Abstract
Schistosoma mansoni is a well-adapted blood-dwelling parasitic helminth, persisting for decades in its human host despite being continually exposed to potential immune attack. Here, we describe in detail micro-exon genes (MEG) in S. mansoni, some present in multiple copies, which represent a novel molecular system for creating protein variation through the alternate splicing of short (≤36 bp) symmetric exons organized in tandem. Analysis of three closely related copies of one MEG family allowed us to trace several evolutionary events and propose a mechanism for micro-exon generation and diversification. Microarray experiments show that the majority of MEGs are up-regulated in life cycle stages associated with establishment in the mammalian host after skin penetration. Sequencing of RT-PCR products allowed the description of several alternate splice forms of micro-exon genes, highlighting the potential use of these transcripts to generate a complex pool of protein variants. We obtained direct evidence for the existence of such pools by proteomic analysis of secretions from migrating schistosomula and mature eggs. Whole-mount in situ hybridization and immunolocalization showed that MEG transcripts and proteins were restricted to glands or epithelia exposed to the external environment. The ability of schistosomes to produce a complex pool of variant proteins aligns them with the other major groups of blood parasites, but using a completely different mechanism. We believe that our data open a new chapter in the study of immune evasion by schistosomes, and their ability to generate variant proteins could represent a significant obstacle to vaccine development.
Schistosomes are blood-dwelling helminth parasites that represent a serious health problem in tropical regions, affecting approximately 200 million people. They can persist in the bloodstream for decades despite being permanently exposed to the host immune system, indicating that they must deploy a very effective system of immune evasion. Despite intensive studies of the host–parasite interaction, how the parasite succeeds in avoiding immune attack is not fully understood. One component involves the production of a relatively inert barrier at the surface of the tegument, the major interface between the parasite and host. This comprises a conventional plasma membrane overlain by a membrane-like secretion, the membranocalyx, which is believed to shield parasite proteins from the immune attack (Skelly and Wilson 2006). In contrast, the other major groups of blood-dwelling parasites, exemplified by Trypanosoma brucei and Plasmodium falciparum, use molecular mechanisms that involve both protein variation and polymorphism, to evade host immune responses. T. brucei relies on switching expression of multicopy genes during the course infection, to produce a unique dominant but immunogenic surface glycoprotein (Taylor and Rudenko 2006). As the host mounts an antibody response to eliminate the parasites from the circulation, a new dominant variant appears in a virtually endless succession of parasite populations. P. falciparum employs clonal variation too, switching the expression of protein isoforms encoded by large gene families, on the surface of infected erythrocytes (Kyes et al. 2007). The parasite also possesses multiple alternative allelic forms at single loci that encode key merozoite antigens (Anders et al. 1993). In both cases, the proteins provoke immune responses that are circumvented by the parasites’ capacity for protein variation.
Micro-exons are very short exons (≤36 base pairs [bp]) that have been described in a few genes (ranging from 0.5% to 1.6% of total genes) from the genome of several organisms; in most cases a single micro-exon was detected per gene (Volfovsky et al. 2003). Micro-exons have been previously described in S. mansoni in a portion of the gene for the antigen 10.3 in which 27-bp exons made up repeats of 81 bp found in tandem (Davis et al. 1988). However, the lack of its genomic sequence prevented a description of its complete structure. Recently, Chalmers et al. (2008) also described a structure containing some micro-exons in the SmVAL-6 gene and showed alternative use of such exons in the generation of transcripts.
Our attention was first drawn to a group of unusual S. mansoni genes of diverse sequence during a proteomic search of schistosomula secretions, which identified peptides with homologies with an EST contig Sm11845 that mapped to a predicted coding region in the v1 genome assembly, now designated Smp_138080. We noted that the structure of this gene uniquely comprised very short internal symmetrical exons. By further analysis of the genome assembly, using the methodology developed to detect single micro-exons (Volfovsky et al. 2003), we found 27 genes with the same organization, which we named micro-exons genes (MEGs). A one-paragraph preliminary description was included in the recent publication of the S. mansoni genome (Berriman et al. 2009). A full account is presented here showing details of how micro-exon genes undergo alternative splicing to generate multiple transcripts, together with proteomic and localization data. These indicate that the multiple protein products generated by MEG transcripts are secreted from glands and epithelia, a situation which would allow them to interact with the host immune system. These data highlight a novel system to create protein variation in a blood-dwelling helminth parasite.
Results
Micro-exon genes from schistosomes represent a novel group of genes with unique features
Analysis of genes differentially expressed in the microarray experiment permitted us to pinpoint a further four gene families with micro-exon structure in the S. mansoni genome (MEG-15–MEG-18), additional to those briefly described by us (Berriman et al. 2009). The nonannotated Smp gene models from the day 3 schistosomulum-germ ball comparisons were extracted from the top table of array results and ordered by fold-difference in expression. Simple visual inspection of the unspliced DNA sequence on the sequence page for each gene at GeneDB, version 2.1, revealed the micro-exon structure. Four genes with list positions 9, 10, 18, and 24, and differences of 33-, 18-, 15-, and 14-fold in expression were identified in this way.
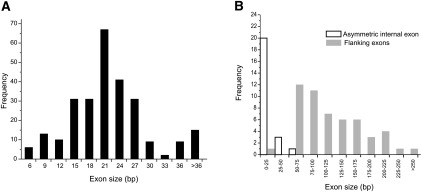
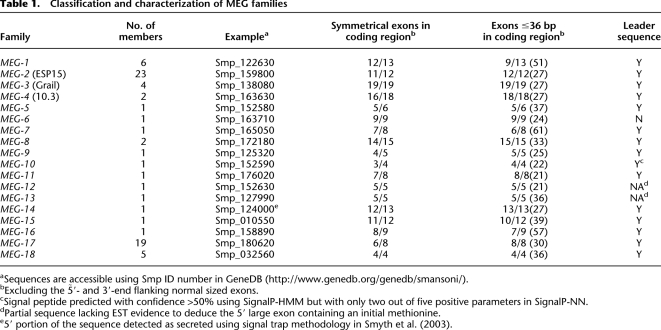
We classified the 31 predicted MEGs by protein sequence homology into 18 families (alignment with E-value lower than 10−4 in a BLASTP using all MEG proteins as database and queries). We then performed a BLASTN search of the S. mansoni genome assembly using the 31 known cDNAs, to find any further family members missed by the Volfovsky method (Volfovsky et al. 2003). This permitted the mapping of exons displaying similar sequences to the previously described MEGs and revealed a total of 72 micro-exon genes, probably an underestimate because the genome assembly is incomplete. The 18 families divide into two groups, 11 with single and seven with multiple members, the gene complement of the latter ranging from 2 to 23 (average = 8.7). The vast majority of micro-exons (∼92%) in all families are symmetrical, ranging in size from 6 to 36 bp (with two outliers at 51 bp), the commonest frequency being 21 bp (Fig. 1A). A small number of micro-exons (24/289) are not symmetrical. The majority of these are near the extremities of the genes, where they code either for part of the signal peptide that is not expected to vary, or near the stop codon, with a minimal disruptive potential for frameshift caused by their alternative splicing (Supplemental Fig. 1). Additionally, there is a small number (17) of larger internal exons ranging from 37 to 61 bp (Fig. 1B). The flanking exons, including untranslated regions (UTRs) deduced from EST sequences, range from 22 to 274 bp (average = 120 bp) (Fig. 1B). However, it is important to note that most of the UTR from 5′-end exons are incompletely sequenced so flanking exon sizes may actually be bigger than currently perceived. These flanking exons do not contain much coding sequence, representing on average only 27% of the total bases coding for amino acids in the genes. Despite their small size, the micro-exons use the same mRNA splicing mechanism as conventional genes, with conserved 5′ and 3′ canonical splicing sites (Supplemental Fig. 2) in the respective introns. Indeed, the 5′ site of the micro-exon introns is even more conserved than that of conventional genes.
Figure 1.
Exon size distribution in S. mansoni micro-exon genes. (A) Histogram of the symmetrical internal exon size distribution. (B) Histogram of asymmetrical internal exon and flanking exon sizes.
All MEGs code for small (6–20 kDa) predicted proteins but BLASTP searches of the NCBI nr database revealed no homology with proteins of known function in other genera. Furthermore, no conserved domains were identified by programs such as Prosite, Interpro, and SMART. However, SignalP analysis predicted an N-terminal leader sequence for almost all MEG proteins (Table 1). The prediction of leader sequence combined with the absence in all but three families of additional membrane spanning regions predicted by HMMTOP lead us to conclude that most of the MEG proteins are secreted.
Table 1.
Classification and characterization of MEG families
aSequences are accessible using Smp ID number in GeneDB (http://www.genedb.org/genedb/smansoni/).
bExcluding the 5- ′- and 3′-end flanking normal sized exons.
cSignal peptide predicted with confidence >50% using SignalP-HMM but with only two out of five positive parameters in SignalP-NN.
dPartial sequence lacking EST evidence to deduce the 5′ large exon containing an initial methionine.
e5′ portion of the sequence detected as secreted using signal trap methodology in Smyth et al. (2003).
We performed an exhaustive search in platyhelminths for homologs of protein sequences encoded by the micro-exon genes. ESTs databases from all platyhelminths deposited at GenBank, plus separate databases for Fasciola hepatica (∼ 25,000 EST contigs), S. haematobium, Echinococcus multilocularis, and E. granulosus (the last three obtained from WTSI: http://www.sanger.ac.uk/Projects/Helminths/) and genome sequences from S. japonicum (Zhou et al. 2009), Schmidtea_mediterranea (The Schmidtea mediterranea Sequencing Consortium), and E. multilocularis (WTSI) were used as databases for BLASTN and BLASTX searches with transcripts and protein products from all 18 MEG families, respectively. We found significant hits only for nine MEG families among S. japonicum ESTs and 11 MEG families among S. haematobium ESTs. Further analysis of the S. japonicum genome indicates that MEGs in this species have similar structure, with small symmetrical exons, to those verified in S. mansoni. Alignment of S. haematobium MEG ESTs with available 454 reads from a preliminary genome sequencing effort (available at http://www.cebio.org/projetos/schistosoma-haematobium-genome) suggest that a similar structure is also present in this parasite, but the fragmentary nature of the sequences currently prevents us from clearly defining the micro-exon structure of any genes.
Evolution of micro-exon genes
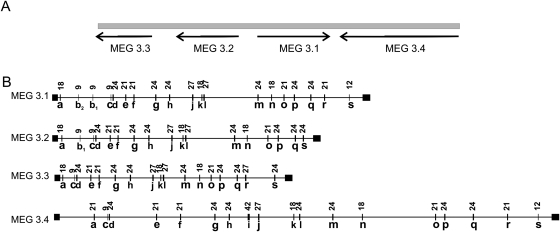
In order to understand better the evolution of micro-exon genes, the MEG-3 family was chosen since it contained multiple closely related members, well described in the assembled genome sequence. Figure 2 illustrates the structure of the four members of this family that form a gene cluster on a chromosome (Fig. 2A). Alignment of the protein products using BLASTP indicates that the first three members have ∼60% identity and 70% positive amino acids, with a fourth more divergent member (MEG-3.4; ∼25% identity and 40% positive). We interpret this organization to be the result of a series of gene duplications, one of them an inversion, with MEG-3.4 the first to diverge and MEG-3.2 and MEG-3.3 being the more closely related elements (Supplemental Fig. 3A). Due to the high similarity between the MEG-3.1, MEG-3.2, and MEG-3.3 gene sequences, we were able to align them to obtain further insights into the evolution of micro-exon genes. The number and size of micro-exons is largely conserved (Fig. 2B), implying a strong evolutionary pressure to maintain structure. Nonetheless, some differences are evident, with acquisition and loss of micro-exons during the history of this family (Supplemental Fig. 3A).
Figure 2.
Structure of micro-exon genes in the MEG-3 family. (A) Schematic representation of the position of four MEG-3 genes on a chromosome. (B) Structure of the four MEG-3 family members. Wide portions represent exons while narrow portions represent introns. Figure is to scale, but exons are depicted three times larger than introns to permit better visualization. Numbers above micro-exons indicate length in base pairs. Letters indicate homologous exons between the different members. Homology between MEG-3.1, MEG-3.2, and MEG-3.3 has been determined by genomic sequence alignment. Proposed homology between MEG-3.4 and the other genes has been determined by protein alignment, the similarity of exon positions in the gene and by their length.
A variation in the length of homologous micro-exons is detectable at the exons s and r (Fig. 2). A more detailed analysis of those sequences shows that variation in length of exon s was caused by two point mutations (Supplemental Fig. 3B). Considering that MEG-3.4 (12 bp in length for the same exon) must represent an ancestral state, it is possible to hypothesize that after its divergence, the original 12-bp exon was transformed into a 24-bp exon by the ancestral introduction of a stronger splicing acceptor site (condition illustrated by MEG-3.3 and MEG-3.2); the ancestral acceptor site was then destroyed by a point mutation in MEG-3.2. However, MEG-3.1 appears to have undergone an additional mutation that caused the loss of the newer stronger splicing signal and reversion of the exon to its former state. Another point mutation is responsible for the conversion of the 21-bp exon r present in MEG-3.1 and MEG-3.4 into a 27-bp exon in MEG-3.3 (Supplemental Fig. 3B).
The most remarkable instance is the gain of two 9-bp exons just after the portion encoding the signal peptide (Fig. 2, exons b1 and b2). After divergence from MEG-3.3, the ancestor of MEG-3.1 and MEG-3.2 must have undergone an insertion (∼1.2 kb) that contained the 9-bp sequence of that exon (G1-I1 and G2-I1 in Supplemental Fig. 4). We could not find any locus in the genome displaying a similar sequence, so we assume that a recombination event must have excised this sequence from its original position and transferred it to the MEG-3 gene. It remains to be determined whether this 9-bp exon was originally part of another micro-exon gene or appeared by chance after integration of this genomic portion into the MEG-3 gene. The second insertion in the MEG-3.1 gene (G1-I2 in Supplemental Fig. 4) has a smaller size (∼400 bp) and a high similarity to G1-I1 and G2-I2 (Supplemental Fig. 4). In addition, this second insertion is flanked by transposons with traits similar to the previously described Sm-alpha retroposon (Ferbeyre et al. 1998), such as the presence of a hammerhead ribozyme, being in that regard similar to the structure detected in some viroids of plants (Flores et al. 1999). We hypothesize that the insertion of such an element into the MEG-3.1 gene may have caused a double-strand break, and during the repair process a portion of the G2-I1 containing the 9-bp exon was copied, which would explain the high similarity between G1-I2 and G2-I1. We can infer that this is a series of exon gains, rather than deletions, because the more divergent ancestral MEG-3.4 contains only one 9-bp exon. In addition, it is possible to note that MEG-3.2 contains one fewer micro-exon near the end of the gene, relative to MEG-3.1 and MEG-3.3 (exon r, Fig. 2), due to a deletion of a portion of ∼1.3 kb.
Transcription of most micro-exon genes is up-regulated in the intramammalian stages of the parasite
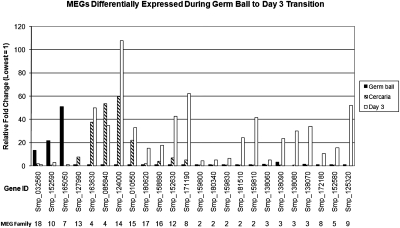
Analysis of all available EST data, by life cycle stage (Supplemental Table 1) indicates that MEGs are mostly transcribed in intramammalian stages of the parasite and in some cases in cercariae and germ balls, but are practically absent from miracidia, except for MEG-18. Probing of the oligonucleotide array with double-stranded cDNA from the three life cycle stages associated with infection of the mammalian host revealed numerous MEG transcripts among the genes showing the greatest change in expression, particularly in the day 3 schistosomula, but also in the germ balls and cercariae (Fig. 3). Only three of the 18 MEG families (MEG-1, MEG-6, and MEG-11) showed no variation in the three life cycle stages. Expression of MEG-1 was low, whereas MEG-6 was high in all three life cycle stages studied; MEG-11 was not detected (data not shown). Transcripts for MEG-7, MEG-10, and MEG-18 were highly expressed in the germ balls relative to the cercariae and schistosomula. It is notable that MEG-7 and MEG-10 were the only two for which ESTs generated from germ balls were found in public databases (Supplemental Table 1). Transcripts for MEG-13 were most highly expressed at the cercarial stage, while both MEG-4 genes were highly expressed in cercariae and schistosomula (up ∼50-fold) relative to the germ ball. All the remaining MEGs were most highly expressed in the schistosomulum, with MEG-8 (both genes), MEG-12 (one gene), MEG-14, MEG-15, and MEG-16 also having appreciable gene expression in cercariae. MEG-2 (six genes), MEG-3 (four genes), MEG-5, and MEG-17 were exclusively up-regulated in the day 3 schistosomulum, relative to the preceding stages. We have also been able to detect high expression of genes from four different MEG families (MEG-2, MEG-3, MEG-17, and MEG-18) in eggs when compared with cercariae (Supplemental Table 2).
Figure 3.
Detection of MEG expression using microarrays. Expression of MEG transcripts detected by hybridization of double-stranded cDNA from intramolluscan germ balls, infective cercariae and day 3 skin-stage schistosomula, to a genome-wide oligonucleotide array. Fold differences in level of expression are plotted relative to the lowest value, irrespective of stage, which is taken as unity. The triple bar groups are arranged by the stage in which highest expression was found and by their MEG family.
Micro-exon genes display stage-dependent alternative splicing
The occurrence of multiple symmetrical exons would allow one or more to be excised without altering the reading frame. MEG organization therefore clearly favors segmentation of virtually the whole coding region into short modules that might be differentially assembled, potentially to produce diverse protein structure. To investigate this possibility, we selected three MEG families (MEG-1, MEG-2, and MEG-3) to explore in detail, for two of which we had protein secretion data. RT-PCR was performed with primers designed for flanking exons to amplify transcripts. For each family, sampling of transcripts showed a diversity of products upon acrylamide gel separation (Supplemental Fig. 5A–C). It is also possible to note considerable specificity in MEG family expression with the absence of transcription of these selected genes in some stages and different amplification profiles between stages. Sequencing of randomly selected clones from the PCR products confirmed the existence of several transcripts for different members of the studied families. Mapping of these to the genome showed they originated by alternative splicing of micro-exons (Fig. 4; Supplemental Figs. 6–8). The presence of 14 cysteines in MEG-3 proteins implies a complex folded structure for the expressed protein and their position represents a highly conserved feature (Supplemental Fig. 9).
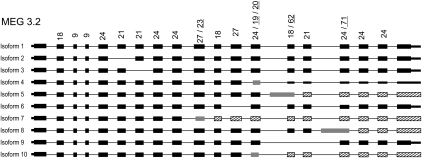
Figure 4.
Alternate splice variants of MEG-3.2. The deduced structure for sequenced transcripts of the MEG-3.2 gene is displayed. Wide lines represent coding region for exons, medium lines represent UTR regions, and narrow lines represent the introns. Exons generated by the use of an alternative splicing site are shown in gray. Coding exons that are being read in a frame different from the most abundant isoform (1) are shown as a hatched box. Exons are shown to scale, but for illustrative purpose intron length is not proportional to size. Numbers above exons indicate exon size, and underlined values are for exons generated by use of alternative splicing sites.
All four genes of the MEG-3 family are actively transcribed with three, 10, three, and two variants detected, respectively. Due to the micro-exon symmetry, in only one of 17 transcripts was an early stop codon introduced by splicing of an asymmetric long exon. Even more remarkable, in four cases asymmetric exons shifted the reading frame generating a novel and extended region (Fig. 4; Supplemental Fig. 10). In the other families studied the effect of alternative splicing of the micro-exon genes on the proteins coded was similar to that observed with MEG-3 (Supplemental Figs. 11, 12).
The impression of the MEG transcription profile gained by visual inspection of the low-resolution acrylamide gels is of numerous bands (Supplemental Fig. 5), whereas counting of sequenced clones suggests that one or a small number of variants dominate (Supplemental Tables 3–5), plus some minor variants. Nevertheless, MEG-1 displayed very distinct RT-PCR profiles when samples derived from adult worms and schistosomula were compared. A tally of clones from the two stages (Supplemental Table 4) revealed that isoforms 15–18 comprised 33% of schistosomula and 0% of adult clones whereas isoforms 2–4 plus 7–14 comprised 16% of adult and 0% of schistosomula clones. This observation suggests a specific stage-dependent mechanism for generation of splice variants, rather than a random process.
MEG-encoded proteins are secreted by migrating schistosomula and mature eggs
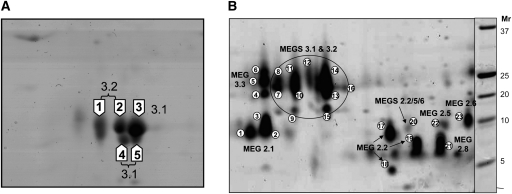
During investigations on the schistosome proteome we identified members of certain MEG families (Mathieson and Wilson 2010; R Curwen, unpubl.). We therefore made a detailed analysis of the relevant preparations, namely, the secretions of schistosomula and mature eggs cultured in vitro. We also examined the tegument surface membranes that comprise the interface between the adult parasite and the host bloodstream. A two-dimensional (2D) electrophoretic separation of the proteins released into culture by 0.5 million skin stage schistosomula between 3 h and 3 d (Supplemental Fig. 13; Fig. 5A) revealed a dominant cluster of gel spots with variable shapes, which are the major secreted protein. (The elongate spots may represent a group of proteins with similar pI and Mr, not resolved by the electrophoresis.) A similar but weaker staining pattern was observed in culture supernatants recovered from lung stage schistosomula between 8 and 10 d (data not shown). Excision of the spots and subjection of their tryptic digests to MALDI TOF/TOF MS revealed that the cluster comprised two members of the MEG-3 family, MEG-3.1 (three spots), and MEG-3.2 (two spots) (Supplemental Table 6). ProQ Emerald staining of gels failed to detect any glycan residues, so it is likely that the cluster represents proteins with small differences in primary sequence. Fainter spots in the cluster did not yield identities, but probably represent scarcer isoforms.
Figure 5.
MEG-encoded proteins are secreted by migrating schistosomula and mature eggs. (A) 2D electrophoretic separation of secretions released from skin-stage schistosomula between 3 h and 3 d of in vitro culture. (B) 2D electrophoretic separation of the less abundant egg-secreted proteins (ESPs) after lectin depletion of the major ESP1-6 proteins. Spot numbers inside the block arrows (A) or circles (B) correspond to the MS data in Supplemental Table 6. The MEG family number is adjacent to each spot or spot group.
Whereas viable schistosomula secrete very little protein when they are developing and migrating through the vasculature, the eggs of approximately the same biomass provide much more material for MS/MS analysis. Nevertheless, accumulation of sufficient egg-secreted protein (ESP) for analysis of the scarce proteins in lectin-depleted samples required the recovery of ∼2.5 million eggs from the livers of 250 mice. A 2D electrophoretic separation of the egg secretions revealed two very dominant spots, comprising >80% of the protein present, which correspond to two previously described egg proteins Omega-1 (ESP1-2) (GenBank DQ013207; Fitzsimmons et al. 2005) and ESP 3-6/IPSE (GenBank AF527011; Schramm et al. 2003) plus a number of fainter spots (Supplemental Fig. 14A; Mathieson and Wilson 2010). The dominant spots (both of which represent non-MEG proteins) stained with both ProQ Emerald and Sypro Ruby, confirming their glycosylated status, but the minor spots stained only with Sypro Ruby. Depletion of the two major glycoproteins by passage down a lectin column, although not complete, produced considerable enrichment of the scarcer spots (Supplemental Fig. 14B). All spots were excised from the gel and their individual tryptic digests subjected to LC separation and electrospray MS/MS to obtain maximum coverage of the peptide composition. The vast majority of spots proved to be MEG proteins, with three members of the MEG-3 (MEG-3.1, MEG-3.2, and MEG-3.3) and five members of the MEG-2 (MEG-2.1, MEG-2.2, MEG-2.5, MEG-2.6, and MEG-2.8) families identified (Fig. 5B; Supplemental Table 6). The MEG-3 members showed a greater diversity of position than in the schistosomular preparations, with pI ranging from 4 to 6 and Mr from 15 to 25 kDa. They occupied both distinct spots and vertical smears on the gel, suggestive of a diversity of isoforms present. However, all peptides identified were encoded by adjacent micro-exons with no examples of exon skipping reaching statistical significance (Supplemental Table 6). The MEG-2 family members had a lower average Mr ranging from 5 to 10 kDa and showed a much wider pI, ranging from 3 to 8 kDa. Nevertheless, the MEG-2 members also showed some diversity of position on the gel, especially MEG-2.1 and MEG-2.2, which reflects the range of isoforms present. Only a single MEG protein, MEG-5, was identified in the tegument membrane preparations from adult worms.
MEG transcripts and proteins are expressed in epithelia and gland cells
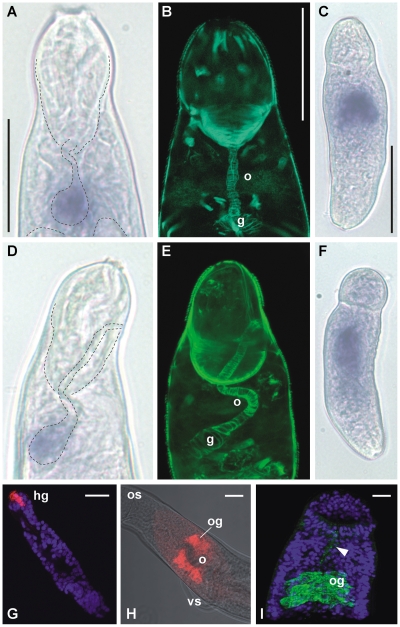
As virtually all MEGs possessed a signal sequence indicating that they encoded either secreted or membrane-anchored proteins, we examined the sites of expression of a small number by WISH and immunocytochemistry. (The mature egg was precluded due to the permeability barrier to antibodies presented by its shell.) Expression of MEG-4.2 and MEG-14 was detected in a very small discrete area of the cercaria (Fig. 6A,D). The distribution of f-actin in smooth muscle fibers revealed that this staining pattern is associated with the gut primordium (Fig. 6B,E). In accord with the microarray data, expression of both MEGs had increased considerably by 3 d of in vitro culture (Fig. 6C,F). However, it was not possible to say whether the precise site lay in the developing gut caecae, the nascent esophageal gland, or both. Expression of a second MEG-4 family member was investigated in adult worms, where much greater resolution is possible. High-level expression of MEG-4.1 was confined to the esophageal gland (Fig. 6H) that lies ventral to the posterior esophagus, into which its secretions empty. This localization was confirmed by the presence of MEG-4.1 protein in the glands and also in the esophageal lumen, detected by specific antibody to a synthetic peptide (Fig. 6I). Finally, expression of MEG-3.2 transcripts was identified in a very discrete area at the extreme anterior of the schistosomulum (Fig. 6G). This lies within the muscular head capsule and corresponds to the head gland, secretions from which are used in skin and intravascular migration.
Figure 6.
Whole-mount in situ hybridization (A,C,D,F–H) and immunocytochemistry (C,E,I) to identify the location of selected MEG gene expression products. (A) Focal expression of MEG-4.2 associated with the gut primordium of the nonfeeding infective cercariae, lying on its back. The fine dashed lines indicate the positions of the muscular head capsule and gut to aid interpretation. (B) The position of the embryonic esophagus and gut in a cercaria with similar orientation, revealed by Phalloidin staining of actin in the surrounding myocytes. (C) Increased expression of MEG-4.2 in the day 3 (skin stage) schistosomulum. (D) Focal expression of MEG-14 in a cercariae lying on its side, with structures outlined as in A. (E) A phalloidin stained day 3 schistosomulum in the same orientation to aid interpretation. Note that myocytes around the gut show only minimal expansion of the organ. (F) Increased expression of MEG-4.2 in the day 3 schistosomulum. (G) MEG-3.2 transcript expression sharply demarcated in the head gland (hg) of a day 10 (lung stage) schistosomulum. (H) Expression of MEG-4.1 in the esophageal gland (og) that envelops the ventral aspect of the esophagus (o). The gland lies adjacent to the ventral sucker (vs) distal to the mouth and oral sucker (os). (I) Confocal projection of MEG-4.1 protein detected in the male esophageal gland (og) using antibody to a synthetic peptide; traces of protein are also present in the esophageal lumen (arrowhead). The nuclei are counterstained with DAPI to reveal body outline. Magnifications: A,B,D,E all approximately the same, bar, 20 μm; C,F, bar, 50 μm; G–I, bars, 20 μm.
Discussion
Here, we characterize a novel class of genes with a unique structure that code for secreted proteins in schistosomes. Previously, micro-exons have largely been described as occurring singly in genes with normal-sized exons (Volfovsky et al. 2003). Apparently, MEGs are restricted to schistosomes, as no homologous proteins could be found in other genera, despite intensive computational searches in public databases that included genomes and EST data from other Platyhelminthes. The absence of homology with proteins from any other organism and the lack of any known protein domain prevented us from computationally assigning any function to this obviously diverse range of proteins, other than their likely secretion from cells.
Our data suggest a mechanism for acquisition and diversification of micro-exons by duplication or recombination of genomic segments containing micro-exons and modification of micro-exon lengths by point mutation events generating new splicing sites. Notably, most of the modifications observed in MEG-3 family gene structure occur at their extremities, suggesting that an evolutionary pressure must exist to restrict disruptive modifications at the core of the protein. In a particular case, the duplication of one exon was apparently associated with the insertion of a retroelement with viroid-like characteristics; such elements are mainly represented in plants but have been previously described in schistosomes (Ferbeyre et al. 1998). It is still unclear if these exon duplications are somehow triggered by events of transposition, but if this is the case the high transcription levels of some retroelements in S. mansoni (DeMarco et al. 2004) may constitute a catalyst for evolutionary modifications of MEG structure. The intricate structure of MEGs is by no means a trivial one to acquire and maintain, and we speculate that some adaptive advantage must arise from such a structure. Moreover, some of the families had an expansion in the number of members as a result of several gene duplication events. Some of these expansions were quite recent, as demonstrated by the strong similarity of the genomic regions for three MEG-3 family members.
The absence of this class of genes in other platyhelminths suggests that a rapid appearance and expansion must have occurred in an ancestor located at a basal position of the Schistosoma genus, thus representing an adaptation to parasitism. Considering that scenario, we hypothesize that the development of micro-exon genes in schistosomes resulted from the expansion of a few ancestral micro-exons that were reiteratively duplicated and mutated in order to generate the multiple micro-exons observed in these genes. Together with gene duplication events, this mechanism could explain the emergence of the numerous families. Considering that similarities between proteins coded by genes of different families are very low, we must assume that mutation rates in MEGs are quite high. In fact, examining the MEG-3 family, the identity observed between the MEG-3.4 protein product and the other members of the family is low (∼26%), despite the very similar gene structure depicted in Figure 2. The assembly of a micro-exon gene from multiple sources of micro-exons combined with the high mutation rates in the small sized exons would create new families that in relatively short time would not resemble any of the parent sources of micro-exons.
RT-PCR experiments of selected MEGs showed that some copies of the genes produce numerous splice variants. Such events provide an explanation for maintenance of small symmetrical exons, since they permit small portions of a transcript be removed without disrupting the reading frame. As a result, we anticipated that translation of such variant transcripts would generate several protein isoforms from each MEG, most of them with very small differences in molecular weight and pI. In fact, in the secretions of eggs and migrating schistosomula we found just that. We detected multiple gel spots that were identified as proteins coded by five copies of MEG-2 and three of MEG-3. Considering the variable appearance of the spots and the resolution of 2D electrophoresis, it is very unlikely that all isoforms were individually separated suggesting that each spot/smear represents one to several isoforms. Trypsinization of a mixture of isoforms contained in a single spot would create a pool of peptides for MS/MS in which the more common peptides would predominate relative to those representing an arrangement less represented in the pool. This may explain why we have so far been unable to identify exon-skipping peptides diagnostic for single splice variants in our mass spectrometric analysis at an acceptable level of statistical significance, detecting only more abundant peptides which represent portions coded by adjacent exons. It is not possible to make a clear correlation between the abundance of the spots in the 2D gel and the count of DNA clones, which suggests that translational controls may be occurring to influence the abundance of proteins. Moreover, cloning of cDNA is not a totally unbiased process and some messages may be under/overrepresented, depending on several factors.
Finally, the MEGs largely encode secreted proteins and we have confirmed for a small number that they are expressed in epithelia and gland cells. Many of the MEG transcripts are up-regulated in stages involved in skin penetration, and intravascular migration during which the parasite gut develops to initiate blood feeding. In the case of the schistosome egg, the MEG-2 and MEG-3 variants are particularly prolific and may be involved in egg transit from the blood vessel to the lumen of the host gut. (In this context, it is important to establish where the rest of the MEG proteins are expressed.) All the above are situations where MEG proteins can interact with the host immune system.
Schistosomes are unusual among parasitic platyhelminths in dwelling in the mammalian bloodstream, where they are bathed in antibodies and complement and surrounded by leukocytes. The generation of variant proteins (and hence antigens) as a strategy to evade host immune responses is a recurrent theme in the analogous protozoan blood parasites Trypanosoma and Plasmodium (Taylor and Rudenko 2006; Kyes et al. 2007). It is thus tempting to conclude that the protein variation we have described in schistosomes is similarly one component of an immune evasion strategy, conferring some benefit on the parasite. However, there are significant differences between schistosomes and the two protozoans. The schistosome variant proteins are produced simultaneously, not sequentially. In addition, there is currently no evidence for the distribution of allelic variants of MEGs across schistosome populations. If the generation of protein variation by expression of MEGs is a strategy for immune evasion, then a different underlying mechanism is implicit in schistosomes. Nevertheless, the extent of MEG transcript and protein variation suggests some form of pressure is exerted to select for micro-exon persistence when it has appeared, potentially by the immune system. The ramifications of MEG diversity in the context of immune evasion are a subject for future work. Individual schistosomes can survive in the bloodstream for >30 yr (Harris et al. 1984) and do not possess intracellular stages that would be protected from host immune attack. It would be expected that strategies to ensure long-term immune evasion by using protein variation in schistosomes would not be the same of those observed in protozoan parasites, where need for long-term immune evasion is less imperative since either elimination of the infection or death of the host occurs on a much shorter time span.
Methods
Biological material
Infective cercariae from a Puerto Rican isolate of S. mansoni were shed from Biomphalaria glabrata snails by exposure to bright light. Adult worms were obtained by portal perfusion of C57BL/6 mice 7 wk after percutaneous infection with 180 cercariae. Eggs were recovered from female NMRI mouse livers by trypsin digestion and then cleaned by washing and sieving in PBS (Ashton et al. 2001). Schistosomula were derived from samples of 0.5 million cercariae, mechanically transformed, and cultured in vitro in 24-well plates for up to 10 d, as required (Dillon et al. 2006). The schistosomula need a protein supplement in the medium to develop normally and retain viability but this is incompatible with proteomic analysis. The problem was solved by short-term culture in medium containing human serum albumin (HSA) at 10 μg/mL concentration, either between 3 h and day 3 or day 8 and day 10. Viability of parasites under these culture conditions was checked by trypan blue exclusion and by visual inspection for lack of motility and ”granular appearance” to indicate death; it was found to be the same as with addition of fetal calf serum over a similar period. A single culture of 0.5 million schistosomula, distributed between eight 24-well plates at 1.5 mL medium per well, yielded ∼300 mL of medium containing 2.9 mg of HSA. This was depleted from the culture supernatant using a cartridge containing POROS perfusion chromatography medium coupled to goat anti-human albumin IgG (a gift from Applied Biosystems), with a binding capacity of 2 mg/mL and a total volume of 1.4 mL. The manufacturer's recommendations not to exceed 50% of the binding capacity of the cartridge were adhered to so a single culture supernatant was split into two or three subsamples, according to its protein content, for HSA depletion, with intervening cartridge regeneration steps. Egg-secreted proteins were collected by incubating mature eggs in serum-free RPMI (GIBCO) for two periods of 3 d with a medium change interposed (Ashton et al. 2001; Mathieson and Wilson 2010). Post-culture viability of eggs was routinely >96%, as assessed by observation of muscular and flame cell activity in unhatched miracidia. Germ ball progenitors of cercariae, derived from daughter sporocysts, were carefully dissected from snail hepatopancreas 22 to 26 d after infection with 40 miracidia per snail.
Microarray experiments
Integral to a study of changes in gene expression associated with infection of the mammalian host we designed a comprehensive oligonucleotide microarray comprising a compilation of the S. mansoni gene predictions on GeneDB (http://www.genedb.org/genedb/smansoni/) together with expressed sequence tag (EST) contig assemblies available on the Wellcome Trust Sanger Institute (WTSI) ftp site (ftp://ftp.sanger.ac.uk/pub/pathogens/Schistosoma/mansoni/) (Manuel 2010), that was manufactured by Roche/Nimblegen. Samples of total RNA were extracted with TRIzol (Invitrogen) following manufacturer's instructions and high-salt solution (0.8 M sodium citrate and 1.2 M NaCl) was added with the isopropanol to remove glycoprotein. Using this protocol, RNA was extracted from three biological replicates of the intramolluscan germ balls, cercariae, schistosomula transformed from cercariae and cultured in vitro for 3 d to the skin stage of development (Dillon et al. 2006), and eggs (Mathieson and Wilson 2010). Double-stranded cDNA was synthesized (without amplification) using a SuperScript II kit (Invitrogen) according to the manufacturer's instructions, labeled with Cy3, and hybridized to the array overnight at 42°C. Two-way comparisons were made of gene expression levels in the three life cycle stages. Fluorescence intensity data were normalized at the probe level, before being averaged for each S. mansoni locus, analyzed using the Bioconductor package (http://www.bioconductor.org/) and values that differed by a fold change of two between stages with an adjusted P < 0.0001 and a B-value > 3 (corresponding to a 95% confidence limit), were taken as significant. Expression of MEGs in eggs was assessed against the level in the cercaria, set at unity.
RT-PCR and cloning of MEG transcripts
Approximately 400 ng of mRNA from 7-d cultivated schistosomula, adult worms, and eggs were treated with 5 U of RQ1 RNase-free DNase (Promega) for 30 min at 37°C. DNase-treated mRNA was separated into two aliquots, one being used for RT-PCR and the other for a no-RT parallel negative control in which the same reagents of the normal reaction were added, except for the reverse transcriptase. Reverse transcription was performed with 50 U of Superscript II (Invitrogen) and 0.5 μg of poly-dT primer in a 20-μL volume for 60 min at 42°C. The PCR step was performed with Advantage II polymerase (Clontech) with buffer supplied by the manufacturer, 1 μL of reverse transcription reaction and 200 nM of each primer using the following program: 94°C (1 min); 35 cycles of 94°C (30 sec), 55°C (30 sec), and 68°C (5 min); final extension of 68°C (5 min). PCR products were submitted to electrophoresis in polyacrylamide gel and stained by silver using a previously described protocol (Sanguinetti et al. 1994). Products were cloned into pGem-T vector (Promega) and resulting clones stocked in TB-ampicillin. Colony PCR was performed using Biolase DNA polymerase (Bioline), 10 pmol of each specific internal primer and the following cycling: 94°C (5 min), plus 40 cycles of 94°C (30 sec), 57°C (45 sec), 72°C (1 min), and final extension of 72°C for 5 min. Resulting products were sequenced by the dideoxyterminator method using primers for both extremities of the clone.
Resulting sequences for each clone were individually assembled using the phredphrap package and were compared against each other after multiple alignment using the ClustalX program (Larkin et al. 2007). Sequences were separated into groups representing each splice variant. One sequence representative of each group was chosen and mapped into the genome sequence for deduction of its structure using the Spidey program (Wheelan et al. 2001), followed by manual curation. A representative sequence for each isoform was submitted to GenBank under accession numbers GU258169–GU258215 and GU258219
Cloning and sequencing of genomic regions of MEG-3
To obtain a complete sequence for the MEG-3 family, PCR from genomic DNA was performed using primers flanking existing gaps in the gene sequences. All primers designed were aligned against the genome sequence from S. mansoni using the BLASTN program and the result verified to assure that they mapped perfectly only to the desired position in the genome. The PCR step was performed with Advantage II polymerase (Clontech) with buffer supplied by the manufacturer, 100 ng of genomic DNA, and 200 nM of each primer using the following program: 94°C (1 min); 35 cycles of 94°C (30 sec), 55°C (30 sec), and 68°C (5 min); final extension of 68°C (5 min). Resulting amplicons were cloned and sequenced. These genomic sequences were deposited in GenBank under the accession numbers GU258216–GU258218.
Proteomics
Sample preparation
Culture medium containing ESPs was gravity-filtered through 0.2-μm filters then concentrated in Vivaspin-20, 5-kDa cutoff, centrifugal concentrators (both Sartorius, Fisher Scientific). Samples containing 75–500 μg of soluble protein were fractionated by standard 2D electrophoresis techniques (Curwen et al. 2004). Culture medium containing ESPs was first replaced by dialysis for 18 h at 4°C into 20 mM Tris-HCl (pH 7.2) (Sigma) using a Slide-A-Lyzer dialysis unit (Pierce), concentrated to less than 20 μL with an Amicon Ultrafree centrifugal concentrator (Millipore) and then diluted in rehydration buffer to a total of 125 μL. All preparations were separated by isoelectric focusing (IEF) on 7-cm, pH 3–10, IEF strips (Amersham Biosystems), under the following conditions: 15 h rehydration, then 500 V (30 min), 1000 V (30 min), and 8000 V (4 h) (at 50 microA per strip). The strips were then reduced, alkylated, and separated by molecular mass on 4%–12% SDS-PAGE gels (Invitrogen). The gels were fixed in 40% methanol/10% acetic acid solution and stained initially with ProQ Emerald for glycans and/or Sypro Ruby for proteins (Invitrogen), according to the manufacturer's instructions, prior to imaging on a Versadoc Imaging System Model 3000 (Bio-Rad), and then with Biosafe Coomassie (Bio-Rad) prior to excision of spots. ESPs of low abundance were enriched using the previously demonstrated affinity of the major protein, IPSE, for Aleuria aurantia lectin (Schramm et al. 2003), as adapted by Mathieson and Wilson (2010). A 460-μg aliquot of crude ESP (∼1 mL) was applied to the lectin column and the effluent (containing lectin-depleted ESP) collected over four column volumes. It was then processed for 2D electrophoretic separation on 7-cm gels as previously described (Mathieson and Wilson 2010).
“Flow-through” fractions from the HSA depletion column were assessed for protein concentration by the Coomassie Plus assay (Pierce), and those containing schistosomular-secreted proteins were combined. They were concentrated ∼30-fold using a Vivaspin 20 (5 kDa cutoff) spin column before desalting prior to separation by 2D electrophoresis.
Tegument membranes were removed from adult worms by a freeze/thaw method and surface membranes enriched by sucrose-gradient centrifugation as previously described (Roberts et al. 1983). They were pelleted by centrifugation at 100g for 30 min at 4°C, resuspended in 0.5 mL of 10 mM Tris, and enriched on a continuous 20%–70% sucrose gradient, with centrifugation at 100,000g for 40 min. The fractions containing alkaline phosphatase activity (an apical membrane marker) were pooled and centrifuged at 100,000g for 1 h to produce a “gradient pellet” highly enriched in surface membranes. Proteins were sequentially extracted from the gradient pellet using a three-step process with reagents of increasing solubilizing power to yield a final pellet (Braschi et al. 2006), which was solubilized in 200 μL of 0.1% SDS, 1% Triton X-100 in 20 mM NH4HCO3, and digested overnight in-solution with trypsin at an enzyme:substrate ratio of ∼1:20. The digested material was first separated by cation exchange and then by reversed phase chromatography and collected directly onto a MALDI plate as previously described (Braschi et al. 2006).
Protein digestion and MALDI-MS/MS
Gel pieces were destained with 40% methanol/10% acetic acid solution, washed in 20 mM ammonium bicarbonate/50% acetonitrile solution, dehydrated in 100% acetonitrile, dried in a Speedvac (Thermo Lifesciences), and digested into peptides overnight at 37°C with 20 μL of 10 μg/mL sequencing grade porcine trypsin per gel piece (Promega). The peptides were applied to C18 ZipTips (Millipore), desalted as per the manufacturer's instructions, and eluted in 5 μL of 80% acetonitrile/0.1% TFA solution. Two microliters of the eluate were mixed with 1 μL of matrix solution (α-cyano-4-hydroxycinnamic acid [Sigma] saturated in a 50% acetonitrile/0.1% TFA solution) and dried onto a MALDI plate.
Tryptic peptides from schistosomular secretions, adult tegument, and the ESP before lectin depletion were analyzed on a 4700 MALDI-TOF/TOF mass spectrometer (Applied Biosystems) operating in positive ion reflector mode. A peptide mass fingerprint was acquired from each spot using 1500 laser shots at 2800–3500 V and the 15 most intense peaks were selected for fragmentation (excluding common tryptic autolysis and human keratin peaks). The MS/MS data were processed by GPS Explorer (Applied Biosystems) and the resulting peak lists exported to Mascot v 2.1 (Matrix Science). The search parameters were set so that only MS/MS fragmentation data with a tolerance <0.2 Da were used. One missed cleavage was allowed for, as were carbamidomethylation modifications to the cysteine and oxidation of the methionine residues. Databases searches were (1) NCBInr; (2) SchistoCDS, an in-house database comprising clusters of all publicly available ESTs (Sm numbers) plus gene predictions from version 1 of the S. mansoni genome database (http://www.genedb.org); (3) Sm genes + ESTs which superseded SchistoCDS when version 4 was loaded into GeneDB; (4) Exon permutations. This last is a theoretical database comprising all possible combinations of exons encoding MEG-3 and some MEG-2 family proteins. Created by Peter Ashton, it was an essential tool to facilitate Mascot searching for exon skipping, which would not be detected by the normal Mascot search algorithm. Conventional matches were only accepted if the confidence interval of the total peptide ion scores was >99.9%.
Electrospray tandem MS
In order to obtain better coverage, peptides derived from lectin-depleted ESPs were subjected to electrospray tandem MS. Aliquots (1–3 μL) of trypsin-digested spots were loaded onto an Ultimate nano-HPLC system (Dionex) equipped with a PepMap C18 trap (Dionex), previously equilibrated and washed with 0.1% (v/v) formic acid, and an Onyx C18 monolithic silica capillary column (Phenomenex). A gradient elution of sample was achieved by the sequence: initial eluant 100% Solvent A (2% [v/v] acetonitrile, 0.1% [v/v] formic acid in H2O) for 3 min, trap switched, a 0%–50% linear gradient of Solvent B (pure acetonitrile, 0.1% [v/v] formic acid) for 20 or 60 min, a 5-min wash at 95% Solvent B, all at a flow rate of 1.2 μL/min and column temperature 60°C. Control and data acquisition were with Chromeleon v6.6 software (Dionex). The HPLC was interfaced with a QSTAR API Pulsar i Hybrid LC/MS/MS System (Applied Biosystems) with a MicroIonSpray source (fitted with a 20-μm ID capillary). Positive ESI MS and MS/MS spectra were acquired over the range 350–1800 m/z using information dependent acquisition with instrument, data acquisition, and analysis controlled by Analyst QS v1.1 software. MS and MS/MS data were searched as for MALDI, above.
Whole-mount in situ hybridization and immunocytochemistry
For whole-mount in situ hybridization (WISH), parasites were fixed and processed as described by Dillon et al. (2007). The preparation of digoxygenin-labeled probes, the conditions for in situ-hybridization, and detection of expression sites were also as previously described (Dillon et al. 2007). The substrates BM purple and Fast Red TR were used for conventional light and confocal microscopy detection, respectively. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) where confocal microscopy was used.
For visualization of the musculature by f-actin, larvae were stained with Alexafluor 488-conjugated phalloidin (Invitrogen, Molecular Probes) and counterstained with DAPI according to the protocol of Bahia et al. (2006). Specimens were fixed for 1 h in 4% paraformaldehyde in phosphate buffered saline (PBS) on ice and then permeabilized for 1 h in PBS, 0.2% gelatin, 0.1% saponin, 0.1% NaN3 at room temperature, before staining, and washed several times in PBS before viewing. For detection of MEG-4.1 in adult worms, an antibody was raised by vaccinating rats with a synthetic peptide (CPKSIADIFLINKPKVP-OH derived from the protein sequence and coupled via the cysteine to carrier ovalbumin), emulsified in Titermax Gold (Sigma-Aldrich), and subsequently boosted at 3 wk with conjugate alone. Whole adult worms were fixed and permeabilized using the protocol described by Mair et al. (2000) prior to reaction with primary antibody, extensive washing, and detection of localization by Alexa-fluor 488 labeled goat-anti rat antibody (Sigma). They were counterstained with DAPI before examination under a LSM 510 Meta confocal microscope (Zeiss).
Acknowledgments
This work was financed by a “young researcher” and “proteomic network of São Paulo” grants from Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP), by the program “Institutos Nacionais de Ciência e Tecnologia” (INCT) from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq/MCT) and FAPESP, and by a postdoctoral fellowship of Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brazil (CAPES). W.M. and S.J.M. were supported by BBSRC postgraduate studentships, R.S.C. by a research grant from the Wellcome Trust, and G.P.D. by NIH grant AI054711-01A2, (P.I. Dr. C King, Case Western Reserve University, Cleveland, OH), both held by R.A.W. The microarray element received core support from the Pathogen Sequencing Group, Wellcome Trust Sanger Institute, Hinxton, Cambridge. We thank Renato Alvarenga for technical assistance. M.B. is supported by the Wellcome Trust grant number WT085775/Z/08/Z.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/genbank) under accession nos. GU258169–GU258219. The microarray data from this study have been submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession nos. GSE22037 and GPL10466. The proteomic data from this study have been submitted to the Proteomics Identification Database (http://www.ebi.ac.uk/pride/) under accession nos. 12831–12859 and 12873–12878.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.100099.109.
References
- Anders RF, McColl DJ, Coppel RL 1993. Molecular variation in Plasmodium falciparum: Polymorphic antigens of asexual erythrocytic stages. Acta Trop 53: 239–253 [DOI] [PubMed] [Google Scholar]
- Ashton PD, Harrop R, Shah B, Wilson RA 2001. The schistosome egg: Development and secretions. Parasitology 122: 329–338 [DOI] [PubMed] [Google Scholar]
- Bahia D, Avelar LG, Vigorosi F, Cioli D, Oliveira GC, Mortara RA 2006. The distribution of motor proteins in the muscles and flame cells of the Schistosoma mansoni miracidium and primary sporocyst. Parasitology 133: 321–329 [DOI] [PubMed] [Google Scholar]
- Berriman M, Haas BJ, LoVerde PT, Wilson RA, Dillon GP, Cerqueira GC, Mashiyama ST, Al-Lazikani B, Andrade LF, Ashton PD, et al. 2009. The genome of the blood fluke Schistosoma mansoni. Nature 460: 352–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braschi S, Curwen RS, Ashton PD, Verjovski-Almeida S, Wilson A 2006. The tegument surface membranes of the human blood parasite Schistosoma mansoni: A proteomic analysis after differential extraction. Proteomics 6: 1471–1482 [DOI] [PubMed] [Google Scholar]
- Chalmers IW, McArdle AJ, Coulson RM, Wagner MA, Schmid R, Hirai H, Hoffmann KF 2008. Developmentally regulated expression, alternative splicing and distinct sub-groupings in members of the Schistosoma mansoni venom allergen-like (SmVAL) gene family. BMC Genomics 9: 89 doi: 10.1186/1471-2164-9-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curwen RS, Ashton PD, Johnston DA, Wilson RA 2004. The Schistosoma mansoni soluble proteome: A comparison across four life-cycle stages. Mol Biochem Parasitol 138: 57–66 [DOI] [PubMed] [Google Scholar]
- Davis RE, Davis AH, Carroll SM, Rajkovic A, Rottman FM 1988. Tandemly repeated exons encode 81-base repeats in multiple, developmentally regulated Schistosoma mansoni transcripts. Mol Cell Biol 8: 4745–4755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarco R, Kowaltowski AT, Machado AA, Soares MB, Gargioni C, Kawano T, Rodrigues V, Madeira AM, Wilson RA, Menck CF, et al. 2004. Saci-1, -2, and -3 and Perere, four novel retrotransposons with high transcriptional activities from the human parasite Schistosoma mansoni. J Virol 78: 2967–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon GP, Feltwell T, Skelton JP, Ashton PD, Coulson PS, Quail MA, Nikolaidou-Katsaridou N, Wilson RA, Ivens AC 2006. Microarray analysis identifies genes preferentially expressed in the lung schistosomulum of Schistosoma mansoni. Int J Parasitol 36: 1–8 [DOI] [PubMed] [Google Scholar]
- Dillon GP, Illes JC, Isaacs HV, Wilson RA 2007. Patterns of gene expression in schistosomes: Localization by whole mount in situ hybridization. Parasitology 134: 1589–1597 [DOI] [PubMed] [Google Scholar]
- Ferbeyre G, Smith JM, Cedergren R 1998. Schistosome satellite DNA encodes active hammerhead ribozymes. Mol Cell Biol 18: 3880–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzsimmons CM, Schramm G, Jones FM, Chalmers IW, Hoffmann KF, Grevelding CG, Wuhrer M, Hokke CH, Haas H, Doenhoff MJ, et al. 2005. Molecular characterization of omega-1: A hepatotoxic ribonuclease from Schistosoma mansoni eggs. Mol Biochem Parasitol 144: 123–127 [DOI] [PubMed] [Google Scholar]
- Flores R, Navarro JA, de la Pena M, Navarro B, Ambros S, Vera A 1999. Viroids with hammerhead ribozymes: Some unique structural and functional aspects with respect to other members of the group. Biol Chem 380: 849–854 [DOI] [PubMed] [Google Scholar]
- Harris AR, Russell RJ, Charters AD 1984. A review of schistosomiasis in immigrants in Western Australia, demonstrating the unusual longevity of Schistosoma mansoni. Trans R Soc Trop Med Hyg 78: 385–388 [DOI] [PubMed] [Google Scholar]
- Kyes SA, Kraemer SM, Smith JD 2007. Antigenic variation in Plasmodium falciparum: Gene organization and regulation of the var multigene family. Eukaryot Cell 6: 1511–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Mair GR, Maule AG, Day TA, Halton DW 2000. A confocal microscopical study of the musculature of adult Schistosoma mansoni. Parasitology 121: 163–170 [DOI] [PubMed] [Google Scholar]
- Manuel SJ 2010. “Patterns of gene expression in Schistosoma mansoni larvae associated with infection of the mammalian host.” PhD thesis, University of York, York, UK [Google Scholar]
- Mathieson W, Wilson RA 2010. A comparative proteomic study of the undeveloped and developed Schistosoma mansoni egg and its contents: The miracidium, hatch fluid and secretions. Int J Parasitol 40: 617–628 [DOI] [PubMed] [Google Scholar]
- Roberts SM, MacGregor AN, Vojvodic M, Wells E, Crabtree JE, Wilson RA 1983. Tegument surface membranes of adult Schistosoma mansoni: Development of a method for their isolation. Mol Biochem Parasitol 9: 105–127 [DOI] [PubMed] [Google Scholar]
- Sanguinetti CJ, Dias Neto E, Simpson AJ 1994. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques 17: 914–921 [PubMed] [Google Scholar]
- Schramm G, Falcone FH, Gronow A, Haisch K, Mamat U, Doenhoff MJ, Oliveira G, Galle J, Dahinden CA, Haas H 2003. Molecular characterization of an interleukin-4-inducing factor from Schistosoma mansoni eggs. J Biol Chem 278: 18384–18392 [DOI] [PubMed] [Google Scholar]
- Skelly PJ, Wilson RA 2006. Making sense of the schistosome surface. Adv Parasitol 63: 185–284 [DOI] [PubMed] [Google Scholar]
- Smyth D, McManus DP, Smout MJ, Laha T, Zhang W, Loukas A 2003. Isolation of cDNAs encoding secreted and transmembrane proteins from Schistosoma mansoni by a signal sequence trap method. Infect Immun 71: 2548–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JE, Rudenko G 2006. Switching trypanosome coats: What's in the wardrobe? Trends Genet 22: 614–620 [DOI] [PubMed] [Google Scholar]
- Volfovsky N, Haas BJ, Salzberg SL 2003. Computational discovery of internal micro-exons. Genome Res 13: 1216–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelan SJ, Church DM, Ostell JM 2001. Spidey: A tool for mRNA-to-genomic alignments. Genome Res 11: 1952–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zheng H, Liu F, Hu W, Wang ZQ, Gang L, Ren S 2009. The Schistosoma japonicum genome reveals features of host-parasite interplay. Nature 460: 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]