Abstract
Mutational screens are an effective means used in the functional annotation of a genome. We present a method for a mutational screen of the mouse X chromosome using gene trap technologies. This method has the potential to screen all of the genes on the X chromosome without establishing mutant animals, as all gene-trapped embryonic stem (ES) cell lines are hemizygous null for mutations on the X chromosome. Based on this method, embryonic morphological phenotypes and expression patterns for 58 genes were assessed, ∼10% of all human and mouse syntenic genes on the X chromosome. Of these, 17 are novel embryonic lethal mutations and nine are mutant mouse models of genes associated with genetic disease in humans, including BCOR and PORCN. The rate of lethal mutations is similar to previous mutagenic screens of the autosomes. Interestingly, some genes associated with X-linked mental retardation (XLMR) in humans show lethal phenotypes in mice, suggesting that null mutations cannot be responsible for all cases of XLMR. The entire data set is available via the publicly accessible website (http://xlinkedgenes.ibme.utoronto.ca/).
X-linked genetic disorders play a major role in the impact of Mendelian traits on human health. The human X chromosome contains only 4% of all annotated genes (1098) (Ross et al. 2005), but nearly 10% of all known Mendelian diseases in the Online Mendelian Inheritance in Man (OMIM) database are linked to the X chromosome (387/3983), as of October 2009. It has been suggested that this may be due to a discovery bias (Ross et al. 2005). Most of these traits disproportionately affect males, where there is only one copy of the mutant X chromosome. Females can often escape severe effects of these mutations because X inactivation renders them functionally mosaic. Alternatively, the X chromosome carrying the mutated allele is preferentially inactivated (Garrick et al. 2006). Of these X-linked diseases, 30% (149/387) are not mapped to a mutation in a specific gene. In addition, embryonic lethal mutations in X-linked genes are thought to underlie the preferential loss of male embryos in many cases of recurrent spontaneous abortion (RSA) (Pegoraro et al. 1997; Lanasa et al. 2001). Clearly, there is a need to better understand the function of genes on the X chromosome in order to link them to the full range of human X-linked disorders.
Comparative analysis of the mouse and human genomes reveals that there is a high degree of conservation and synteny between the X chromosomes, with 90% of known genes (629/699) conserved (Ross et al. 2005). The ability to genetically manipulate the genome in mouse embryonic stem cells (mESC) makes it possible to generate mouse models of human X-linked disorders. However, phenotypic analysis of such mutations to date has been slow, because of the frequent difficulties in generating high-percentage chimeras with hemizygous XY mESCs and transmitting the mutant X-linked allele through the germ line. There are 211 targeted mutant phenotypes on the mouse X chromosome reported in the Mouse Genome Database (http://www.informatics.jax.org/phenotypes.shtml), 50 of which are associated with a human disorder. However, the efforts of the International Gene Trap Consortium (IGTC) (Nord et al. 2006) and the International Knockout Mouse Consortium (IKMC) (International Mouse Knockout Consortium 2007) have placed a growing resource of mutant mESC lines in the public domain. As of January 2010, 473 of the genes on the mouse X chromosome are now associated with either a gene trap (379 genes) or targeted mutant cell line. As a means of identifying novel disease genes, the mouse gene trap (GT) resource can be mined for insertions in genes with human X-linked orthologs that have yet to be associated with developmental processes in either mice or humans.
Using a straightforward approach of generating and analyzing mESC-derived embryos, we have added embryonic phenotypic and functional data to almost 10% of syntenic genes on the mouse X chromosome. This rapid and cost-effective screening method has identified 19 (13 novel to mouse and human) X-linked mutations that cause developmental defects in the mouse and are therefore likely to cause human congenital disease. These embryonic abnormalities occur during gastrulation, neurulation, and organogenesis and include phenotypes such as neural tube closure defects, axis truncation, cardia bifida, and heart failure, all leading to early embryonic lethality. Moreover, the beta-galactosidase (lacZ) reporter gene introduced by the GT vector insertion provided gene expression information of value in predicting gene function.
Two of the embryonic lethal phenotypes resulted from mutations in the genes BCL6 interacting corepressor (Bcor) and the Wnt-ligand modifier porcupine homolog (Porcn). Interestingly, after the start of our screen, these genes were associated with X-linked diseases in humans: oculofaciocardiodental syndrome (OFCD) (OMIM:300166; BCOR) (Ng et al. 2004) and focal dermal hypoplasia (FDH) (OMIM:305600; PORCN) (Grzeschik et al. 2007; Wang et al. 2007). In both cases symptomatic human females survive into adulthood, but fully hemizygous null males are never observed (Ng et al. 2004; Grzeschik et al. 2007; Wang et al. 2007). We show that GT mutants for Porcn fail to complete gastrulation, while Bcor GT mutants have severely affected heart development. This provides proof-of-principle of the utility of mutagenic screens in the mouse to identify candidate genes for human disease.
Recent resequencing of the coding exons of the human X chromosome identified few novel candidates for X-linked mental retardation (XLMR), indicating that XLMR may be due to mutations in gene regulatory regions rather than coding regions. We observe that many of the known XLMR genes assessed in our screen have lethal phenotypes when null in the mouse, suggesting that human XLMR may be caused by hypomorphic rather than null mutations. To facilitate disease candidate data mining between human and mouse phenotypes, expression patterns, and genetic loci, we have compiled our data set into an online relational database (http://xlinkedgenes.ibme.utoronto.ca/), along with human data from OMIM.
Results
The genotype-driven screen
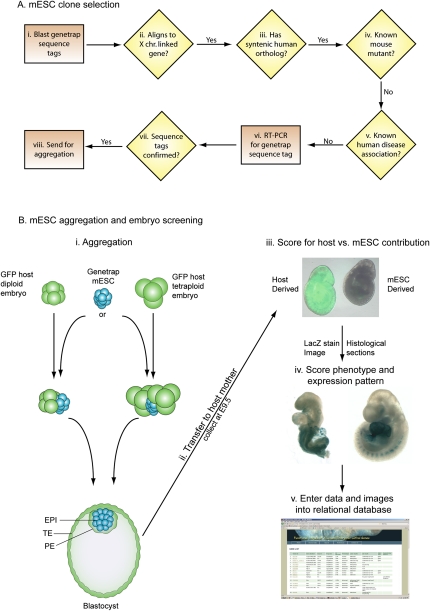
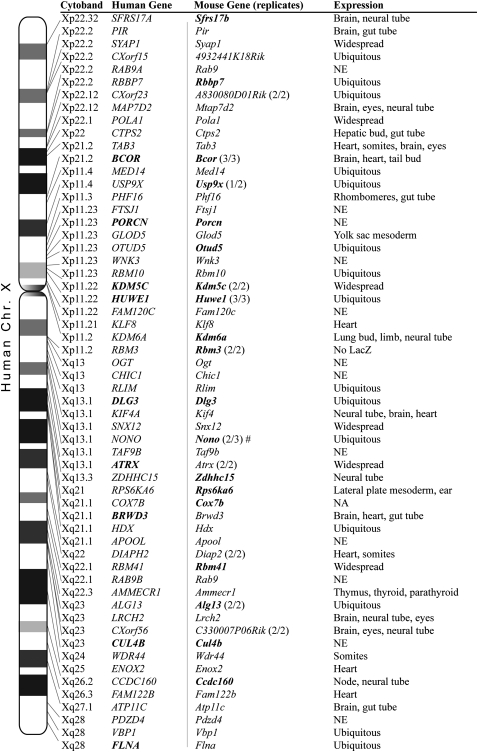
The protocol for choosing GT mESC lines is summarized in Figure 1A and detailed in the Methods section. Briefly, sequence tags for trapped genes were aligned against transcripts that localize to the X chromosome. Those that aligned were selected based on deleting >50% of the gene (exceptions were made if no alternate traps were available). Finally, genes were filtered for reports of mutagenesis in the mouse or linkage to a human disease as of September 2004, when the screen began. Two genes that are linked to human diseases, Atrx and Flna, were included as positive and negative controls. Null mutations in ATRX have no embryonic lethality in humans, while null mutations in FLNA cause embryonic male lethality. A total of 68 GT cell lines representing 58 genes were aggregated with wild-type embryos to generate mESC-derived embryos for phenotypic analysis (Table 1; Fig. 2). While the focus of the screen was to identify novel human disease candidates, several human disease alleles were mapped during the course of the screen (Fig. 2, human genes in bold).
Figure 1.
An overview of the screening method. (A) mESC GT clone selection involved blasting of sequence tags to transcripts from the X chromosome (i). All BLAST (Altschul et al. 1990) hits >95% identical were then compared with the human X chromosome (ii). Those genes with a syntenic homolog were then filtered for any known mouse mutants and known links to human disease (iv,v). GT lines were ordered and quality controlled by RT-PCR to confirm the sequence tag prior to aggregation (vii,viii). (B) Two different aggregation techniques (i) were used with similar results and efficiency (Table 1). mESCs were aggregated with either one diploid or two tetraploid host embryos and cultured overnight. After overnight culture, the colony of mESC preferentially forms the epiblast of the embryo and displaces the host cells to the primitive endoderm and trophectoderm of the blastocyst. mESCs contribute only to the epiblast (EPI) that will generate the embryo proper, while the host (all tetraploid and a large proportion of diploid) embryos generate the primitive endoderm (PE) and the trophectoderm (TE). Aggregated embryos were transferred to pseudopregnant mothers and collected at E9.5 (ii). Dissected embryos were scored for mESC contribution by observing GFP fluorescence (iii). The embryo on the left is GFP positive and has diploid host embryo contribution, while the embryo on the right is GFP negative and is fully mESC-derived (iii). Fully mESC-derived embryos were fixed and stained for beta-galactosidase (lacZ) activity. After staining, the embryos were digitally imaged and scored for expression and phenotype (iv). All digital images and aggregation data were entered into a relational database (v).
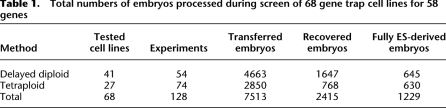
Table 1.
Total numbers of embryos processed during screen of 68 gene trap cell lines for 58 genes
Figure 2.
Linkage to human homologs and summary of expression and phenotype. The human orthologs of the mouse GT genes are aligned to their cytoband on an ideogram of the human X chromosome. Also shown are the names of human and mouse homologs and a brief summary of the observed expression pattern of the beta-galactosidase reporter gene in the embryos. (Bold mouse genes) Gene names for GT lines that gave a phenotype. Eleven mouse genes were tested by two or more GT lines and are indicated in parentheses next to the mouse gene name as a ratio of lines with similar expression or phenotype. The phenotype for the gene Nono was confirmed by inverting the GT and restoring Nono expression and normal phenotype in aggregated embryos (#). (Bold human genes) Names associated with human disease or syndromes when mutated.
Two different aggregation techniques were used to generate embryos that were derived completely from the GT mESC lines: aggregation with tetraploid (Nagy et al. 1990) and diploid (Nagy et al. 1990; Poueymirou et al. 2007) eight-cell stage embryos, with the exception that we aggregated rather than injected cells under the zona (see Methods; Fig. 1B). While the two techniques produce comparable numbers of mESC-derived embryos, tetraploid aggregation requires twice as many host embryos as diploid aggregation, since two embryos are aggregated with a clump of mESCs. We noted similar rates of embryo implantation and mESC-derived embryos regardless of genetic background or source of cell line (Supplemental Table 1). At mouse mid-gestation stage (embryonic day [E9.5]; equivalent to human week 5), the fully mESC-derived embryos were fixed and stained for lacZ expression (Fig. 1B), then imaged and assessed for phenotype and expression pattern. At this stage, the basic body plan is laid down, patterning of the nervous system is apparent, and organogenesis has begun. It thus provides an effective developmental stage to compare human and mouse development, gene expression patterns, and mouse mutant phenotypes relevant to human congenital disease. All image and text information has been placed in a relational database (Fig. 1B). The entire data set includes results from 128 different aggregation experiments that generated over 1200 fully mESC-derived embryos (Table 1).
Phenotypes and gene expression patterns
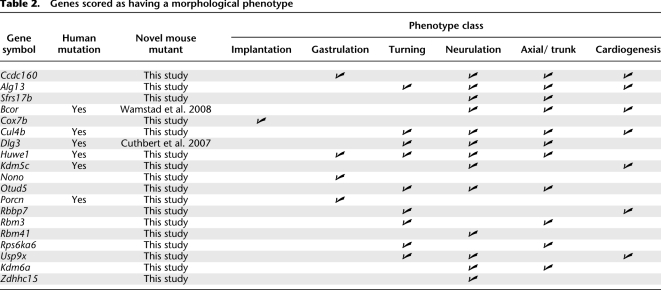
Figure 2 summarizes the genes analyzed in our screen and their phenotypes and expression patterns. In total, 26 cell lines representing 19 genes were scored as having a phenotype in the mouse embryo by E9.5 (Fig. 2, mouse genes in bold). Of these 19 phenotypes, 17 are novel mouse phenotypes where no mutants have been reported in the literature. Thirteen of the novel mouse mutants have no reported mutations in human homologs, making them candidate human disease loci. The remaining 38 genes had no obvious morphological phenotype by E9.5. Aggregation of the parental lines E14 and R1 by either tetraploid or eight-cell-stage diploid embryos produced healthy and normal embryos at E9.5 (data not shown).
For assessment of phenotypes we considered the developmental processes completed or in progress by E9.5, then classified the observed phenotypes into several broad categories (Table 2). The majority of phenotypes fell into the classes of turning, neurulation, cardiogenesis, and axial growth (i.e., somitogenesis and posterior axis elongation). Several genes were scored with a gastrulation phenotype (i.e., formation of the basic body plan and three germ layers), and showed arrested development or altered morphology. Only one GT mESC line did not yield any gastrulation-stage embryos, possibly because the mutated gene, cytochrome c oxidase subunit VIIb, produces an enzyme for general energy metabolism of cells, although mESC are viable (Table 2; Cox7b).
Table 2.
Genes scored as having a morphological phenotype
We also scored expression of the lacZ GT reporter in the embryos. Of the lines with embryonic phenotypes, 15 had expression of lacZ at the stages observed, with six restricted and nine ubiquitous expression patterns (Fig. 2, expression information and database). Three cell lines had no lacZ staining: Cul4b, Rbm3 (no lacZ reporter), and Porcn (insertion before the start codon). Of the GT mutations without a phenotype at E9.5, 13 had restricted expression patterns, four were widespread, nine were ubiquitous, and 10 had no expression (Fig. 2). Lack of a phenotype at E9.5 does not preclude later effects, which were beyond the scope of this study, but the observation of expression restricted to particular tissues at E9.5 can be used to focus future phenotypic analysis of mutant mouse lines.
To assess the robustness of the screening method, we aggregated at least two independent lines for 11 genes (Fig. 2, replicates). In 10 out of 11 cases the resulting replicates had highly similar phenotypes (Fig. 2). Several of the GT lines used in this screen contained conditional multipurpose alleles that can be inverted using site-specific recombination to restore wild-type function (Schnutgen et al. 2005). The loss-of-function Nono GT mESC line presented a strong gastrulation phenotype, with developmental arrest at E7.5. We confirmed specificity of the phenotype by Cre-mediated GT inversion, rescue of gene function, and restoration of Nono protein to normal levels (Supplemental Fig. 1). Furthermore, embryos derived from inversion-rescued GT lines were phenotypically normal at E9.0 (n = 10; Supplemental Fig. 1). Although we did not test all GT lines similarly, the results of the redundant lines, the high correlation of phenotype and lacZ reporter expression, and the inversion of the GT demonstrate that the screen was robust. There is high confidence that the observed phenotypes are the results of the GT mutations and not nonspecific effects associated with particular mESC clones.
Relationship of embryonic phenotypes to human disease genes
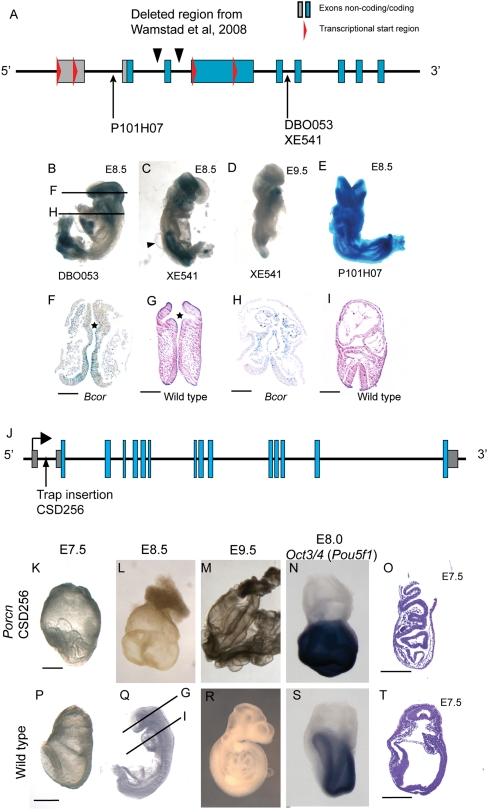
Although our screen was intended to focus on genes with no known phenotype in mouse or human, several human homologs were linked to human diseases during the course of our screen. Two of the GT lines assayed in our screen recapitulated known human diseases with embryonic lethality in male embryos: Bcor and Porcn. BCOR is a transcriptional repressor that has been described to affect levels of histone H3 K4 trimethylation (activated gene mark) (Fan et al. 2009) and physically interact with histone demethylases (Sanchez et al. 2007). The human homolog of mouse Bcor is associated with OFCD (OMIM:300166) (Ng et al. 2004). We generated fully mESC-derived mutant embryos from three different GT lines of Bcor: one in intron 1c (P101H07) and two in intron 5 (DB0053 and XE541, the latter was used in the study by Wamstad et al. 2008) (Fig. 3A). Based on cap-analysis gene expression (CAGE) tag analysis of the Bcor locus, there are two alternate transcriptional start sites in the first exon and two more in the fourth exon (Fig. 3A; Kawaji et al. 2006). Thus, a GT in the first intron would not trap all alternative transcripts, but the two GTs inserted after exon 5 would, possibly causing a more severe phenotype (Fig. 3A). To test this idea, we analyzed and compared all three Bcor GT lines.
Figure 3.
Analysis of Bcor and Porcn early embryo phenotypes. (A) Diagram of Bcor gene locus with locations of gene trap alleles, the conditional targeted allele as reported by Wamstad et al. (2008), and transcriptional start sites (red triangles). (B–E) mESC-derived embryos from three different Bcor GTs stained for lacZ expression. (B) Embryo from DB0053 cell line, used for histological analysis in F and H. Note the small head and heart, and truncated tail bud compared with controls (Q,R). (C) E8.5 mESC-derived embryo from XE541 cell line. Note the similar appearance and expression to DB0053 line. Also shown is a cyst (arrowhead) on the dorsal region of the embryo flanking the neural tube. (D) E9.5 Bcor mutant embryo stained for lacZ expression; of note is the severe posterior truncation, and small head and heart compared with wild-type controls (Q,R). (E) E8.5 embryo from P101H07 cell line exhibits exencephaly. (F) Transverse histological section of Bcor mutant embryo (B) head showing poor formation of optic pits (star), compared with an equivalent region from a wild-type stage-matched embryo (G). (H) Transverse histological section through heart field of Bcor mutant embryo (C) compared with equivalent region in stage-matched embryo (I). Bcor mutants (H) have a small symmetrical heart compared with the looped asymmetrical heart of controls (I). Also, Bcor mutants have an open neural tube compared with control embryos. (F–I) Scale bar, 100 μm. (J) Schematic of Porcn gene locus showing exons and GT insertion in first intron. (K–N) Porcn fully mESC-derived embryos. (P–S) Stage-matched wild-type embryos. (K) At E7.5, the Porcn null epiblast is highly folded and accumulates in the amniotic cavity compared to wild-type embryos (P). (L) At E8.5, Porcn mutants have no head, heart, allantois, or somite structures compared with stage-matched controls (Q). (M) At E9.5, Porcn mutant embryos consist of a yolk sac and small cyst-like structures, compared with wild-type controls that have a fully formed embryo body axis (R). (N) At E7.75 head-fold stage, Porcn mutant embryos fail to down-regulate Pou5f1 compared with stage-matched controls (S). (O) Hematoxylin and eosin-stained sagittal section along the presumed anterior posterior axis of Porcn mutants reveals details of highly folded epiblast and epiblast vesicles compared with the smooth epiblast of controls at E7.75 (T). Porcn mutants had no morphological node apparent in any of the sections from two mutant E7.5 embryos examined; also, the visceral endoderm does not appear to be displaced from the embryonic region of Porcn mutants. (K,P,O,T) Scale bar, 200 μm.
Bcor GT lines DB0053 and XE541 both show lethality by E9.5 with apparent fusion of the ventral forebrain, greatly reduced and malformed cardiac tissue, and severe posterior truncation with failure of chorioallantoic fusion (cf. Fig. 3B,C,D and Q,R). Consistent with the phenotype, lacZ reporter staining was observed strongly in the head, heart, and tail bud at E8.5 (Fig. 3B,C). Embryos derived from both GT lines showed variable numbers of cysts along the dorsal region flanking the neural tube and occasionally in the head region (Fig. 3C; data not shown). Histological sections through the head revealed the eyes to be either delayed in formation or missing completely (cf. Fig. 3F and G, star). Histological sections through the heart showed a bilateral symmetrical heart, compared with the looped asymmetrical heart of similarly staged wild-type controls (cf. Fig. 3H and I). At this level of analysis it is not clear if this is a result of failed looping or cardia bifida. These results clearly indicate that different GT mESC lines give reliable and consistent embryonic phenotypes.
GT line P101H07 displayed a less severe phenotype with an open head and delayed cardiac folding, but a more elongated tail region, proper chorioallantoic fusion, and normal optic cup anlage (Fig. 3E; data not shown). These results demonstrate that a GT line is a valuable resource to mutagenize splice-specific and promoter-specific isoforms, and that this can generate tissue-specific phenotypes with different levels of severity.
We aggregated a GT cell line for Porcn for which the human homolog is associated with FDH (OMIM:305600) (Grzeschik et al. 2007; Wang et al. 2007). The PORCN gene encodes a membrane-bound O-acyl transferase (MBOAT) that is involved in the acyl modification of Wnt ligands, which is important for long-range signaling (Tanaka et al. 2000). The GT insertion for Porcn lies between the first and second exons before the translational start codon (Fig. 3J), truncating the entire coding region of the transcript (as confirmed by RT-PCR; data not shown). Porcn null embryos were observed from E7.5 to E9.5 (Fig. 3K–M), but embryonic development arrested during gastrulation, with no recognizable anterior–posterior axis based on morphological landmarks, such as head fold or allantois (cf. Fig. 3K and P). Histological sections at E7.75 (Fig. 3O,T) revealed that the mutant epiblast accumulated in the amniotic cavity, and there was a marked absence of the allantois (data not shown).
Epiblast overgrowth may be a result of a failure of the epiblast to differentiate. To assess the differentiation state of the Porcn mutant epiblast, we assayed the RNA expression of Pou5f1 (Oct3/4) as a marker of pluripotency by whole mount in situ hybridization. Compared with E8.0 controls, where Pou5f1 was sufficiently down-regulated and epiblast cells started to differentiate (Fig. 3S), Porcn GT mutants showed prolonged expression indicating a lack of differentiation (Fig. 3N). This is very similar to Wnt3 (Liu et al. 1999) and beta-catenin (Morkel et al. 2003) mutants, which fail to undergo EMT and lack mesoderm and endoderm. The Porcn GT mutant embryos are phenotypically similar to Wnt3 mutants (the earliest Wnt phenotype), which supports accumulating evidence that Porcn is required for the secretion of WNT. More detailed analysis of anterior–posterior axis patterning genes is needed to confirm this phenotype. Our observations provide insight into the cause of male embryonic lethality in humans with PORCN mutations.
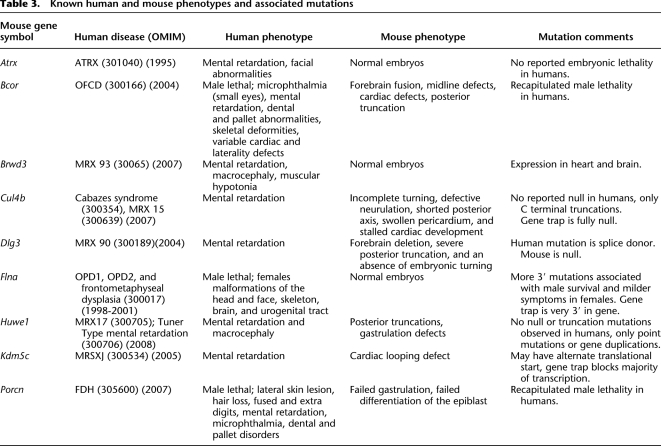
There were seven additional known human X-linked disease genes where human mutations are viable, but for which we report an embryonic phenotype; four of these are described here for the first time (Brwd3, Cul4b, Huwe1, and Kdm5c (formerly Jarid1c); Table 3), while the others have been previously published (Atrx, Dlg3, Flna; Table 3). In many cases the GT line was a null allele, while no null alleles have been found in the human pathologies (Table 3). For example, mutations in CUL4B are associated with Cabezas syndrome (OMIM:300354) and mental retardation (MRX 15, OMIM:300639). We analyzed a GT resulting in a null mutation for Cul4b and observed incomplete turning, defective neurulation, shortened posterior axis, swollen pericardium, and stalled cardiac development. In humans with Cabezas syndrome (CUL4B), all reported mutations are C-terminal truncations. As the null mutation has not been observed in humans, this strongly suggests that a null allele may also cause early gestational lethality in humans.
Table 3.
Known human and mouse phenotypes and associated mutations
Novel early embryonic lethal genes
We observed 17 novel embryonic mutant phenotypes in X-linked genes that exhibited a wide range of defects and had no previous mutation reported in mice (Table 2). As noted above, 13 of these have no reported mutations in human homologs and therefore represent novel candidates for human disease loci. We present a focused morphological assessment of three candidates, one from each phenotype class of cardiac development, neurulation, and axis formation.
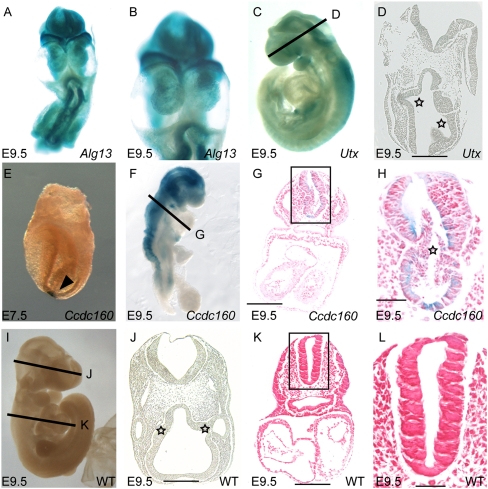
Of the genes giving cardiac defects, the mutation in Alg13 was the most striking, causing cardia bifida where the two heart primordia remain separate and form two hearts on either side of midline (Fig. 4A,B). This cardiac defect is typically due to defects in foregut development resulting in failure of the cardiac precursors to meet at the midline. ALG13 is a soluble cytosolic protein recruited to the endoplasmic reticulum (ER) to form a heterodimer with the ER membrane protein ALG14 (Gao et al. 2005). Together, they generate a UDP-GlcNAc glycosyltransferase that catalyzes the second sugar addition in the synthesis of the dolichol-linked oligosaccharide precursor in N-linked glycosylation (Gao et al. 2005). The exact enzymatic targets of the ALG13/14 complex are unknown, but several protein ligands (i.e., WNTs, SHH) and transmembrane receptors require N-linked glycosylation for proper function.
Figure 4.
Novel embryonic phenotypes. (A,B) Alg13 mutants have widespread expression of the lacZ reporter, are severely developmentally delayed, and have cardia bifida. (C) Kdm6a mutant embryos have an open head and lacZ reporter expression in the head and neural tube. (D) Histological analysis of heads of Kdm6a mutants shows a fused forebrain and uneven, delayed optic pits (stars), compared with a similar region in a stage-matched control embryo (J). (E) Ccdc160 mutant embryos have expression in the node at E7.5. (F) At E9.5, expression of Ccdc160 has become highly restricted to the developing brain and neural tube. (G) Histological analysis of an E9.5 Ccdc160 mutant embryo revealed the presence of cells in the lumen of the neural tube; this was observed in three out of three embryos examined (data not shown). (H) Closer view of the neural tube in G highlighting the trapped cells (star). (I) A wild-type E9.5 embryo has a single folded heart and closed head. (J) Histological section of the head of an E9.5 wild-type embryo showing segmented developing brain and symmetrical optic pits (stars). (K) Histological section though a wild-type E9.5 embryo showing normal number of developing chambers and a closed neural tube with an empty lumen (star). (L) Higher magnification view of neural tube in K.
In Kdm6a (formerly Utx) mutant embryos we observed restricted GT lacZ reporter gene expression in the neuroectoderm of the brain and neural tube at E9.5 (Fig. 4C). The embryos showed exencephaly in the mid- and hindbrain region. Histological sections through the head showed that the forebrain vesicle fails to fuse in the rostral region and the optic pits are asymmetrical as compared to control embryos (cf. Fig. 4D and J, stars). KDM6A has been identified as a histone H3 K27 demethylase involved in epigenetic regulation of gene expression (Lee et al. 2007). Kdm6a also escapes X inactivation in both the mouse and human (Greenfield et al. 1998).
Expression of Ccdc160 in mutant embryos was first observed in the node at E7.5 (Fig. 4E; Supplemental Fig. 2A,E) and in the neural tube at E9.5 (Fig. 4F). At E9.5, Ccdc160 mutant embryos had a severe truncation of the posterior body axis (cf. Fig. 4F and I). Histological analysis of lacZ expression revealed highly restricted expression in the ventral neural tube (Fig. 4G,H). The neural epithelium was disrupted bilaterally in three out of three embryos examined (cf. Fig. 4G,H and K,L; data not shown). The posterior truncation and other defects may result from the expression and function of Ccdc160 in the node, which gives rise to the notochord and is required for dorsal–ventral patterning of the neural tube as well as trunk and tail organizers (Yamanaka et al. 2007). Moreover, a weak and transient expression of the lacZ reporter was observed in the late streak at E7.5 (Supplemental Fig. 2B–D), which suggests a role for Ccdc160 in posterior mesoderm development. The gene codes for a small protein of 323 amino acids with no known domains aside from a putative coiled-coil domain that is a common feature of centrosome/basal body proteins (Andersen et al. 2003). Interestingly, Ccdc160 is expressed in regions of high Shh signaling (i.e., node and ventral neural tube), which in mammals depends on primary cilia. Genomic BLAST analysis showed that the gene Ccdc160 and its homologs are found only in mammals, including opossum and platypus, perhaps indicating that Ccdc160 is an evolutionary adaptation to mammalian development.
Online database
Data on aggregation efficiency and images of mESC-derived embryos are available on a publicly accessible website (http://xlinkedgenes.ibme.utoronto.ca/). The database is searchable by gene name or can be browsed. We have integrated our image and text data of these mutant embryos with the OMIM database. As mentioned in the introduction, there are over 140 suspected X-linked diseases that have not been mapped to a specific gene. To facilitate data mining we have extracted these OMIM entries and entered them into our database, placing all mouse GTs for orthologous human genes within the defined linkage regions for OMIM entries on the human X chromosome. Constraints can be placed on either the expression pattern or phenotype of the mouse homologues to find those similar to the human disease or syndrome. Phenotype data and images of analyzed cell lines have been deposited with MGI (http://www.informatics.jax.org). MGI allele IDs for each of the characterized lines can be found in Supplemental Table 2.
Discussion
Using publicly available GT insertion mESC lines and a rapid and cost-effective phenotypic assay based on generating entirely mESC-derived embryos, we have added functional annotation to almost 10% of mouse syntenic X-linked genes and provided an online resource to link mouse phenotype and expression information to human disease.
It has been suggested that the observation of a higher percentage of Mendelian genetic diseases being linked to the X chromosome is a discovery bias (Ross et al. 2005) based on the ease of detection of X-linked phenotypes in males and, in some cases, in X-inactivation mosaic females. In our unbiased screen for embryonic phenotypes in X-linked GT mutations in the mouse, we have detected about 32% (19/58) with an early developmental phenotype. This is in line with other GT-based mutagenesis studies that focused on the autosomes, where 33% (Mitchell et al. 2001) and 37% (Friedrich and Soriano 1991) of genes tested presented phenotypes. This assessment does come with the caveat that we have included both recessive and dominant phenotypes of the previous gene trap screens. We justify this by the fact that our screen is a hemizygous null so we cannot discriminate recessive from dominant mutations in our screen. Thus, we conclude from our analysis that autosomal and X-chromosomal-lethal phenotypes occur at similar rates.
Importantly, as a proof-of-principle we confirmed early male embryo lethality in two mouse models, Bcor and Porcn, where lethality in humans was only suspected. BCOR mutations are associated with human OFCD (OMIM 300166) (Ng et al. 2004). Human males do present with OFCD when BCOR has a hypomorphic mutation such that some functional protein is made (Hilton et al. 2009). Bcor mutants have previously been generated in the mouse and, although no hemizygous null males were observed postnatal, there was no assessment of the embryonic phenotype (Wamstad et al. 2008). We clearly demonstrate that Bcor has an essential function during embryonic development and we offer an explanation for male embryonic lethality.
Mutations in PORCN are associated with human FDH (OMIM:305600) (Grzeschik et al. 2007; Wang et al. 2007). The majority of patients are female, and only males with mosaic spontaneous somatic mutations are viable, indicating that hemizygous null males die during gestation (Grzeschik et al. 2007; Wang et al. 2007). Patients present with lateral skin lesion and hair loss, fused and extra digits, mental retardation, microphthalmia, and dental and palate disorders (Grzeschik et al. 2007; Wang et al. 2007). Our data demonstrate that Porcn mutants phenocopy known Wnt/beta-catenin mutants and that Porcn is essential for epiblast differentiation, as Pou5f1 expression was not down-regulated. This embryo morphology is very similar to Wnt3 mutants, the earliest of Wnt mutant phenotypes (Liu et al. 1999), which fail to undergo EMT and lack mesoderm and endoderm. Taken together, our observations support accumulating evidence that Porcn is required for the secretion of functional Wnt proteins and provide a cause/mechanism for male embryonic lethality in humans. From the analysis of Bcor and Porcn we conclude that our screen can identify embryonic phenotypes that can give insight into key developmental pathways associated with postnatal disease.
Although the screen was designed to identify mid-gestation lethal mutations, we also observed many embryos with a normal morphology, even though the trapped genes showed clear expression by lacZ reporter at and before E9.5. It is possible that loss of function of these genes could be compensated by the coexpression of close paralogs or that the phenotype may not be overtly apparent at this stage. Of the genes that were observed to have cardiac expression, for example, many were in the trabecular cells, which are only just developing at E9.5. Defects in trabecular formation may not be phenotypically or even morphologically apparent until later in gestation. An example of this was Klf8, which had expression in the left side ventricle and outflow tract. Interestingly, the human homolog KLF8 maps to the locus of the human syndrome TARPS (Kurpinski et al. 2003) that has left side cardiac defects. Unfortunately, sequencing of the coding regions of KLF8 in two families did not reveal any mutations in the coding regions; however, mutations in the promoter or enhancers have not been tested (L Biesecker, pers. comm.).
Another common class was genes with expression in the developing brain; these could be associated with XLMR, which would not be apparent until postnatal time points. For example, Brwd3, which was associated with XLMR by another group during the course of our study (Field et al. 2007), has lacZ reporter expression in the brain and neural tube. Expression does not have to be restricted; an example of this from our screen is Dlg3, where in humans mutations in DLG3 are associated with mental retardation (Tarpey et al. 2004). Aggregation results from mESC line with a gene-trap insertion in Dlg3 (clone P038A02) revealed ubiquitous expression of lacZ and showed a range of phenotype severity. Embryo phenotypes for Dlg3 mutants ranged from morphologically normal to failure of embryonic turning (five out of 18), including rare cases with forebrain deletion (one out of 18). This GT phenotype has been confirmed using a Dlg3 KO mouse line in an inbred background (C van Campenhout, P Giallonardo, C Gloeckner, A Eitelhuber, M Gegg, S Grant, D Krappmann, M Ueffing, and H Lickert, in prep.). The mouse phenotype indicates a critical role for Dlg3 in neural induction and that non-null mutations could cause more subtle developmental defects leading to cognitive deficiencies. The partial penetrance of this phenotype suggests that alleles of other genes could modify the penetrance of the mutant phenotype.
A large-scale resequencing of the X chromosome in families with XLMR found few novel genes with exonic mutations that could be associated with mental retardation (Tarpey et al. 2009). A possible explanation is that the causative mutations are in regulatory elements or splicing control regions; a difficult prospect for sequence analysis. This is indirectly supported by our observation that some human XLMR genes had lethal phenotypes when null in the mouse (Cul4b, Dlg3, Huwe1, Kdm5c; Table 3). Thus, it is possible that null mutations give rise to early embryo lethality, but tissue-specific loss of gene expression caused by mutations in regulatory regions may cause XLMR. We suggest that mutagenic screens in the mouse, such as this current study, may be used to identify genes with expression patterns or phenotypes consistent with a function in brain or neural development. These genes would be candidates for more focused resequencing efforts.
The screening method used here does not rely on the generation of founder animals, extensive crossing schemes, or mapping of mutations. Instead, preselection and testing of candidate disease genes can be carried out in a very short time frame. Generation of mESC-derived embryos was first reported by aggregation of mESC with tetraploid embryos (Nagy et al. 1990), a technique also used in part of our screen. However, a subsequent report suggested that laser-assisted injection of mESCs under the zona pellucida of diploid eight-cell embryos could also generate mESC-derived mice at an efficient rate (Poueymirou et al. 2007). In this study, we used an aggregation method with diploid eight-cell embryos and were able to generate completely mESC-derived embryos with similar success to aggregation with tetraploid embryos. Eliminating the need for electrofusion or laser-assisted injection simplified the method and dramatically reduced labor and cost of the assay. The majority of GT lines were generated using a vector that only traps genes expressed in mESCs, thus limiting the genes available for analysis. However, newer GT lines from the Center for Modeling Human Disease node of the IGTC are designed to trap all genes independent of expression (Stanford et al. 2006). This and the international functional genomic effort to generate targeted mutations in all genes (International Mouse Knockout Consortium 2007) are adding new X-linked mutations to the mouse mutant resource. In principle, using this screening method in combination with newer mutant mESC resources should make it possible to assess the embryonic phenotype of every gene on the mouse X chromosome. While this goal would require the coordinated efforts of several research centers, it would represent a major contribution to functional analysis of the X chromosome.
Overall, the integration of our data set with the OMIM data set will produce an important resource for both the biological and clinical communities to identify the molecular causes of human genetic disease. The data set will also expand our understanding of the role of the X chromosome during embryonic development.
Methods
Selection and verification of ES lines
A multistep approach was used to select all trapped genes on the X chromosome. First, sequence tags for all available GT lines were obtained from the IGTC database. These tags were then blasted against a library of mRNA sequences with chromosomal and genetic locations from the mouse. BLAST (Altschul et al. 1990) scores had to be below E−30 sequences and have >95% identity over the span of the tag to be considered a match to a gene. The next selection criterion was that the trap deleted >50% of the gene. From this list, mouse genes with synteny to the human X chromosome were selected. Finally, any gene that had been mutated in mice or was known to be associated with a human disease was removed to generate a final list of genes for screening. In some cases, multiple cell lines per gene were used to test the specificity of observed phenotypes as well as the differences in insertion position and phenotype.
Confirmation of the GT integration site, as mapped by the German Gene Trap Consortium (GGTC), BayGenomics via Mutant Mouse Regional Resource Center (MMRRC), or Center for Modeling Human Disease (CMHD) GT node, was done by RT-PCR. Primers were designed in an exon directly before the trap, or, if no appropriate primer was available, an exon further upstream was used. A primer was also placed in the GT as supplied by the GT resource group. Lastly, a primer was placed in an exon 3′ of the trap to test for the presence of normal transcripts. In cases where the PCR fragment size was not close to the expected size, the PCR product was sequenced to confirm the PCR product. mRNA from wild-type cells was used as a negative control for the trapped transcript and as a positive control for the wild-type transcript. All primers are listed in Supplemental Table 3. RNA was isolated from a 6-cm dish of mESCs using TRIzol (Invitrogen) and dissolved in 30 μL of DEPC water. Two micrograms of total RNA was used for reverse transcription, which was performed with the SuperScript II Reverse Transcriptase kit (Invitrogen) and primed with random octamers. From a final reverse transcription reaction volume of 20 μL, 1 μL was used for subsequent PCR analysis. PCR extension times and annealing temperatures were optimized for each transcript. Each line was tested for mycoplasm infection and confirmed negative prior to aggregation.
Culture of ES lines
Cell lines were thawed and expanded on a feeder layer of Mitomycin C-inactivated mouse embryonic fibroblasts (MEFs) using standard protocols (Joyner 2000). mESC media contained high glucose DMEM supplemented with 15% FBS (tested to support generation of chimeras; Hyclone), 1000 U/mL LIF, 2 mM GlutaMAX (Invitrogen), 1 mM Na pyruvate (Invitrogen), 0.1 mM nonessential amino acids (Invitrogen), 50 U/mL penicillin and streptomycin (Invitrogen), and 0.1 mM 2-mercaptoethanol (Invitrogen). Cells were fed daily and passaged every 2 d. Each cell line was tested for mouse pathogens (Research Animal Diagnostics Laboratory [RADIL]) prior to use for aggregation.
Aggregation assays
Tetraploid embryos
Embryos at the two-cell stage were collected at E1.5 from superovulated ICR (Harlan) or BDF1 (Charles River) mice mated with ICR, BDF1, or B5/EGFP [Tg(CAG-EGFP)B5Nagy] (Hadjantonakis et al. 1998), eYFP (7AC5/EYFP) [Tg(CAG-EYFP)7AC5Nagy] (Hadjantonakis et al. 2002), or DsRed*MST [Tg(CAG-DsRed*MST)1Nagy] (Vintersten et al. 2004) males and electrofused to generate tetraploid embryos (Nagy 2003). The fused, tetraploid embryos were cultured in microdrops of KSOM with amino acids (Millipore) under mineral oil at 37°C in 94% air/6% CO2. Twenty-four hours after fusion the tetraploid embryos that developed to four-cell stage embryos were used for aggregation. Zonae pellucidae of the embryos were removed by treatment with acid Tyrode's solution (Sigma). mESCs were briefly trypsinized to form clumps of loosely connected cells, and clumps of eight to 15 mESCs were placed between two zona-free tetraploid embryos in depression wells made in the plastic dish. These aggregates were cultured overnight in KSOM microdrops before morulae and blastocysts were transferred into the uteri of 2.5-d pseudopregnant recipients.
Diploid embryos
Superovulated ICR (Harlan) female mice were mated with male mice homozygous for a ubiquitously expressed transgene coding for eGFP (B5/EGFP) [Tg(CAG-EGFP)B5Nagy] (Hadjantonakis et al. 1998), eYFP (7AC5/EYFP) [Tg(CAG-EYFP)7AC5Nagy] (Hadjantonakis et al. 2002), or DsRed*MST [Tg(CAG-DsRed*MST)1Nagy] (Vintersten et al. 2004). Embryos were collected at day E1.5 and cultured overnight in microdrops of KSOM with aminoacids (Millipore) under mineral oil at 37°C in 94% air/6% CO2 until the uncompacted eight-cell stage. The next morning each embryo was aggregated with a clump of eight to 15 mES cells in depression wells and cultured one more night in KSOM microdrops, then morulae and blastocysts were transferred into the uteri of pseudopregnant females at E2.5 as described (Nagy 2003).
Phenotypic analysis
Embryos from diploid aggregations were dissected at day E9.5 and screened for host embryo contribution. Only embryos with no host contribution, aside from the typical hindgut contribution, were used (Kwon et al. 2008; Burtscher and Lickert 2009). An initial morphological assessment determined if there was any obvious phenotypic difference between the mESC-derived embryos, chimeric, and fully host-derived embryos. Embryos were fixed and processed for lacZ staining. After lacZ staining the embryos were again assessed for staining patterns and morphology.
Embryos from tetraploid aggregations were screened in two stages. In the first stage mESC were aggregated with ICR or BDF1 tetraploid embryos, dissected at E7.5, E8.5, and E9.5, and compared with controls to assess any morphological defect. ES cell clones that gave either patchy lacZ staining (an indication of chimerism with host cells) or constant phenotypes were validated in a second screen. In the second screen the ES cells were again aggregated with tetraploid embryos, but using CD-1 females that ubiquitously expressed a fluorescent marker (dsRed*MST, eGFP, or eYFP) to exclude tetraploid cell contribution to the embryo.
For lacZ, embryos to be stained (E6.5–E11.5) were fixed in PBS (pH 7.3) containing 0.02% NP-40, 5 mM EGTA (pH 8.0), 2 mM MgCl2, 1% formaldehyde, and 0.2% glutaraldehyde for 30 min at room temperature, washed three times with 0.02% NP-40 in PBS, and then placed in X-gal staining solution (PBS at pH 7.3 containing 0.02% NP-40, 2 mM MgCl2, 5 mM K3[Fe(CN)6], 5 mM K4[Fe(CN)6] × 6H2O, 0.01% sodium deoxycholate, 1 mg/mL X-gal). Staining was carried out in the dark, overnight at room temperature. After staining, samples were washed in PBS and stored in 4% paraformaldehyde at 4°C. Whole-0mount in situ hybridization was performed as described previously (Tamplin et al. 2008).
All mouse mutant embryos were digitally imaged regardless of lacZ staining pattern or body morphology. Image data were stored in a relational database. Embryos were imaged in PBS on a stereo fluorescence microscope (Leica MZ16F) and attached digital camera (QIMAGING MicroPublisher 5.0RTV); raw images were processed with Volocity5 software or a Zeiss SteREO Lumar.V12 microscope using an AxioCam MRc5 camera.
Embryos were sectioned by the Center for Modeling Human Disease core histology facility at the Toronto Center for Phenogenomics. Slides of embryo sections were imaged on a ZEISS scanner (MIRAX SCAN), and single images were captured in MIRAX VIEWER 1.11 software and processed in Adobe Photoshop CS2.
Database construction
An internal database was constructed with File Maker Pro 8.5. All data were manually entered and curated in the database. Tables were then exported from the internal database to construct the online version using MySQL, Java, Php, and HTML.
Acknowledgments
We thank Jorge Cabezas, Suzanne MacMaster, and Sandra Tondat from the Toronto Center for Phenogenomics; and Patrizia Giallonardo, Adrianne Tasdemir, and Susanne Weidemann for technical support in generating aggregation embryos. We also thank Valerie Prideaux for assistance with FileMakerPro and Christine To for programming assistance in the generation of the online version of the phenotype database. We thank Les Biesecker for sequencing of KLF8 in carriers of TARPS at our request. We thank Thomas Kislinger and Chi-Chung Hui for critical reading of the manuscript and Jean Cox for editorial assistance. This work was supported in part by the March of Dimes (grant no. 6-FY06-364 and 6-FY08-315) to J.R. H.L. is supported by the German research foundation and the Helmholtz society. W.W. and R.K. are funded by the NGFN-Plus (FKZ: 01GS0858/01GS0823) and by the EU, FP6 (EUCOMMEUCOMM: LSHM-CT-2005-018931).
Footnotes
[Supplemental material is available online at http://www.genome.org. Phenotype data and images of analyzed cell lines have been submitted to Mouse Genome Informatics (http://www.informatics.jax.org) under the allele IDs listed in Supplemental Table 2.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.105106.110.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ 1990. Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M 2003. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426: 570–574 [DOI] [PubMed] [Google Scholar]
- Burtscher I, Lickert H 2009. Foxa2 regulates polarity and epithelialization in the endoderm germ layer of the mouse embryo. Development 136: 1029–1038 [DOI] [PubMed] [Google Scholar]
- Cuthbert PC, Stanford LE, Coba MP, Ainge JA, Fink AE, Opazo P, Delgado JY, Komiyama NH, O'Dell TJ, Grant SG 2007. Synapse-associated protein 102/dlgh3 couples the NMDA receptor to specific plasticity pathways and learning strategies. J Neurosci 27: 2673–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z, Yamaza T, Lee JS, Yu J, Wang S, Fan G, Shi S, Wang CY 2009. BCOR regulates mesenchymal stem cell function by epigenetic mechanisms. Nat Cell Biol 11: 1002–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field M, Tarpey PS, Smith R, Edkins S, O'Meara S, Stevens C, Tofts C, Teague J, Butler A, Dicks E, et al. 2007. Mutations in the BRWD3 gene cause X-linked mental retardation associated with macrocephaly. Am J Hum Genet 81: 367–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich G, Soriano P 1991. Promoter traps in embryonic stem cells: A genetic screen to identify and mutate developmental genes in mice. Genes Dev 5: 1513–1523 [DOI] [PubMed] [Google Scholar]
- Gao XD, Tachikawa H, Sato T, Jigami Y, Dean N 2005. Alg14 recruits Alg13 to the cytoplasmic face of the endoplasmic reticulum to form a novel bipartite UDP-N-acetylglucosamine transferase required for the second step of N-linked glycosylation. J Biol Chem 280: 36254–36262 [DOI] [PubMed] [Google Scholar]
- Garrick D, Sharpe JA, Arkell R, Dobbie L, Smith AJ, Wood WG, Higgs DR, Gibbons RJ 2006. Loss of Atrx affects trophoblast development and the pattern of X-inactivation in extraembryonic tissues. PLoS Genet 2: e58 doi: 10/1371/journal.pgen.0020058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield A, Carrel L, Pennisi D, Philippe C, Quaderi N, Siggers P, Steiner K, Tam PP, Monaco AP, Willard HF, et al. 1998. The UTX gene escapes X inactivation in mice and humans. Hum Mol Genet 7: 737–742 [DOI] [PubMed] [Google Scholar]
- Grzeschik KH, Bornholdt D, Oeffner F, Konig A, del Carmen Boente M, Enders H, Fritz B, Hertl M, Grasshoff U, Hofling K, et al. 2007. Deficiency of PORCN, a regulator of Wnt signaling, is associated with focal dermal hypoplasia. Nat Genet 39: 833–835 [DOI] [PubMed] [Google Scholar]
- Hadjantonakis AK, Gertsenstein M, Ikawa M, Okabe M, Nagy A 1998. Generating green fluorescent mice by germline transmission of green fluorescent ES cells. Mech Dev 76: 79–90 [DOI] [PubMed] [Google Scholar]
- Hadjantonakis AK, Macmaster S, Nagy A 2002. Embryonic stem cells and mice expressing different GFP variants for multiple non-invasive reporter usage within a single animal. BMC Biotechnol 2: 11 doi: 10.1186/1472-6750-2-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton E, Johnston J, Whalen S, Okamoto N, Hatsukawa Y, Nishio J, Kohara H, Hirano Y, Mizuno S, Torii C, et al. 2009. BCOR analysis in patients with OFCD and Lenz microphthalmia syndromes, mental retardation with ocular anomalies, and cardiac laterality defects. Eur J Hum Genet 17: 1325–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Knockout Mouse Consortium 2007. A mouse for all reasons. Cell 128: 9–1317218247 [Google Scholar]
- Joyner AL 2000. Gene targeting: A practical approach Oxford University Press, Oxford [Google Scholar]
- Kawaji H, Kasukawa T, Fukuda S, Katayama S, Kai C, Kawai J, Carninci P, Hayashizaki Y 2006. CAGE Basic/Analysis Databases: The CAGE resource for comprehensive promoter analysis. Nucleic Acids Res 34: D632–D636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurpinski KT, Magyari PA, Gorlin RJ, Ng D, Biesecker LG 2003. Designation of the TARP syndrome and linkage to Xp11.23-q13.3 without samples from affected patients. Am J Med Genet A 120A: 1–4 [DOI] [PubMed] [Google Scholar]
- Kwon GS, Viotti M, Hadjantonakis AK 2008. The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Dev Cell 15: 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanasa MC, Hogge WA, Kubik CJ, Ness RB, Harger J, Nagel T, Prosen T, Markovic N, Hoffman EP 2001. A novel X chromosome-linked genetic cause of recurrent spontaneous abortion. Am J Obstet Gynecol 185: 563–568 [DOI] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R 2007. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318: 447–450 [DOI] [PubMed] [Google Scholar]
- Liu P, Wakamiya M, Shea MJ, Albrecht U, Behringer RR, Bradley A 1999. Requirement for Wnt3 in vertebrate axis formation. Nat Genet 22: 361–365 [DOI] [PubMed] [Google Scholar]
- Mitchell KJ, Pinson KI, Kelly OG, Brennan J, Zupicich J, Scherz P, Leighton PA, Goodrich LV, Lu X, Avery BJ, et al. 2001. Functional analysis of secreted and transmembrane proteins critical to mouse development. Nat Genet 28: 241–249 [DOI] [PubMed] [Google Scholar]
- Morkel M, Huelsken J, Wakamiya M, Ding J, van de Wetering M, Clevers H, Taketo MM, Behringer RR, Shen MM, Birchmeier W 2003. Beta-catenin regulates Cripto- and Wnt3-dependent gene expression programs in mouse axis and mesoderm formation. Development 130: 6283–6294 [DOI] [PubMed] [Google Scholar]
- Nagy A 2003. Manipulating the mouse embryo. A laboratory manual Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Nagy A, Gocza E, Diaz EM, Prideaux VR, Ivanyi E, Markkula M, Rossant J 1990. Embryonic stem cells alone are able to support fetal development in the mouse. Development 110: 815–821 [DOI] [PubMed] [Google Scholar]
- Ng D, Thakker N, Corcoran CM, Donnai D, Perveen R, Schneider A, Hadley DW, Tifft C, Zhang L, Wilkie AO, et al. 2004. Oculofaciocardiodental and Lenz microphthalmia syndromes result from distinct classes of mutations in BCOR. Nat Genet 36: 411–416 [DOI] [PubMed] [Google Scholar]
- Nord AS, Chang PJ, Conklin BR, Cox AV, Harper CA, Hicks GG, Huang CC, Johns SJ, Kawamoto M, Liu S, et al. 2006. The International Gene Trap Consortium Website: A portal to all publicly available gene trap cell lines in mouse. Nucleic Acids Res 34: D642–D648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro E, Whitaker J, Mowery-Rushton P, Surti U, Lanasa M, Hoffman EP 1997. Familial skewed X inactivation: A molecular trait associated with high spontaneous-abortion rate maps to Xq28. Am J Hum Genet 61: 160–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poueymirou WT, Auerbach W, Frendewey D, Hickey JF, Escaravage JM, Esau L, Dore AT, Stevens S, Adams NC, Dominguez MG, et al. 2007. F0 generation mice fully derived from gene-targeted embryonic stem cells allowing immediate phenotypic analyses. Nat Biotechnol 25: 91–99 [DOI] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, et al. 2005. The DNA sequence of the human X chromosome. Nature 434: 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C, Sanchez I, Demmers JA, Rodriguez P, Strouboulis J, Vidal M 2007. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics 6: 820–834 [DOI] [PubMed] [Google Scholar]
- Schnutgen F, De-Zolt S, Van Sloun P, Hollatz M, Floss T, Hansen J, Altschmied J, Seisenberger C, Ghyselinck NB, Ruiz P, et al. 2005. Genomewide production of multipurpose alleles for the functional analysis of the mouse genome. Proc Natl Acad Sci 102: 7221–7226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford WL, Epp T, Reid T, Rossant J 2006. Gene trapping in embryonic stem cells. Methods Enzymol 420: 136–162 [DOI] [PubMed] [Google Scholar]
- Tamplin OJ, Kinzel D, Cox BJ, Bell CE, Rossant J, Lickert H 2008. Microarray analysis of Foxa2 mutant mouse embryos reveals novel gene expression and inductive roles for the gastrula organizer and its derivatives. BMC Genomics 9: 511 doi: 10.1186/1471-2164-9-511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Okabayashi K, Asashima M, Perrimon N, Kadowaki T 2000. The evolutionarily conserved porcupine gene family is involved in the processing of the Wnt family. Eur J Biochem 267: 4300–4311 [DOI] [PubMed] [Google Scholar]
- Tarpey P, Parnau J, Blow M, Woffendin H, Bignell G, Cox C, Cox J, Davies H, Edkins S, Holden S, et al. 2004. Mutations in the DLG3 gene cause nonsyndromic X-linked mental retardation. Am J Hum Genet 75: 318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey PS, Smith R, Pleasance E, Whibley A, Edkins S, Hardy C, O'Meara S, Latimer C, Dicks E, Menzies A, et al. 2009. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet 41: 535–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vintersten K, Monetti C, Gertsenstein M, Zhang P, Laszlo L, Biechele S, Nagy A 2004. Mouse in red: Red fluorescent protein expression in mouse ES cells, embryos, and adult animals. Genesis 40: 241–246 [DOI] [PubMed] [Google Scholar]
- Wamstad JA, Corcoran CM, Keating AM, Bardwell VJ 2008. Role of the transcriptional corepressor Bcor in embryonic stem cell differentiation and early embryonic development. PLoS ONE 3: e2814 doi: 10.1371/journal.pone.002814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Reid Sutton V, Omar Peraza-Llanes J, Yu Z, Rosetta R, Kou YC, Eble TN, Patel A, Thaller C, Fang P, et al. 2007. Mutations in X-linked PORCN, a putative regulator of Wnt signaling, cause focal dermal hypoplasia. Nat Genet 39: 836–838 [DOI] [PubMed] [Google Scholar]
- Yamanaka Y, Tamplin OJ, Beckers A, Gossler A, Rossant J 2007. Live imaging and genetic analysis of mouse notochord formation reveals regional morphogenetic mechanisms. Dev Cell 13: 884–896 [DOI] [PubMed] [Google Scholar]