening in multiple distinct laboratory locations. Although the error rate was at first much higher than in the TEDDY PT, most of the errors occurred in the first year of the VQPT. Performance in the VQPT program improved over time, and in several of the latest quarterly challenges, all the laboratories received a perfect score. Combined with the highly accurate PT results from TEDDY, our experience documents the technical feasibility of population-based public health newborn screening, which could provide the early identification of T1DM risk essential for subsequent prediction and intervention strategies.25
Abbreviations
- CDC
Centers for Disease Control and Prevention
- DBS
dried blood spot
- DKA
diabetic ketoacidosis
- FDR
first-degree relative
- HLA
human leukocyte antigen
- MHC
major histocompatibility complex
- NBS
newborn bloodspot screening
- PT
proficiency testing
- T1DM
type 1 diabetes mellitus
- TEDDY
The Environmental Determinants of Diabetes in the Young
- VQPT
voluntary quarterly proficiency testing
References
- 1.Kiviniemi M, Hermann R, Nurmi J, Ziegler AG, Knip M, Simell O, Veijola R, Lövgren T, Ilonen J, TEDDY study group A high-throughput population screening system for the estimation of genetic risk for type 1 diabetes: An application for the TEDDY (The Environmental Determinants of Diabetes in the Young) study. Diabetes Technol Ther. 2007;9(5):460–472. doi: 10.1089/dia.2007.0229. [DOI] [PubMed] [Google Scholar]
- 2.Berger M, Stassen HH, Köhler K, Krane V, Mönks D, Wanner C, Hoffmann K, Hoffmann MM, Zimmer M, Bickeböller H, Lindner TH. Hidden population substructures in an apparently homogeneous population bias association studies. Eur J Hum Genet. 2006;14(2):236–244. doi: 10.1038/sj.ejhg.5201546. [DOI] [PubMed] [Google Scholar]
- 3.Hagopian WA, Lernmark A, Rewers MJ, Simell OG, She JX, Ziegler AG, Krischer JP, Akolkar B. TEDDY--The Environmental Determinants of Diabetes in the Young: an observational clinical trial. Ann NY Acad Sci. 2006;1079:320–326. doi: 10.1196/annals.1375.049. [DOI] [PubMed] [Google Scholar]
- 4.TEDDY Study Group The Environmental Determinants of Diabetes in the Young (TEDDY) Study. Ann NY Acad Sci. 2008;1150:1–13. doi: 10.1196/annals.1447.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.TEDDY Study Group The Environmental Determinants of Diabetes in the Young (TEDDY) study: study design. Pediatr Diabetes. 2007;8(5):286–298. doi: 10.1111/j.1399-5448.2007.00269.x. [DOI] [PubMed] [Google Scholar]
- 6.Dantonio P, Meredith N, Earley M, Cordovado S, Callan WJ, Rollin D, Morris D, Vogt RF, Hannon WH. A screening system for detecting genetic risk markers of type 1 diabetes in dried blood spots. Diabetes Technol Ther. 2006;8(4):433–443. doi: 10.1089/dia.2006.8.433. [DOI] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute (CLSI) Blood collection on filter paper for newborn screening programs; approved standard. Fifth ed. CLSI document LA4-A5. Wayne: Clinical and Laboratory Standards Institute; 2007. [Google Scholar]
- 8.Rewers M, Bugawan TL, Norris JM, Blair A, Beaty B, Hoffman M, McDuffie RS, Jr, Hamman RF, Klingensmith G, Eisenbarth GS, Erlich HA. Newborn screening for HLA markers associated with IDDM: Diabetes Autoimmunity Study in the Young (DAISY) Diabetologia. 1996;39(7):807–812. doi: 10.1007/s001250050514. [DOI] [PubMed] [Google Scholar]
- 9.Buzzetti R, Galgani A, Petrone A, Del Buono ML, Erlich HA, Bugawan TL, Lorini R, Meschi F, Multari G, Pozzilli P, Locatelli M, Bottazzo G, Di Mario U. Genetic prediction of type 1 diabetes in a population with low frequency of HLA risk genotypes and low incidence of the disease (the DIABFIN study) Diabetes Metab Res Rev. 2004;20(2):137–143. doi: 10.1002/dmrr.426. [DOI] [PubMed] [Google Scholar]
- 10.Sjöroos M, Ilonen J, Reijonen H, Lövgren T. Time-resolved fluorometry based sandwich hybridisation assay for HLA-DQA1 typing. Dis Markers. 1998;14(1):9–19. doi: 10.1155/1998/350145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wion E, Brantley M, Stevens J, Gallinger S, Peng H, Glass M, Hagopian W. Population-wide infant screening for HLA-based type 1 diabetes risk via dried blood spots from the public health infrastructure. Ann NY Acad Sci. 2003;1005:400–403. doi: 10.1196/annals.1288.067. [DOI] [PubMed] [Google Scholar]
- 12.Patterson CC, Dahlquist GG, Gyürüs E, Green A, Soltész G, EURODIAB Study Group Incidence trends for childhood type 1 diabetes in Europe during 1989-2003 and predicted new cases 2005-20: a multicentre prospective registration study. Lancet. 2009;373(9680):2027–2033. doi: 10.1016/S0140-6736(09)60568-7. [DOI] [PubMed] [Google Scholar]
- 13.Dabelea D. The accelerating epidemic of childhood diabetes. Lancet. 2009;373(9680):1999–2000. doi: 10.1016/S0140-6736(09)60874-6. [DOI] [PubMed] [Google Scholar]
- 14.Cardwell CR, Stene LC, Joner G, Cinek O, Svenesson J, Goldacre MJ, Parslow RC, Pozzilli P, Brigis G, Stoyanov D, Urbonaite B, Sipetić S, Schober E, Ionescu-Tirgoviste C, Devoti G, de Beaufort CE, Buschard K, Patterson CC. Caesarean section is associated with an increased risk of childhood-onset type 1 diabetes mellitus: a meta-analysis of observational studies. Diabetologia. 2008;51(5):726–735. doi: 10.1007/s00125-008-0941-z. [DOI] [PubMed] [Google Scholar]
- 15.Lernmark B, Lynch K, Lernmark A. Cord blood islet autoantibodies are related to stress in the mother during pregnancy. Ann NY Acad Sci. 2006;1079:345–9. doi: 10.1196/annals.1375.053. [DOI] [PubMed] [Google Scholar]
- 16.Harris SS. Vitamin D in type 1 diabetes prevention. J Nutr. 2005;135(2):323–325. doi: 10.1093/jn/135.2.323. [DOI] [PubMed] [Google Scholar]
- 17.Sanjeevi CB, Schatz DA, Atkinson MA. Immunology of diabetes V: from bench to bedside. New York Academy of Science; 2008. [PubMed] [Google Scholar]
- 18.Fung EY, Smyth DJ, Howson JM, Cooper JD, Walker NM, Stevens H, Wicker LS, Todd JA. Analysis of 17 autoimmune disease-associated variants in type 1 diabetes identifies 6q23/TNFAIP3 as a susceptibility locus. Genes Immun. 2009;10(2):188–191. doi: 10.1038/gene.2008.99. [DOI] [PubMed] [Google Scholar]
- 19.Concannon P, Chen WM, Julier C, Morahan G, Akolkar B, Erlich HA, Hilner JE, Nerup J, Nierras C, Pociot F, Todd JA, Rich SS. Type 1 Diabetes Genetics Consortium. Genome-wide scan for linkage to type 1 diabetes in 2,496 multiplex families from the Type 1 Diabetes Genetics Consortium. Diabetes. 2009;58(4):1018–1022. doi: 10.2337/db08-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida K, Corper AL, Herro R, Jabri B, Wilson IA, Teyton L. The diabetogenic mouse MHC class II molecule I-Ag7 is endowed with a switch which modulates TCR affinity. J Clin Invest. 2010;120(5):1578–1590. doi: 10.1172/JCI41502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ilonen J, Sjöroos M, Knip M, Veijola R, Simell O, Akerblom HK, Paschou P, Bozas E, Havarani B, Malamitsi-Puchner A, Thymelli J, Vazeou A, Bartsocas CS. Estimation of genetic risk for type 1 diabetes. Am J Med Genet. 2002;115(1):30–36. doi: 10.1002/ajmg.10341. [DOI] [PubMed] [Google Scholar]
- 22.Rewers A, Klingensmith G, Davis C, Petitti DB, Pihoker C, Rodriguez B, Schwartz ID, Imperatore G, Williams D, Dolan LM, Dabelea D. Presence of diabetic ketoacidosis at diagnosis of diabetes mellitus in youth: the Search for Diabetes in Youth study. Pediatrics. 2008;121(5):e1258–e1266. doi: 10.1542/peds.2007-1105. [DOI] [PubMed] [Google Scholar]
- 23.Barker JM, Goehrig SH, Barriga K, Hoffman M, Slover R, Eisenbarth GS, Norris JM, Klingensmith GJ, Rewers M, DAISY Study. Clinical characteristics of children with diagnosed with type 1 diabetes through intensive screening and follow-up. Diabetes Care. 2004;27(6):1399–1404. doi: 10.2337/diacare.27.6.1399. [DOI] [PubMed] [Google Scholar]
- 24.Herold KC, Gitelman S, Greenbaum C, Puck J, Hagopian W, Gottlieb P, Sayre P, Bianchine P, Wong E, Seyfert-Margolis V, Bourcier K, Bluestone JA, Immune Tolerance Network ITN007AI Study Group Treatment of patients with new onset type 1 diabetes with a single course of anti-CD3 mAb Teplizumab preserves insulin production for up to 5 years. Clin Immunol. 2009;132(2):166–173. doi: 10.1016/j.clim.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ludvigsson J, Faresjö M, Hjorth M, Axelsson S, Chéramy M, Pihl M, Vaarala O, Forsander G, Ivarsson S, Johansson C, Lindh A, Nilsson NO, Aman J, Ortqvist E, Zerhouni P, Casas R. GAD treatment and insulin secretion in recent-onset type 1 diabetes. N Engl J Med. 2008;359(18):1909–120. doi: 10.1056/NEJMoa0804328. [DOI] [PubMed] [Google Scholar]



