Abstract
In this review, we discuss several important issues concerning the discovery of protein biomarkers for complex human diseases, with a focus on type 1 diabetes. Serum or plasma is the first choice of specimen due to its richness in biological information and relatively easy availability. It is a challenging task to comprehensively characterize the serum/plasma proteome because of the large dynamic range of protein concentration. Therefore, sample pretreatment is required in order to explore the low- to medium-abundance proteins contained in serum/plasma. In this regard, enrichment of low-abundance proteins using random hexapeptide library beads has distinct advantages over the traditional immune-depletion methods, including higher efficiency, higher binding capacity, and lower cost. In-depth mining of serum/plasma proteome using different separation techniques have also been evaluated and are discussed in this review. Overall, the shotgun proteomics—multidimensional separation of digested peptides followed by mass spectrometry analysis—is highly efficient and therefore has become a preferred method for protein biomarker discovery.
Keywords: depletion, enrichment, mass spectrometry, protein biomarker, proteomics, type 1 diabetes mellitus
Introduction
Biomarkers are useful to type 1 diabetes mellitus (T1DM) for a number of purposes, including disease prediction, understanding disease mechanism, monitoring response to therapy, and risk assessment for diabetic complications.1–3 Type 1 diabetes mellitus in humans may be preventable by avoiding those factors that trigger the disease process (primary prevention) or by use of therapy that modulates the destruction of islet cells before the onset of clinical symptoms (secondary prevention). Accurate prediction is vital for secondary prevention so that therapy can be given to those individuals who are very likely to develop the disease or those who will benefit from the therapy. The prediction for any disease is dependent on three parameters, which must be carefully assessed for predictive tests to be clinically useful: sensitivity, specificity, and positive predictive value. Specificity is important if a disease marker is to be used to identify individuals either for counseling or for therapy to prevent the disease. Research by many investigators since the 1980s has identified several useful biomarkers for T1DM prediction,4,5 including genetic risk factors, immunological markers (such as islet cell autoantibodies), and metabolic markers. These markers have provided a good foundation for T1DM prediction and prevention, yet are far from being perfect due to low specificity or arrival late in the disease stage. It is believed that genetic susceptibility is a prerequisite for the development of T1DM; however, not all genetically predisposed individuals do develop clinical disease. The vast majority (~90%) of T1DM patients develop autoantibodies against pancreatic β cells before the clinical onset. Although the time period between the appearance of autoantibodies and clinical onset varies greatly, it usually takes years for the clinical disease to occur. Furthermore, only a proportion of autoantibody-positive (AbP) individuals will progress to clinical diabetes. This lengthy asymptomatic period, from genetic predisposition to prediabetes marked by autoimmunity (autoantibodies and cellular immunity) and finally to clinical disease, provides excellent opportunities for disease prevention. However, prevention for human T1DM is still not available today, partly because we cannot precisely predict the disease and accurately assess risk for the high-risk population and partly because the etiology of the disease is potentially very heterogeneous and poorly understood. Therefore, prevention tailored for the whole at-risk population may not be effective and personalized prevention strategies based on one’s own risk, and etiology may prove to be more efficient. To achieve these ambitious goals, biomarkers for the disease process are urgently needed for both risk assessment and, more importantly, for tailoring and monitoring therapies. Compared with genetic biomarkers, protein biomarkers are the direct executors for all aberrant genetic changes. Proteomic analysis of the protein/peptide expression, modifications, and interactions for T1DM will lead to indispensable contribution for the disease prediction and prognosis. We believe that current proteomics study for T1DM needs to pay attention to several issues. First, various biological specimens will be selected for different diseases and for various purposes. For example, in T1DM protein biomarker discovery, serum/plasma probably is the most suitable specimen because of the high richness in biological information and relatively easy availability compared with other biological fluids or solid specimens. Obviously, advanced technology to conquer the challenges with serum analysis is another very important issue. Last but not least, reasonable sample size plays a critical role in T1DM biomarker evaluation, which has been neglected in many studies. In this review, combining our own research results, we focus on using mass spectrometry (MS)-based proteomic technologies for T1DM biomarker discovery in human serum.
Biomarker Discovery Using Multidimensional Protein Identification Technology
While most of the initial efforts in proteomics have focused on protein identification, MS-based technology developments have provided useful platforms for the study of quantitative changes in protein components. Quantitative analysis of the global serum proteome is an essential step for understanding the molecular changes associated with the disease progression and onset. Several methods are widely used to generate global quantitative protein profiles, including two-dimensional (2D) gel electrophoresis followed by MS analysis, stable isotope labeling-based quantification, MS signal intensity-based quantification, and intact protein-based quantification (see review6 and reports7,8 for details).
Comprehensive analysis of the serum/plasma proteome is a challenging task due to its extraordinary complexity and high multidimensionality of its components.9,10 It is therefore unrealistic that any one analytical technique would be well suited to address all protein complexities. Desirable objectives include extending the detection, quantification and identification to low-abundance proteins, assessment of protein distribution among cells and subcellular structures, as well as assessment of posttranslational modifications (PTMs). As a result, various schemes are currently being implemented to reduce the complexity of biological samples prior to analysis by MS.1 Two main categories of methodologies have gained popularity for normalization of serum/plasma protein content, e.g., depletion of high-abundance proteins using antibodies11 and enrichment of low-abundance proteins.12
Depletion-Based Strategy
This strategy is based on utilizing immobilized antibodies against selective high-abundance proteins to concentrate the low-abundance proteins in a column chromatography format. This method has been in use for decades, and an increasing number of major proteins can be depleted using this approach.13,14 However, the application of this method is hindered by the limited binding capacity of immobilized antibodies, limited antibody availability, and high cost.15
Enrichment-Based Approaches
A different strategy to normalize serum/plasma protein content is through binding to a random hexapeptide library that is conjugated to small beads to enrich all the proteins on the column. The technology was commercialized under the name of ProteoMiner (PM) by Bio-Rad Laboratories (Hercules, CA). The core concept of the PM kit is the use of a large, highly diverse bead-based library of combinatorial hexapeptide ligands, which provide significant binding diversities (~206) for proteins. When complex samples are applied to the beads, high-abundance proteins usually saturate the binding sites on the beads and the unbound (excess) proteins are removed by washing, while medium- and low-abundance proteins usually do not saturate their specific binding sites, leading to the increase of their relative concentration.16 This process can significantly reduce the dynamic range of protein concentrations while maintaining representation of many proteins in the original samples.12
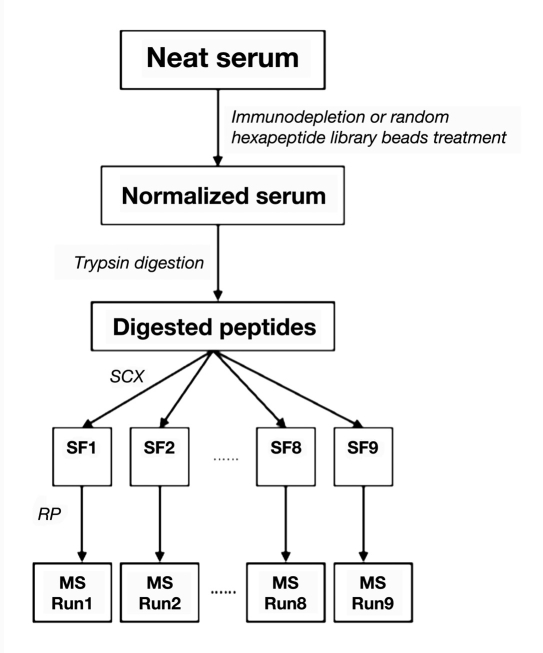
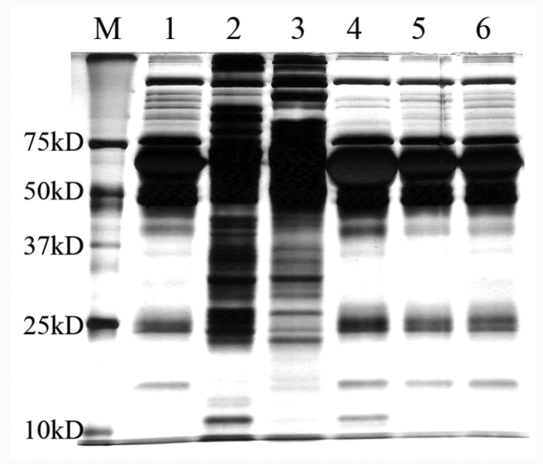
Our lab used a multidimensional-protein-identification-technology-based serum biomarker study platform, as presented in ,Figure 1, to compare the efficiencies of these two normalization methods for serum samples. Multiple pools of serum samples are generated, each pool containing serum from 10 individuals. A pool of serum sample is normalized using the MARS-14 (Agilent Technologies, Santa Clara, CA) and/or the PM kit (Biorad). The differences in the protein content for these two approaches were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) by separating 40 μg of proteins and further compared to neat serum loaded under the same conditions (,Figure 2).
Figure 1.
Workflow of the proteomics-based biomarker discovery. SCX, strong cation exchange chromatography.
Figure 2.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis images of serum proteins before and after depletion of major proteins or affinity enrichment. Lane M, molecular weight marker; lanes 1 and 6, neat serum; lane 2, bound fraction after enrichment by the PM kit; lane 3, flow-through fraction after depletion by MARS-14 column; lane 4, depleted fraction by MARS-14 column; lane 5, flow-through fraction after PM processing.
Compared with neat serum samples (lanes 1 and 6 of ,Figure 2), both PM (lane 2) and MARS-14 (lane 3) greatly decreased the concentration of major proteins to enriched proteins with medium to low abundance as indicated by the disappearance of major proteins and significantly increased number of protein bands after processing. The undesired fractions from each method, namely, bound proteins on MARS-14 column (lane 4) and the flow through of the PM kit (lane 5), are found very similar to neat serum. More protein bands appear to be obtained with PM (lane 2) than MARS-14 (lane 3), with a broader distribution of MWs for the proteins, contrary to the notion that these two lanes should be similar. To better assess the performance of these two approaches, 5 μg of normalized serum fractions from each method (bound fraction for PM and flow-through fraction for MARS-14) as well as neat serum with equal protein amount were further analyzed by a 2D shotgun proteomic approach consisting of online 2D chromatographic separation and a linear ion-trap MS (LTQ). Database searches were then performed to generate protein identification results at 1% false positive rate, according to a previously described method.17 Due to the analytical incompleteness of shotgun proteomics,18 we replicated the whole procedure six times and compared the results on the total unique protein identified from these six runs for each of the three samples (neat serum, MARS-14, and PM), overlap between protein identifications within these six runs, number of immunoglobulins and major proteins, common identifications among the three samples and between any two samples.
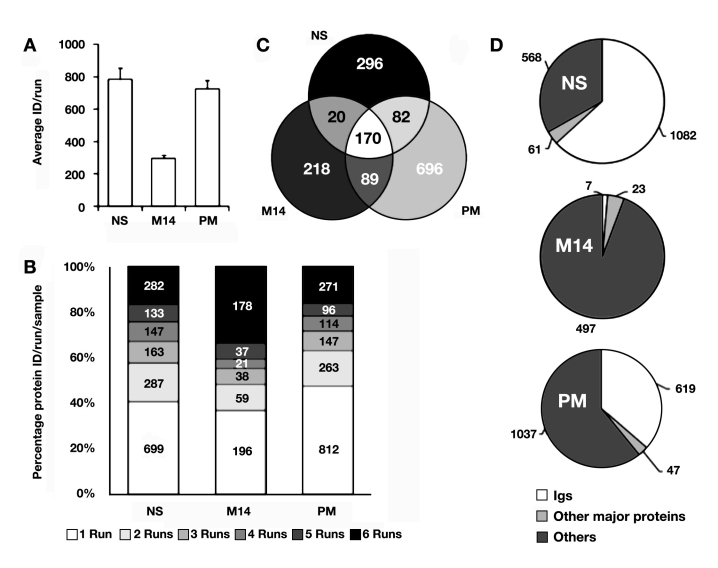
An average of 785, 724, and 294 proteins are confidently identified in each run from neat serum, PM, and MARS-14 normalized serum, respectively (,Figure 3A). A total of 1711, 1703, and 527 unique protein identifications were obtained from neat serum, PM, and MARS-14, respectively, when all six runs were combined together (Figures 3B and 3C). Detailed examination of the protein identifications in each of the three datasets revealed that a large portion of protein identifications for neat serum belonged to immunoglobulin (1082 out of 1711; ,Figure 3D). As expected, only a small number of identifications from MARS-14 was immunoglobulin (7 out of 527; ,Figure 3D). A moderate portion of protein identifications from PM belonged to immunoglobulin (619 out of 1703; ,Figure 3D).After subtracting immunoglobulin proteins and 14 other major proteins (e.g., albumin, transferrin, and haptoglobin), the number of proteins identified from neat serum and MARS-14 treatment were similar (568 versus 497), while PM was able to identify a much larger number of proteins (1037; ,Figure 3D). The number of protein identifications from neat serum was larger than the commonly expected results. The better-than-expected results were probably due to the combination of high separation and identification power of our 2D shotgun proteomic platform. After PM enrichment, 1037 proteins could be identified from serum, indicating that affinity enrichment is an excellent method for the analysis of complex proteomes by MS. The relatively large number of immunoglobulin molecules after PM enrichment is actually expected, because the diverse set of peptides in the PM kit can bind to the variable regions of immunoglobulin. The presence of large numbers of immunoglobulin molecules does not pose a serious problem for identifying other proteins, because the total abundance of the immunoglobulins is relatively low, despite their sequence diversity.
Figure 3.
Protein identification results for neat serum, MARS-14-treated serum, and PM-treated serum. (A) Average unique protein number per run; (B) overlap of protein identification among replicate runs; (C) overlap of protein identification among the three samples; (D) classification of protein identification. ID, identification; NS, neat serum; M14, MARS-14.
The poor performance of the affinity-depletion method was initially to our surprise but should be expected. MARS-14 treatment, like all other depletion methods, is designed to remove the intended proteins. The primary reason for this poor performance may be due to two nonalternative problems: (1) the emergence of new major proteins after the depletion of the initial major proteins and (2) loss of proteins due to codepletion. To test the first possibility, we examined the major proteins that were identified by each of the three methods (,Table 1). The original major serum proteins are infrequently found among proteins identified with either MARS-14 or PM, indicating that both methods are highly efficient in eliminating the major proteins. However, the samples processed with MARS-14 or PM contain several proteins with >100 peptide counts, an indication of their relatively high protein abundance.7,8 For MARS-14, none of the newly emerged major proteins except apolipoprotein AI (129 ± 15 peptides) belongs to the 14 major serum proteins that the kit is designed to deplete, indicating the high depletion efficiency of the column. The newly emerged major proteins include apolipoprotein B-100, complement C4-A, ceruloplasmin, and several others, and they can interfere with the identification of low-abundance proteins in the processed samples. Similarly, most of the abundant serum proteins (e.g., albumin, IgG, antitrypsin, transferring, and haptoglobin) were greatly reduced, while a few major serum proteins (apolipoprotein AI and complement C3) were still present in high abundance after normalization with PM. Furthermore, a group of proteins, including complement component 4 and its binding protein, was preferably enriched by PM. The selective enrichment was probably due to the presence of large numbers of binding peptides in the peptides library.12
Table 1.
Major Proteins Identified in Neat Serum or after Normalization
| Protein name | Peptide number (ratio to total peptide number) | ||
|---|---|---|---|
| Neat serum | MARS-14 | PM | |
| Albumina | 3513 ± 327 (37.40%) | 86 ± 36 (1.48%) | 296 ± 33 (3.69%) |
| Iga | 1790 ± 214 (19.06%) | 3 ± 2 (0.05%) | 619 ± 87 (7.71%) |
| Antitrypsina | 5 ± 1 (0.05%) | 0 | 5 ± 2 (0.06%) |
| Transferrina | 364 ± 116 (3.88%) | 0 | 15 ± 4 (0.19%) |
| Haptoglobina | 105 ± 26 (1.12%) | 0 | 22 ± 3 (2.74%) |
| Fibrinogena | 4 ± 2 (0.04%) | 1 ± 1 (0.02%) | 16 ± 3 (0.20%) |
| Alpha-2-macroglobulina | 318 ± 37 (3.39%) | 7 ± 3 (0.12%) | 61 ± 8 (0.76%) |
| Alpha-1-acid glycoproteina | 45 ± 7 (0.48%) | 0 | 2 ± 2 (0.02%) |
| Apolipoprotein AIa | 142 ± 9 (1.51%) | 129 ± 15 (2.22%) | 222 ± 50 (2.77%) |
| Apolipoprotein AIIa | 46 ± 8 (0.49%) | 50 ± 10 (0.86%) | 37 ± 8 (0.46%) |
| A Chain A, Human Complement Component C3a | 159 ± 18 (1.69%) | 2 ± 1 (0.03%) | 630 ± 162 (7.85%) |
| B Chain B, Human Complement Component C3a | 108 ± 14 (1.15%) | 24 ± 4 (0.41%) | 374 ± 46 (4.66%) |
| Transthyretina | 14 ± 3 (0.15%) | 0 | 65 ± 15 (0.81%) |
| Vitamin D-binding protein | 85 ± 6 (0.91%) | 327 ± 22 (5.62%) | 30 ± 3 (0.37%) |
| Ceruloplasmin precursor | 70 ± 3 (0.75%) | 482 ± 75 (8.29%) | 162 ± 28 (2.02%) |
| Complement C4-A precursor | 108 ± 13 (1.15%) | 229 ± 23 (3.94%) | 653 ± 134 (8.14%) |
| Hemopexin | 50 ± 15 0.53%) | 624 ± 49 (10.74%) | 5 ± 2 (0.06%) |
| Apolipoprotein B-100 precursor | 154 ± 17 (1.64%) | 306 ± 22 (5.26%) | 204 ± 66 (2.54%) |
| C4 binding protein, α chain precursor | 41 ± 2 (0.44%) | 57 ± 7 (0.98%) | 158 ± 18 (1.97%) |
| Fibronectin precursor | 45 ± 6 (0.48%) | 101 ± 16 (1.74%) | 312 ± 40 (3.87%) |
Top 14 serum proteins that the MARS-14 kit is designed to deplete.
We also investigated the proteins bound to the MARS-14 column using 2D shotgun proteomic analysis. As expected, 13 of the 14 major serum proteins (except fibrinogen) were found in the bound fraction. Interestingly, 167 other proteins were also identified in the bound fraction (data not shown). Fifteen of the 167 proteins belong to the same family of proteins targeted by the antibodies in the MARS-14 kit or interacted with the targeted proteins (e.g., apolipoprotein and complement components). The presence of 152 additional proteins bound to MARS-14 column may be a result of nonspecific binding to the antibodies or to the resin. Another possibility is that proteins like cytokines and similar proteins need a carrier protein for their transport in serum and are therefore codepleted with albumin.19,20 Similar observations have been reported on a different MS platform when serum was used as a starting material for the biomarker discovery in other diseases/conditions. Sihlbom and colleagues21 showed that using peptide library beads to process human plasma was superior to antibody depletion through separately combining these two technologies with surface-enhanced laser desorption and ionization time-of-flight (SELDI-TOF) MS and 2D difference gel electrophoresis (DIGE; 1100 versus 675 spots), yet both techniques (SELDI-TOF MS and 2D DIGE) suffered from the inability of directly getting protein identification information. Dwivedi and associates22 suggest that PM may provide a better basis for probing lower-abundance species in serum samples as compared to IgY antibodies-based depletion column (IgY-12, ProteomeLab, Beckman Coulter, CA), while accessing the reproducibility of PM beads.
These observations also raised the possibility that such libraries might be useful in the preparation of samples for quantitative and comparative proteomic analysis. Hartwig and coworkers,23 using spiking experiments together with 2D gels, showed that the concentration of the spiked bacteria proteome (into serum), reflected by the maintained proportional spot intensities, was not altered by PM treatment. There are also studies on the reproducibility of the PM kit using SDS-PAGE,15 SELDI-TOF MS,21 and isobaric tag for relative and absolute quantitation +2D high-performance liquid chromatography (HPLC)-MS,22 and all reached good results. The overall results of these studies indicated that there is enough redundancy in each column to bind to a given protein, although each of the PM columns is predicted to contain a nonidentical repertoire of hexapeptides on the PM beads. However, in order to have enough redundancy of PM columns, relatively large volumes of PM resins are required, and therefore the standard protocol of PM columns needs 1 ml of serum/plasma. The large serum/plasma volume could be a limiting factor for the application of PM beads on precious samples (e.g., clinical samples), and reducing both the volumes of PM beads and serum/plasma samples would be a rational attempt. Yet, in this case, the reproducibility of the PM treatment has not been reported and needs to be further evaluated.
Intact Protein-Based Approaches for Biomarker Discovery for Type 1 Diabetes Mellitus
The high complexity of eukaryotic proteome requires initial prefractionation, and separation of proteins/peptides before MS are mandatory to most proteomic studies. Two strategies for the characterization of complex proteomes focused on separating either intact proteins or digested peptides. The first strategy involves processing the protein samples on a traditional 2D gel system or higher-dimensional intact protein separation systems that use multiple in-solution protein separation techniques, such as isoelectric focusing (IEF) coupled with liquid chromatography and followed by SDS-PAGE.24–26 Mass spectrometry is then used in offline mode to analyze the much simplified peptide mixture digested from protein fractions containing a single or limited number of proteins after separation. This approach of multidimensional separation has a better resolution but is more labor intensive compared to 2D electrophoresis.
The multidimensionality and the complexity of the proteome at a given time cannot be resolved using a single universal proteomic platform. The choice of appropriate proteomic methods for a specific project can be difficult, because no comprehensive comparison of the different approaches has been undertaken in the same setting. Our lab has compared the performance of three proteomic platforms, including a shotgun method based on 2D HPLC electrospray ionization MS (platform 1), a three-dimensional (3D) intact protein separation method (platform 2), and a combination of intact protein separation and shotgun method (platform 3).
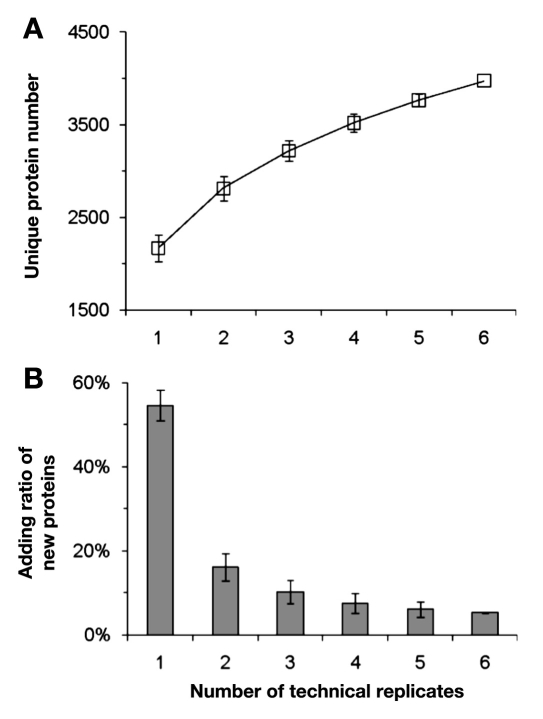
Using our platform 1, we were able to reliably identify over 2000 proteins in each run from total cellular extract (DC2.4; ,Figure 4A). The shotgun approach was found to identify a diverse set of proteins in each run, suggesting that multiple replicate runs for a single sample are required for a comprehensive characterization of complex cellular proteome. The data indicated that the number of unique proteins increased with each replicate run, but the total number of identified unique proteins begins to decrease more rapidly after three to four replicates (,Figure 4). With five to six replicate runs, we can generally identify ~4000 unique proteins from a total DC2.4 cell extract (,Figure 4).
Figure 4.
Shotgun analysis of the total DC2.4 proteome. (A) Total number of unique proteins identified by different number of replicates; (B) percentage of new proteins identified by each consecutive replicate analysis. This was calculated by dividing the newly identified proteins in a single run by total identified proteins in all six replicate runs.
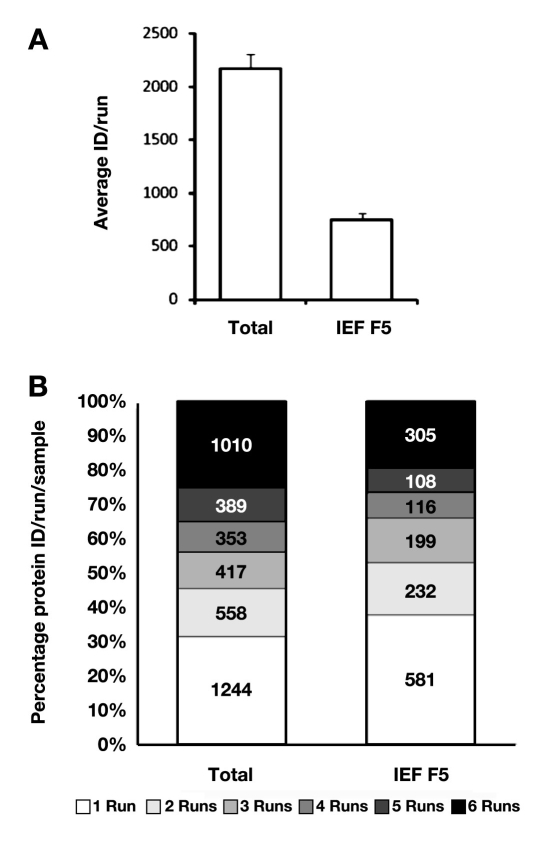
Furthermore, prefractionation of intact proteins before shotgun analysis has the potential to increase the number of identifiable proteins. Based on our results, prefraction-ation of the DC2.4 proteome into five fractions using liquid phase IEF (platform 3) may double the total number of identifiable proteins (,Figure 5); however, this approach increases the analysis time by four-fold. Therefore, one must balance the cost and benefit between the total number of proteins identified and the total time required to complete the analysis when deciding whether or not to prefractionate before shotgun proteomic analysis.
Figure 5.
Protein identification results for total DC 2.4 proteome and IEF fraction V of DC 2.4 proteome. (A) Average unique protein number per run; (B) overlap of protein identification among replicate runs. ID, identification.
We further compared data from the 2D shotgun analyses (platform 1) and a 3D intact protein separation system (platform 2) composed of liquid-phase IEF, reverse phase (RP)-HPLC and SDS-PAGE. Typically, several hundred proteins from each IEF fraction are resolved into unique bands after RP-HPLC and SDS-PAGE. Overall, 7000–8000 protein bands were found from all five IEF fractions. After eliminating potentially redundant protein bands due to tailing effect of RP-HPLC (identical protein on adjacent lanes), we estimated that ~2500 unique bands could be resolved for the DC2.4 proteome.
The shotgun proteomics approach described earlier is relatively simple and can be set up for the analysis of a relatively large number of samples compared to the liquid-phase IEF, RP-HPLC, and SDS-PAGE approach. It has the potential to identify large numbers of proteins from very complex proteomes after employing proper prefractionation/separation prior to MS analysis. Other investigators have also compared the performances of different prefractionation methods prior to shotgun proteomic analysis and provided good suggestions on the selection of prefractionation methods. Wang and colleagues27 compared the performance of gel-based liquid chromatography (GeLC)-MS/MS (one-dimensional protein + one-dimensional peptide separation) method and a 3D method that adds a liquid IEF step before the GeLC-MS/MS analysis and found that the latter detected more unique peptides (32,216 versus 25,641) and proteins (3486 versus 2850) than the sum of four repetitive GeLC-MS/MS analysis. Fang and associates28 systematically compared the performances of the three most popular upstream fractionation methods prior to MS analysis, including strong cation exchange chromatography, IEF, and SDS-PAGE, both at protein level and peptide level, and concluded that, for maximal proteome coverage, SDS-PAGE is very clearly the most effective method tested, with more than 90% of the entire dataset found. But when considering the amount of material recovered after each fractionation procedure, solution-based IEF and strong cation exchange chromatography performed similarly, with approximately 80% of the input being recovered.
After using these various techniques on serum samples from T1DM patients and normal controls, our initial analyses have successfully identified 50 different serum proteins that are differentially expressed between T1DM patients and controls. Most of these 50 proteins have known functions and can be grouped into six functional categories: innate immunity, inflammation/oxidation, lymphocyte activation/proliferation, lymphocyte trafficking/infiltration, immunoglobulin, and other disease. The functional annotation clearly indicates that these proteins are highly relevant to the pathogenesis of T1DM. We have also validated several protein biomarker candidates identified using these approaches. These results are being prepared for publication elsewhere. Other researchers have been trying to find protein biomarkers in serum/plasma for diabetes. But most of the candidate proteins identified are limited to proteins of relatively high abundance in human serum/plasma, such as albumin,29 transferring,29 apolipoprotein,29,30 transthyretin,29 and C-reactive protein,31,32 which, based on our results, are apparently due to the limited mining of serum proteome mainly caused by inadequate pretreatment of serum samples.
Biomarkers Based on Multivariate Models
While proteins or genes may be used individually in predicting disease, the accuracy of prediction can be significantly improved by using multiple markers simultaneously in multivariate statistical models for prediction. Indeed, multivariate models may be the only hope for highly specific and sensitive biomarkers, as any single marker is unlikely to prove acceptable for many complex diseases. Development of multivariate models requires solving two statistical issues: (1) selecting an optimal subset of markers—a single multivariate model—from all available sets of variables with which to make predictions and (2) predicting the phenotypic/disease statuses based on the selected subset of markers. The subset of markers that would be selected depends on the method used for predicting disease status, and prediction of disease status can be accomplished by classifying subjects into known classes.
We have previously applied multivariate analysis to the serum proteomic data generated by SELDI-TOF to identify predictive models based on multiple proteins.33 We identified nine models based on normal kernel discriminant analysis, each of which utilized 25 peaks (total peaks used = 176), and each had 10–25% error rates in classifying AbP and T1DM subjects based on leave-one-out cross validation. As the misclassified subjects in one model may be “correctly” classified in a different model, the “final” classification for a subject is the averaged outcome using plurality voting, which classify the AbP subjects with 92.8% accuracy (specificity) and the T1DM subject with 90.2% accuracy (sensitivity). These results show that the identified models hold promise in accurately classifying AbP and T1DM subjects.
Conclusions
Innovations in biological MS have made it a desirable tool for proteomic analysis. The combination of separation science and biological MS has become the current workhorse in proteomics. These technologies are continuing to evolve to meet the needs of high-sensitivity and high-throughput requirements. Biological samples subjected to proteomic analysis consist of three major types: (1) tissues, (2) cell populations, and (3) biological fluids. A common feature of biological samples is their extraordinary complexity, which is a result of the high multidimensionality of their protein constituents. These proteins differ in their cellular and subcellular distribution; their occurrence in complexes; their charge, molecular mass, and hydrophobicity; and their expressed levels and PTMs. As a result, any single analytical technique would not be sufficient to analyze all proteins in complex proteomes. The results presented here demonstrate the validity of various schemes to reduce the complexity of biological samples prior to analysis by MS to improve proteomic analyses. Despite these advances in proteomic technology, this technology still needs improvement in several areas, including extending the detection, quantification, and identification to low-abundance proteins; assessment of protein distribution among cells and subcellular structures; and assessment of PTM.
Abbreviations
- 2D
two dimensional
- 3D
three dimensional
- AbP
autoantibody-positive
- DIGE
difference gel electrophoresis
- GeLC
gel-based liquid chromatography
- HPLC
high-performance liquid chromatography
- IEF
isoelectric focusing
- PM
ProteoMiner
- PTM
posttranslational modification
- RP
reverse phase
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SELDI-TOF
surface-enhanced laser desorption and ionization time-of-flight
- T1DM
type 1 diabetes mellitus
References
- 1.Jacobs JM, Adkins JN, Qian WJ, Liu T, Shen Y, Camp DG, 2nd, Smith RD. Utilizing human blood plasma for proteomic biomarker discovery. J Proteome Res. 2005;4(4):1073–1085. doi: 10.1021/pr0500657. [DOI] [PubMed] [Google Scholar]
- 2.Simpson RJ, Bernhard OK, Greening DW, Moritz RL. Proteomics-driven cancer biomarker discovery: looking to the future. Curr Opin Chem Biol. 2008;12(1):72–77. doi: 10.1016/j.cbpa.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 3.Li F, Fang Y, Xiao ZQ, Chen ZC. Clinical proteomics: the application of proteomics in the research of clinical medicine. Prog Biochem Biophys. 2006;33(1):5–9. [Google Scholar]
- 4.Purohit S, She JX. Biomarkers for type 1 diabetes. Int J Clin Exp Med. 2008;1(2):98–116. [PMC free article] [PubMed] [Google Scholar]
- 5.Sosenko JM, Krischer JP, Palmer JP, Mahon J, Cowie C, Greenbaum CJ, Cuthbertson D, Lachin JM, Skyler JS, Diabetes Prevention Trial-Type 1 Study Group A risk score for type 1 diabetes derived from autoantibody-positive participants in the diabetes prevention trial-type 1. Diabetes Care. 2008;31(3):528–533. doi: 10.2337/dc07-1459. [DOI] [PubMed] [Google Scholar]
- 6.Maurya P, Meleady P, Dowling P, Clynes M. Proteomic approaches for serum biomarker discovery in cancer. Anticancer Res. 2007;27(3A):1247–1255. [PubMed] [Google Scholar]
- 7.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;4(10):1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Asara JM, Christofk HR, Freimark LM, Cantley LC. A label-free quantification method by MS/MS TIC compared to SILAC and spectral counting in a proteomics screen. Proteomics. 2008;8(5):994–999. doi: 10.1002/pmic.200700426. [DOI] [PubMed] [Google Scholar]
- 9.Anderson L. Candidate-based proteomics in the search for biomarkers of cardiovascular disease. J Physiol. 2005;563(Pt 1):23–60. doi: 10.1113/jphysiol.2004.080473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobs JM, Adkins JN, Qian WJ, Liu T, Shen Y, Camp DG, 2nd, Smith RD. Utilizing human blood plasma for proteomic biomarker discovery. J Proteome Res. 2005;4(4):1073–1085. doi: 10.1021/pr0500657. [DOI] [PubMed] [Google Scholar]
- 11.Gong Y, Li X, Yang B, Ying W, Li D, Zhang Y, Dai S, Cai Y, Wang J, He F, Qian X. Different immunoaffinity fractionation strategies to characterize the human plasma proteome. J Proteome Res. 2006;5(6):1379–1387. doi: 10.1021/pr0600024. [DOI] [PubMed] [Google Scholar]
- 12.Righetti PG, Boschetti E, Lomas L, Citterio A. Protein Equalizer Technology: the quest for a “democratic proteome.”. Proteomics. 2006;6(14):3980–3992. doi: 10.1002/pmic.200500904. [DOI] [PubMed] [Google Scholar]
- 13.Brand J, Haslberger T, Zolg W, Pestlin G, Palme S. Depletion efficiency and recovery of trace markers from a multiparameter immunodepletion column. Proteomics. 2006;6(11):3236–3242. doi: 10.1002/pmic.200500864. [DOI] [PubMed] [Google Scholar]
- 14.Echan LA, Tang HY, Ali-Khan N, Lee K, Speicher DW. Depletion of multiple high-abundance proteins improves protein profiling capacities of human serum and plasma. Proteomics. 2005;5(13):3292–3303. doi: 10.1002/pmic.200401228. [DOI] [PubMed] [Google Scholar]
- 15.Sennels L, Salek M, Lomas L, Boschetti E, Righetti PG, Rappsilber J. Proteomic analysis of human blood serum using peptide library beads. J Proteome Res. 2007;6(10):4055–4062. doi: 10.1021/pr070339l. [DOI] [PubMed] [Google Scholar]
- 16.Boschetti E, Lomas L, Citterio A, Righetti PG. Romancing the “hidden proteome,” Anno Domini two zero zero seven. J Chromatogr A. 2007;1153(1-2):277–290. doi: 10.1016/j.chroma.2007.01.136. [DOI] [PubMed] [Google Scholar]
- 17.Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods. 2005;2(9):667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 18.Wilkins MR, Appel RD, Van Eyk JE, Chung MC, Görg A, Hecker M, Huber LA, Langen H, Link AJ, Paik YK, Patterson SD, Pennington SR, Rabilloud T, Simpson RJ, Weiss W, Dunn MJ. Guidelines for the next 10 years of proteomics. Proteomics. 2006;6(1):4–8. doi: 10.1002/pmic.200500856. [DOI] [PubMed] [Google Scholar]
- 19.Liu T, Qian WJ, Mottaz HM, Gritsenko MA, Norbeck AD, Moore RJ, Purvine SO, Camp DG, 2nd, Smith RD. Evaluation of multiprotein immunoaffinity subtraction for plasma proteomics and candidate biomarker discovery using mass spectrometry. Mol Cell Proteomics. 2006;5(11):2167–2174. doi: 10.1074/mcp.T600039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yocum AK, Yu K, Oe T, Blair IA. Effect of immunoaffinity depletion of human serum during proteomic investigations. J Proteome Res. 2005;4(5):1722–1731. doi: 10.1021/pr0501721. [DOI] [PubMed] [Google Scholar]
- 21.Sihlbom C, Kanmert I, Bahr H, Davidsson P. Evaluation of the Combination of Bead Technology with SELDI-TOF-MS and 2-D DIGE for detection of plasma proteins. J Proteome Res. 2008;7(9):4191–4198. doi: 10.1021/pr800340c. [DOI] [PubMed] [Google Scholar]
- 22.Dwivedi RC, Krokhin OV, Cortens JP, Wilkins JA. Assessment of the reproducibility of random hexapeptide peptide library-based protein normalization. J Proteome Res. 2010;9(2):1144–1149. doi: 10.1021/pr900608z. [DOI] [PubMed] [Google Scholar]
- 23.Hartwig S, Czibere A, Kotzka J, Passlack W, Haas R, Eckel J, Lehr S. Combinatorial hexapeptide ligand libraries (ProteoMiner): an innovative fractionation tool for differential quantitative clinical proteomics. Arch Physiol Biochem. 2009;115(3):155–160. doi: 10.1080/13813450903154224. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Hanash S. Intact-protein based sample preparation strategies for proteome analysis in combination with mass spectrometry. Mass Spectrom Rev. 2005;24(3):413–426. doi: 10.1002/mas.20018. [DOI] [PubMed] [Google Scholar]
- 25.Horn A, Kreusch S, Bublitz R, Hoppe H, Cumme GA, Schulze M, Moore T, Ditze G, Rhode H. Multidimensional proteomics of human serum using parallel chromatography of native constituents and microplate technology. Proteomics. 2006;6(2):559–570. doi: 10.1002/pmic.200500142. [DOI] [PubMed] [Google Scholar]
- 26.Assiddiq BF, Williamson JC, Snijders AP, Cook K, Dickman MJ. Multidimensional liquid phase protein separations in conjunction with stable isotope labelling for quantitative proteomics. Proteomics. 2007;7(21):3826–3834. doi: 10.1002/pmic.200700367. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Chang-Wong T, Tang HY, Speicher DW. Comparison of extensive protein fractionation and repetitive LC-MS/MS analyses on depth of analysis for complex proteomes. J Proteome Res. 2010;9(2):1032–1040. doi: 10.1021/pr900927y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang Y, Robinson DP, Foster LJ. Quantitative analysis of proteome coverage and recovery rates for upstream fractionation methods in proteomics. J Proteome Res. 2010;9(4):1902–1912. doi: 10.1021/pr901063t. [DOI] [PubMed] [Google Scholar]
- 29.Sundsten T, Eberhardson M, Göransson M, Bergsten P. The use of proteomics in identifying differentially expressed serum proteins in humans with type 2 diabetes. Proteome Sci. 2006;4:22. doi: 10.1186/1477-5956-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HJ, Cho EH, Yoo JH, Kim PK, Shin JS, Kim MR, Kim CW. Proteome analysis of serum from type 2 diabetics with nephropathy. J Proteome Res. 2007;6(2):735–743. doi: 10.1021/pr060489g. [DOI] [PubMed] [Google Scholar]
- 31.Cho WC, Yip TT, Chung WS, Leung AW, Cheng CH, Yue KK. Differential expression of proteins in kidney, eye, aorta, and serum of diabetic and non-diabetic rats. J Cell Biochem. 2006;99(1):256–268. doi: 10.1002/jcb.20923. [DOI] [PubMed] [Google Scholar]
- 32.Riaz S, Alam SS, Akhtar MW. Proteomic identification of human serum biomarkers in diabetes mellitus type 2. J Pharm Biomed Anal. 2010;51(5):1103–1107. doi: 10.1016/j.jpba.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 33.Purohit S, Podolsky R, Schatz D, Muir A, Hopkins D, Huang YH, She JX. Assessing the utility of SELDI-TOF and model averaging for serum proteomic biomarker discovery. Proteomics. 2006;6(24):6405–6415. doi: 10.1002/pmic.200600420. [DOI] [PubMed] [Google Scholar]