Abstract
The liver and pancreas share a common origin and coexpress several transcription factors. To gain insight into the transcriptional networks regulating the function of these tissues, we globally identify binding sites for FOXA2 in adult mouse islets and liver, PDX1 in islets, and HNF4A in liver. Because most eukaryotic transcription factors bind thousands of loci, many of which are thought to be inactive, methods that can discriminate functionally active binding events are essential for the interpretation of genome-wide transcription factor binding data. To develop such a method, we also generated genome-wide H3K4me1 and H3K4me3 localization data in these tissues. By analyzing our binding and histone methylation data in combination with comprehensive gene expression data, we show that H3K4me1 enrichment profiles discriminate transcription factor occupied loci into three classes: those that are functionally active, those that are poised for activation, and those that reflect pioneer-like transcription factor activity. Furthermore, we demonstrate that the regulated presence of H3K4me1-marked nucleosomes at transcription factor occupied promoters and enhancers controls their activity, implicating both tissue-specific transcription factor binding and nucleosome remodeling complex recruitment in determining tissue-specific gene expression. Finally, we apply these approaches to generate novel insights into how FOXA2, PDX1, and HNF4A cooperate to drive islet- and liver-specific gene expression.
The liver and pancreas develop from a common progenitor pool in the foregut endoderm (Zaret 2008; Zaret and Grompe 2008). Many of the transcription factors initially expressed within the endoderm remain expressed in both the liver and pancreas and function to maintain homeostasis in the adult tissues (Jensen 2004; Zaret 2008; Zaret and Grompe 2008). For example, FOXA2 is essential for both pancreas and liver development (Lee et al. 2005; Gao et al. 2008). FoxA proteins, which share homology with histone H5, function as pioneer factors and can bind compact chromatin and open local chromatin structures (Cirillo et al. 2002; Sekiya et al. 2009). FOXA2 regulates the expression of several transcription factors, including Pdx1 (Gao et al. 2008) and the nuclear hormone receptor Hnf4a (Bailly et al. 2001). PDX1 is essential for pancreas development (Jonsson et al. 1994) and is required for the expression of insulin and several other genes critical to pancreas development and function (Ohlsson et al. 1993; Ahlgren et al. 1998; Gerrish et al. 2001; Svensson et al. 2007). HNF4A is not essential for the early phases of liver development, but controls many genes that are vital for adult liver functions, and is considered a central regulator of hepatocyte differentiation and function (Hayhurst et al. 2001; Battle et al. 2006).
Recent genome-wide analyses of transcription factor–DNA association in mammalian genomes show that most transcription factors bind thousands of sites (Robertson et al. 2007; Chen et al. 2008; Marson et al. 2008; Wederell et al. 2008; Hoffman and Jones 2009; Zheng et al. 2009). It has been suggested that sites that are less occupied are nonfunctional (Li et al. 2008); however, there is no clear boundary between highly and poorly occupied sites, and no methods are available that can identify which sites are functional. To interpret genome-wide transcription factor localization data, it is therefore essential to develop novel methods that can discriminate functionally active binding events from inactive ones.
Cis-regulatory regions are associated with diverse histone modifications that include acetylations and methylations of histone tails (Bernstein et al. 2006; Roh et al. 2006; Barski et al. 2007; Heintzman et al. 2007; Robertson et al. 2008; Wang et al. 2008). Lineage-specific combinations of histone marks at enhancers correlate with lineage-specific enhancer function (Heintzman et al. 2009), suggesting that specific histone modifications can be used to predict the functionality of cis-regulatory loci. For example, active genes are associated with the mono- and tri-methylation of lysine 4 of histone H3 (H3K4me1 and H3K4me3) (Wang et al. 2008). H3K4me1 occurs at cis-regulatory regions both distal and proximal to transcriptional start sites (TSSs), while H3K4me3 is typically regionally restricted to TSS-proximal loci (Robertson et al. 2008), making H3K4me1 useful in predicting locations of lineage-specific enhancers (Heintzman et al. 2009).
To gain insight into how FOXA2, PDX1, and HNF4A regulate distinct transcriptional programs in the liver and pancreas, we used chromatin immunoprecipitation coupled with flow cell sequencing (ChIP-seq) to identify genome-wide locations of loci bound by FOXA2 in adult mouse islets and liver, by PDX1 in islets, and by HNF4A in liver. Furthermore, to discriminate functional binding events, we used ChIP-seq to determine the genome-wide enrichment profiles of H3K4me1 and H3K4me3.
Results
Genome-wide identification of FOXA2-, PDX1-, and HNF4A-occupied loci in adult islets and liver
We used ChIP-seq to globally identify loci occupied by FOXA2 in adult mouse islets and liver, PDX1 in islets, and HNF4A in liver. The antibodies used produced only the expected bands in Western blots (Supplemental Fig. S1), suggesting that our data reflect specific protein–DNA interactions. Aligning the DNA sequence reads to the reference genome identified regions enriched for immunoprecipitated fragments, called “peaks” (Supplemental Fig. S2). We thresholded the peaks to discriminate statistically significant sites from background, identifying 7189 FOXA2- and 13,448 PDX1-occupied loci in islets, as well as 12,494 HNF4A-occupied loci in liver (Supplemental Table S1). Reprocessing previously published FOXA2 ChIP-seq data from liver (Wederell et al. 2008) with current methods identified 10,701 occupied loci (Robertson et al. 2008). Comparing these data with literature reported sites and performing validations using ChIP-qPCR confirmed the high quality of the data (Supplemental Figs. S3, S4).
To test whether the identified sites represent direct DNA–protein interactions and to determine the in vivo binding potential of the factors, we used de novo motif discovery (see Supplemental Methods; Supplemental Figs. S5–S8; Li 2009). In islets and liver, we identified FOXA2-like motifs in 72% and 75% of the FOXA2-enriched regions, respectively. These motifs were highly similar to each other (E-value 1.1 × 10−16), were similar to the known FOXA2 motif (E-values 5.2 × 10−12 and 5.6 × 10−16, respectively) (Wederell et al. 2008), and were centrally concentrated around sites of maximal enrichment (Supplemental Figs. S5, S6). We identified a PDX1-like motif in 45% of enriched regions that was similar to the TRANSFAC PDX1 motif (M00463; E-value 9.6 × 10−7) and was moderately centrally concentrated. PDX1 can interact and bind DNA as a heterodimer with PBX1 (Swift et al. 1998), and we also identified a PDX1:PBX1 heterodimer-like motif in 42% the PDX1-enriched regions (Supplemental Fig. S7), which was also moderately centrally concentrated. PDX1 and/or PDX1:PBX1-like motifs were found in 62% of the PDX1-enriched sites. The derived HNF4A motif was very similar to the HNF4A TRANSFAC motif (M01031, E-value 0.0), was identified in 92% of the enriched regions, and was also centrally concentrated (Supplemental Fig. S8). These results confirm that the majority of identified sites contain the expected binding sequences for FOXA2, PDX1, and HNF4A.
Transcription factor occupied loci display three patterns of H3K4 methylation
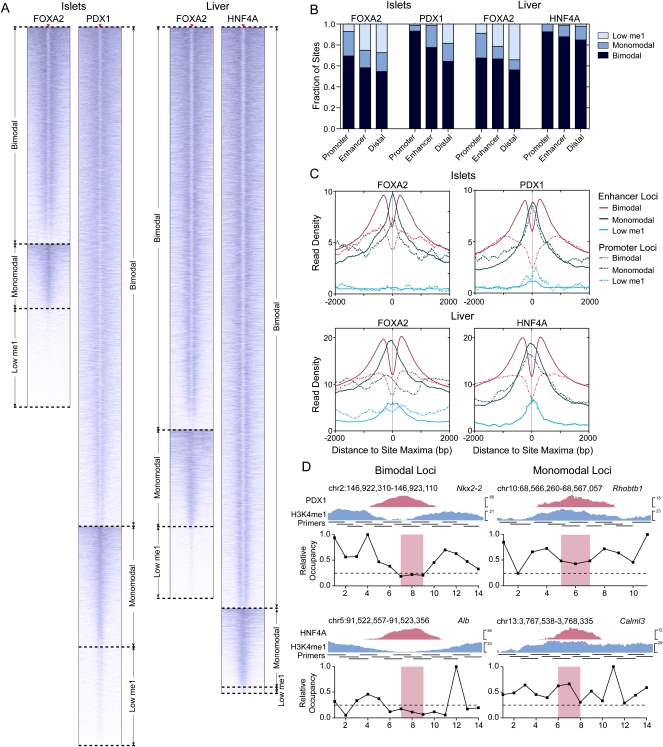
As histone H3 lysine 4 mono-methylation (H3K4me1) and tri-methylation (H3K4me3) are commonly associated with functional cis-regulatory regions (Barski et al. 2007; Heintzman et al. 2007; Robertson et al. 2008), we asked whether these marks could be used to discriminate functionally active FOXA2-, PDX1-, and HNF4A-occupied loci. For this we generated H3K4me1 and H3K4me3 enrichment profiles in ±2-kb regions around the transcription factor peak maxima in islets and liver. Unexpectedly, we found three classes of H3K4me1 enrichment profiles associated with the identified loci (Fig. 1A; Supplemental Fig. S9). For each transcription factor, the majority of promoter, enhancer, and distal loci (Fig. 1B) had a bimodal H3K4me1 profile, with peaks of H3K4me1 enrichment on either side of the identified sites (Fig. 1C), indicating the presence of H3K4me1-marked nucleosomes flanking these sites (Heintzman et al. 2007; Robertson et al. 2008). The second most common H3K4me1 profile had a monomodal shape, with a peak of H3K4me1 enrichment co-occurring at the identified transcription factor binding sites, suggesting the presence of a central H3K4me1-marked nucleosome, or nucleosomes, at the transcription factor binding site. For FOXA2, in both islets and liver, and for PDX1, >10% of loci were associated with a low, nonsignificant level of H3K4me1, whereas only 1% of HNF4A sites were in this category. Similar profile classes were identified using the H3K4me3 data (Supplemental Fig. S10), although the fraction of loci associated with a significant level of this modification was considerably lower as H3K4me3 is primarily localized to promoter regions (Robertson et al. 2008).
Figure 1.
H3K4me1 profiles segregate discrete classes of transcription factor occupied loci. (A) Heatmaps of H3K4me1 read density in ±2-kb regions centered on FOXA2 (islets or liver), PDX1, and HNF4A peak maxima. Peak max locations are indicated by red triangles, with flanking H3K4me1 read density plotted horizontally in blue for each site. H3K4me1 read density is represented by the intensity of blue in the heatmaps: (dark blue) high; (light blue) low read density; (white) minimal or no H3K4me1. (B) Fractions of bimodal, monomodal, or low H3K4me1 promoter, enhancer, or distal (>50 kb from any RefSeq TSS) loci in each library. (C) Mean H3K4me1 enrichment profiles associated with promoter and enhancer loci in each site class in the FOXA2 (islets or liver), PDX1, and HNF4A peak sets. Peak maxima are centered at 0. (D) Nucleosome mapping by MNase-qPCR confirms the presence of nucleosomes. (Left) Flanking selected bimodal sites; (right) immediately at or adjacent to transcription factors binding locations at monomodal sites. UCSC Genome Browser screenshots of representative PDX1 (top panel) and HNF4A (bottom panels) loci are shown, and the locations of the primers used in the MNase-qPCR are indicated. The panels beneath the browser screenshots show MNase-qPCR results, with regions of relative enrichment indicative of nucleosome positions. The red-highlighted regions indicate the primer pairs flanking identified motif locations and represent the location of transcription factor binding; the dashed lines, at a relative occupancy of 0.25, are shown as a reference.
To ensure that the bimodal, monomodal, and low H3K4me1 profile classes identified are not the result of ChIP-seq processing artifacts, we generated a set of high-confidence profile class calls. For this we calculated Pearson correlations between the H3K4me1 profiles of individual loci with mean bimodal, monomodal, or low H3K4me1 profiles, and used a minimum correlation threshold of 0.35 to identify loci that strongly conformed to the mean profiles (Supplemental Fig. S11). The distribution of these loci into the three classes, and the enrichment profiles obtained, were similar to those obtained using the whole data set (Supplemental Fig. S12). Next, to verify that bimodal and monomodal sites were a result of differences in nucleosome distributions, and not of H3K4me1 enrichment differences, we mapped nucleosome positions at representative enhancer loci (Figs. 1D and 4H below). These data indicate the presence of flanking nucleosomes at bimodal sites and of central nucleosomes at monomodal sites, confirming that the H3K4me1 bimodal and monomodal profiles are largely a result of nucleosome distribution differences. In sum, these data indicate that H3K4me1 profiles can be used to classify transcription factor occupied loci into three distinct subsets: those with a bimodal, a monomodal, or a low H3K4me1 profile.
Figure 4.
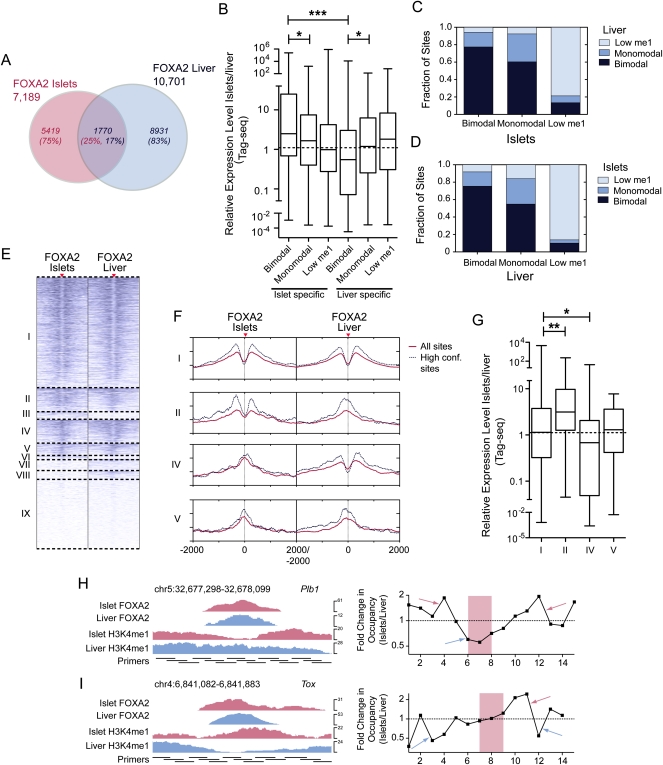
Tissue-specific FOXA2 binding and nucleosome occupancy determine tissue-specific target gene expression. (A) Venn diagram representing the proportion of sites shared between the FOXA2 islet and liver peak sets. (B) Box-whisker plot of the relative expression level (islets/liver) of expressed genes with associated islet- or liver-specific FOXA2 bimodal, monomodal, or low H3K4me1 sites. The fraction of FOXA2 sites shared in islets and liver that are bimodal, monomodal, or low H3K4me1 in the (C) FOXA2 islet or (D) liver peak sets. (E) A heatmap of H3K4me1 profiles in islets and liver for shared FOXA2 sites. Sites in group I are bimodal in both tissues; group II sites are bimodal in islet and monomodal in liver; groups III and VI–IX sites have low H3K4me1 (i.e., below the profile's global FDR threshold); group IV sites are islet monomodal and liver bimodal; and group V sites are islet and liver monomodal. (F) Profiles of average H3K4me1 read density for the shared site groups. Solid red lines are the average red densities for all peaks in a category, while dotted lines represent data from the high-confidence subset. (G) Box-whisker plot of the relative expression level (islets/liver) of expressed genes associated with peaks in groups I, II, IV, and V. Nucleosome mapping by MNase-qPCR confirms that nucleosome occupancy at transcription factor loci is altered at loci that are bimodal in islets and monomodal in liver (H), and vice versa (I). (Left) ChIP-seq enrichment profiles of the assessed loci, with the locations of the primers used for MNase-qPCR. (Right) Fold changes in nucleosome enrichment in islets/liver, calculated from MNase-qPCR results. Red arrows indicate regions that were specifically enriched for nucleosomes in islets, while blue arrows indicate regions that were specifically enriched for nucleosomes in liver. The red-highlighted regions indicate the primer pairs flanking identified FOXA2 binding motif locations and represent the relative location of FOXA2 binding. In B and G differences in expression were assessed using a Kruskall-Wallace non-parametric test with a Dunn's comparison. (*) P < 0.05; (***) P < 0.001.
Bimodal and monomodal sites are concentrated in transcriptionally active regions
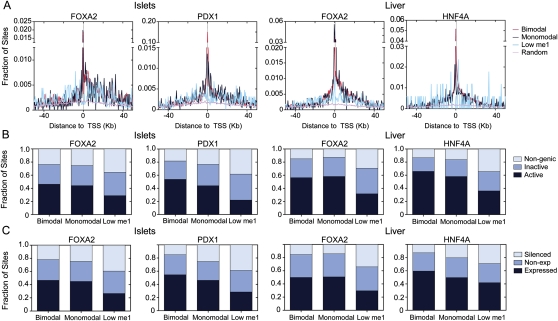
To determine the relevance of transcription factor occupied loci with bimodal, monomodal, or low H3K4me1 profiles in determining whether a gene is expressed, we plotted the distances of bimodal, monomodal, or low H3K4me1 sites to the nearest RefSeq TSS. Loci with a bimodal or monomodal profile frequently occurred proximal to TSSs, while low H3K4me1 loci showed no obvious enrichment at proximal locations (Fig. 2A). To test whether the transcription factor locus classes were associated with active transcription, we generated islet and liver Tag-seq (Morrissy et al. 2009) data and identified RefSeq genes that were expressed (tag count >5) in these tissues. A similar fraction of bimodal and monomodal loci occurred in genomic regions with actively transcribed genes (identified here by overlapping the regulatory regions ±50 kb from the TSSs of expressed genes). Loci in the low H3K4me1 class occurred significantly more often (P < 1.5 × 10−8) in genomic regions that contain no active genes (identified as contiguous regions of the genome lacking expressed genes formed by overlapping the regulatory regions ±50 kb from the TSSs of non-expressed genes), or in non-genic regions (Fig. 2B). Similarly, we asked whether bimodal, monomodal, or low H3K4me1 loci had different levels of association, directly, with expressed versus non-expressed genes. For this we grouped RefSeq genes into three categories: expressed (tag count >5), non-expressed (H3K4me3 marks at the promoter but tag count <5), and silenced (no H3K4me3 at the promoter and tag count <5). Low H3K4me1 loci were significantly more frequently (P < 0.01) associated with silenced genes (Fig. 2C) than either bimodal or monomodal loci, which were equally associated with expressed, non-expressed, and silenced genes. In sum, these results indicate that bimodal and monomodal loci more commonly occur in regions of the genome that would be expected to be involved in active gene regulation, than loci with a low H3K4me1, but that none of these locus types is sufficient to activate gene expression.
Figure 2.
Bimodal and monomodal transcription factor occupied loci have a similar genomic distribution. (A) Distributions of distance to the closest transcriptional start site (dTSS) for transcription factor occupied loci within 50 kb of a RefSeq gene TSS. (B) Fractions of bimodal, monomodal, or low H3K4me1 sites in transcriptionally active, inactive, or non-genic regions. (C) Proportion of bimodal, monomodal, or low H3K4me1 sites associated with an expressed (tag count >5), non-expressed (H3K4me3 marks at the promoter, tag count <5), or silenced (no H3K4me3 at the promoter, tag count <5) genes.
Bimodal sites are functionally active
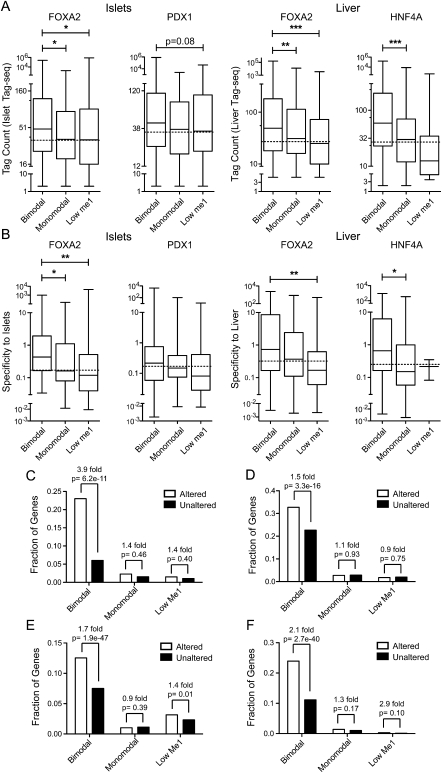
Since none of the classes of transcription factor occupied loci were deterministic of whether a gene was active or not, we assessed whether they were able to modulate the expression levels of target genes. We found that genes with associated bimodal loci had significantly higher expression (P < 0.001) than genes with associated monomodal or low H3K4me1 loci (Fig. 3A). As FOXA2, PDX1, and HNF4A have tissue-restricted expression, we predicted that genes regulated by these factors would also show tissue-restricted expression. Using expression data from 203 SAGE libraries (Siddiqui et al. 2005; Hoffman et al. 2008), we found that genes associated with bimodal loci were significantly more tissue-specific (P < 0.001) than genes associated with monomodal or low H3K4me1 loci (Fig. 3B).
Figure 3.
Bimodal sites regulate target gene expression. Box-whisker plots of islet or liver (A) Tag-seq counts or (B) specificity against 202 other mouse SAGE libraries, for expressed genes associated with a bimodal, monomodal, or low H3K4me1 site in the FOXA2 (islet or liver), PDX1, or HNF4A peak sets. Expressed genes have a tag count >5 in the particular Tag-seq library. Statistically significant differences were detected using a Kruskall-Wallis non-parametric test with a Dunn's multiple comparison correction, with a P < 0.05 threshold. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. Fractions of RefSeq genes with bimodal, monomodal, or low H3K4me1sites whose expression was significantly altered or not in (C) PancChip 6.1 (http://www.betacell.org/ma/) comparisons of Foxa2 deficient (Foxa2loxP/loxP, Pdx1-CreERT2, tamoxifen treated) versus control (Foxa2loxP/loxP, tamoxifen treated) islets (Gao et al. 2007). (D) Tag-seq analysis of islets treated with siRNA's targeting Pdx1 versus with siRNA's targeting Ppib. (E) Agilent 4×44k array comparisons of Foxa2 deficient (Foxa2loxP/loxP, Alfp-Cre) versus wild-type livers (Bochkis et al. 2009). (F) Agilent Whole Mouse Genome Microarray comparisons of Hnf4a deficient (Hnf4aloxP/loxP, Alb-Cre) versus wild-type livers (Holloway et al. 2008). P-values were determined using Fisher's exact test.
To further confirm these results, we compared the association of bimodal, monomodal, and low H3K4me1 sites with genes whose expression was significantly altered by adult-specific disruption of Foxa2, Pdx1, or Hnf4a. For this we used available microarray data from adult-specific knockout mouse models of Foxa2 in islets (Gao et al. 2007), Foxa2 in liver (Bochkis et al. 2009), and Hnf4a in liver (Holloway et al. 2008). Because no equivalent data were available from adult-specific knockout models of Pdx1, we suppressed Pdx1 using siRNAs in islets and used Tag-seq to identify genes significantly altered by Pdx1 suppression as compared to islets treated with siRNAs targeting Ppib (negative control). We identified 409 RefSeq genes up-regulated (P < 0.001, based on Audic-Claverie statistics, and >1.5-fold change) in the siPdx1 library as compared to the siPpib library, and 979 RefSeq genes as down-regulated (including Ins1, Ins2, Slc2a2, etc). From the analysis of these four data sets, we found that genes altered by Foxa2 or Hnf4a disruption, or Pdx1 suppression, were enriched in bimodal loci, but not in monomodal or low H3K4me1 loci (Fig. 3C–F). Interestingly, both up- and down-regulated genes were enriched in bimodal sites for PDX1 (1.5-fold, P = 9.7 × 10−16 up-regulated; 1.4-fold, P = 4.1 × 10−3 down-regulated), and for FOXA2 in islets (2.1-fold, P = 5.6 × 10−3 up-regulated; 5.6-fold, P = 5.6 × 10−12 down-regulated) and in liver (1.7-fold, P = 1.9 × 10−29 up-regulated; 1.6-fold, P = 1.4 × 10−26 down-regulated); whereas for HNF4A only down-regulated genes were enriched in bimodal sites (2.9-fold, P = 1.7 × 10−62). To further validate the functionality of bimodal sites, we used RT-qPCR to determine the expression levels of 41 different genes with an associated bimodal FOXA2 peak in islets treated with siRNA targeting Foxa2 (Supplemental Fig. S13). The expression levels of 33 (80%, P < 0.05) of the target genes were altered by the suppression of Foxa2, with nearly equal numbers showing increased versus decreased expression, while none of the genes without an associated peak (Neg1-3) were altered. Together, these data argue that bimodal loci are functionally active in regulating target gene expression.
Tissue-specific FOXA2 binding and nucleosome displacement determine target gene expression
FOXA1 recruitment has been suggested to drive cell-type-specific target gene expression in prostate cancer and breast cancer cell lines (Lupien et al. 2008). We wanted to determine whether this is the case for FOXA2 in islets and liver in vivo, and, additionally, to assess the relevance of the nucleosome occupancy of FOXA2-bound loci in regulating the lineage-specific expression of FOXA2 target genes. For this, we assessed the relationship between FOXA2 bimodal, monomodal, and low H3K4me1 sites with target gene expression in islets and liver. First, we identified 1770 FOXA2 sites common to both tissues, 5419 sites unique to islets, and 8931 unique to liver (Fig. 4A). We found that genes with islet-specific bimodal sites had substantially higher expression in islets (P < 0.001) than genes with liver-specific bimodal sites, and vice versa (Fig. 4B). As well, genes with islet- or liver-specific bimodal sites had significantly (P < 0.05) higher tissue-specific expression than genes with islet- or liver-specific monomodal sites (Fig. 4B). For the 1770 loci bound by FOXA2 in both islets and liver, the majority of loci that were bimodal (>75%) or low H3K4me1 (>80%) had the same profile in both tissues (Fig. 4C,D). On the other hand, >55% of the monomodal loci in islets or liver were bimodal in the alternative tissue (Fig. 4C–E). Assessing the H3K4me1 profiles of individual loci in our high-confidence data confirmed these results (Fig. 4F), and similar results were obtained for loci in both promoters and enhancers (Supplemental Fig. S14A). Correlating this with the expression of genes associated with these loci indicated that genes with loci that were bimodal in both tissues were, on average, equally expressed in both tissues (Fig. 4G). In contrast, genes with loci that were bimodal only in islets had significantly higher expression in islets (P < 0.01); while genes with loci that were bimodal only in liver had significantly higher expression in liver (P < 0.05). To verify that these site transitions were a result of nucleosome occupancy changes, we mapped nucleosome positions, in islets and liver, of representative enhancer loci that were bimodal in one tissue and monomodal in the other. These data show enrichment of nucleosomes flanking the loci in the tissue in which they were identified as having a bimodal profile, and of central nucleosomes at, or immediately adjacent to, the transcription factor binding site, in the tissue in which they were identified as having a monomodal profile (Fig. 4H,I). This confirms that the lineage-specific profile class changes identified are, at least in part, due to changes in nucleosome occupancy, although the replacement of H3 with H3.3 histones in central nucleosomes at monomodal sites may also be a contributing factor, as H3.3 is known to be enriched at transcription factor binding sites (Jin et al. 2009; He et al. 2010). Regardless, our data confirm that bimodal sites are functionally active and indicate the significance of both tissue-specific transcription factor binding and nucleosome displacement in determining tissue-specific target gene expression.
Nucleosome occupancy of transcription factor bound loci can be altered during development or in response to signaling events
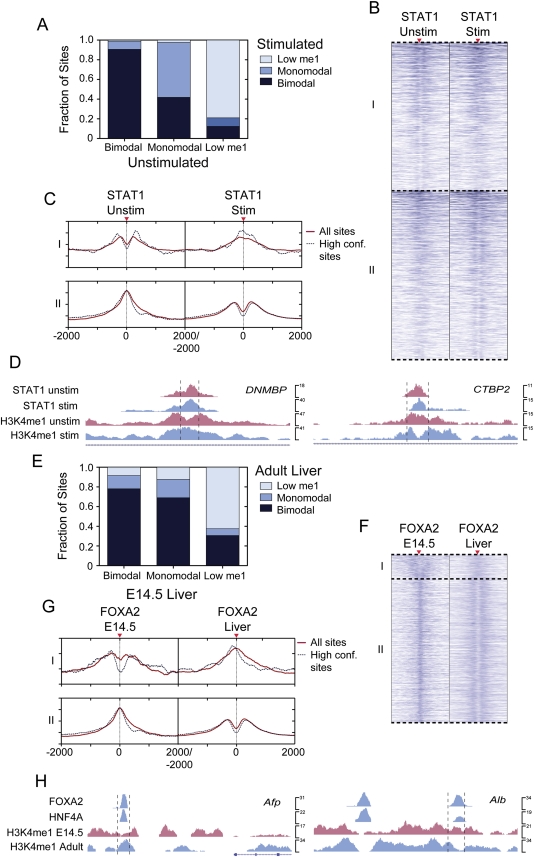
It is becoming recognized that the nucleosome occupancy of promoters and enhancers can be dynamically altered to control their function (Hogan et al. 2006; Shivaswamy et al. 2008; Ramirez-Carrozzi et al. 2009; He et al. 2010); however, the relevance of this to the control of transcription factor occupied loci has not been reported. Therefore, to determine whether the regulated nucleosome occupancy of transcription factor bound loci may be a more general mechanism for controlling locus activity, we first asked whether signaling events could regulate the nucleosome occupancy of such loci. For this, we used STAT1 binding and H3K4me1 localization data from IFNG-stimulated and unstimulated HeLa cells (Robertson et al. 2007, 2008). Of the 17,136 loci that were bound by STAT1 in both cell states, most were in the same H3K4me1 class in both conditions (Fig. 5A). However, >40% of the monomodal STAT1 occupied loci in unstimulated cells were bimodal in IFNG-stimulated cells (Fig. 5A–C). Also, ∼8% of the bimodal STAT1-occupied loci in unstimulated cells were monomodal in IFNG-stimulated cells (Fig. 5A–C). Similar results to these were obtained when we assessed loci only in promoter or enhancer regions (Supplemental Fig. S14B). Loci associated with DNMBP and CTBP2 undergo such transitions (Fig. 5D).
Figure 5.
H3K4me1-marked nucleosome occupancy regulates transcription factor bound cis-regulatory site function. Analysis of H3K4me1 transitions for transcription factor occupied loci during (A–D) an IFNG-stimulated signaling event, or (E–H) liver development. (A) STAT1 site class transitions in the IFNG-stimulated and unstimulated shared peaks, or (E) FOXA2 adult occupied loci in E14.5 hepatocytes and adult liver. Categories I and II in the heatmaps in B and F represent bimodal–monomodal and monomodal–bimodal transitions for STAT1 and FOXA2 sites, respectively. Average H3K4me1 enrichment profiles of category I or II sites are plotted in red, with dotted lines showing the average profiles for high confidence peaks. UCSC mm8 Genome Browser screenshots of representative loci that are bound by (D) STAT1 in the IFNG-stimulated and unstimulated cells that transition from bimodal to monomodal or vice versa, or (H) by FOXA2 and HNF4A in adult liver but transition from bimodal to monomodal or vice versa. The dotted lines in D and H mark 1-kb regions.
To determine whether a similar process controls the activity of transcription factor occupied loci during development, we compared the H3K4me1 profiles of FOXA2-occupied loci from E14.5 hepatocytes and adult liver. Of the FOXA2 binding sites in adult liver, 90.5% were associated with H3K4me1 at E14.5. The majority of loci that were bimodal in E14.5 hepatocytes were also bimodal in the adult (Fig. 5E), as were the majority of E14.5 monomodal sites (Fig. 5E–G), while a smaller fraction (13%) of loci that were bimodal at E14.5 were monomodal in the adult (Fig. 5E–G). As we found for STAT1, assessing only loci in promoters or enhancer regions gave similar results (Supplemental Fig. S14C). As an example, a locus ∼4 kb upstream of alpha fetoprotein (Afp) was bimodal in E14.5 hepatocytes but monomodal in adult liver (Fig. 5H). Consistent with this, Afp expression was high in developing hepatocytes but turned off in the adult liver. In contrast, a locus ∼10 kb upstream of albumin (Alb) was monomodal in E14.5 hepatocytes but bimodal in adult liver (Fig. 5H), corresponding to higher Alb expression in adult liver. Together, these data clearly indicate that signaling events and developmental programs can dynamically regulate the nucleosome occupancy of transcription factor bound loci in promoters and enhancers.
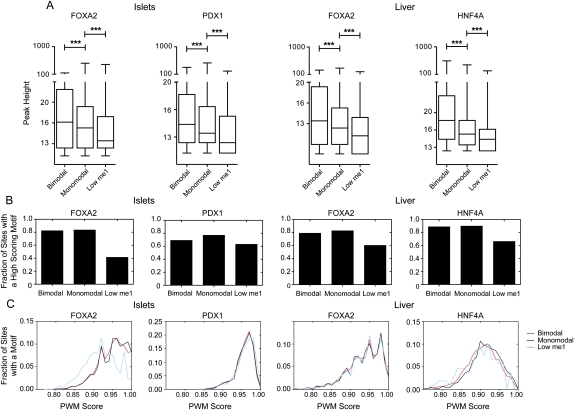
Transcription factor bound loci with bimodal H3K4me1 profiles are more highly occupied
To determine whether the different classes of transcription factor occupied loci are associated with different levels of transcription factor occupancy, we compared the peak heights, or enrichment levels, of sites in each class. Bimodal sites were significantly (P < 0.001) more enriched than monomodal sites, and monomodal sites were significantly (P < 0.01) more enriched than low H3K4me1 sites (Fig. 6A). As higher enrichment levels imply that a larger fraction of cells in the ChIP experiments contained a bound locus, this suggests that bimodal loci are more highly occupied than monomodal loci, which, in turn, are more occupied than loci with low H3K4me1. We next assessed whether these occupancy differences correlated with underlying differences in the fraction of loci containing high-scoring sequence motifs. We found that equivalent fractions of bimodal and monomodal loci, but a moderately reduced fraction of low H3K4me1 loci, contained an appropriate motif (Fig. 6B). The distribution of motif scores was also similar for the different H3K4me1 profile classes (Fig. 6C), although some reduction in the motif score distribution for low H3K4me1 FOXA2 islet sites was found. We then compared the peak heights from FOXA2 sites bound in both islets and liver. We found that sites that were bimodal in both tissues typically had similar peak heights (Supplemental Fig. S15). Sites that were bimodal in one tissue and monomodal in the other, however, were significantly taller in the tissue containing the bimodal site. These data indicate that the lower enrichment of monomodal sites over bimodal sites cannot be explained by underlying differences in motif presence or strength and suggest that further histone modifications and/or nucleosome displacement are necessary for full binding.
Figure 6.
Site modality is independent of motif presence or score. (A) Box-whisker plots of bimodal, monomodal, or low FOXA2, PDX1, or HNF4A peak heights. Statistical significance between groups was detected using a Kruskall-Wallis non-parametric test with a Dunn's post-test, with a P < 0.05 threshold. (***) P < 0.001. (B) Fraction of peaks in the bimodal, monomodal, or low site classes in the FOXA2 (islet and liver), PDX1, HNF4A peak sets that contain a high-scoring binding motif. (C) Frequency distribution of PWM scores for the three site classes for each TF peak set.
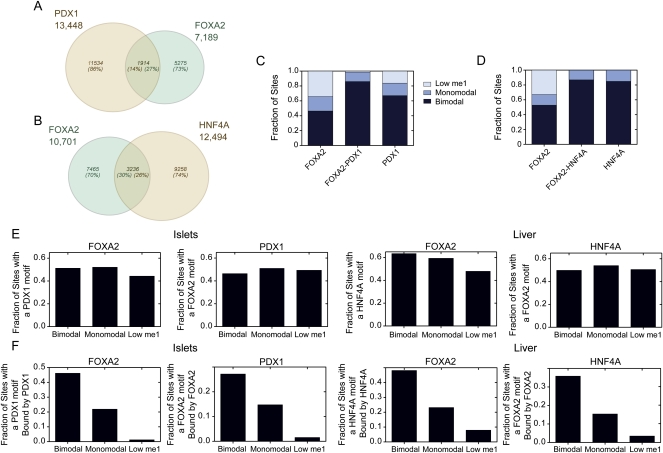
Secondary co-occupying transcription factors ensure nucleosome displacement
Previously, it was shown that differential recruitment of ESR1 and AR in breast and prostate cancer cell lines, respectively, correlates with the accessibility and functionality of FOXA1 sites (Eeckhoute et al. 2009). Based on this, we wondered whether co-binding of tissue-specific transcription factors would be more associated with loci in a bimodal state. To address this we identified 1914 loci in islets that were co-bound by both FOXA2 and PDX1, and 3236 loci in liver that were co-bound by FOXA2 and HNF4A (Fig. 7A,B). The fraction of co-bound loci in a bimodal state was increased as compared to loci singly bound by FOXA2 or PDX1 in islets, or FOXA2 in liver (Fig. 7C,D). Next, to determine whether the co-binding of secondary transcription factors was affected by H3K4me1 class type we assessed, in islets, the fraction of FOXA2-bound loci with a PDX1 motif and the fraction of PDX1-bound loci with a FOXA2 motif; and in liver, the fraction of FOXA2-bound loci with an HNF4A motif and the fraction of HNF4A-bound loci with a FOXA2 motif. In each case, a roughly equivalent fraction of bimodal, monomodal, or low H3K4me1 loci contained a motif for putative co-binding transcription factor (Fig. 7E). We then determined the fraction of loci with an appropriate motif for a collaborating transcription factor, that were actually bound by that factor, that is, the fraction of FOXA2-bound loci with a PDX1 motif bound by PDX1, and so on. We found that a reduced fraction of monomodal loci with a appropriate motif for a putative co-binding transcription factor were actively bound by that factor as compared to bimodal loci, and the fraction of actively bound, motif-containing, low H3K4me1 loci was reduced even further (Fig. 7F). Consistent with previous reports (Adams and Workman 1995), these results indicate that binding of a collaborating transcription factor is highly correlated with nucleosome-free DNA, but also indicates that the ability of collaborating factors to bind loci with appropriate motifs is determined by additional levels of control. Thus, the presence of collaborating transcription factors alone is insufficient to regulate the nucleosome status and the cis-regulatory activity of a transcription factor occupied loci. These data further implicate the ability of an occupied locus to recruit chromatin-remodeling complexes as a primary mechanism of locus control.
Figure 7.
Co-factors bind few of their available motifs at monomodal or low H3K4me1 sites. Venn diagrams showing the number of loci shared by (A) PDX1 and FOXA2 in islets or (B) FOXA2 and HNF4A in the liver. (C) Fraction of co-bound islet sites (FOXA2–PDX1) versus FOXA2 (FOXA2) only, or PDX1 (PDX1) only sites that have bimodal, monomodal, or low H3K4me1 profiles. (D) Fraction of co-bound liver sites (FOXA2–HNF4A) versus FOXA2 (FOXA2) only or HNF4A (HNF4A) only sites that have bimodal, monomodal, or low H3K4me1 profiles. (E) Fraction of bimodal, monomodal, or low H3K4me1 sites in the FOXA2 (islet and liver), PDX1, and HNF4A peak sets that contain a motif for the indicated transcription factors. (F) The fraction of sites that contain a motif for a potential TF binding partner (FOXA2 with PDX1 in islets and FOXA2 with HNF4A in liver) actually co-bound in the bimodal, monomodal, or low H3K4me1 peak sets.
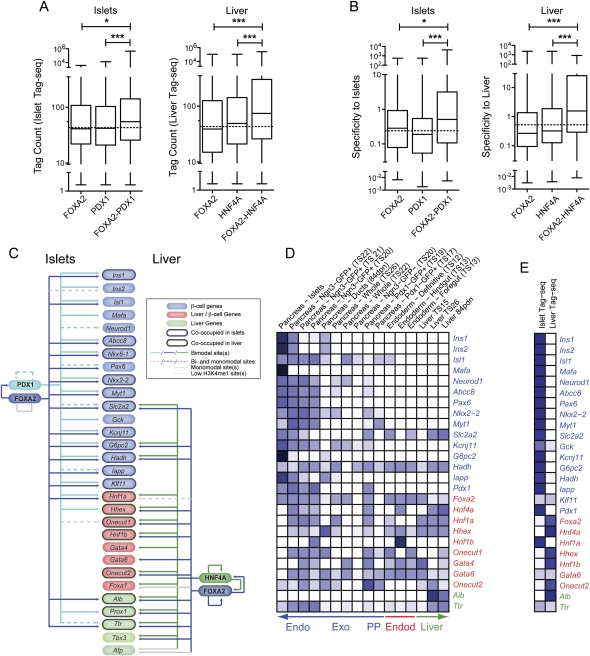
Cooperative binding at bimodal sites drives tissue-specific gene expression
We next sought to use our ability to detect functional bimodal sites to gain insight into how FOXA2 and PDX1, and FOXA2 and HNF4A cooperate to drive transcriptional networks in islets and liver. We first asked if bimodal loci co-bound by FOXA2 and PDX1 in islets, and those co-bound by FOXA2 and HNF4A in liver, had higher expression or tissue specificity than loci singly bound by FOXA2, PDX1, or HNF4A alone (Fig. 8A). In fact, genes with sites co-occupied by FOXA2 and PDX1 had a median tag count ∼1.4× higher in islets than genes with a FOXA2 or PDX1 singly bound site (P < 0.01), while genes with associated sites co-occupied by FOXA2 and HNF4A sites had a median tag count ∼1.7× higher in liver than genes associated with singly bound loci (P < 0.001). These results indicate that site co-occupancy increases the level of activation conferred to target genes. We next determined whether this increase in expression translated into increased tissue specificity. Genes with FOXA2–PDX1 co-occupied sites had a median specificity to islets ∼2.4× higher than genes with a FOXA2 or PDX1 singly bound site (P < 0.001) (Fig. 8B), and genes with a FOXA2–HNF4A co-occupied site in liver had a median specificity ∼5.2× higher than genes with a FOXA2 or HNF4A site alone (P < 0.001). These data indicate that site co-occupancy plays a significant role in conferring tissue-specific target gene expression.
Figure 8.
Co-occupancy at bimodal sites drives tissue-specific gene expression. Box-whisker plots of islet or liver (A) Tag-seq counts or (B) specificity against 202 other mouse SAGE libraries, for expressed genes associated with bimodal sites either singly occupied by FOXA2, PDX1, or HNF4A, or co-occupied by FOXA2–PDX1 in islets, or FOXA2–HNF4A in liver. Statistically significant differences were detected using a Kruskall-Wallis non-parametric test with a Dunn's multiple comparison correction, with a P < 0.05 threshold. (*) P < 0.05; (***) P < 0.001. Bimodal FOXA2, PDX1, and/or HNF4A sites are commonly associated with islet and liver critical genes. (C) A schematic representing the presence of bimodal, monomodal, and low H3K4me1 FOXA2 (blue) and PDX1 (aqua) sites in islets (left), and of FOXA2 (blue) and HNF4A (green) sites in liver (right) associated with genes critical to islets (blue ovals), islets and liver (red ovals), or liver (green ovals) specification and/or function. The presence of one or more FOXA2, PDX1, or HNF4A bimodal sites associated with a gene is indicated by a solid colored line. The presence of one or more bimodal and one or more monomodal sites associated with a gene is indicated by a dotted colored line; the presence of monomodal sites only by a solid gray line, and of low H3K4me1 sites only by a dotted gray line. (D,E) Heatmaps of the relative expression levels (tag counts) of the indicated factors essential for proper pancreas and/or liver development or function in (D) SAGE libraries derived from endoderm, developing pancreas and liver, and adult pancreas and liver tissues, or (E) in the adult islet or liver Tag-seq libraries. Darker blue shades represent higher relative expression levels. Unambiguous tags above minimum count thresholds (four of the SAGE libraries and six for the Tag-seq libraries) for Foxa1, Nkx6-1, Prox1, Tbx3, and Afp could not be identified in the SAGE libraries.
To gain insight into the biological significance of these results, we asked if genes with co-occupied sites were enriched for association with specific KEGG pathway terms, as compared with genes with singly bound sites. Focusing on expressed genes, we found that genes with FOXA2–PDX1 co-occupied sites in islets were more frequently associated with the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways “Neuroactive ligand–receptor interaction” (5.5-fold increase, P = 2.35 × 10−5), “Arginine and proline metabolism” (4.6-fold increase, P = 5.15 × 10−4), and “Maturity onset diabetes of the young” (3.7-fold increase, P = 9.39 × 10−3) than singly bound sites. Genes with FOXA2–HNF4A co-occupied sites in liver were more frequently associated with the KEGG pathways “PPAR signaling pathway” (3.8-fold increase, P = 5.30 × 10−4), “Fatty acid metabolism” (3.1-fold increase, P = 7.11 × 10−3), and “Arginine and proline metabolism” (7.2-fold increase, P = 5.92 × 10−4).
We next asked how these factors cooperate to drive the expression of genes that play essential roles in islet and/or liver specification or function. We found bimodal co-occupied FOXA2–PDX1 sites associated with 11 of the 17 genes assessed that are specifically involved in islet function or development, with no such sites associated with genes involved in islet and liver development, or with genes involved in liver development or function only. In contrast, bimodal co-occupied FOXA2–HNF4A sites were associated with one of the 17 genes specifically involved in islet function or development, five of eight sites associated with genes involved in islet and liver development, and three of five genes involved in only liver development or function (Fig. 8C). These results correlate well with the expression profiles of these genes (Fig. 8D,E), which show that genes involved specifically in islet function or development tend to have high relative expression in islets and no expression in endoderm or liver, while genes involved in both islet and liver development tend to be expressed only in developing pancreas tissues and not in adult islets, but are commonly expressed in adult liver. Together these data implicate FOXA2 and PDX1 collaborations in initiating and regulating the expression of genes involved in beta-cell function, and FOXA2 and HNF4A collaborations in maintaining the expression in the adult liver of genes that were initially coexpressed in the developing pancreas and liver.
Discussion
We identified between 7189 and 13,448 DNA-binding sites for FOXA2, PDX1, and HNF4A in islets and/or liver in the adult mouse. Based on previous studies, we anticipated that many of these loci would be inactive. To discriminate functionally active loci, we compared our transcription factor binding data with localization data for H3K4me1 and H3K4me3, which are associated with active genes and transcription factor binding sites (Barski et al. 2007; Heintzman et al. 2007; Robertson et al. 2008). We found that H3K4me1 enrichment profiles discriminate three classes of transcription factor occupied loci: those with a bimodal profile, a monomodal profile, or with little or no associated H3K4me1. Bimodal sites appear to be flanked by a pair of H3K4me1-marked nucleosomes, and similarly H3K4me2-marked nucleosomes have been found to flank FOXA1 sites in LNCaP cells (He et al. 2010). In contrast, monomodal sites appear to contain centrally located H3K4me1-marked nucleosomes, indicating that transcription factor binding at these sites occurs within nucleosome-associated DNA. We are unable to determine the positioning or presence of nucleosomes at sites with little or no associated H3K4me1.
As both bimodal and monomodal loci are found within regions of the genome undergoing active transcription, are near expressed genes, and as they are typically associated with H3K4me1 and/or H3K4me3 (Barski et al. 2007; Wang et al. 2008), we suggest that both bimodal and monomodal sites generally occur within open chromatin. While neither of these locus types appears able to induce genes to be expressed, several lines of evidence indicate that bimodal loci can regulate associated expressed genes. For instance, bimodal sites are associated with genes that are in general significantly more highly expressed and tissue-specific. Also, genes altered by disruption or suppression of Foxa2, Pdx1, or Hnf4a were enriched in bimodal sites. Furthermore, genes associated with loci occupied by FOXA2 in both islets and liver, but that were bimodal in one tissue and monomodal in the other, had increased expression in the tissue containing the bimodal locus. Together, these results indicate that bimodal loci can modulate gene expression levels and that H3K4me1 profiles provide an effective method to discriminate such loci.
It is known that some yeast transcription factors can interact with nucleosomal DNA in vitro and at promoters in vivo (Adams and Workman 1995; Albert et al. 2007; Koerber et al. 2009), possibly due to binding at DNA sequences found within nucleosome exit or entry points, or at sequences exposed due to rotational phasing on the nucleosome surface (Albert et al. 2007; Jiang and Pugh 2009; Koerber et al. 2009). However, in mammalian systems, the ability to interact with nucleosomal DNA has been considered one of the defining features of pioneer transcription factors, such as the FOXA proteins (Cirillo et al. 1998, 2002; Sekiya and Zaret 2007; Sekiya et al. 2009). FOXA1 is the only mammalian transcription factor that has previously been shown to interact directly with nucleosomal DNA in vivo (Chaya et al. 2001). Here, we show that 10%–20% of FOXA2, PDX1, HNF4A, and STAT1 sites occur within DNA associated with H3K4me1-marked nucleosomes. Although we find that such loci are functionally inactive, they are often converted to an active nucleosome-flanked state by lineage, signaling, or developmental-stage-specific factors, regardless of whether they are in a promoter or enhancer, and likely represent loci that are poised for activity. Likewise, we find that signaling and developmental-stage-specific factors can induce nucleosomes to re-form, or re-position, inactivating loci while transcription factor occupancy is maintained. These data, taken together with our expression analyses, indicate that tissue-specific gene expression can be dictated by tissue-specific transcription factor binding and by the tissue-specific recruitment of nucleosome-remodeling complexes (e.g., SWI/SNF) (Hogan et al. 2006; Schwabish and Struhl 2007) to transcription factor occupied loci. Transcription factor binding (Gutierrez et al. 2007) and the presence of appropriate histone modifications (Hogan et al. 2006; Jiang and Pugh 2009) have been implicated as mechanisms for targeting nucleosome-remodeling complexes to specific sites in the genome. Our data, though, suggest that neither transcription factor binding nor the presence of H3K4me1 is sufficient to recruit such complexes to the monomodal loci identified here.
Although similar fractions of the transcription factors assessed were bound at monomodal loci, they showed significantly different abilities to bind loci with low H3K4me1 levels. Given their genomic distribution and lack of associated H3K4me1, we propose that low H3K4me1 loci usually occur within closed chromatin. In support of this, our data show that these loci are typically functionally inactive and not co-occupied, in agreement with previous studies (Lupien et al. 2008; Eeckhoute et al. 2009). These sites may represent pioneer factor activity. FOXA2 is a well-established pioneer factor (Sekiya et al. 2009), while HNF4A and PDX1 are not. Consistent with this, a substantial fraction of FOXA2 sites are in the low H3K4me1 class, while virtually no HNF4A sites are in this class. In contrast, a considerable fraction of PDX1 sites are in the low H3K4me1 class. PDX1 does, though, interact with PBX1 (Liu et al. 2001; Oliver-Krasinski et al. 2009), which can bind closed chromatin (Berkes et al. 2004), and it is possible that these sites represent pioneer activity of one of PDX1's binding partners. Thus, although our data suggest that the ability of mammalian transcription factors to interact with nucleosome-associated DNA is widespread, the ability to do so may rely on the presence of H3K4me1 or other “active” histone marks, and pioneer activity may be required for a transcription factor to associate with sites within regions of the genome devoid of such marks (Sekiya and Zaret 2007).
Using our ability to discriminate functional sites, we show that genes associated with bimodal sites that are bound by PDX1, FOXA2, or HNF4A alone are not strongly tissue-specific, despite the fact that these transcription factors themselves have relatively restricted expression profiles. On the other hand, we find that genes with associated FOXA2–PDX1 or FOXA2–HNF4A co-occupied bimodal sites are highly tissue-specific. These data globally demonstrate how transcription factors that have diverse expression profiles can function cooperatively at cis-regulatory loci to drive highly tissue-specific target gene expression. Consistent with this, we find that FOXA2–PDX1 and FOXA2–HNF4A co-occupied sites are enriched for genes involved in islet- and liver-specific functions, respectively. For example, FOXA2–PDX1 co-occupied sites are enriched for genes involved in maturity onset diabetes of the young, such as Nkx2-2, Ins1, Iapp, and Slc2a2. Similarly, FOXA2–HNF4A co-occupied sites are enriched in genes involved in PPAR signaling and fatty acid metabolism, both of which are important components of liver function. It is interesting that both FOXA2–PDX1 co-occupied sites in islets and FOXA2–HNF4A co-occupied sites in liver are enriched for genes involved in arginine and proline metabolism. Arginine is a precursor of nitric oxide (NO), which plays critical roles in controlling insulin secretion from beta-cells, beta-cell apoptosis, and islet inflammation, as well as in controlling liver regeneration, inflammation, and blood flow (Schmidt et al. 1992; Shimabukuro et al. 1997; Wu and Morris 1998; Adeghate and Parvez 2000; Rockey and Shah 2004). Thus, our data suggest a novel role for FOXA2–PDX1 and FOXA2–HNF4A collaborations in controlling these processes by directly regulating genes involved in arginine metabolism.
Our data indicate that mammalian transcription factors commonly bind sites within DNA associated with H3K4me1-marked nucleosomes in vivo, and that the regulated gain and loss of such nucleosomes at transcription factor occupied loci controls their function. Extending this, we demonstrate that by considering both the H3K4me1-marked nucleosome occupancy and H3K4me1 levels of transcription factor occupied loci, we can discriminate loci that are functionally active from those poised to become active, and from those that reflect pioneer-like activity. By using this approach to discriminate active binding sites, we show that FOXA2–PDX1 and FOXA2–HNF4A co-occupancy at bimodal sites drives islet- and liver-specific gene expression, respectively. These advances should greatly enhance the utility of global transcription factor binding data in generating biological insights.
Methods
ChIP-seq library generation and identification of enriched regions
For each islet ChIP experiment, islets from at least 10 adult (8–10 wk old) ICR (for transcription factor libraries) or C57BL6/J (for histone methylation libraries) mice were purified by collagenase digestion and gradient centrifugation, with subsequent hand picking. Perfused liver or purified E14.5 hepatocytes were obtained from C57BL6/J, and ChIP was performed as described (Wederell et al. 2008) using 3 μg of anti-FOXA2 (Santa Cruz), anti-PDX1 (Upstate), anti-HNF4A (Santa Cruz), anti-H3K4me1 (Santa Cruz), anti-H3K4me3 (Santa Cruz), or normal rabbit IgG (Santa Cruz). DNA from triplicate pooled ChIP experiments was purified by 8% PAGE to obtain 100–300-bp fragments and sequenced on an Illumina 1G sequencer as previously reported (Robertson et al. 2007).
To identify enriched regions, Illumina 1G 36-bp-long sequence reads were aligned to the genome (mm8) using Eland and directionally extended to 200 bp in length. Clusters of overlapping extended reads were defined as enriched regions as described in Robertson et al. (2008). Enriched regions were thresholded using a false discovery rate of 0.01, and refined further by trimming at 20% of their maximal height, in order to remove low height flanks, and separate composite regions. Regions that overlapped sites identified using an islet or liver input control sample, as appropriate, were removed.
H3K4me1 or me3 enrichment profiles were obtained by quantifying the number of sequence reads occurring in 5-bp bins ±2 kb from loci of maximum enrichment for the FOXA2, PDX1, or HNF4A enriched region data sets. Sites with low H3K4me1 levels were first discriminated using a cumulative distribution plot of the total number of H3K4me1 sequence reads in each ±2-kb window (Supplemental Fig. S9A). Subsequently, bimodal and monomodal sites were discriminated based on whether the H3K4me1 profiles formed a stereotypical peak–trough–peak shape or not (Supplemental Fig. S9B).
Association of transcription factor enriched regions to genes
Enriched regions were associated with genes by identifying the closest annotated RefSeq gene within 50 kb. Enriched regions >50 kb from any annotated RefSeq gene were not associated with any gene, using alternative site-gene association distance gave similar results (Supplemental Fig. S16). Enriched regions were associated with only the closest gene. While some binding sites may regulate several neighboring genes, this is likely not the general case, and, while the approach described would miss such interactions, it eliminates a large number of incorrect associations. Sites were considered to be in promoter regions if they fell within 2 kb of a RefSeq TSS, within a enhancer if they fell between 2 kb and 50 kb from a RefSeq TSS, and distal if they were >50 kb from a RefSeq TSS.
To identify overlapping and co-occupied sites, locations of sites of maximal enrichment were compared for FOXA2 sites from islets and liver; FOXA2 and PDX1 sites in islets, FOXA2 and HNF4A sites in liver (Supplemental Fig. S17). Sites were considered to be overlapping if the distance between their locations of maximal enrichment was <100 bp. For STAT1, unstimulated and stimulated shared sites were identified per Robertson et al. (2007).
Generation and analysis of Tag-seq and SAGE libraries
Total RNA was isolated from hand-picked islets and liver using TRIzol, and RNA quality was assessed using a Bioanalyser (Agilent). DNase I–treated total RNA was used to construct Tag-seq libraries, which were sequenced to a depth of 7,481,000 tags for islet and 9,510,780 for liver. For the islet library, cDNA was prepared according to the SAGE-Lite method, with 20 cycles of amplification. Both samples were processed using a modified LongSAGE protocol (Morrissy et al. 2009). Seventeen-base-pair SAGE tags were extracted and mapped to the NCBI Build 36 (mm8) reference mouse genome. Tags mapping to more than one transcript in a sense direction, mapping only in an antisense direction, or not mapping were eliminated. Tag counts in the libraries ranged from 1 (∼1 × 10−7% of the library) to 867,668 (∼12% of the library for Ins2 in islets). Tags with a count of <6 in both libraries were considered to be not expressed. For genes producing more than one tag, the tag counts were added. Seven thousand ninety-eight (7098) unique RefSeqs were identified in islets, and 6098 in liver. Four thousand eight hundred and ten (4810) RefSeq transcripts were expressed in both libraries. The counts of the identified RefSeq transcripts were compared in the islet and liver libraries to obtain their relative expression levels.
All SAGE libraries were generated as part of the Mouse Atlas of Gene Expression project (Siddiqui et al. 2005; Hoffman et al. 2008). Specificity of a gene to a library was calculated using the following equation:
where Mr is the ratio of the counts of the tag in the library of interest over the mean of the counts of the tag in all other libraries, Ac is the absolute count of the tag in the library of interest, and Lc is the number of libraries the tag is found in.
Identification of genes altered by Pdx1 suppression
Single-cell suspensions were generated from hand-picked islets and plated at 100,000 cells per well. Cells were cultured overnight at 37°C, 5.2% CO2 in RPMI media containing 10% FBS and 1% L-glutamine. ON-TARGETplus siRNAs pools targeting Pdx1 or Pbip (Dharmacom) were combined with siGLO transfection indicator and used to transfect the cultured islets with DharmaFECT4 (Dharmacon) transfection reagent according to the manufacturer's recommendation. Transfected cells were cultured for 48 h and subsequently subjected to FACS sorting to isolate siGLO-positive cells. After confirmation of Pdx1 knockdown, the material collected from three separate experiments was pooled and used for Tag-seq-lite library construction as described above. The counts of identified RefSeq genes were subsequently compared between the libraries generated using siRNA's targeting Pdx1 or Ppib. RefSeq genes were considered significantly altered if their counts were significantly different (P < 0.001, based on Audic-Claverie statistics) in the siPdx1 library as compared to the siPpib library and if the change was >1.5-fold.
Nucleosome mapping by MNase-qPCR
To isolate nuclei from hand-picked islets and from livers, we treated single-cell suspensions of the tissues with cell lysis buffer (10 mM Tris-HCl pH 8, 10 mM NaCl, 3 mM MgCl2, 0.5% Igepal [w/v], protease inhibitor) to obtain nuclei pellets. These pellets were subsequently re-suspended in 250 μL of MNase digestion buffer (0.32 M sucrose, 50 mM Tris-HCl pH 7.5, 4 mM MgCl2, 1 mM CaCl2, protease inhibitor), homogenized by passaging through a 26G needle, and treated with 3 μL of MNase (Sigma; 1 U/5 μL) for 7 min at 37°C. MNase digestion was halted by addition of EDTA to 10 mM. One milliliter of lysis buffer (1% SDS [w/v], 5 mM EDTA, 50 mM Tris-HCl pH 8, protease inhibitor) was added and the tubes incubated for 60 min on ice with vortexing at 10-min intervals. Tubes were centrifuged, and the chromatin-containing supernatant was removed to a new tube. The MNase-digested chromatin was then purified by phenol/chloroform with ethanol precipitation and run on a 2% gel. The mononucleosome-sized (140–220 bp) fragments were then excised from the gel and column-purified (QIAGEN). Next, the mononucleosomal DNA obtained was characterized by qPCR. For each loci (∼800 bp), a tiling series of qPCR primers was used, each producing a product between 85 and 95 bp as outlined in Figure 1D and Figure 4, H and I. To determine the enrichment level for each primer pair, Ct values obtained from the mononucleosomal DNA were normalized against values obtained using genomic DNA and compared to a reference primer pair that was selected for each locus.
Motif discovery and identification of high-confidence positive weight matrix (PWMs)
Motif discovery analyses were performed using an extended version of the GADEM package (Li 2009) in which a PWM was used as the starting point for each run. Results are reported for FOXA2, HNF4A, and PDX1 motifs (see Supplemental Methods). The 10-mer FOXA2, 11-mer PDX1, and 13-mer HNF4A sequence motifs were then used to identify enriched TF regions with high-confidence PWMs. We scanned the reported PWMs in TF-enriched regions with a custom Java algorithm, largely as described in Wederell et al. (2008). When an enriched region contained multiple sites and scores, we retained only the highest score.
Acknowledgments
We acknowledge the Genome Sciences Centre sequencing, bioinformatics, and SAGE library construction teams, and Joanne Johnson and Amanda Kotzer for project management. M.A.M., P.A.H., and S.J.M.J. are Senior Scholars of the Michael Smith Foundation for Health Research. L.L. was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Z01-ES-101765). Funding was provided by Genome Canada, Genome British Columbia, the Juvenile Diabetes Research Foundation and the Canadian Institutes for Health Research, with infrastructure support provided by the British Columbia Cancer Foundation.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra) under accession no. SRA008281 and are also available for download at http://www.bcgsc.ca/data/histone-modification.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.104356.109.
References
- Adams CC, Workman JL 1995. Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Mol Cell Biol 15: 1405–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeghate E, Parvez SH 2000. Nitric oxide and neuronal and pancreatic beta cell death. Toxicology 153: 143–156 [DOI] [PubMed] [Google Scholar]
- Ahlgren U, Jonsson J, Jonsson L, Simu K, Edlund H 1998. Beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev 12: 1763–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446: 572–576 [DOI] [PubMed] [Google Scholar]
- Bailly A, Torres-Padilla ME, Tinel AP, Weiss MC 2001. An enhancer element 6 kb upstream of the mouse HNF4Alpha1 promoter is activated by glucocorticoids and liver-enriched transcription factors. Nucleic Acids Res 29: 3495–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Battle MA, Konopka G, Parviz F, Gaggl AL, Yang C, Sladek FM, Duncan SA 2006. Hepatocyte nuclear factor 4alpha orchestrates expression of cell adhesion proteins during the epithelial transformation of the developing liver. Proc Natl Acad Sci 103: 8419–8424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell 14: 465–477 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Bochkis IM, Schug J, Rubins NE, Chopra AR, O'Malley BW, Kaestner KH 2009. Foxa2-dependent hepatic gene regulatory networks depend on physiological state. Physiol Genomics 38: 186–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaya D, Hayamizu T, Bustin M, Zaret KS 2001. Transcription factor FoxA (HNF3) on a nucleosome at an enhancer complex in liver chromatin. J Biol Chem 276: 44385–44389 [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega V, Wong E, Orlov Y, Zhang W, Jiang J 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Cirillo LA, McPherson CE, Bossard P, Stevens K, Cherian S, Shim EY, Clark KL, Burley SK, Zaret KS 1998. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J 17: 244–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS 2002. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell 9: 279–289 [DOI] [PubMed] [Google Scholar]
- Eeckhoute J, Lupien M, Meyer CA, Verzi M, Shivdasani R, Liu XS, Brown M 2009. Cell-type selective chromatin remodeling defines the active subset of FOXA1-bound enhancers. Genome Res 19: 372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N, White P, Doliba N, Golson ML, Matschinsky FM, Kaestner KH 2007. FOXA2 controls vesicle docking and insulin secretion in mature beta cells. Cell Metab 6: 267–279 [DOI] [PubMed] [Google Scholar]
- Gao N, Lelay J, Vatamaniuk MZ, Rieck S, Friedman JR, Kaestner KH 2008. Dynamic regulation of PDX1 enhancers by Foxa1 and FOXA2 is essential for pancreas development. Genes Dev 22: 3435–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrish K, Cissell MA, Stein R 2001. The role of hepatic nuclear factor 1 alpha and PDX-1 in transcriptional regulation of the pdx-1 gene. J Biol Chem 276: 47775–47784 [DOI] [PubMed] [Google Scholar]
- Gutierrez JL, Chandy M, Carrozza MJ, Workman JL 2007. Activation domains drive nucleosome eviction by SWI/SNF. EMBO J 26: 730–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ 2001. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol 21: 1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. 2010. Nucleosome dynamics define transcriptional enhancers. Nat Genet 42: 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman BG, Jones SJ 2009. Genome-wide identification of DNA–protein interactions using chromatin immunoprecipitation coupled with flow cell sequencing. J Endocrinol 201: 1–13 [DOI] [PubMed] [Google Scholar]
- Hoffman BG, Zavaglia B, Witzsche J, Ruiz de Algara T, Beach M, Hoodless PA, Jones SJ, Marra MA, Helgason CD 2008. Identification of transcripts with enriched expression in the developing and adult pancreas. Genome Biol 9: R99 doi: 10.1186/gb-2008-9-6-r99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan GJ, Lee CK, Lieb JD 2006. Cell cycle-specified fluctuation of nucleosome occupancy at gene promoters. PLoS Genet 2: e158 doi: 10.1371/journal.pgen.0020158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway MG, Miles GD, Dombkowski AA, Waxman DJ 2008. Liver-specific hepatocyte nuclear factor-4alpha deficiency: Greater impact on gene expression in male than in female mouse liver. Mol Endocrinol 22: 1274–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J 2004. Gene regulatory factors in pancreatic development. Dev Dyn 229: 176–200 [DOI] [PubMed] [Google Scholar]
- Jiang C, Pugh BF 2009. Nucleosome positioning and gene regulation: Advances through genomics. Nat Rev Genet 10: 161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G 2009. H3.3/H2A.Z double variant containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet 41: 941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson J, Carlsson L, Edlund T, Edlund H 1994. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 371: 606–609 [DOI] [PubMed] [Google Scholar]
- Koerber RT, Rhee HS, Jiang C, Pugh BF 2009. Interaction of transcriptional regulators with specific nucleosomes across the Saccharomyces genome. Mol Cell 35: 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Friedman JR, Fulmer JT, Kaestner KH 2005. The initiation of liver development is dependent on Foxa transcription factors. Nature 435: 944–947 [DOI] [PubMed] [Google Scholar]
- Li L 2009. GADEM: A genetic algorithm guided formation of spaced dyads coupled with an EM algorithm for motif discovery. J Comput Biol 16: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, et al. 2008. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol 6: e27 doi: 10.1371/journal.pbio.0060027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, MacDonald RJ, Swift GH 2001. DNA binding and transcriptional activation by a PDX1.PBX1b.MEIS2b trimer and cooperation with a pancreas-specific basic helix–loop–helix complex. J Biol Chem 276: 17985–17993 [DOI] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M 2008. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132: 958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. 2008. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134: 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissy AS, Morin RD, Delaney A, Zeng T, McDonald H, Jones S, Zhao Y, Hirst M, Marra MA 2009. Next-generation tag sequencing for cancer gene expression profiling. Genome Res 19: 1825–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson H, Karlsson K, Edlund T 1993. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J 12: 4251–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver-Krasinski JM, Kasner MT, Yang J, Crutchlow MF, Rustgi AK, Kaestner KH, Stoffers DA 2009. The diabetes gene PDX1 regulates the transcriptional network of pancreatic endocrine progenitor cells in mice. J Clin Invest 119: 1888–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi V, Braas D, Bhatt D, Cheng CS, Hong C, Doty K, Black J, Hoffmann A, Carey M, Smale ST 2009. A Unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 138: 114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. 2007. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods 4: 651–657 [DOI] [PubMed] [Google Scholar]
- Robertson AG, Bilenky M, Tam A, Zhao Y, Zeng T, Thiessen N, Cezard T, Fejes AP, Wederell ED, Cullum R, et al. 2008. Genome wide relationship between histone H3 lysine 4 mono- and tri-methylation and transcription factor binding. Genome Res 18: 1906–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockey DC, Shah V 2004. Nitric oxide biology and the liver: Report of an AASLD research workshop. Hepatology 39: 250–257 [DOI] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Cui K, Zhao K 2006. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci 103: 15782–15787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HH, Warner TD, Ishii K, Sheng H, Murad F 1992. Insulin secretion from pancreatic B cells caused by L-arginine-derived nitrogen oxides. Science 255: 721–723 [DOI] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K 2007. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol Cell Biol 27: 6987–6995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T, Zaret KS 2007. Repression by Groucho/TLE/Grg proteins: Genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol Cell 28: 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T, Muthurajan UM, Luger K, Tulin AV, Zaret KS 2009. Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev 23: 804–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimabukuro M, Ohneda M, Lee Y, Unger RH 1997. Role of nitric oxide in obesity-induced beta cell disease. J Clin Invest 100: 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR 2008. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol 6: e65 doi: 10.1371/journal.pbio.0060065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui AS, Khattra J, Delaney AD, Zhao Y, Astell C, Asano J, Babakaiff R, Barber S, Beland J, Bohacec S, et al. 2005. A mouse atlas of gene expression: Large-scale digital gene-expression profiles from precisely defined developing C57BL/6J mouse tissues and cells. Proc Natl Acad Sci 102: 18485–18490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson P, Williams C, Lundeberg J, Rydén P, Bergqvist I, Edlund H 2007. Gene array identification of Ipf1/PDX1−/− regulated genes in pancreatic progenitor cells. BMC Dev Biol 7: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift GH, Liu Y, Rose SD, Bischof LJ, Steelman S, Buchberg AM, Wright CV, MacDonald RJ 1998. An endocrine-exocrine switch in the activity of the pancreatic homeodomain protein PDX1 through formation of a trimeric complex with PBX1b and MRG1 (MEIS2). Mol Cell Biol 18: 5109–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wederell ED, Bilenky M, Cullum R, Thiessen N, Dagpinar M, Delaney A, Varhol R, Zhao Y, Zeng T, Bernier B, et al. 2008. Global analysis of in vivo FOXA2-binding sites in mouse adult liver using massively parallel sequencing. Nucleic Acids Res 36: 4549–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Morris SM Jr 1998. Arginine metabolism: Nitric oxide and beyond. Biochem J 336: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS 2008. Genetic programming of liver and pancreas progenitors: Lessons for stem-cell differentiation. Nat Rev Genet 9: 329–340 [DOI] [PubMed] [Google Scholar]
- Zaret KS, Grompe M 2008. Generation and regeneration of cells of the liver and pancreas. Science 322: 1490–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Zhao K, Mehler MF 2009. Profiling RE1/REST-mediated histone modifications in the human genome. Genome Biol 10: R9 doi: 10.1186/gb-2009-10-1-r9 [DOI] [PMC free article] [PubMed] [Google Scholar]