Abstract
Fanconi anemia is a human cancer predisposition syndrome caused by mutations in thirteen Fanc genes. The disorder is characterized by genomic instability and cellular hypersensitivity to chemicals that generate DNA interstrand crosslinks (ICLs). A central event in the activation of the Fanconi anemia pathway is the mono-ubiquitylation of the FANCI-FANCD2 complex, but how this complex confers ICL resistance remains enigmatic. We make use of a cell-free system to show that the FANCI-FANCD2 complex is required for replication-dependent ICL repair. Removal of FANCD2 from extracts inhibits nucleolytic incisions near the ICL as well as translesion DNA synthesis past the lesion. Reversal of these defects requires ubiquitylated FANCI-FANCD2. Our results show that multiple steps of the essential S phase ICL repair mechanism fail when the Fanconi anemia pathway is compromised.
Cells derived from Fanconi anemia (FA) patients are hypersensitive to agents that induce ICLs and exhibit ICL-induced chromosomal instability(1, 2). Eight FANC proteins form a nuclear “core complex”, which mono-ubiquitylates the FANCI-FANCD2 complex after DNA damage (3–5). Ubiquitylated FANCI-FANCD2 is recruited to the chromatin where it co-localizes with DNA repair factors (6, 7). Mutation of the ubiquitin acceptor site in FANCD2 prevents FANCI-FANCD2 chromatin binding and sensitizes cells to ICL inducing agents (4). Although the FA pathway might play a minor role in ICL repair during the G1 phase of the cell cycle (8, 9), its primary function is exerted in S phase (6, 10–12). These results suggest that ubiquitylated FANCI-FANCD2 controls ICL repair during DNA replication, but the underlying mechanism is unknown.
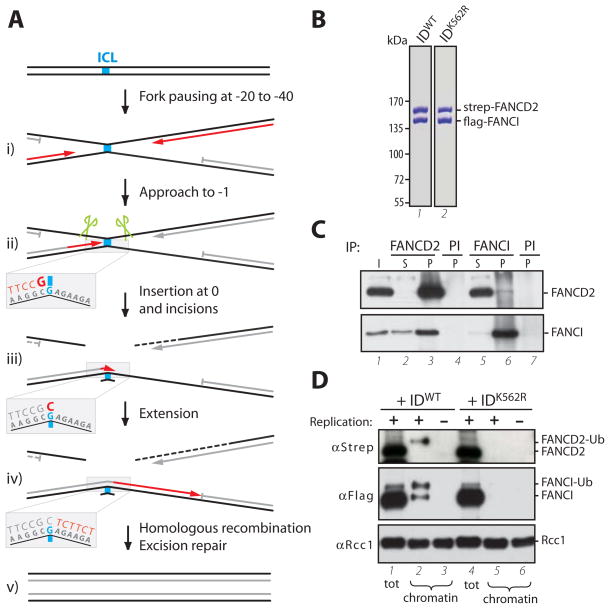
Xenopus egg extracts support replication-dependent repair of a plasmid-born cisplatin ICL (pICL) [(12), Fig. S1A]. Initially, two replication forks converge on the ICL, with their leading strands stalling 20–40 nucleotides from the lesion (Fig. 1A, i). One of the two leading strands then approaches the ICL, stalling again 1 nucleotide from the crosslinked base (the “−1” position; Fig. 1A, ii). Subsequently, incisions on the parental strand uncouple the crosslink (Fig. 1A, ii, green scissors) and lesion bypass occurs in two steps. First, a nucleotide is inserted across from the damaged template base (‘insertion’; Fig. 1A, iii), after which the strand is extended beyond the ICL in a DNA polymerase ζ-dependent manner (‘extension’; Fig. 1A, iv). The final steps in repair are thought to involve excision repair and/or homologous recombination (Fig. 1A, v).
Fig. 1.
(A) Schematic representation of lesion bypass in ICL repair (12). (B) Purified FANCI-FANCD2WT and FANCI-FANCD2K562R stained with Coomassie blue. (C) Reciprocal co-immunoprecipitation of xlFANCI and xlFANCD2 from Xenopus egg extract. Input (I) and supernatant (S) (0.2 μl extract), or precipitated proteins (P, from 1 μl extract) were blotted for FANCI and FANCD2. PI: pre-Immune serum. (D) Replication-dependent binding of FANCI-FANCD2 to damaged chromatin. Crosslinked sperm chromatin was replicated in undepleted extracts supplemented with FANCI-FANCD2WT or FANCI-FANCD2K562R (310 nM). Chromatin-bound fractions (from 2 μl extract) or total extract (0.2 μl), were analyzed by Western blotting with anti-strep-tag (to visualize recombinant FANCD2), anti-FLAG-tag (recombinant FANCI), and anti-RCC1 (loading control) antibodies. Where indicated, replication was inhibited with Geminin. Note that only ubiquitylated FANCD2 binds chromatin, while both ubiquitylated and unubiquitylated FANCI bind (see also (14)).
To examine the function of the Xenopus FANCI-FANCD2 complex in ICL repair, we co-expressed Xenopus FANCI (Fig. S2) and FANCD2 (10) in insect cells and purified a stable 1:1 FANCI-FANCD2 complex (Fig. 1B and S3A). Using antibodies to FANCI (Fig. S3B) and FANCD2 (12), we showed that in Xenopus egg extracts, FANCI and FANCD2 interact (Fig. 1C), and the proteins undergo replication-dependent mono-ubiquitylation and chromatin-binding, both of which are enhanced by the presence of an ICL (Fig. S3C-E) (10). We also purified FANCI-FANCD2K562R, in which the ubiquitin acceptor lysine in FANCD2 is mutated to arginine (Fig. 1B). Unlike FANCI-FANCD2WT, FANCI-FANCD2K562R did not bind to chromatin (Fig. 1D, compare lanes 2 and 5). In summary, Xenopus FANCI-FANCD2 binds chromatin dependent on DNA damage, FANCD2 ubiquitylation, and DNA replication.
To investigate whether FANCD2 is required for ICL repair, >95% of FANCD2 was immunodepleted from Xenopus egg extracts, which resulted in ~75% co-depletion of FANCI (Fig. S4A), but no defect in pICL replication (Fig. S4B). pICL repair efficiency was determined by measuring the regeneration of a SapI restriction site that coincides with the crosslink (Fig. 2A and S5A). In mock-depleted extracts, 15–24% of the replicated DNA became cleavable by SapI after 150–220 minutes (Fig. 2B and Fig. S6). SapI site regeneration is not 100% efficient due to significant destruction of the incised sister chromatid (12), incomplete removal of the unhooked ICL (Fig. S5B)(12), and possibly some mutagenic lesion bypass events. In contrast, in FANCD2-depleted extracts, regeneration of SapI cleavable products was reduced on average 14-fold (Fig. 2B and Fig. S6). The residual SapI products might arise from incomplete FANCD2 depletion or FANCD2-independent ICL repair. Addition of recombinant FANCI-FANCD2 (Fig. 1B) rescued the repair defect (Fig. 2B and Fig. S6) ruling out that FANCD2 depletion non-specifically inactivated repair. In contrast, FANCI-FANCD2K562R, which does not undergo ubiquitylation (Fig. S4C) or bind chromatin (Figure 1D), did not rescue repair (Fig. 2B and Fig. S6), demonstrating a role for ubiquitylated FANCI-FANCD2 in replication-coupled ICL repair. A recent study showed that FANCL depletion reduces origin-dependent ICL repair in Xenopus egg extracts, but the defect was minor (30%) and not rescued with recombinant proteins (8). Our experiments thus represent the first demonstration that the Fanconi anemia pathway is integral to replication-dependent ICL repair, and together with previous observations (13), provide powerful evidence that Fanconi anemia is a bona fide DNA repair disorder.
Fig. 2.
FANCD2 and its ubiquitylation are required for ICL repair. (A) Sequence surrounding the ICL and relevant restriction sites of pICL. (B) pICL (2.3 ng/μl) was replicated in mock-depleted extract, FANCD2-depleted extract (ΔFANCD2) or ΔFANCD2 extract supplemented with 375 nM FANCI-FANCD2WT or FANCI-FANCD2K562R, and repair efficiency was plotted. For primary data and calculation of repair efficiency, see Fig. S5A.
The ATR signaling pathway confers ICL resistance in vertebrate cells (14, 15), and replication of pICL in Xenopus egg extracts activates ATR as measured by chk1 and Rad1 phosphorylation [(12) and Fig. S1B and Fig. S7]. However, the repair defects we observed in FANCD2-depleted extracts were not due to defective checkpoint activation (Fig S7).
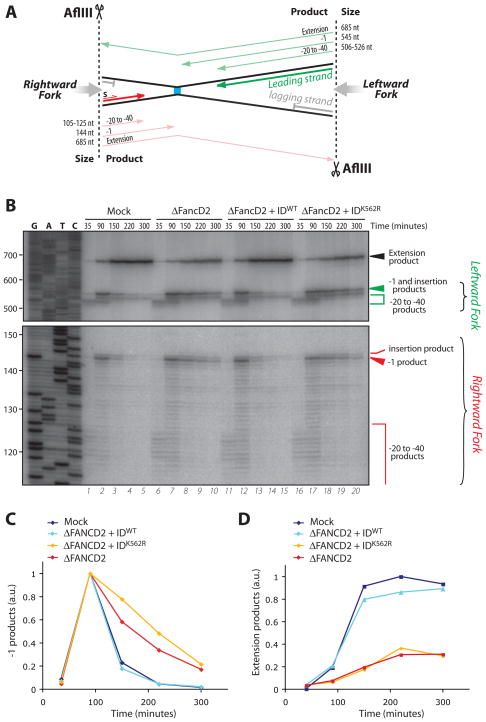
To address which step in ICL repair is dependent on the FANCI-FANCD2 complex, we replicated pICL in mock-depleted or FANCD2-depleted extract and performed lesion bypass analysis. To this end, DNA samples were withdrawn at various times, digested with AflIII, which cuts on either side of the ICL (Fig. 3A), and nascent strands were examined. In mock-depleted extract, the leading strands of the leftward and the rightward forks initially stalled 20–40 nt from the ICL (Fig. 3B, lane 1, green and red brackets), as previously described [(12) and Figure 1A]. Then, one of the two forks resumed synthesis and stalled again one nucleotide before the crosslinked base, causing a peak of “−1” products at ~90 minutes (Fig. 3B, lane 2, green and red arrowheads). Finally, insertion of a nucleotide across from the adducted base followed by extension resulted in formation of a 685 nt extension product (Fig. 3B, lanes 3–5, black arrowhead). In FANCD2-depleted extracts, there was a marked persistence of the −1 product and a reduction in extension products (Fig. 3B, lanes 6–10; for quantification, see Fig. 3, C and D, and Fig. S8). Although the low level of SapI site regeneration in the absence of FANCD2 generally plateaued by 150 minutes (Fig. 2B and Fig. S6, red graphs), extension products continued to accumulate at a low rate (Figure 3D and Fig. S8, red graphs), suggestive of a FANCD2-independent, error-prone lesion bypass process. The defects observed in FANCD2-depleted extracts were fully rescued by recombinant FANCI-FANCD2WT but not by FANCI-FANCD2K562R (Fig. 3, B to D, and Fig. S8). In the samples lacking functional FANCI-FANCD2, the level of −1 products did eventually decrease (Fig. 3, B and C, and Fig. S8). However, since the extension products never accumulated to more than ~30% of the mock-depleted samples (Fig. 3D and Fig. S8), we infer that this decline of −1 products is primarily due to the high susceptibility of stalled products to degradation (12). We conclude that the absence of functional FANCI-FANCD2 inhibits nucleotide insertion opposite the crosslinked base. This contrasts with the effect of DNA pol ζ immunodepletion, which arrests lesion bypass immediately after the insertion step (Figure S9;(12)).
Fig. 3.
Insertion of a nucleotide across from the damaged base is compromised in FANCD2-depleted extracts (A) Schematic representation of leading strand intermediates generated after pICL digestion with AflIII. (B) Samples from the reactions described in Fig. 2B were digested with AflIII, separated on a sequencing gel alongside a ladder derived from extension of primer S (S, in panel A) on pControl, and visualized via autoradiography. Nascent strands generated by the rightward (red) and leftward (green) replication forks are indicated to the right and illustrated in (A). −1 and extension products observed in (B) were quantified and graphed in (C) and (D), respectively.
An important event in ICL repair is thought to be incision of the parental strand on either side of the lesion (15)(Fig. 1A). To examine whether FANCI-FANCD2 is required for incisions, DNA repair intermediates were digested with HincII (Fig. 2A) and analyzed by denaturing gel electrophoresis and Southern blotting. Dual incisions surrounding the ICL are expected to convert the high molecular weight, parental X-shaped molecule into a 5.6 kb linear product (Figure 4A). As expected, in mock-depleted extract, we observed a time-dependent decrease of X-shaped molecules and a concomitant increase in linear species (Fig. 4B, lanes 4–9; quantified in Fig. 4, C and D). Nascent strands stalled at the ICL (Fig. 4A, top, grey strands) were detected as 2.3 and 3.3kb arm fragments (Fig. 4B), and then declined over time due to lesion bypass and resection. As we reported previously (12), this assay revealed that the majority of incisions occur after forks reached the −1 position (Fig. S10C).
Fig. 4.
ICL-proximal incisions are compromised in FANCD2-depleted extracts. (A) Schematic representation of predicted fragments generated by HincII digestion of pICL, before and after dual incisions and lesion bypass. Parental strands in black, nascent strands in grey. (B) pICL (2.5 ng/μl) was replicated in mock-depleted extract, FANCD2-depleted extract (ΔFANCD2) alone or supplemented with 386 nM FANCI-FANCD2WT. Samples were digested with HincII and separated on a denaturing agarose gel. Both parental and nascent strands are detected by Southern blotting. Unreplicated pControl and pICL were optionally digested with the indicated enzymes and used as size markers for arm/linear species and X-shaped molecules, respectively (lanes 1–3). Note the small amount of linear products in lane 3 (3% of the total), which represents contaminating non-crosslinked plasmids. X-structures and linear species observed in (B) were quantified and graphed in (C) and (D), respectively.
In the absence of FANCD2, incisions were severely inhibited, as seen from the persistence of X-shaped species and a severe delay in the accumulation of linear molecules (Fig. 4B, lanes 10–15; quantified in Fig. 4, C and D). Incisions and the accumulation of linear species were fully rescued by FANCI-FANCD2WT (Fig. 4D). Next, we repeated the incision-assay using pICL that had been pre-labeled with 32P-α-dATP via nick translation, which allowed us to specifically visualize the parental strands. This assay confirmed that incisions are inhibited in the absence of FANCD2, and showed that the defect is rescued by FANCI-FANCD2WT but not by FANCI-FANCD2K562R (Fig. S11).
Finally, we addressed the precise timing of FANCI-FANCD2 ubiquitylation. As shown in Fig. S12C, FANCI and FANCD2 ubiquitylation correlated with the arrival of leading strands at the −1 position, consistent with a role for the activated FANCI-FANCD2 complex in the insertion and incision steps, which occur after forks reach the −1 position ((12), Fig. S10 and S12).
Using a chemically homogeneous cisplatin ICL and a bona fide repair assay, we show that the Fanconi anemia pathway is required for replication-coupled ICL repair. These results indicate that the accumulation of FA cells in late S phase after exposure to ICLs (11) is due to defective replication-dependent repair. We further demonstrate that FANCI-FANCD2 must be ubiquitylated and thereby bind chromatin to support repair, suggesting that its role in this process is direct. In the absence of FANCI-FANCD2, incisions near the ICL are severely inhibited. In addition, insertion of a nucleotide across from the damaged template base is compromised, consistent with genetic epistasis between TLS polymerases and the FA pathway (16), and reduced damage-dependent mutagenesis in FA cells (17). Although we cannot presently determine whether the insertion or incision occurs first during ICL repair, it is widely envisioned that incisions must precede insertion (15). In this view, FANCI-FANCD2 might directly promote incisions and thereby affect TLS indirectly (Fig. S13A). For example, FANCI-FANCD2, which contains no apparent nuclease domains, could promote dual incisions by recruiting the Slx4 nuclease complex to the lesion (18). However, we cannot rule out the converse scenario in which TLS precedes incisions (Fig. S13B). In this case, the primary function of FANCI-FANCD2 might be to promote TLS. An attractive model is that ubiquitylated FANCI-FANCD2 facilitates insertion by recruiting the TLS polymerase REV1 via its ubiquitin binding domains. Finally, FANCI-FANCD2 might directly control both the incision and insertion steps (Fig. S13C). Future experiments will be required to distinguish between these possibilities.
Supplementary Material
Acknowledgments
This work was supported by NIH grant GM62267 and Leukemia and Lymphoma Society Award 1406-07 to J.C.W., New York State of Science, Technology & Academic Research (NYSTAR) Faculty Development Award C040069 and Swiss Cancer League grant OCS-01413-08-2003 to O.D.S., and a Dutch Cancer Society fellowship to P.K.. A.S. was supported by T32CA09216 to the Pathology Department at the Massachusetts General Hospital and by the Burroughs Wellcome Fund. S.J.E. is an investigator of the Howard Hughes Medical Institute. We thank Maureen Hoatlin for the Xenopus FANCD2 cDNA, Karlene Cimprich for phospho-Rad1 antibody, and the Walter lab for feedback. M.R. thanks Stefan Jentsch and the Max Planck Institute for generous support.
References
- 1.D’Andrea AD, Grompe M. Nat Rev Cancer. 2003 Jan;3:23. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 2.Patel KJ, Joenje H. DNA Repair (Amst) 2007 Jul 1;6:885. doi: 10.1016/j.dnarep.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Alpi AF, Pace PE, Babu MM, Patel KJ. Mol Cell. 2008 Dec 26;32:767. doi: 10.1016/j.molcel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Higuera I, et al. Mol Cell. 2001 Feb;7:249. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 5.Smogorzewska A, et al. Cell. 2007 Apr 20;129:289. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taniguchi T, et al. Blood. 2002 Oct 1;100:2414. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Andreassen PR, D’Andrea AD. Mol Cell Biol. 2004 Jul;24:5850. doi: 10.1128/MCB.24.13.5850-5862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben-Yehoyada M, et al. Mol Cell. 2009 Sep 11;35:704. doi: 10.1016/j.molcel.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen X, et al. Mol Cell. 2009 Sep 11;35:716. doi: 10.1016/j.molcel.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sobeck A, et al. Mol Cell Biol. 2006 Jan;26:425. doi: 10.1128/MCB.26.2.425-437.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akkari YM, et al. Mol Genet Metab. 2001 Dec;74:403. doi: 10.1006/mgme.2001.3259. [DOI] [PubMed] [Google Scholar]
- 12.Raschle M, et al. Cell. 2008 Sep 19;134:969. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howlett NG, et al. Science. 2002 Jul 26;297:606. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 14.Ishiai M, et al. Nat Struct Mol Biol. 2008 Nov;15:1138. doi: 10.1038/nsmb.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W. Nat Rev Genet. 2007 Oct;8:735. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 16.Niedzwiedz W, et al. Mol Cell. 2004 Aug 27;15:607. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 17.Mirchandani KD, McCaffrey RM, D’Andrea AD. DNA Repair (Amst) 2008 Jun 1;7:902. doi: 10.1016/j.dnarep.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein HL, Symington LS. Cell. 2009 Jul 10;138:20. doi: 10.1016/j.cell.2009.06.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.