Abstract
KLF1 regulates a diverse suite of genes to direct erythroid cell differentiation from bipotent progenitors. To determine the local cis-regulatory contexts and transcription factor networks in which KLF1 operates, we performed KLF1 ChIP-seq in the mouse. We found at least 945 sites in the genome of E14.5 fetal liver erythroid cells which are occupied by endogenous KLF1. Many of these recovered sites reside in erythroid gene promoters such as Hbb-b1, but the majority are distant to any known gene. Our data suggests KLF1 directly regulates most aspects of terminal erythroid differentiation including production of alpha- and beta-globin protein chains, heme biosynthesis, coordination of proliferation and anti-apoptotic pathways, and construction of the red cell membrane and cytoskeleton by functioning primarily as a transcriptional activator. Additionally, we suggest new mechanisms for KLF1 cooperation with other transcription factors, in particular the erythroid transcription factor GATA1, to maintain homeostasis in the erythroid compartment.
Erythropoiesis, the production of red blood cells (erythrocytes), is characterized by an intricate coordination between proliferation and differentiation of hematopoietic stem cells into mature enucleated cells (Koury et al. 2002). As differentiation proceeds, there are well-described changes in cell surface protein expression (e.g., CD71 and Ter119), reduction in cell size, progressive hemoglobinization, and nuclear condensation which finally results in extrusion of the nucleus, RNA, and mitochondria (Cantor and Orkin 2002; Zhang et al. 2003; Testa 2004; Fraser et al. 2007). During erythropoiesis there is also an activation of red cell–specific biochemical pathways to harness energy in the absence of mitochondria (i.e., anaerobic glycolysis) and to generate antioxidant small molecules to protect the red cell against oxidant stressors. The coordination of this complex biology and biochemistry remains poorly understood.
A relatively small number of transcription factors are critical for erythropoiesis (Cantor and Orkin 2002); two of the best understood are GATA1 and KLF1 (formerly known as EKLF), which bind to GATA (5′-WGATAR-3′) and extended CACC (5′-CCN-CNC-CCN-3′) DNA motifs in the promoters, enhancers, and locus control regions (LCRs) of many erythroid genes (Weiss and Orkin 1995; Perkins 1999). Our current understanding of GATA1- and KLF1-regulated genes has in the most part arisen from gene knockout studies (Pevny et al. 1991; Nuez et al. 1995; Perkins et al. 1995; Fujiwara et al. 1996), expression profiling experiments (Welch et al. 2004; Drissen et al. 2005; Hodge et al. 2006; Pilon et al. 2008), and gene-by-gene promoter-reporter studies. These approaches are likely to have elucidated only a small subset of the true direct target genes for these factors. This has become particularly evident through recent global studies of in vivo GATA1 binding that show it underpins extensive erythroid gene regulatory networks (Cheng et al. 2009; Fujiwara et al. 2009; Yu et al. 2009).
KLF1 is the founding member of a family of 17 transcription factors in mammals, which is defined by the presence of three highly similar C2H2 type zinc fingers at the C terminus (Miller and Bieker 1993; van Vliet et al. 2006). Expression of Klf1 is remarkably restricted to erythroid cells and their precursors (Miller and Bieker 1993) and is critical in erythroid versus megakaryocyte lineage choice (Starck et al. 2003; Siatecka et al. 2007; Tallack and Perkins 2010). Klf1−/− mice die from anemia by E15.5, with severe defects in differentiation, hemoglobinization, enucleation, and membrane-cytoskeleton organization of red blood cells (Nuez et al. 1995; Perkins et al. 1995). Direct transcriptional target genes of KLF1 are few to date, but loss of some are likely to explain these phenotypes, at least in part. These KLF1 target genes include Hbb-b1 (beta major globin), Ahsp, Epb4.9 (also known as Dematin), Klf3 (also known as Bklf), E2f2, and Cdkn2c (also known as p18INK4c) (Miller and Bieker 1993; Nuez et al. 1995; Perkins et al. 1995; Drissen et al. 2005; Hodge et al. 2006; Pilon et al. 2006, 2008; Funnell et al. 2007; Keys et al. 2007; Tallack et al. 2007, 2009).
At present, the best prediction of KLF1 binding site preference has arisen from the crystal structure of the three zinc fingers of a related protein, ZIF268 (also known as EGR1 or KROX24), bound to is cognate DNA motif (Feng et al. 1994). However, the extent to which this reflects the true in vivo binding of KLF1 is not known. Descriptions of transcription factor binding sites determined in vitro have been shown in some cases to be information poor compared with those defined in vivo, as is the case for the tumor suppressor protein TRP53 (Wei et al. 2006).
The aim of this study was to gain a better understanding of how KLF1 regulates erythropoiesis through determination of the sites bound by KLF1 in vivo. By employing chromatin immunoprecipitation (ChIP) in primary fetal liver erythroid cells, coupled with high-throughput DNA sequencing, we have described the transcriptional networks that are underpinned by KLF1 in erythropoiesis. Additionally, by de novo motif discovery and motif enrichment analysis we have provided insights into the cooperation of KLF1 with other transcription factors in vivo. In particular, we describe that KLF1 often works with GATA1 under specific rules of engagement. Together, these results shed new light on how erythropoiesis is programmed and how KLF1 functions in vivo.
Results
KLF1 occupies CACC motifs located proximally and distally to transcription start sites
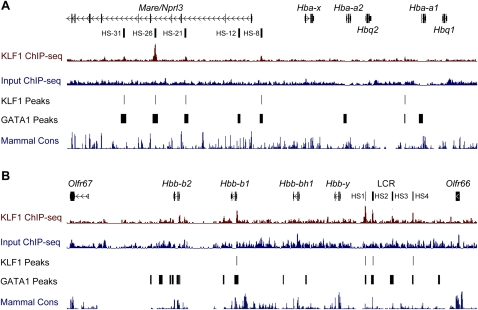
In order to broaden our understanding of how KLF1 regulates erythropoiesis, we set out to determine all of the binding sites occupied by endogenous KLF1 in vivo. We performed ChIP-seq on E14.5 fetal liver, a rich source of definitive erythroid cells at various stages of maturation, to uncover a comprehensive list of direct KLF1 target genes (Supplemental Table S1). DNA recovered from KLF1 ChIP was used to construct fragment libraries and sequenced with the SOLiD next-generation sequencing platform (Applied Biosystems). We generated 17,438,921 and 47,356,494 uniquely mapping ChIP-seq reads to the mm9 genome for KLF1 ChIP-seq and Input DNA libraries, respectively, and performed various peak calling approaches to estimate that between 945 and 1380 sites are bound by KLF1 in vivo (Supplemental Table S1).
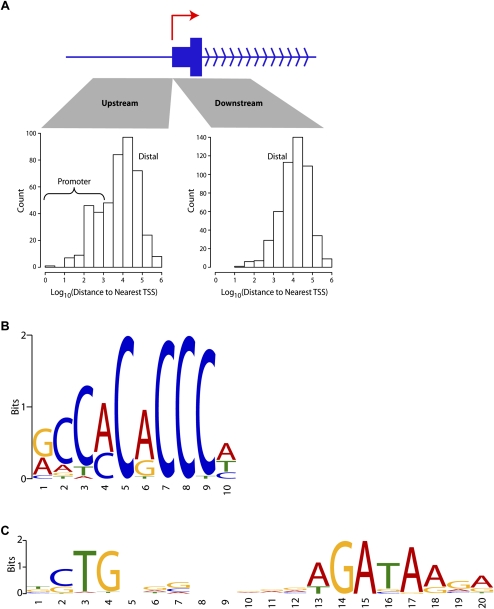
Many (∼16%) of these in vivo KLF1 binding sites occur in the vicinity (within 1 kb) of transcription start sites (TSSs), but the majority (∼52%) are located at distances of >10 kb from any known TSS (Fig. 1A). A list of the most highly occupied sites (based on greatest peak height) reveals well-known direct target genes which are critical for erythroid function such as Nprl3 (also known as Mare) (alpha-globin LCR), Alad, Gypc, and Ermap/Scianna (Table 1). However, many of these highly occupied sites are located within or close to genes with no previously described erythroid function or are located at vast genomic distances from any known gene (Table 1; Fig. 1A).
Figure 1.
KLF1 ChIP-seq reveals a role in both proximal and distal gene regulation and a refined in vivo binding site motif. (A) Characteristics of KLF1 binding with respect to transcriptional start sites (TSSs). Distances between all KLF1 ChIP-seq peak sites and the nearest mm9 UCSC Genome Browser annotated TSS were determined and sorted into upstream and downstream sets as shown. Distances are plotted as a histogram on a Log10 scale (e.g., 3 represents 1000 bp). (B) KLF1 in vivo consensus binding site predicted by the motif discovery algorithm MEME 4.3 and presented as a sequence LOGO. (C) Analysis of KLF1 ChIP-seq peaks with MEME 4.3 also uncovered the motif shown here which strikingly resembles the motif for murine TAL1(SCL)-GATA1 from the JASPAR CORE database (TOMTOM E-value = 6.6 × 10−11).
Table 1.
Highest ranking KLF1 ChIP-seq peaks
aThe 6820431F20Rik peak appears to be an artifact of chromatin conformation. This is further discussed in Supplemental Figure S1.
In order to evaluate the quality of our peak calls we measured the frequency of CACC motif occurrences in the peak sequences. KLF1 is known to bind DNA via a CACC motif, so the peak sequences should contain a nonrandomly high number of CACC motif occurrences, if the peak calls are valid. In order to obtain an unbiased estimate of CACC motif frequency, we used the independently derived CACC motif for KLF7 (Newburger and Bulyk 2009), whose DNA-binding domain is highly similar to that of KLF1. Using an independently derived CACC motif avoids the problem of circularity that could arise if we instead employed a CACC motif derived using de novo motif discovery on our peak sequences. We measured average motif affinity, using the program AMA (Buske et al. 2010), rather than simply counting the number of sequences that had at least one CACC motif occurrence at a specified score threshold. By calculating average motif affinity, we more accurately model the thermodynamic interaction of the transcription factor with the DNA. Thus, we account for sequences that have a high ChIP-seq signal due to multiple weak CACC motif occurrences.
By performing a q-value analysis (Storey and Tibshirani 2003) applied to the AMA scores of binding regions, we estimated that 75% of sequences are bound directly via the CACC motif. Applying the same analysis to our 330 strongest binding regions, we estimate that 95% of such sequences are bound via the CACC motif. The fact that 95% of the strongest peaks are estimated to be directly bound via the CACC motif indicates the high quality of the ChIP-seq data. The 75% statistic does not imply 25% of the sequences are noise, as CACC motif presence is only one factor contributing to binding. In particular, cooperative transcription factor binding (T Whitington, MC Frith, and TL Bailey, in prep.) and chromatin accessibility (Whitington et al. 2009) are critical to in vivo occupancy.
By interrogating the same 330 strongest occupied sites using the de novo pattern recognition algorithm MEME, we have refined the KLF1 in vivo binding site preference (Fig. 1B) to 5′-R-CCM-CRC-CCN-3′ (where R represents A or G and M represents A or C), thus providing additional specificity to the “N” positions previously predicted from in vitro studies (Feng et al. 1994). This site is very similar to the KLF7 binding site deposited in UniPROBE (Newburger and Bulyk 2009) and the KLF4 binding site as determined by ChIP-seq in ES cells (Chen et al. 2008) as we would expect based upon the similarity of the amino acid sequences at the DNA-binding domains of most KLFs. This refined KLF1 binding motif (Fig. 1B) appears in the majority of our 945 KLF1 ChIP-seq peak sequences (572/945 with a P-value threshold of 0.0001).
De novo motif discovery using MEME also uncovered a GATA-type motif in KLF1 ChIP-seq peak regions (Fig. 1C). This motif is almost identical to the TAL1(SCL)-GATA1 motif from the JASPAR CORE database (TOMTOM E-value of 6.6 × 10−11). This GATA-type motif (Fig. 1C) was frequently observed in KLF1 ChIP-seq peak sequences (279/945 with a P-value threshold of 0.0001). This result suggested an interaction between KLF1 and the TAL1(SCL)-GATA1 complex at erythroid gene promoters and other regulatory elements might be common.
KLF1 directly regulates the production of the alpha-globin and beta-globin subunits of hemoglobin
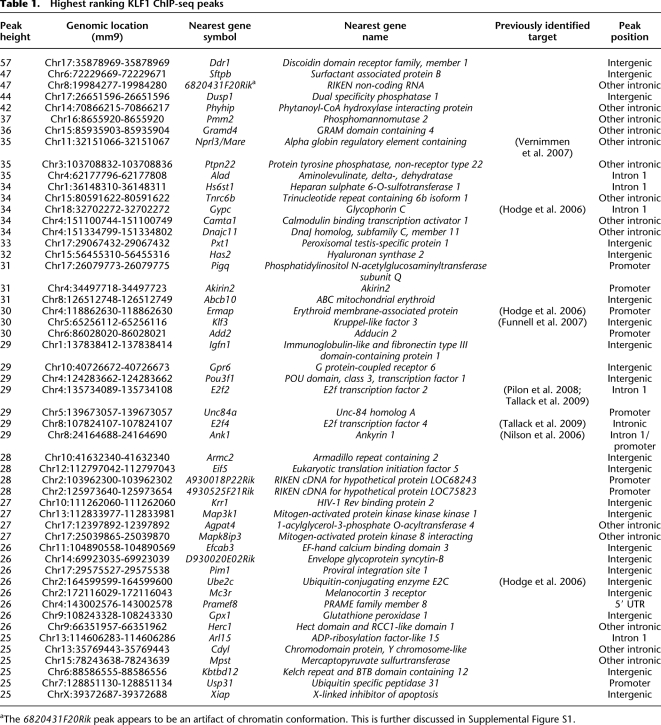
We expected KLF1 ChIP-seq peaks would be found at the proximal promoters of genes within the beta-globin gene cluster (e.g., Hbb-b1) (Im et al. 2005; Keys et al. 2007) and within the beta-globin LCR (Im et al. 2005; Shyu et al. 2006). Indeed, we observed KLF1 binding at the Hbb-b1 promoter and at HS1, HS2, HS4, and HS3 (weakly) of the beta-globin LCR (as defined in Hardison et al. 1997) (Fig. 2B). In each of these cases there is concomitant occupancy of GATA1, as determined by ChIP-seq in G1-ER cells (Cheng et al. 2009), suggesting the two factors cooperate at each of these cis-regulatory elements (Fig. 2B). We also determined KLF1 occupancy within the alpha-globin gene cluster at sites corresponding to HS-31, HS-26, HS-21, HS-8, and an intergenic site between the Hbq2 (also known as F830116E18Rik) and Hba-a1 genes (Fig. 2A). KLF1 occupancy at HS-26 has been observed previously (Vernimmen et al. 2007); however, this is the first report of occupancy at additional DNase I hypersensitive sites in the alpha-globin gene cluster. Again, we observed concomitant occupancy of GATA1 at all of these sites consistent with cooperation between these two factors in regulation of the alpha-globin gene cluster (Fig. 2A). In particular, the peak of KLF1 binding at HS-26 (corresponding to HS-40 in humans) has a high tag count suggesting strong affinity (Table 1). These observations suggest KLF1 is a transcriptional regulator of both the alpha- and beta-globin loci and may coordinate the balanced production of globin chains. It also provides further validation of our ChIP-seq data set and peak calling specificity.
Figure 2.
KLF1 binds the mouse alpha- and beta-globin loci at proximal promoters and locus control regions. (A) An image of the murine alpha-globin gene cluster and the upstream locus control region regulatory element (HS-31 to HS-8) from the UCSC Genome Browser. Genes and DNase I hypersensitivity (HS) sites are represented in the top track. KLF1 ChIP-seq signal (maroon) and Input ChIP-seq signal (blue) tracks are also shown together with KLF1 peak calls (KLF1 Peaks). An additional track describing GATA1 ChIP-seq peak calls from Cheng et al. (2009) is shown (GATA1 Peaks) together with a mammalian conservation track (Mammal Cons). (B) An image of the murine beta-globin gene cluster and locus control region flanked by olfactory receptor genes from the UCSC Genome Browser. Genes and DNase I HS sites are represented in the top track. Other tracks are presented as for A.
Heme synthesis and iron procurement are directly coordinated by KLF1
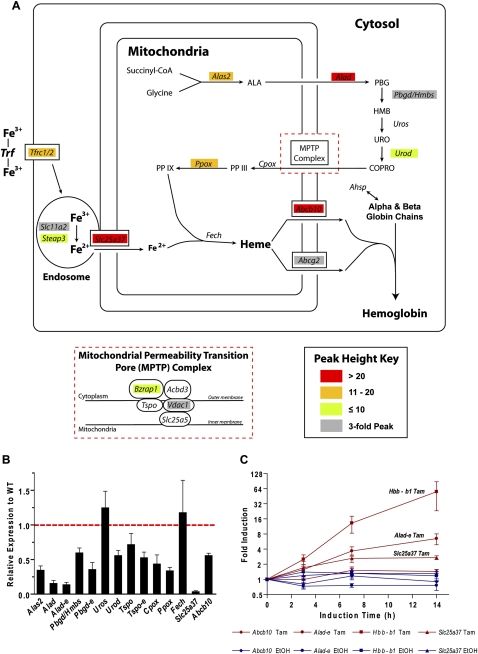
Our study demonstrated KLF1 occupancy at many (∼2/3) of the genes that are required to produce enzymes of the heme synthesis pathway (Ponka 1997), or those required for iron procurement (Fig. 3A; Dunn et al. 2007). Remarkably the majority of the genes in the heme synthesis pathway have KLF1 ChIP-seq peaks at their promoters, within introns (often the first intron) or in neighboring intergenic regions (Fig. 3A; Supplemental Fig. S2; Supplemental Table S2). In order to confirm that occupancy coincided with expression of each of these genes, we performed qRT-PCR of some of these heme synthesis–related genes in wild-type (WT) versus Klf1−/− fetal liver (Fig. 3B). All of the genes that we tested with the exception of Uros and Fech, which do not contain KLF1 ChIP-seq peaks, have decreased expression in Klf1−/− fetal liver compared with WT. Some of these genes appear to produce erythroid specific transcripts (e.g., Alad-e, Hmbs/Pbgd-e, Tspo-e) which are also specifically reduced at the mRNA level in Klf1−/− erythroid tissue (Fig. 3B).
Figure 3.
KLF1 directly regulates synthesis of heme and procurement of iron. (A) Schematic diagram of an erythroid cell simplistically depicting the pathways of heme synthesis, iron procurement, heme export, and hemoglobin assembly. Genes are shown in italics at their point of involvement in the pathways. The components of the mitochondrial permeability transition pore (MPTP) complex are shown in the inset for clarity. Genes in the figure are color coded as shown in the peak height key according to peak height from KLF1 ChIP-seq. (B) Relative gene expression in Klf1−/− fetal liver (compared with WT) for many of the genes depicted in A. Gene expression as determined by qRT-PCR is normalized to the housekeeping gene Hprt and shown for Klf1−/− fetal liver relative to WT (set to 1 as shown by the red dotted line). Data are presented as mean ± SEM. n ≥ 4 in each instance. (C) Relative gene expression in 4-hydroxytamoxifen (Tam)–induced K1-ER cells at 0, 3, 7, and 14 h following induction. Gene expression has been determined by qRT-PCR and normalized to the housekeeping gene Hprt. Induced gene expression (Tam, maroon) is shown together with the corresponding vehicle control (EtOH, blue) on a Log2 scale. Data are presented as mean ± SEM, n = 4.
KLF1 also appears to regulate importation of coproporphyrinogen III (COPRO) into the mitochondria where ferrous iron can be safely incorporated into heme (Fig. 3A). This process is regulated by a recently described multiprotein complex known as the mitochondrial permeability transition pore (MPTP) complex (Fig. 3A, inset; Papadopoulos et al. 2006). KLF1 binds with GATA1 to the promoters of two genes, Bzrap1 and Vdac1, which encode proteins that contribute to the MPTP (Fig. 3A; Supplemental Fig. S2).
Many of the genes required for iron transport have been discovered only recently, and more are likely to await discovery (Dunn et al. 2007). We have known for many years that surface expression of the transferrin receptor (Tfrc, CD71) on erythroid cells is reduced in the absence of Klf1 in a gene dosage-dependent manner, suggesting it might be a direct target of KLF1 (Hodge et al. 2006). We therefore asked whether KLF1 directly regulates either of the two transferrin receptor genes (Tfrc1 [also known as Tfrc] and Tfrc2) and other genes involved in trafficking of iron from the receptor to the heme molecule inside mitochondria (Fig. 3A). Indeed we found KLF1 ChIP-seq peaks in the vicinity of Tfrc1, the mitochondrial iron transporter mitoferrin (Slc25a37), and the Slc11a2 and Steap3 genes which encode proteins involved in iron transport and its reduction in the endosome (Fig. 3A). Additionally, Slc25a37 is markedly down-regulated in the Klf1−/− fetal liver (Fig. 3B).
The final step in the assembly of the hemoglobin molecule requires a fully constructed iron containing heme group to be transported outside of the mitochondria and coordinated with the appropriate globin protein chains (Fig. 3A). KLF1 additionally appears to control this process by direct regulation of the Abcb10 (ABC-me) and Abcg2 genes that produce heme exporter proteins (Fig. 3A). Previous studies have also described KLF1 as a critical regulator of the gene encoding the alpha-globin chaperone protein Ahsp (Pilon et al. 2006; Keys et al. 2007). Unfortunately the Ahsp gene is missing from the mm8 and mm9 genome assemblies, preventing its inclusion in the comprehensive list of KLF1 ChIP-seq target genes reported in this study.
We chose to validate that the heme synthesis–related genes with strong KLF1 ChIP-seq peaks (red boxes, Fig. 3A) were directly induced by KLF1 in a Klf1−/− erythroid cell line with tamoxifen-inducible KLF1 activity (Fig. 3C; Coghill et al. 2001). This system facilitates testing of direct chromatin-dependent induction of gene expression and is more physiological than reporter assays with plasmids (Coghill et al. 2001; Brown et al. 2002). We compared the induction of these genes (Alad, Slc25a37, and Abcb10) to that of beta-globin (Hbb-b1). We found beta-globin to be strongly induced (∼50-fold by 14 h) by tamoxifen in these cells as expected (Keys et al. 2007). We also found Alad-e (erythroid specific transcript of Alad) and Slc25a37 to be rapidly induced by tamoxifen, albeit at levels much lower than for beta-globin (approximately sixfold and approximately threefold, respectively). Expression of Abcb10 was only mildly induced by tamoxifen in these cells (∼1.5-fold), although these cells do not fully differentiate as erythroid cells do in vivo. These observations confirm that KLF1 is a critical direct regulator of many genes acting throughout the production of hemoglobin, from heme synthesis to iron procurement to the assembly of hemoglobin itself.
KLF1 regulates erythroid cell survival, proliferation, and integrity by direct regulation of specific target genes
The site of additional KLF1 ChIP-seq peaks provided us with an interesting insight into other processes that were likely to be controlled by KLF1 activity. We found peaks within the genes Bcl2l1 (Bcl-X), Xiap, and Pim1, which are known to be anti-apoptotic, prosurvival components of erythroid cells (Supplemental Fig. S3). We also confirmed our previous identification of E2f2 as a direct KLF1 target gene via an enhancer located in the first intron (Tallack et al. 2009); however, we were unable to identify KLF1 binding at the gene promoter (Supplemental Fig. S4). Additionally we determined a region of strong KLF1 occupancy in intron 5 of the E2f4 gene (Supplemental Fig. S4). We previously could not identify KLF1 binding to this region in K1-ER cells that have been retrovirally immortalized (Tallack et al. 2009). However, the use of primary tissue for ChIP-seq in this case provides evidence that E2f4 is indeed a direct KLF1 target gene. Further KLF1 target genes that play roles in the maintenance and integrity of the erythroid cell membrane and cytoskeleton such as the Ermap (Ye et al. 2000), glycophorin C (Gypc), and ankyrin 1 (Ank1) (Nilson et al. 2006) genes were uncovered (Fig. 4B; Supplemental Fig. S4). Also, KLF1 occupies the Fn3k, Slc2a4, and Pigq genes (Table 1; Fig. 4B) suggesting a direct role in sugar metabolism and protein modification such as glycosylation and GPI biosynthesis. In short, our study shows that KLF1 directly regulates virtually all processes necessary for the production and survival of erythroid cells.
Figure 4.
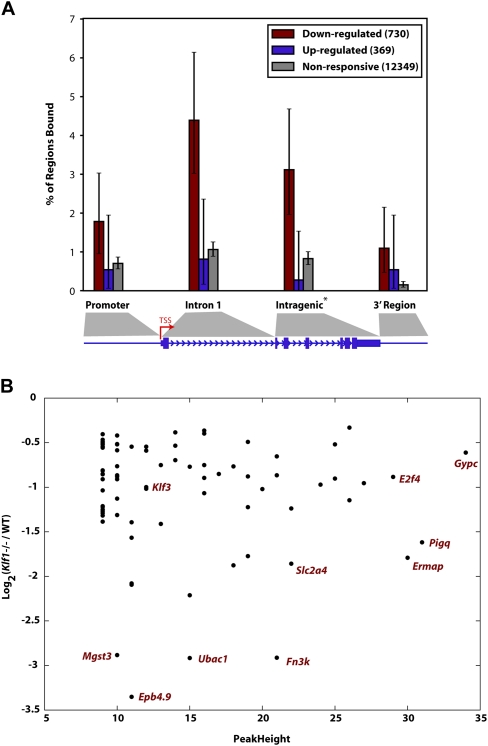
KLF1 acts primarily as a direct activator of target genes and not exclusively via promoter regions. (A) Each of the genes from the Hodge et al. (2006) expression profiling study were classified as “down-regulated” in Klf1−/−, “up-regulated” in Klf1−/−, or nonresponsive, when compared with WT expression. The plot shown represents the fraction of regions in the genes from each of these groups containing a KLF1 ChIP-seq peak binned into categories as shown in the cartoon underneath where the size of the promoter and 3′ region are each 2 kb. Each error bar represents the 95% confidence interval for the given fraction, assuming the number of regions with a peak follows a binomial distribution with N equal to the number of regions of that class. Note genes consisting of a single exon do not contribute to the intragenic region calculation. (B) A plot describing the relationship between fold change in expression in Klf1−/− fetal liver compared with WT for genes that are “bound” by KLF1 within the gene or 2 kb on either side. The down-regulated in Klf1−/− fetal liver gene set is shown (i.e., genes represented by maroon bars in A). The genes represented by some of the points are shown for clarity.
KLF1 acts primarily as a transcriptional activator in vivo and not exclusively via promoter regions
Our qRT-PCR experiments only permitted testing of a small fraction of the set of KLF1-bound genes for KLF1-dependent changes in expression. Thus, we compared our KLF1 ChIP-seq data with the KLF1 expression profiling data of Hodge et al. (2006), to elucidate the relationship between KLF1 binding and gene activation or repression, on a global scale. Microarray-probed genes were divided into three groups: “down-regulated” in the Klf1−/− fetal liver (inferred to be KLF1 activated, 730 genes), “up-regulated” in the Klf1−/− fetal liver (inferred to be KLF1 repressed, 369 genes), or “nonresponsive” to loss of Klf1 (12,349 genes). We recorded the presence of KLF1 ChIP-seq peaks in the promoter (within 2 kb of TSS), first intron, intragenic region, and proximal downstream region (within 2 kb of transcript end) of each gene (shown by the cartoon in Fig. 4A). Figure 4A displays a barplot indicating the fraction of regions that are estimated as bound, for each (responsiveness, region) combination. A much larger fraction of “down-regulated” genes are bound by KLF1, compared with the “up-regulated” or “nonresponsive” genes. This supports the view that KLF1 primarily activates gene expression. In contrast, there is no significant difference between the fraction of “up-regulated” genes bound and the fraction of “nonresponsive” genes bound, for any gene interval. Thus, we conclude that KLF1 rarely functions as a direct transcriptional repressor in vivo. The genes that are bound by KLF1 and “down-regulated” (64 genes) in Figure 4A (maroon bars) provide a very high confidence subset of direct KLF1 target genes for future studies. These are provided in Supplemental Table S3.
We were not surprised to observe that a high proportion of the “down-regulated” genes (presumably activated by KLF1) were not bound within their promoter region but were in fact bound at sites within the first intron or at other intragenic regions (Fig. 4A). For example, Alas2, Alad, Hmbs, Urod, Abcg2, Slc11a2, Bcl2l1, and Ank1 all have KLF1 binding sites in the first intron (Supplemental Figs. S2, S3, and S4 and Supplemental Table S4). This result supports the notion that KLF1 might be activating transcription of target genes at alternative erythroid specific promoters (either known or supported by mRNA, EST, or CAGE data, Supplemental Table S4) in many cases, or via intronic enhancer elements, as for E2f2 and E2f4 (Tallack et al. 2009).
We also investigated the possibility that the strength or frequency of KLF1 binding (peak height) to the regulatory element of a gene might relate to the degree to which that gene is activated by KLF1. We hypothesized that the genes which are most “down-regulated” in the absence of KLF1 might contain the strongest peaks. However, we found no relationship between strength of binding and degree of responsiveness to KLF1 (Fig. 4B). Indeed some of the most “down-regulated” genes have only modest peak heights (e.g., Epb4.9, Fig. 4B). Likewise, some of the strongest peaks occur in genes that are only modestly “down-regulated” in the Hodge et al. (2006) data set (e.g., Gypc, Fig. 4B). However, in many cases it is difficult to assign a peak to a TSS, since for distal peaks the nearest TSS is often not the true target gene. Nevertheless, our study clearly shows that KLF1 binding leads to activation of target gene expression, but the degree to which gene activation occurs depends on many additional factors besides the binding of this single transcription factor.
KLF1 and GATA1 co-occupy many sites in the erythroid genome in a distinct nonrandom configuration
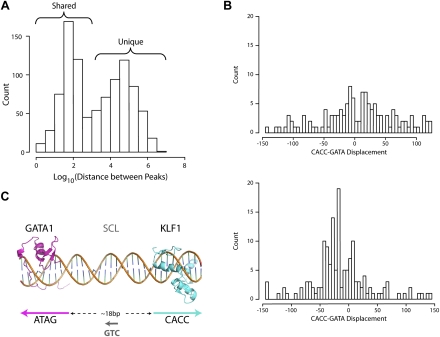
Previous work has suggested cooperation between KLF1 and GATA1 in erythroid gene regulation (Merika and Orkin 1995; Gregory et al. 1996). However, KLF1 has not been found with GATA1 containing protein complexes in vivo (Rodriguez et al. 2005), and we have never been able to coimmunoprecipitate KLF1 and GATA1 in the absence of DNA. Nevertheless, to investigate a functional DNA-dependent interaction between KLF1 and GATA1 at a global level we took advantage of a recent ChIP-seq study performed by Cheng et al. which defined the full complement of GATA1 binding sites in G1-ER cells (Cheng et al. 2009). We determined the distance to the nearest GATA1 ChIP-seq peak for all of our KLF1 ChIP-seq peaks by performing a liftOver of GATA1 ChIP-seq peaks onto the mm9 genome (see Methods). A histogram of these distances shows that ∼48% of our KLF1 ChIP-seq peaks have a corresponding GATA1 ChIP-seq peak within 1 kb (Log103) (Fig. 5A, Shared), suggesting in vivo cooperation between the two factors is likely. GATA1 is known to function together with the transcription factor SCL/TAL1 (hereafter SCL) to form an activating complex (Tripic et al. 2009). Our de novo motif discovery (Fig. 1C) suggested that a complex containing KLF1, GATA1, and SCL might be present in erythroid cells. However, it was interesting to find that by incorporating in vivo erythroid ChIP-seq data for SCL (Wilson et al. 2009) into our analysis we find that KLF1-GATA1 and SCL-GATA1 cobound regions show very little overlap in vivo (Fig. 6; Venn diagram). The SCL ChIP-seq data set may not be comprehensive; nevertheless most SCL-bound sites are not bound by KLF1.
Figure 5.
Correlation with GATA1 ChIP-seq reveals extensive co-occupancy by KLF1 and GATA1 throughout the erythroid genome. (A) A histogram of the distance between all KLF1 ChIP-seq peaks and the nearest GATA1 ChIP-seq peak as determined by Cheng et al. (2009) plotted on a Log10 scale as for Figure 1A. The bimodal distribution of distances indicates shared and unique binding sites for KLF1 as shown. (B) Histogram plots of the distance between the KLF1 motif and GATA1 motifs in the same (same strand, top) and opposing (opposite strand, bottom) directions for the KLF1 ChIP-seq peaks. (C) A model for KLF1-GATA1 cooperation based on the data presented in B. Crystal structures of the GATA1 DNA-binding zinc fingers and Zif268 (representative of KLF1) imposed onto a DNA backbone are shown in pink and blue, respectively. The preferred directions and spacing of the motifs for cooperation is shown. The hypothesized existence of SCL between the two factors binding to a “CTG” motif is indicated in gray.
Figure 6.
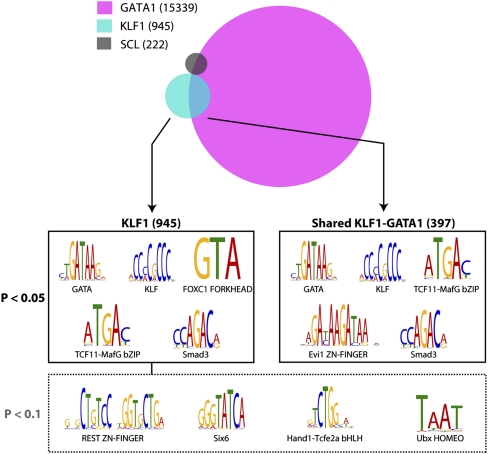
Analysis of KLF1- and GATA1-bound genomic regions reveals potential interacting transcription factor partners. A schematic representation of the overlap between KLF1, GATA1 (Cheng et al. 2009) and SCL (Wilson et al. 2009) erythroid ChIP-seq datasets defines two major groups of KLF1 binding sites, total binding sites (945), and shared KLF1-GATA1 binding sites (397). Interrogation of binding site sequences using the Clover algorithm identifies the motifs shown as being overrepresented in the groups shown at 0.05 and 0.1 P-value significance levels.
Although KLF1 and GATA1 can act synergistically at artificial promoters in reporter assays (Merika and Orkin 1995), co-occupancy of KLF1 and GATA1 in vivo does not necessarily indicate cooperation. So we next sought to determine if there were particular “rules of engagement” that might provide evidence for such a functional interaction. We used the algorithm SpaMo (T Whitington, MC Frith, and TL Bailey, in prep.) to determine whether CACC and GATA motifs exhibited any preferred spatial configuration in the KLF1 ChIP-seq peak sequences. SpaMo produces two histograms indicating the frequency of each possible displacement between the two input motifs. These histograms (one for same strand motif hits, one for opposite strand motif hits) clearly demonstrate an enrichment for CACC-GATA motif displacement at 15–45 bp apart on opposite strands (Fig. 5B). SpaMo did not detect enriched spacing for any non-GATA motifs in our set of 259 input motifs. This supports the veracity of the observed CACC-GATA enrichment in KLF1 ChIP-seq peak sequences.
The most highly represented displacement between CACC and GATA motifs is at a distance of 18 bp apart on opposite strands. We used this information to construct a 3D model of cooperative binding, by placing the crystal structures for the zinc finger domains of GATA1 (pink) and Zif268 (a surrogate for KLF1, blue) onto a short segment of DNA (Fig. 5C). We have no knowledge of the structure of the non–DNA-binding domains of either KLF1 or GATA1, but we speculate that the non–DNA-binding domains of KLF1 and GATA1 as well as cofactors are likely to exist in the center of the space between the CACC and GATA sites (Fig. 5C). Based on our de novo motif discovery analysis with MEME, which recovered an SCL-GATA1 site in KLF1 ChIP-seq peak regions (Fig. 1C) and the preferential orientations of CACC and GATA motifs, we suggest that SCL is likely to be one factor at the center of the KLF1-GATA1 erythroid complex in a small subset of erythroid cis-regulatory modules (Fig. 5C).
Transcription factor partners for KLF1
Our discovery of an interaction between KLF1 and GATA1 at a large number of genomic sites prompted us to ask what other factors might also participate at these sites, and more importantly, what interactions might be at play for KLF1 exclusive sites (i.e., those with no GATA1 binding). We utilized the motif enrichment analysis tool Clover to interrogate all KLF1-bound genomic locations as well as KLF1-GATA1 cobound genomic locations for overrepresentation of specific known DNA motifs (see Methods). We found an overrepresentation of GATA and KLF motifs in both the total KLF1-bound regions and cobound regions as expected (Fig. 6). More interestingly we found enrichment of an AP-1–type motif (represented by TCF11-MafG bZIP [Johnsen et al. 1998]) and Smad (represented by Smad3) motifs in both types of regions (Fig. 6). Overrepresentation of a number of other motifs was also observed, including a FORKHEAD motif (FOXC1 FORKHEAD) and a homeobox half site motif, TAAT (Ubx HOMEO, Fig. 6). Taken together, these results suggest that KLF1 may interact in transcriptional complexes that also include proteins capable of binding to AP-1, Smad, and homeobox motifs (see Discussion).
Discussion
We have described in this study a broad repertoire of direct KLF1 target genes that regulate virtually all processes associated with erythropoiesis. Depending on the specificity of our peak calling we declare between 945 and 1380 in vivo KLF1 binding sites in erythroid cells. We have deliberately favored high specificity over sensitivity in our peak calling approach to reduce the risk of declaring false-positive KLF1 binding sites. Thus, much greater sequencing depth is likely to find a small number of additional lower affinity sites, but will mostly resequence the same high affinity sites. These declared binding sites describe the majority of the genome wide transcriptional regulatory activities of KLF1 during erythropoiesis.
In the vicinity of our declared KLF1 ChIP-seq peaks we identified by de novo motif discovery a SCL-GATA1 binding motif suggesting that these two factors might interact with KLF1 to drive the appropriate expression of erythroid genes (Fig. 1C). In addition, by using the newly developed tool SpaMo we identified a set of specific constraints governing the positioning between CACC and GATA motifs in KLF1 ChIP-seq peaks (Fig. 5B,C). SCL is likely to bind between KLF1 and GATA1 in some instances, yet when we consider the overlap between KLF1-GATA1 shared peaks and SCL-GATA1 cobound regions it is only minimal (Fig. 6). We believe that the high specificity and relatively low sensitivity of the SCL ChIP-seq data set might partly explain for this apparent contradiction, whereby a less stringent peak calling approach in the SCL ChIP-seq data set would likely lead to a greater number of peaks and more significant overlap with KLF1 ChIP-seq peaks. Nevertheless, many SCL-GATA1–bound regions in the genome function independently of KLF1 and vice versa.
Additionally, our study suggests there is a significant advantage in using primary tissue rather than engineered cell lines for identifying all sites of binding in vivo. For example, we previously conducted ChIP experiments in an immortalized Klf1−/− erythroid cell line with inducible KLF1 activity (K1-ER cells) and were unable to find any binding of KLF1 at the E2f4 gene locus (Tallack et al. 2009). In direct contrast we describe here that the E2f4 gene locus is robustly bound by KLF1 in primary fetal liver cells at a site located within intron 5 (Supplemental Fig. S4). This is likely due to KLF1 occupying different regions of the genome as an erythroid precursor differentiates into a mature late normoblast. Both E2F2 and E2F4 have roles in erythroid differentiation, perhaps balancing proliferation with differentiation (Kinross et al. 2006; Dirlam et al. 2007). Such dynamic changes in occupancy are sometimes difficult to observe in most cell lines. This is an example of a binding event we missed in K1-ER, and would probably miss in MEL cell lines, that we believe highlights the advantage of performing ChIP-seq experiments in primary tissue when it is possible.
We also describe a set of cis-elements we have associated with KLF1 target genes (Table 1) that significantly overlaps with that previously postulated from expression profiling experiments (Fig. 4) (Drissen et al. 2005; Hodge et al. 2006; Pilon et al. 2008). This includes not only genes we might have expected to find such as those responsible for the production of hemoglobin (Figs. 2, 3) and the integrity and deformability of the red cell (Supplemental Fig. S4), but also genes required for cell survival and nuclear condensation like Bcl2l1, Xiap, and Pim1 (Supplemental Fig. S3). Thus it appears that KLF1 (together with GATA1 50% of the time) regulates virtually all of the processes at work to produce and maintain erythroid cells.
In addition to the discovery of a new suite of KLF1 target genes, we have also produced evidence to suggest unique modes of gene regulation by KLF1. We noticed that the vast majority of KLF1 binding sites (peaks) are located at large genomic distances from known transcription start sites (Fig. 1A). Many of these sites have high tag counts (strongly bound) and many are co-occupied by GATA1. KLF1 has recently been described as a critical looping factor particularly in the context of the beta-globin locus (Drissen et al. 2004). It has also been shown very recently that KLF1 is critical in the formation of nuclear transcription factories consisting of RNA pol II and highly transcribed erythroid genes (Schoenfelder et al. 2010). These studies strongly suggest that KLF1 is not only capable of, but necessary for, long-distance gene regulation within erythroid cells. We suggest many of the in vivo binding sites for KLF1 that are found at vast distances from any known gene might function in chromatin looping, or be involved in recruiting genomic regions into transcription factories, and may not function as canonical enhancers. However, we cannot be sure that these binding sites are not functional as very distant enhancers. In such cases it is very difficult to know which gene(s) might be responsive to such a putative enhancer, making functional testing extremely difficult.
We also determined that for direct KLF1 target genes, regulation of transcription was frequently occurring via binding at intragenic regions, the first intron in particular (Fig. 4A; Supplemental Figs. S2–S4). Some of these instances are previously reported alternative erythroid-specific promoters, others have supporting EST evidence for alternative promoters (Supplemental Table S4), but many have not yet been discovered. Erythroid cells have a unique metabolic demand and appear to have coevolved a unique set of promoters together with KLF1 so that the coding outputs of many genes in general biochemical pathways can be specifically tailored to the particular metabolic needs of erythroid cells. Further studies describing the comprehensive transcriptional landscape of erythroid cells will need to be performed to validate this hypothesis.
In addition to the KLF1-GATA1 interaction, we have uncovered enrichment of several transcription factor DNA-binding motifs in the vicinity of KLF1 occupied sites that suggest the involvement of transcription factors capable of binding to AP-1 like (TCF11-MafG bZIP), Smad (Smad3), and Homeobox (Ubx HOMEO) motifs (Fig. 6). The AP-1-like motif (TCF11-MafG bZIP [Johnsen et al. 1998]) is most likely bound by bZIP heterodimers like p45-NFE2/p18-MAF which are important in erythropoiesis (Andrews et al. 1993; Kotkow and Orkin 1995; Johnsen et al. 1998). Interestingly a member of the Smad family of transcription factors, Smad5, is partly responsible for the induction of Klf1 expression in erythroid cells (Lohmann and Bieker 2008). Smad5 continues to be expressed in erythroid cells and is critical for the proper expansion of erythroid progenitors in the fetal liver and in stress erythropoiesis (Porayette and Paulson 2008). In addition, our data suggest KLF1 cooperates directly with “ventral” Smads such as with Smad1/5/8 to mediate erythroid expansion in response to BMP4 signaling. This hypothesis is consistent with experiments performed in Xenopus which show the KLF1-like protein, Klf17 (also known as Neptune), is essential for BMP4 induction of red blood cell formation (Huber et al. 2001). In short, we suggest Smad5 activates KLF1 gene expression and then works with KLF1 in the nucleus to regulate the erythroid response to stress.
In summary, our study has described the in vivo DNA-binding activities of KLF1 by performing primary erythroid tissue ChIP-seq. We have revealed a complex interaction between KLF1 and GATA1 that is responsible for regulating the expression of erythroid genes, and we provide future challenges and considerations for the interpretation of ChIP-seq studies particularly with regard to erythropoiesis.
Methods
Primary fetal liver ChIP
ChIP assays were performed as previously described (Hodge et al. 2006) using a rabbit polyclonal antibody raised against the N terminus of KLF1 (Perkins et al. 1995; Keys et al. 2007) in Klf3−/− or wild-type (WT) fetal livers collected from E14.5 mouse embryos. At this stage of development the fetal liver is entirely erythroid and composed of cells at various stages of maturation from proerythroblasts to late enucleating normoblasts. Klf3−/− fetal livers were utilized in addition to WT due to anticipated stronger KLF1 ChIP occupancy based on a recently proposed model of competition between KLF1 and KLF3 (BKLF) for CACC box occupancy in vivo (Eaton et al. 2008). In brief, ChIP assays were performed on pools of four homogenized E14.5 fetal livers (∼8 × 107 cells) and validated by quantitative real-time PCR as previously described to determine occupancy at an E2f2 enhancer (E2f2-i1en) and the Ahsp promoter (Keys et al. 2007; Tallack et al. 2009).
ChIP-seq library construction
The SOLiD System 2.0 workflow for Lower Input/Lower Complexity DNA fragment library preparation (Applied Biosystems) was followed to prepare libraries containing 80–130 bp of ChIP DNA (or Input DNA) flanked by the appropriate adaptors. A detailed description of the method is provided in the Supplemental Methods.
SOLiD chemistry sequencing
SOLiD ChIP-seq DNA fragment libraries were sequenced by SOLiD system 2.0 and SOLiD system 3.0 chemistries to produce DNA sequence reads of 35 and 50 nucleotides (nt), respectively. Sequencing reads were mapped to the mm9 mouse genome using the mapreads algorithm (Applied Biosystems). In order to maximize the number of mappable reads we removed the final 5 nt for unmapped reads in a recursive strategy until the reads reached 25 nt in length, similar to an approach previously described (Cloonan et al. 2009). We recovered 17,438,921 and 47,356,494 unique reads (defined by a unique sequencing read start site) that mapped exactly once to the mm9 genome for the KLF1 ChIP-seq and Input DNA libraries, respectively.
KLF1 ChIP-seq peak detection
We employed the same method used by Chen et al. (2008) in order to declare a set of KLF1-bound regions for downstream analysis. When generating the tag intensity profile used in peak declaration, we added a count of one to each position up to 100 bp 3′ from the start of each tag. To be declared a peak, a genomic position must pass both a tag intensity threshold and a fold-change threshold. We applied a false discovery rate (FDR) threshold of 1%, which resulted in a tag intensity profile threshold of nine. We applied negative control tag library fold-change threshold of 5.0 at the center of each declared peak, yielding the final set of 945 declared peaks. We additionally declared a less stringent set of 1380 peaks by applying a fold-change threshold of 3.0 (Supplemental Table S1).
Motif analyses
We used several motif analysis tools to study the relationship between KLF1 binding and transcription factor DNA-binding sequence specificity.
Average motif affinity (AMA) (Buske et al. 2010) was used to estimate the fraction of regions around peaks that have a nonrandomly strong KLF1 affinity as predicted by a CACC box PWM.
The de novo motif discovery tool MEME (Bailey 2002) was used to determine KLF1 DNA-binding sequence specificity without prior information.
We used the tool Clover (Frith et al. 2004) to measure enrichment of known motifs in sets of KLF1 ChIP-seq peak regions of interest, in order to identify motifs of possible co-binding transcription factors. This was necessary in order to detect enrichments that are invisible to de novo discovery, due to the inherently large search space of de novo discovery.
We used SpaMo (T Whitington, MC Frith, and TL Bailey, in prep.) to detect significant spacing relationships between known motifs.
For the methodology and parameters employed when running these programs, refer to the Supplemental Methods.
Inference of KLF1-DNA-GATA1 ternary complex structure
The enriched motif spacing information inferred using SpaMo allowed us to predict the likely structure of the corresponding KLF1-DNA-GATA1 ternary complex, using existing transcription factor-DNA X-ray crystallography structures. See Supplemental Methods for an explanation of this approach.
Comparing KLF1 binding and transcriptional response to perturbation of KLF1
To investigate the relationship between in vivo KLF1 binding and transcriptional response to pertubation of Klf1, we compared our ChIP-seq data with Klf1−/− expression profiling data from primary fetal liver (Hodge et al. 2006). Refer to Supplemental Methods for an explanation of this analysis.
K1-ER cell line induction
K1-ER cells (Coghill et al. 2001) were maintained and treated with 4-hydroxytamoxifen (Tam; Sigma) or vehicle control (EtOH) as previously described (Keys et al. 2007). The assessment of induced specific changes in gene expression was performed as described below.
Gene expression profiling by qRT-PCR
Fetal liver or K1-ER cell line RNA was isolated and used to produce cDNA as described in the Supplemental Methods. Gene expression profiling was performed by qRT-PCR using specific primer sets as described in the Supplemental Methods and Supplemental Table S6.
Acknowledgments
This work was supported by an Australian Research Council Discovery Grant (DP0770471/ACP) and a grant from the Cancer Council Queensland (519718/ACP). M.R.T. and T.W. were the recipients of an Australian Postgraduate Award. N.C. is the recipient of an Australian Research Council Postdoctoral Fellowship. S.M.G. is the recipient of an Australian NH&MRC Senior Research Fellowship.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE20478.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.106575.110.
References
- Andrews NC, Erdjument-Bromage H, Davidson MB, Tempst P, Orkin SH 1993. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature 362: 722–728 [DOI] [PubMed] [Google Scholar]
- Bailey TL 2002. Discovering novel sequence motifs with MEME. Curr Protoc Bioinformatics Chapter 2: Unit 2.4. [DOI] [PubMed] [Google Scholar]
- Brown RC, Pattison S, van Ree J, Coghill E, Perkins A, Jane SM, Cunningham JM 2002. Distinct domains of erythroid Kruppel-like factor modulate chromatin remodeling and transactivation at the endogenous beta-globin gene promoter. Mol Cell Biol 22: 161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buske FA, Bodén M, Bauer DC, Bailey TL 2010. Assigning roles to DNA regulatory motifs using comparative genomics. Bioinformatics 26: 860–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor AB, Orkin SH 2002. Transcriptional regulation of erythropoiesis: An affair involving multiple partners. Oncogene 21: 3368–3376 [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Wu W, Kumar SA, Yu D, Deng W, Tripic T, King DC, Chen KB, Zhang Y, Drautz D, et al. 2009. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res 19: 2172–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N, Xu Q, Faulkner GJ, Taylor DF, Tang DT, Kolle G, Grimmond SM 2009. RNA-MATE: A recursive mapping strategy for high-throughput RNA-sequencing data. Bioinformatics 25: 2615–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghill E, Eccleston S, Fox V, Cerruti L, Brown C, Cunningham J, Jane S, Perkins A 2001. Erythroid Kruppel-like factor (EKLF) coordinates erythroid cell proliferation and hemoglobinization in cell lines derived from EKLF null mice. Blood 97: 1861–1868 [DOI] [PubMed] [Google Scholar]
- Dirlam A, Spike BT, Macleod KF 2007. Deregulated E2f-2 underlies cell cycle and maturation defects in retinoblastoma null erythroblasts. Mol Cell Biol 27: 8713–8728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W 2004. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev 18: 2485–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen R, von Lindern M, Kolbus A, Driegen S, Steinlein P, Beug H, Grosveld F, Philipsen S 2005. The erythroid phenotype of EKLF-null mice: Defects in hemoglobin metabolism and membrane stability. Mol Cell Biol 25: 5205–5214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn LL, Rahmanto YS, Richardson DR 2007. Iron uptake and metabolism in the new millennium. Trends Cell Biol 17: 93–100 [DOI] [PubMed] [Google Scholar]
- Eaton SA, Funnell AP, Sue N, Nicholas H, Pearson RC, Crossley M 2008. A network of Kruppel-like Factors (Klfs). Klf8 is repressed by Klf3 and activated by Klf1 in vivo. J Biol Chem 283: 26937–26947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng WC, Southwood CM, Bieker JJ 1994. Analyses of beta-thalassemia mutant DNA interactions with erythroid Kruppel-like factor (EKLF), an erythroid cell-specific transcription factor. J Biol Chem 269: 1493–1500 [PubMed] [Google Scholar]
- Fraser ST, Isern J, Baron MH 2007. Maturation and enucleation of primitive erythroblasts during mouse embryogenesis is accompanied by changes in cell-surface antigen expression. Blood 109: 343–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Fu Y, Yu L, Chen JF, Hansen U, Weng Z 2004. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res 32: 1372–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH 1996. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci 93: 12355–12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, O'Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH 2009. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell 36: 667–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funnell AP, Maloney CA, Thompson LJ, Keys J, Tallack M, Perkins AC, Crossley M 2007. Erythroid Kruppel-like factor directly activates the basic Kruppel-like factor gene in erythroid cells. Mol Cell Biol 27: 2777–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RC, Taxman DJ, Seshasayee D, Kensinger MH, Bieker JJ, Wojchowski DM 1996. Functional interaction of GATA1 with erythroid Kruppel-like factor and Sp1 at defined erythroid promoters. Blood 87: 1793–1801 [PubMed] [Google Scholar]
- Hardison R, Slightom JL, Gumucio DL, Goodman M, Stojanovic N, Miller W 1997. Locus control regions of mammalian beta-globin gene clusters: Combining phylogenetic analyses and experimental results to gain functional insights. Gene 205: 73–94 [DOI] [PubMed] [Google Scholar]
- Hodge D, Coghill E, Keys J, Maguire T, Hartmann B, McDowall A, Weiss M, Grimmond S, Perkins A 2006. A global role for EKLF in definitive and primitive erythropoiesis. Blood 107: 3359–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber TL, Perkins AC, Deconinck AE, Chan FY, Mead PE, Zon LI 2001. neptune, a Kruppel-like transcription factor that participates in primitive erythropoiesis in Xenopus. Curr Biol 11: 1456–1461 [DOI] [PubMed] [Google Scholar]
- Im H, Grass JA, Johnson KD, Kim SI, Boyer ME, Imbalzano AN, Bieker JJ, Bresnick EH 2005. Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc Natl Acad Sci 102: 17065–17070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen O, Murphy P, Prydz H, Kolsto AB 1998. Interaction of the CNC-bZIP factor TCF11/LCR-F1/Nrf1 with MafG: Binding-site selection and regulation of transcription. Nucleic Acids Res 26: 512–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys JR, Tallack MR, Hodge DJ, Cridland SO, David R, Perkins AC 2007. Genomic organisation and regulation of murine alpha haemoglobin stabilising protein by erythroid Kruppel-like factor. Br J Haematol 136: 150–157 [DOI] [PubMed] [Google Scholar]
- Kinross KM, Clark AJ, Iazzolino RM, Humbert PO 2006. E2f4 regulates fetal erythropoiesis through the promotion of cellular proliferation. Blood 108: 886–895 [DOI] [PubMed] [Google Scholar]
- Kotkow KJ, Orkin SH 1995. Dependence of globin gene expression in mouse erythroleukemia cells on the NF-E2 heterodimer. Mol Cell Biol 15: 4640–4647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koury MJ, Sawyer ST, Brandt SJ 2002. New insights into erythropoiesis. Curr Opin Hematol 9: 93–100 [DOI] [PubMed] [Google Scholar]
- Lohmann F, Bieker JJ 2008. Activation of Eklf expression during hematopoiesis by Gata2 and Smad5 prior to erythroid commitment. Development 12: 2071–2082 [DOI] [PubMed] [Google Scholar]
- Merika M, Orkin SH 1995. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Kruppel family proteins Sp1 and EKLF. Mol Cell Biol 15: 2437–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller IJ, Bieker JJ 1993. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol Cell Biol 13: 2776–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newburger DE, Bulyk ML 2009. UniPROBE: An online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res 37: D77–D82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilson DG, Sabatino DE, Bodine DM, Gallagher PG 2006. Major erythrocyte membrane protein genes in EKLF-deficient mice. Exp Hematol 34: 705–712 [DOI] [PubMed] [Google Scholar]
- Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F 1995. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature 375: 316–318 [DOI] [PubMed] [Google Scholar]
- Papadopoulos V, Baraldi M, Guilarte TR, Knudsen TB, Lacapere JJ, Lindemann P, Norenberg MD, Nutt D, Weizman A, Zhang MR, et al. 2006. Translocator protein (18kDa): New nomenclature for the peripheral-type benzodiazepine receptor based on its structure and molecular function. Trends Pharmacol Sci 27: 402–409 [DOI] [PubMed] [Google Scholar]
- Perkins A 1999. Erythroid Kruppel like factor: From fishing expedition to gourmet meal. Int J Biochem Cell Biol 31: 1175–1192 [DOI] [PubMed] [Google Scholar]
- Perkins AC, Sharpe AH, Orkin SH 1995. Lethal beta-thalassemia in mice lacking the erythroid caccc-transcription factor Eklf. Nature 375: 318–322 [DOI] [PubMed] [Google Scholar]
- Pevny L, Simon MC, Robertson E, Klein WH, Tsai SF, D'Agati V, Orkin SH, Costantini F 1991. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature 349: 257–260 [DOI] [PubMed] [Google Scholar]
- Pilon AM, Nilson DG, Zhou D, Sangerman J, Townes TM, Bodine DM, Gallagher PG 2006. Alterations in expression and chromatin configuration of the alpha hemoglobin-stabilizing protein gene in erythroid Kruppel-like factor-deficient mice. Mol Cell Biol 26: 4368–4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon AM, Arcasoy MO, Dressman HK, Vayda SE, Maksimova YD, Sangerman JI, Gallagher PG, Bodine DM 2008. Failure of terminal erythroid differentiation in EKLF-deficient mice is associated with cell cycle perturbation and reduced expression of E2F2. Mol Cell Biol 28: 7394–7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponka P 1997. Tissue-specific regulation of iron metabolism and heme synthesis: Distinct control mechanisms in erythroid cells. Blood 89: 1–25 [PubMed] [Google Scholar]
- Porayette P, Paulson RF 2008. BMP4/Smad5 dependent stress erythropoiesis is required for the expansion of erythroid progenitors during fetal development. Dev Biol 317: 24–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez P, Bonte E, Krijgsveld J, Kolodziej KE, Guyot B, Heck AJ, Vyas P, de Boer E, Grosveld F, Strouboulis J 2005. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J 24: 2354–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. 2010. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet 42: 53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu YC, Wen SC, Lee TL, Chen X, Hsu CT, Chen H, Chen RL, Hwang JL, Shen CK 2006. Chromatin-binding in vivo of the erythroid kruppel-like factor, EKLF, in the murine globin loci. Cell Res 16: 347–355 [DOI] [PubMed] [Google Scholar]
- Siatecka M, Xue L, Bieker JJ 2007. Sumoylation of EKLF promotes transcriptional repression and is involved in inhibition of megakaryopoiesis. Mol Cell Biol 27: 8547–8560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck J, Cohet N, Gonnet C, Sarrazin S, Doubeikovskaia Z, Doubeikovski A, Verger A, Duterque-Coquillaud M, Morle F 2003. Functional cross-antagonism between transcription factors FLI-1 and EKLF. Mol Cell Biol 23: 1390–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallack MR, Perkins AC 2010. Megakaryocyte-erythroid lineage promiscuity in EKLF null mouse blood. Haematologica 95: 144–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallack MR, Keys JR, Perkins AC 2007. Erythroid Kruppel-like factor regulates the G1 cyclin dependent kinase inhibitor p18INK4c. J Mol Biol 369: 313–321 [DOI] [PubMed] [Google Scholar]
- Tallack MR, Keys JR, Humbert PO, Perkins AC 2009. EKLF/KLF1 controls cell cycle entry via direct regulation of E2f2. J Biol Chem 284: 20966–20974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa U 2004. Apoptotic mechanisms in the control of erythropoiesis. Leukemia 18: 1176–1199 [DOI] [PubMed] [Google Scholar]
- Tripic T, Deng W, Cheng Y, Zhang Y, Vakoc CR, Gregory GD, Hardison RC, Blobel GA 2009. SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood 113: 2191–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet J, Crofts LA, Quinlan KG, Czolij R, Perkins AC, Crossley M 2006. Human KLF17 is a new member of the Sp/KLF family of transcription factors. Genomics 87: 474–482 [DOI] [PubMed] [Google Scholar]
- Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR 2007. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J 26: 2041–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. 2006. A global map of p53 transcription-factor binding sites in the human genome. Cell 124: 207–219 [DOI] [PubMed] [Google Scholar]
- Weiss MJ, Orkin SH 1995. GATA transcription factors: Key regulators of hematopoiesis. Exp Hematol 23: 99–107 [PubMed] [Google Scholar]
- Welch JJ, Watts JA, Vakoc CR, Yao Y, Wang H, Hardison RC, Blobel GA, Chodosh LA, Weiss MJ 2004. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood 104: 3136–3147 [DOI] [PubMed] [Google Scholar]
- Whitington T, Perkins AC, Bailey TL 2009. High-throughput chromatin information enables accurate tissue-specific prediction of transcription factor binding sites. Nucleic Acids Res 37: 14–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NK, Miranda-Saavedra D, Kinston S, Bonadies N, Foster SD, Calero-Nieto F, Dawson MA, Donaldson IJ, Dumon S, Frampton J, et al. 2009. The transcriptional program controlled by the stem cell leukemia gene Scl/Tal1 during early embryonic hematopoietic development. Blood 113: 5456–5465 [DOI] [PubMed] [Google Scholar]
- Ye TZ, Gordon CT, Lai YH, Fujiwara Y, Peters LL, Perkins AC, Chui DH 2000. Ermap, a gene coding for a novel erythroid specific adhesion/receptor membrane protein. Gene 242: 337–345 [DOI] [PubMed] [Google Scholar]
- Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, et al. 2009. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell 36: 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Socolovsky M, Gross AW, Lodish HF 2003. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: Functional analysis by a flow cytometry-based novel culture system. Blood 102: 3938–3946 [DOI] [PubMed] [Google Scholar]